Abstract
Background:
Little is known about the effect of a minimum number of slides required in generating image datasets used to build generalizable machine-learning (ML) models. In addition, the assumption within deep learning is that the increased number of training images will always enhance accuracy and that the initial validation accuracy of the models correlates well with their generalizability. In this pilot study, we have been able to test the above assumptions to gain a better understanding of such platforms, especially when data resources are limited.
Methods:
Using 10 colon histology slides (5 carcinoma and 5 benign), we were able to acquire 1000 partially overlapping images (Dataset A) that were then trained and tested on three convolutional neural networks (CNNs), ResNet50, AlexNet, and SqueezeNet, to build a large number of unique models for a simple task of classifying colon histopathology into benign and malignant. Different quantities of images (10–1000) from Dataset A were used to construct >200 unique CNN models whose performances were individually assessed. The performance of these models was initially assessed using 20% of Dataset A's images (not included in the training phase) to acquire their initial validation accuracy (internal accuracy) followed by their generalization accuracy on Dataset B (a very distinct secondary test set acquired from public domain online sources).
Results:
All CNNs showed similar peak internal accuracies (>97%) from the Dataset A test set. Peak accuracies for the external novel test set (Dataset B), an assessment of the ability to generalize, showed marked variation (ResNet50: 98%; AlexNet: 92%; and SqueezeNet: 80%). The models with the highest accuracy were not generated using the largest training sets. Further, a model's internal accuracy did not always correlate with its generalization accuracy. The results were obtained using an optimized number of cases and controls.
Conclusions:
Increasing the number of images in a training set does not always improve model accuracy, and significant numbers of cases may not always be needed for generalization, especially for simple tasks. Different CNNs reach peak accuracy with different training set sizes. Further studies are required to evaluate the above findings in more complex ML models prior to using such ancillary tools in clinical settings.
Keywords: Carcinoma, colon, convolutional neural network, machine learning
INTRODUCTION
Microscopic evaluation of histopathology slides by humans remains the gold standard for most pathology diagnoses. At the same time, the field of pathology has a 50-year history of developing computer-based image analysis techniques[1] to potentially augment human interpretation. Concurrent advances in the availability of whole-slide imaging and deep-learning platforms have spurred an increased interest in the computer analysis of histopathology images.[2,3] As such, a new generation of tools may soon be available in the pathologist's diagnostic arsenal.
Current deep-learning platforms are superior to previous histopathology image classification techniques.[2,3,4] Contemporary approaches often utilize a transfer-learning approach whereby an existing convolutional neural network (CNN) which has been optimized for image recognition is fine-tuned by further training with domain-specific images. This technique has shown success in general image recognition[5,6] as well as histopathologic tasks, as demonstrated in one of our recent studies.[7] In particular, recent success has been noted for detecting lymph node metastasis,[4] as well as classifying non-small cell lung cancer,[8] breast pathology,[9] colorectal polyps,[10] and gastric carcinoma,[11] to name a few.
Ideally, classification models should be generalizable, that is, equally effective at classifying histopathologic images regardless of which laboratory prepares the images. Generalizability refers to the model's ability to make accurate predictions on new previously unseen test sets (i.e., Dataset B in our study) that are distinct and completely outside of our original training/initial testing dataset.[12] Unfortunately, although these deep-learning approaches may be very helpful in yielding relatively accurate results, most deep-learning platforms have an inherent “black box” element whereby it is not clear exactly how the models they create come to be.[2] This makes the process more challenging when the need to optimize certain models may arise. In addition, this is also problematic for the goal of developing generalizable models, as it is not clear which features a model in using within the CNN to make its predictions.
Herein, we describe the creation and evaluation of 231 different unique models for the distinction of carcinoma from nonneoplastic colonic tissue. Our goal was to build the simplest discriminating models from the fewest number of slides used to acquire a dataset of overlapping images. This study enabled us to evaluate the correlation of our initial validation accuracy to each model's generalization accuracy. This approach also allowed us to empirically identify the optimal number of images that were required to build the most generalizable model for this very simple histopathologic task: classification into clearly benign and clearly malignant histopathology. Comparing these unique machine-learning (ML) model variations in this study has yielded some interesting results and a degree of transparency into these “black box” platforms that may assist others, especially when the number of histopathology resources within their study may be limited.
METHODS
Data collection
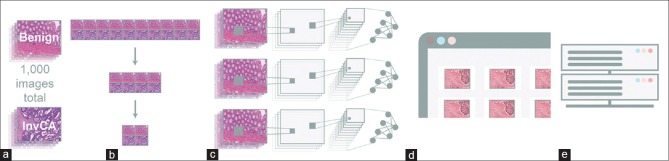
Institutional Review Board approval (ID number 1286225-1) was acquired. One thousand portable network graphics (PNG) partially overlapping images (500 invasive colonic adenocarcinomas [with evidence of submucosal invasion] and 500 normal colonic tissues) were obtained at two magnifications (×40 and × 100) from 10 unique cases (5 colonic carcinoma slides and 5 normal colonic tissue slides) using screen capture techniques from Aperio whole-slide scanned images (Leica Biosystems Aperio XT, Buffalo Grove, IL, USA). Since the goal of the project was to build the simplest binary classification possible, for distinguishing carcinoma from normal colonic tissue, proliferative lesions such as but not limited to tubular adenomas and other polyps were excluded from the training dataset (Dataset A). The carcinoma slides selected included those with histologic variations, including well-differentiated, moderately differentiated, and poorly differentiated subtypes, as well as those with mucinous differentiation. The nonneoplastic colonic tissue showed no evidence of dysplasia, tubular adenoma, malignancy, or other proliferative lesions but included certain nonneoplastic histologic features, such as inflammation, as part of the training set [Figure 1a].
Figure 1.
(a) Dataset A included 1000 images acquired through ten slides (five benign colon and five invasive colon carcinomas). (b) From the above 1000 images from Dataset A, seven distinct image training set categories (1000 images, 500 images, 200 images, 100 images, 50 images, 30 images, and 10 images) were constructed to assess the significance of the number of training images on their respective model's accuracy. (c) A transfer-learning approach was employed to retrain the three distinct well established convolutional neural networks noted above in building models that could distinguish colonic carcinoma from normal colonic tissue. (d) The model's accuracy was then assessed through two distinct data sets “Internal Validation” is based on Dataset A's 20% of the images that were kept outside of the training phase and used for the first validation accuracy measure while the “External Validation” test set is based on Dataset B which was completely unknown to our trained images (taken from a variety of public domain sources) and used to assess each model's generalizability. (e) The performance parameters of the individual models were then compared, contrasted, and statistically analyzed
Data preparation
From the above 1000 images (Dataset A), seven distinct image training subset categories (1000 images, 500 images, 200 images, 100 images, 50 images, 30 images, and 10 images) were constructed to assess the significance of the number of training images on their respective model's accuracy [Figure 1b]. Each subset was composed of equal proportions of benign and malignant images, and each subset was inclusive of the next largest iteration (i.e., the 500 image subsets contained all of the other image subset files [200, 100, 50, 30, and 10], while the 30 image subsets contained all of the 10 image subset files, and so on). Each of these subsets was then trained on three distinct well-established CNNs (ResNet50,[13] SqueezeNet, and AlexNet[14]) to build its respective ML models. In addition, within each model, 11 unique models were constructed to quantitate their respective accuracies and to assess the statistical significance for each subset and respective variables. Hence, a total of 231 unique, optimized ML models were constructed to compare and contrast their performance. The 231 models generated were based on 7 unique datasets (datasets of 1000 images, 500 images, 200 images, 100 images, 50 images, 30 images, and 10 images) using 3 CNN models (ResNet50, AlexNet, and SqueezeNet) that were used to build 11 separate models (7 × 3 × 11 = 231).
Machine-learning and deep-learning models
A transfer-learning approach was employed to retrain the three distinct, well-established CNNs described above in building models that could distinguish colonic carcinoma from normal colonic tissue [Figure 1c]. This approach preserves the core aspects of the original CNN while allowing subsequent fine tuning to render it specific to the task at hand. During this retraining process with our new input images (colon carcinoma versus normal colon), the training performance was measured by cross-entropy loss function to display the learning progress of our new model.
In the training mode, the parameter that initially internally measures this task during training only is the model's calculated “training accuracy,” which calculates the percentage of accurately labeled images on the training batch using a random 5% of the images within the training set. The training steps used in the transfer-learning process were done through the Turi Create library framework, which utilized the ResNet50 and the SqueezeNet CNNs, while the AlexNet CNN was retrained in the TensorFlow platform. The final weights and the number of optimized steps were saved based on the protocol buffer. As part of the training process, all of the images in the training sets were resized to fixed dimensions (224 × 224 pixels) and were set to the default regularization.
Following this initial validation, a “validation accuracy” (herein known as the “internal validation”) was then calculated, which involves testing the above-trained model on a held-out subset of the images (not included in the training phase). In our study, 80% of the images from Dataset A or its dataset subsets were used in the training batch, while 20% of the images were reserved for the “internal validation” accuracy testing step. To then further test the generalization capability of each model, a secondary external set of test images (Dataset B) from a variety of outside public domain sources completely unknown to our training set (herein known as the “external validation”) was used to test each model's true generalizability [Figure 1d and e]. This “external validation” test set was attained through a Fatkun batch process. The images were initially treated as unknowns and were subsequently reclassified as invasive colonic carcinoma or normal colonic tissue by our three board-certified pathologists prior to being used in our testing phase. Images without 100% concordance among the pathologists were excluded from the analysis. This resulted in 50 images within the external validation test set (Dataset B).
In addition to the above 50 colon external test images (Dataset B), test images from breast and prostate histopathology, collected for a prior study evaluating joint photographic experts group (JPEG) versus PNG images in histopathology, were also used to test the specificity of our colon ML models.[7] The external images collected for the prostate and breast were similarly collected and assessed as the colonic images used in this study. Eighty-two breast external test images and seventy prostate external test images were used for this task. This allowed us to test the best colon ML models against these other tissue subtypes to assess our colon ML model's specificity. This approach also validated the generalizability assessment of our models and allowed us to note any potential overfitting phenomena within our ML models.
Statistical measures
The performance parameters of our individual models were then statistically analyzed as follows. Accuracy, precision, sensitivity, and specificity were compared between different numbers of training set images using ANOVA models. These models included effects for the number of images, ML model, and the interaction between the number of images and ML model. Pairwise comparisons between all time points and all ML models were adjusted for multiple testing using the Tukey honestly significant difference method. Aggregate accuracy for each group of 11 models was calculated as the average ± the standard error of the mean. Analyses were conducted using R, version 3.5.1 (R Core Team, 2018, R Core Team: The R Foundation & GNU's Free Software Foundation, Inc. Boston, MA).
RESULTS
”Internal Validation” image test set (based on the 20% held-out test set from Dataset A or its subsets)
The models, when compared based on the number of images within their training phase and their respective neural networks, showed relatively similar performance parameters with respect to their “internal validation” accuracy. The performance differences noted within the models were mainly with the extreme high (1000) and extreme low training sets (<50), [Figure 2]. Specifically, the ResNet50's most accurate models were obtained at the 1000 training image sets with a mean accuracy (range) of 98.8% (98.1%–99.6%). The AlexNet models were able to obtain near 100% accuracy with as few as 50 images in their training set, while the SqueezeNet models achieved their highest accuracy with the 500 training image set with a mean (range) of 96.9% (95.7%–97.9%).
Figure 2.
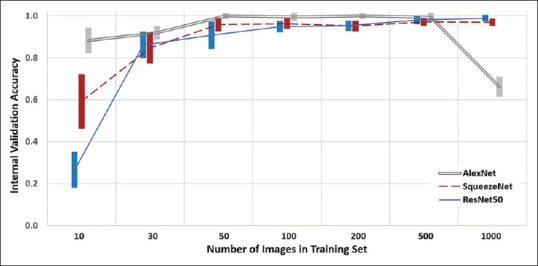
Effect of the size of training set and the respective model's classification accuracy on their internal validation image test set (the 20% held-out images from Dataset A). Mean ± standard error of the mean, for each group
Within the AlexNet models, only the performance of the 1000 training image models was significantly different (inferior) to the other training set sizes (P < 0.001 for all). For both the ResNet50 and SqueezeNet models, only the performance of the 10 training image models was significantly different (inferior) to the other training set sizes (P < 0.001 for all). With a training set of 10 images, the performance of the AlexNet models was superior to SqueezeNet, which was superior to ResNet50 (P < 0.001 for all). With a training set of 1000 images, the performance of the AlexNet models was inferior to the performance of the SqueezeNet and ResNet50 platforms (P < 0.001 for both). When using 30–500 training images, the validation performance of each platform was similar.
“External Validation (Generalization)” image test set (based on Dataset B)
Unlike the internal validation study, the external validation test set was used to assess the generalization of each model, revealing marked differences between the models’ accuracies [Figure 3]. The generalization accuracy of the models was dependent on the number of images within their training set and their respective neural network platform. Overall, the ResNet50 models performed the best with the highest generalization accuracy obtained by the 200 training image models with a mean (range) of 98.0% (94.0%–100%). Similar accuracy was obtained with 500 images, but accuracy with 10–100 and 1000 training images was inferior (P < 0.03 for all).
Figure 3.
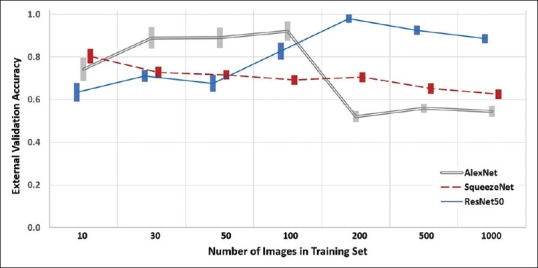
Effect of the size of training set and the respective model's classification accuracy based on Dataset B, the external validation test set (veneralization). Mean ± standard error of the mean, for each group
The highest generalization accuracy for the AlexNet models was obtained by the 100 training image models, with a mean (range) of 92.1% (61.0%–100%). Similar accuracy was obtained with 30 and 50 images, but accuracy with 10 and 200–1000 training images was inferior (P < 0.001 for all).
For SqueezeNet, the highest generalization accuracy was obtained by the models using 10 training images with a mean (range) of 80.4% (66.0%–92.0%). Similar accuracy was obtained with 30 and 50 images, but accuracy with 100–1000 training images was inferior (P < 0.03 for all).
In summary, for 200–1000 training images, ResNet50 is superior to SqueezeNet which is superior to AlexNet (P < 0.03 for all). With 100 training images, AlexNet is superior to ResNet50, which is superior to SqueezeNet (P < 0.01 for all). Notably, with 10 training images, the generalization accuracy of AlexNet and SqueezeNet models is similarly superior to ResNet50 models (P < 0.01 for both). For 30 or 50 training images, ResNet50 and SqueezeNet models are similar to both inferior to AlexNet (P < 0.001 for both).
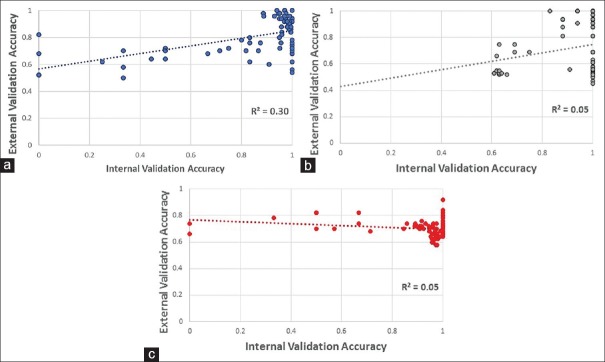
The performance of the models against internal validation test images was a significant but weak predictor of that model's ability to generalize novel colonic tissue images; the exception was ResNet50, which showed the strongest correlation. For all models taken together, the correlation (r2) was weak (r2 = 0.08). However, for models based on their distinct neural networks, the correlation was variable (r2 = 0.30, 0.05, and 0.05 for ResNet50, AlexNet, and SqueezeNet, respectively). The strongest correlation was found in the ResNet50-generated models [Figure 4].
Figure 4.
Correlation between internal validation accuracy (based on the held back 20% of the images from Dataset A and its subsets) and external validation accuracy (based on Dataset B) with regression line for (a) ResNet50, (b) AlexNet, and (c) SqueezeNet. ResNet50 showed the strongest correlation (R2 = 0.3) between the internal and external validation test set accuracies for their respective models. Both ResNet50 and AlexNet showed a similar slope
In addition, the model that performed best at categorizing novel images of colon tissue from Dataset B (ResNet50 using 200 training images, accuracy 98%) was not as successful at classifying H and E-stained images of other tissue types (prostate and breast tissue datasets). Specifically, our best performing colon histology model was also used to distinguish benign breast and prostate images from breast and prostate carcinoma (from our separate prostate and breast external test datasets), which were shown to be unsuccessful (compared to colon test sets) with accuracies of 52% and 54% when tested on our prostate and breast test sets, respectively. This further supported the specificity of our models, and the validity of their respective external validation test set results on Dataset B.
DISCUSSION
In this study, surprisingly, we were able to show how partially overlapping images from a limited number of slides (5 carcinomas and 5 normal colons) may generate generalizable models for simple tasks. In addition, we found that the size of the training set had a profound impact on the ability of models to accurately generalize. Seemingly paradoxically, larger training sets resulted in worse accuracy for the classification of novel (external validation) test images. We propose that overfitting is occurring when using the larger number of training images, as even CNNs are not immune to this effect.[15,16] In essence, the models become better and better at classifying the internal validation images at the expense of incorrectly classifying the external novel images.
Further, to assess the specificity of each CNN to classify colonic histopathology, images from benign breast and prostate, and images from breast and prostate carcinoma,[7] were also tested on our best-performing colon ML models which supported their specificity and generalizability. These external images from the prostate and breast were similary collcected and assessed as the colonic images that were used in this study. Our best performing model performed extremely well on the colonic dataset (Dataset B) while performing abysmally when attempting to classify prostate and breast tissue test sets, with accuracies of 54% and 52%, respectively. Taken together, excellent performance on novel colon histology images (external validation test set, Dataset B) and poor performance on other tissue types suggest that the models are functioning based on true differentiating characteristics and not based on an idiosyncratic finding in the training set.
As others delve into deep-learning pathology model development, the current study demonstrates important principles that should be considered: first, the choice of neural network matters. Others have also shown that CNNs, whether developed from scratch or already optimized for image analysis with domain-specific transfer learning, vary widely in performance even when using similar base techniques and platforms.[4,17] In this study, there was a clear performance difference between AlexNet, ResNet50, and SqueezeNet. This may or may not hold true for other digital pathology deep-learning tasks. For example, as seen in our dataset, the AlexNet or SqueezeNet platform might be preferred with very limited training sets, while ResNet50 may be a better choice for larger sets of data. Its worth noting that these small training sets, with 10 training images, most likely reflect the base, naïve (untrained), and CNN model's performance. Importantly, all of the CNNs could achieve impressive levels of both internal and external accuracies using a limited number of cases and controls (10 slides). This suggests that massive training slide sets may not always be necessary to create generalizable models for simple tasks (e.g., binary classification tasks). Additional study is needed to determine which platform, if any, is routinely better than others for histology image analysis from other anatomic sites, other histologic variations, and for classification beyond dichotomization (e.g., proliferative lesions, preneoplasia, dysplasia, etc.,).
Second, the choice of model selection should not depend solely on the accuracy against the internal validation image test set. In this study, all platforms approached 100% in their internal validation accuracy as the training image set reached 500–1000 images. However, when tested on the novel (external validation) image set (Dataset B), significant differences were detected in the performance of each platform. Indeed, the correlation between internal validation performance and accuracy against external validation images was poor for two of the three neural networks. ResNet50 showed the best correlation (r2 = 0.30) for these two parameters. Especially striking is the number of models that achieved 100% internal validation accuracy but the widely variable performance against novel (external validation) test images [Figure 4].
While it may seem that even 1000 training images are not that large of a dataset in the age of “big data,” one must also remember that the deep-learning platforms used in this study were not naïve. They had been previously highly trained for general image recognition tasks (nonhistopathology). Though a unique finding within digital pathology, at least one other medical study noted a degradation of model performance with seemingly small training sets when using pretrained CNNs.[18] Further study is needed to determine the optimal number of training images and will likely depend on the deep-learning platform, tissue site of origin, and number/complexity of classifications needed.
Transfer learning and ML in recent publications have shown incredible results in identifying subtypes of carcinoma and specific morphologic features.[19,20] These methods showcase that these tools have clinical utility within the laboratory. Depending on the ground truth, these models are used for classification, prediction of immunohistochemical results, molecular markers, and even survival may be predicted from initial H and E slides. However, it is common in many of these studies to use internal datasets for the validation of these models. While this may be appropriate under certain circumstances, this method can also potentially introduce significant bias based on the performing characteristics specific to each laboratory. To minimize such bias, our study's external validation (generalization) was purposely based on external images from variable sources.
Further models that include proliferative colon lesions (e.g., adenomatous polyps) and additional carcinoma subtypes are needed to build deep-learning models that are impactful for pathologists’ day-to-day practice. The classification scheme in this study was purposefully simple to test the aforementioned variables, such as CNN and training image quantity. Although we have shed some light, additional studies to bring further transparency to the process are warranted to make the creation of generalizable models more intentional.
While we may be far from a future wherein these deep-learning models render primary diagnoses, a future in which these models optimize workflow or provide quality assurance review is much closer. This depends on following a systematic approach to develop deep-learning models that are accurate, reproducible, and most importantly, generalizable.
Financial support and sponsorship
The statistical analysis in the project described was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant number UL1 TR001860. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2020/11/1/5/278925
REFERENCES
- 1.Mendelsohn ML, Kolman WA, Perry B, Prewitt JM. Morphological analysis of cells and chromosomes by digital computer. Methods Inf Med. 1965;4:163–7. [PubMed] [Google Scholar]
- 2.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J Pathol Inform. 2016;7:29. doi: 10.4103/2153-3539.186902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med Image Anal. 2016;33:170–5. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proc. Proc Conference on Computer Vision and Pattern Recognition. 2014:580–7. [Google Scholar]
- 6.Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, et al. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics. 2017;18:281. doi: 10.1186/s12859-017-1685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AD, Graff JP, Darrow M, Borowsky A, Olson KA, Gandour-Edwards R, et al. Impact of pre-analytical variables on deep learning accuracy in histopathology. Histopathology. 2019;75:39–53. doi: 10.1111/his.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24:1559–67. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandomkar Z, Brennan PC, Mello-Thoms C. MuDeRN: Multi-category classification of breast histopathological image using deep residual networks. Artif Intell Med. 2018;88:14–24. doi: 10.1016/j.artmed.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, et al. Deep learning for classification of colorectal polyps on whole-slide images. J Pathol Inform. 2017;8:30. doi: 10.4103/jpi.jpi_34_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2–13. doi: 10.1016/j.compmedimag.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad Pathol. 2019;6:2374289519873088. doi: 10.1177/2374289519873088. doi: 10.1177/eCollection 2019 Jan-Dec. Review. PubMed PMID: 31523704; PubMed Central PMCID: PMC6727099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He KZ, Ren S, Sun J. Deep Residual Learning for Image Recognition. arXiv. 2015 151203385. [Google Scholar]
- 14.Krizhevsky AS, Hinton GE. Image net classification with deep convolutional neural networks. Commun ACM. 2017;60:84–90. [Google Scholar]
- 15.Bilbao I, Bilbao J. Overfitting Problem and the Over-training in the Era of Data: Particularly for Artificial Neural Networks. Cairo, Egypt: IEEE; 2017. [Google Scholar]
- 16.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–58. [Google Scholar]
- 17.Veta M, van Diest PJ, Willems SM, Wang H, Madabhushi A, Cruz-Roa A, et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med Image Anal. 2015;20:237–48. doi: 10.1016/j.media.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Shin Y, Balasingham I. Automatic polyp frame screening using patch based combined feature and dictionary learning. Comput Med Imaging Graph. 2018;69:33–42. doi: 10.1016/j.compmedimag.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Sha L, Osinski BL, Ho IY, Tan TL, Willis C, Weiss H, et al. Multi-field-of-view deep learning model predicts nonsmall cell lung cancer programmed death-ligand 1 status from whole-slide hematoxylin and eosin images. J Pathol Inform. 2019;10:24. doi: 10.4103/jpi.jpi_24_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maleki S, Zandvakili A, Gera S, Khutti SD, Gersten A, Khader SN. Differentiating noninvasive follicular thyroid neoplasm with papillary-like nuclear features from classic papillary thyroid carcinoma: Analysis of cytomorphologic descriptions using a novel machine-learning approach. J Pathol Inform. 2019;10:29. doi: 10.4103/jpi.jpi_25_19. [DOI] [PMC free article] [PubMed] [Google Scholar]