Abstract
Acetoin is a potential platform compound for a variety of chemicals. Bacillus licheniformis MW3, a thermophilic and generally regarded as safe (GRAS) microorganism, can produce 2,3-butanediol with a high concentration, yield, and productivity. In this study, B. licheniformis MW3 was metabolic engineered for acetoin production. After deleting two 2,3-butanediol dehydrogenases encoding genes budC and gdh, an engineered strain B. licheniformis MW3 (ΔbudCΔgdh) was constructed. Using fed-batch fermentation of B. licheniformis MW3 (ΔbudCΔgdh), 64.2 g/L acetoin was produced at a productivity of 2.378 g/[L h] and a yield of 0.412 g/g from 156 g/L glucose in 27 h. The fermentation process exhibited rather high productivity and yield of acetoin, indicating that B. licheniformis MW3 (ΔbudCΔgdh) might be a promising acetoin producer.
Keywords: acetoin, Bacillus licheniformis, metabolic engineering, 2, 3-butanediol dehydrogenase, 2, 3-butanediol
Introduction
Acetoin (3-hydroxy-2-butanone) is widely used as a common additive in food industry and a building block for various chemicals (Liu et al., 2011; Xiao and Lu, 2014; Yang et al., 2017). It was listed as one of 30 platforms compound whose development and utilization was given priority by the US Department of Energy in 2004 (Xiao and Lu, 2014). Currently, commercially available acetoin is primarily produced from fossil feedstocks by chemical methods involving radical reactions and severe environmental pollution. With increasing concerns about production cost and environment protection, microbial fermentation production of acetoin has attracted extensive attention in recent years (Xiao and Lu, 2014; Yang et al., 2017).
Many native or recombinant microorganisms, including the genera Serratia, Klebsiella, Saccharomyces, Gluconobacter, and Bacillus, have abilities to produce acetoin (Sun et al., 2012; Wang et al., 2013, 2017; Bae et al., 2016; Jia et al., 2017). Among these species, Bacillus species which can produce various industrial products have become the research hotspot for acetoin production (Zhang Y. et al., 2013; Fan et al., 2018; Liu et al., 2019). Acetoin is an intermediate of 2,3-butanediol fermentation-pathway (Ji et al., 2011; Yang and Zhang, 2019). Except a few native acetoin producers, most of the Bacillus strains produce 2,3-butanediol but not acetoin as their major product (Fu et al., 2016; Dai et al., 2019). Thus, different metabolic engineering strategies were proposed to redistribute the carbon flux to acetoin in various Bacillus strains. For example, Zhang et al. successfully improved acetoin production by Bacillus subtilis through disruption of acetoin reductase (also called as 2,3-butanediol dehydrogenase) and expression of a water-forming NADH oxidase. Acetoin at a concentration of 56.7 g/L was produced with a yield of 0.378 g/g and a productivity of 0.639 g/[L h] (Zhang et al., 2014b).
Bacillus licheniformis, a generally regarded as safe (GRAS) strain with the characteristics of strong sugar consumption and fast growth rate, is a promising chassis strain for acetoin production. Natural B. licheniformis strains usually produce a mixture of (2R,3R)-2,3-butanediol and meso-2,3-butanediol isomers (Li L. et al., 2014; Qi et al., 2014; Qiu et al., 2016). In this study, two 2,3-butanediol dehydrogenases encoding genes in B. licheniformis MW3 were deleted and an engineered strain B. licheniformis MW3 (ΔbudCΔgdh) was constructed. The recombinant strain B. licheniformis MW3 (ΔbudCΔgdh) can produce 64.2 g/L acetoin within 27 h with a productivity of 2.378 g/[L h] and a yield of 0.412 g/g. Considering its desirable characteristics, B. licheniformis MW3 (ΔbudCΔgdh) may be a promising alternative for fermentative production of acetoin.
Materials and Methods
Enzymes and Chemicals
FastPfu DNA polymerase and T4 DNA ligase were purchased from TransGen Biotech (China) and Thermo Fisher (United States), respectively. Restriction enzymes were purchased from Thermo Fisher (United States). Polymerase chain reaction (PCR) primers were provided by Sangon (Shanghai, China). Racemic acetoin, diacetyl and 2,3-butanediol were purchased from Sigma. (2R,3R)-2,3-butanediol (98.0%), (2S,3S)-2,3-butanediol (99.0%), and meso-2,3-butanediol (98.0%) were purchased from ACROS (The Kingdom of Belgium). NADH was purchased from Roche (United States). Yeast extract (FM902) was a kind gift from Angel Yeast Co., Ltd. (Yichang, Hubei, China). All other chemicals were of analytical grade and commercially available.
Bacterial Strains, Media, and Plasmids
All the strains and plasmids used in this study are listed in Supplementary Table S1. The plasmids pKVM1-ΔbudC and pKVM1-Δgdh based on the temperature sensitive plasmid pKVM1 were used for gene knockout in B. licheniformis MW3 (Waschkau et al., 2008; Rachinger et al., 2013; Ge et al., 2016). Lysogenic broth (LB) medium was used for cultivation of Escherichia coli and B. licheniformis. NB medium (8 g/L Nutrient Broth, Difco) supplemented with 50 mM glucose was used for selection of B. licheniformis containing pKVM1. Ampicillin was used at a concentration of 100 μg/mL for the selection of E. coli. Erythromycin (5 μg/mL) and polymyxin B (40 μg/mL) were used for the selection of B. licheniformis. X-Gal was added at a concentration of 40 μg/mL for blue-white screening.
Gene Knockout in B. licheniformis MW3
E. coli S17-1 λpir was used as donor strain to allow the conjugal transfer of plasmids pKVM1-ΔbudC and pKVM1-Δgdh into B. licheniformis MW3. After growth in liquid NB/glucose medium with erythromycin at 30°C, serial dilutions and spreading on NB/glucose plates with erythromycin and X-gal and incubation at 42°C, one of blue colonies of B. licheniformis MW3 with plasmid integration by homologous recombination was picked, inoculated in NB/glucose medium without antibiotic, and cultivated at 30°C for at least two passages. After dilutions and spreading on NB/glucose with X-gal, white colonies in which plasmids used to knockout were cured resulting from a second recombination event were picked. PCR was used to verify the disruption event of gene budC and gdh using primer pairs ΔbudC-f/ΔbudC-r and Δgdh-f/Δgdh-r (Supplementary Table S2), respectively.
Enzyme Activity Assays
To measure 2,3-butanediol dehydrogenase activities in B. licheniformis MW3 and B. licheniformis MW3 (ΔbudCΔgdh), the cells of the strains were grown for 12 h, harvested, washed twice and resuspended in 67 mM phosphate buffer (pH 7.4), and disrupted by sonication with a Sonics sonicator (500 W; 20 kHz). Cell debris was removed through centrifugation (12,000 g, 15 min) and the resulting supernatants were used as the crude extracts for enzyme activity assays.
Activities of 2,3-butanediol dehydrogenase were spectrophotometrically assayed by measuring the change in absorbance at 340 nm corresponding to the oxidation of NADH or reduction of NAD+ at 30°C using a UV/visible spectrophotometer (Ultrospec 2100 pro, Amersham Biosciences, United States). For the reduction reaction, the reaction solution contains 0.2 mM of NADH and 5 mM of acetoin or diacetyl in 67 mM phosphate buffer (pH 7.4). For oxidation reactions, the reaction solution contains 10 mM (2R,3R)-2,3-butanediol, (2S,3S)-2,3-butanediol, and meso-2,3-butanediol and 1 mM NAD+ in 67 mM phosphate buffer (pH 7.4). One unit of activity was defined as the amount of enzyme that consumed or formed 1 μmol of NADH per min. The protein concentration in crude extract was measured by the Lowry method, with bovine serum albumin as the standard (Hartree, 1972).
Batch and Fed-Batch Fermentations
B. licheniformis MW3 was maintained on LB agar slants at 4°C. A loop of cells from the fully grown slant was inoculated into 100 mL of LB in 500-mL Erlenmeyer flasks and incubated at 50°C on a rotary shaker at 180 r/min for 12 h to prepare the seed culture. Then, the seed culture was inoculated (5%, v/v) into the bioreactors for acetoin production.
Batch and fed-batch fermentations were conducted in a 1-L bioreactor (Infors AG, Bottmingen, Switzerland) with 0.8 L initial medium and a 5-L bioreactor (BIOSTAT B, B. Braun Biotech International GmbH, Germany) with 4 L initial medium, respectively. The seed culture was inoculated into the fermentation medium containing about 70 g/L glucose; 1 g/L triammonium citrate; 12 g/L yeast extract; 2 g/L K2HPO4⋅3H2O; 6.5 g/L sodium acetate; 0.25 g/L MgSO4⋅7H2O; 0.0225 g/L FeSO4⋅7H2O; 0.0075 g/L ZnSO4⋅7H2O; and 0.0038 g/L MnSO4⋅H2O. The cultivation was carried out at 50°C, stirring at 500 r/min, and airflow at 1.0 vvm (volume air per volume broth per minute). The pH was maintained at 7.0 by automatic addition of 6 M acetic acid and 6 M NaOH using a program-controlled peristaltic pump. Samples were collected periodically to determine the biomass, concentrations of glucose, 2,3-butanediol, and acetoin.
Analytical Methods
Samples were centrifuged at 12,000 g for 10 min and the concentration of glucose was measured enzymatically using a bio-analyzer (SBA-40D, Shandong Academy of Sciences, China) after diluting to an appropriate concentration. The concentrations of 2,3-butanediol and acetoin were analyzed by gas chromatography (GC; GC2014c, Shimadzu) with capillary GC columns (AT. SE-54, inside diameter, 0.32 mm; length, 30 m; Chromatographic Technology Center, Lanzhou Institute of Chemical Physics, China) as described previously (Zhang L. et al., 2016).
Results and Discussion
Construction of the Strain B. licheniformis MW3 (ΔbudCΔgdh)
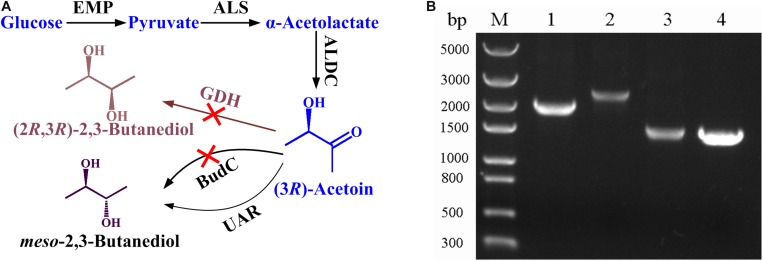
B. licheniformis MW3 is a GRAS and thermophilic strain which can efficiently produce meso-2,3-butanediol and (2R,3R)-2,3-butanediol with a simple stereoisomer formation mechanism (Ge et al., 2016). As shown in Figure 1A, glucose is converted to pyruvate by glycolytic pathway, then α-acetolactate synthase (ALS) condenses two molecules of pyruvate to generate one molecules of α-acetolactate, and then α-acetolactate is transformed into one molecules of acetoin by α-acetolactate decarboxylase (ALDC). Two stereospecific 2,3-butanediol dehydrogenases, (2R,3R)-2,3-butanediol dehydrogenase (encoded by gdh) and meso-2,3-butanediol dehydrogenase (encoded by budC), are responsible for the production of (2R,3R)-2,3-butanediol and meso-2,3-butanediol, respectively (Ge et al., 2016). To construct an acetoin producing strain based on B. licheniformis MW3, both (2R,3R)-2,3-butanediol dehydrogenase and meso-2,3-butanediol dehydrogenase encoding genes should be deleted. The temperature sensitive plasmid pKVM1 was utilized as described previously. The mutant strain B. licheniformis MW3 (ΔbudCΔgdh) was selected and the result in Figure 1B shows that the PCR using the primer pairs generated products of the expected sizes.
FIGURE 1.
Construction of B. licheniformis MW3 (ΔbudCΔgdh) for acetoin production. (A) Technology roadmap for acetoin production from glucose by B. licheniformis MW3 (ΔbudCΔgdh). ALS, α-acetolactate synthase; ALDC, α-acetolactate decarboxylase; BudC, meso-2,3-butanediol dehydrogenase; GDH, (2R,3R)-2,3-butanediol dehydrogenase; UAR, unidentified acetoin reductase (based on the results of gas chromatograph in Supplementary Figure S1, this enzyme produces meso-2,3-butanediol as the reduction product of acetoin). (B) Analysis of PCR fragments to confirm budC and gdh disruption. Lane M: molecular mass standard (DNA/HindIII); Lane 1: PCR product of budC amplified with B. licheniformis MW3 genomic DNA as the template; Lane 2: PCR product of gdh amplified with B. licheniformis MW3 genomic DNA as the template; Lane 3: PCR product of budC amplified with B. licheniformis MW3 (ΔbudCΔgdh) genomic DNA as the template; Lane 4: PCR product of gdh amplified with B. licheniformis MW3 (ΔbudCΔgdh) genomic DNA as the template. The PCRs were performed with primer pairs ΔbudC-f/ΔbudC-r and Δgdh-f/Δgdh-r, respectively.
Activities of 2,3-Butanediol Dehydrogenases in B. licheniformis MW3 (ΔbudCΔgdh)
The activities of (2R,3R)-2,3-butanediol dehydrogenase and meso-2,3-butanediol dehydrogenase in B. licheniformis MW3 (ΔbudCΔgdh) were assayed. As shown in Supplementary Table S3, (2S,3S)-2,3-butanediol oxidation activity was rather low in both B. licheniformis MW3 and B. licheniformis MW3 (ΔbudCΔgdh). Both (2R,3R)-2,3-butanediol and meso-2,3-butanediol oxidation activities decreased in B. licheniformis MW3 (ΔbudCΔgdh). Especially, decrease of acetoin reduction activity occurred after deletion of (2R,3R)-2,3-butanediol dehydrogenase and meso-2,3-butanediol dehydrogenase encoding genes. Taking advantage of its low acetoin reduction activity, acetoin produced by ALDC might not be reduced to 2,3-butanediol and the strain B. licheniformis MW3 (ΔbudCΔgdh) was used for acetoin production in the subsequent experiments.
Batch Fermentation of Acetoin by B. licheniformis MW3 (ΔbudCΔgdh)
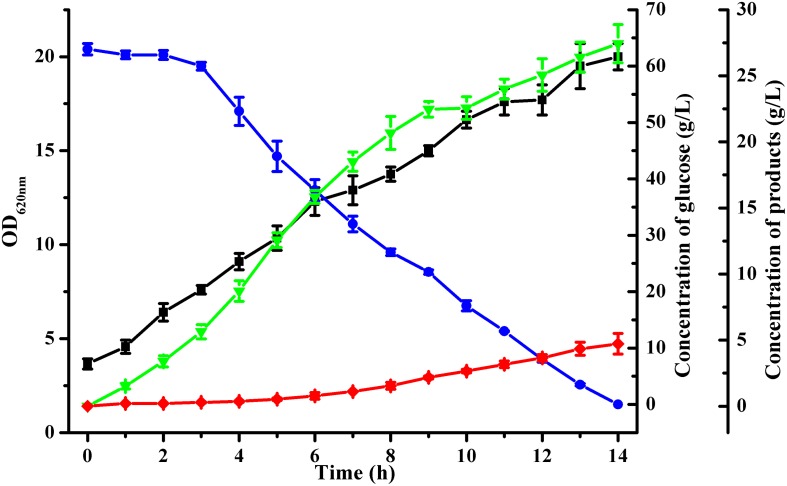
Batch fermentation was conducted in a 5-L bioreactor with 4 L initial medium to analyze acetoin production by B. licheniformis MW3 (ΔbudCΔgdh). As shown in Figure 2, inactivation of both budC and gdh led to a high acetoin accumulation in B. licheniformis MW3 (ΔbudCΔgdh). This strain consumed 63 ± 1 g/L glucose in 14 h and produced 27.4 ± 1.45 g/L acetoin, with a yield of 0.435 ± 0.023 g/g (Figure 2). Although the growth of B. licheniformis MW3 (ΔbudCΔgdh) was slightly lower than that of B. licheniformis MW3, this strain can produce acetoin instead of 2,3-butanediol as its major product (Supplementary Table S4).
FIGURE 2.
Time-course of batch fermentation by B. licheniformis MW3 (ΔbudCΔgdh).  , Glucose;
, Glucose;  , OD620nm;
, OD620nm;  , 2,3-butanediol;
, 2,3-butanediol;  , acetoin. The experiment was conducted in a 1-L bioreactor (Infors AG, Bottmingen, Switzerland) with 0.8 L initial medium with an initial glucose concentration of 70 g/L approximately. Cultivation was carried out at 50°C and an initial pH of 7.0. The pH was maintained at 7.0 by automatic addition of 6 M acetic acid and 6 M NaOH. The agitation speed was 500 r/min and the aeration rate was 1 vvm. Data were the means ± SDs from three parallel experiments.
, acetoin. The experiment was conducted in a 1-L bioreactor (Infors AG, Bottmingen, Switzerland) with 0.8 L initial medium with an initial glucose concentration of 70 g/L approximately. Cultivation was carried out at 50°C and an initial pH of 7.0. The pH was maintained at 7.0 by automatic addition of 6 M acetic acid and 6 M NaOH. The agitation speed was 500 r/min and the aeration rate was 1 vvm. Data were the means ± SDs from three parallel experiments.
Fed-Batch Fermentation of Acetoin by B. licheniformis MW3 (ΔbudCΔgdh)
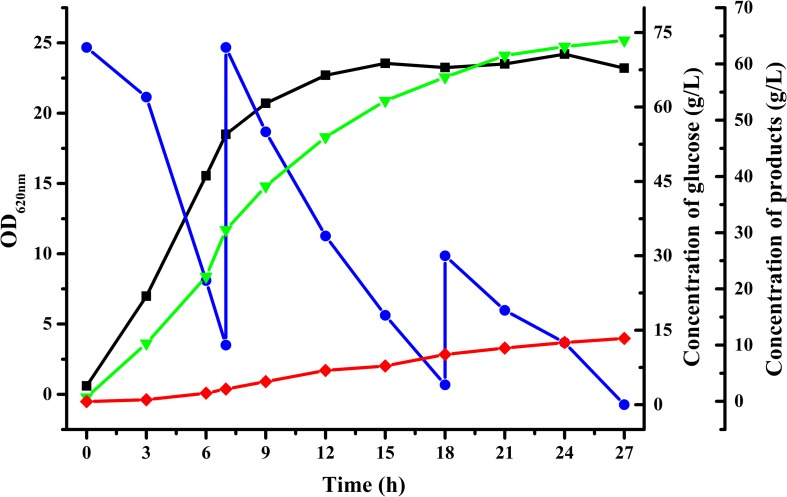
To achieve higher acetoin concentrations, fed-batch fermentation was carried out with an initial glucose concentration of about 70 g/L using B. licheniformis MW3 (ΔbudCΔgdh). As shown in Figure 3, 64.2 g/L acetoin from 156 g/L glucose was obtained in 27 h by B. licheniformis MW3 (ΔbudCΔgdh). The yield of acetoin was 84.1% of the theoretical value and the average productivity was 2.378 g/[L h]. The carbon flux channeled into the acetoin biosynthetic might be further enhanced since there were still 2,3-butanediol accumulated during the fermentation (Figure 3 and Supplementary Figure S1). Toxicity of acetoin can hinder its higher production by various microorganisms. It was reported that an approximate 30% cell growth of B. licheniformis will be inhibited with 40 g/L exogenous acetoin (Yuan et al., 2019). Thus, production of 2,3-butanediol may be due to induction of an unidentified acetoin reductase or meso-2,3-butanediol dehydrogenase to resistant the toxicity of acetoin at high concentrations. Higher production of acetoin by B. licheniformis may be accomplished by searching the undiscovered acetoin reductase or overexpressing NADH oxidase, which could lead to prevention of NADH-dependent reduction of acetoin.
FIGURE 3.
Time-course of fed-batch fermentation by B. licheniformis MW3 (ΔbudCΔgdh).  , Glucose;
, Glucose;  , OD620nm;
, OD620nm;  , 2,3-butanediol;
, 2,3-butanediol;  , acetoin. The experiment was conducted in a 5-L bioreactor (BIOSTAT B, B. Braun Biotech International GmbH, Germany) with 4 L initial medium with an initial glucose concentration of 70 g/L approximately. Cultivation was carried out at 50°C and an initial pH of 7.0. The pH was maintained at 7.0 by automatic addition of 6 M acetic acid and 6 M NaOH. The agitation speed was 500 r/min and the aeration rate was 1 vvm. The fed-batch fermentation was conducted by feeding glucose solution when the residual glucose in the fermentation was lower than 10 g/L. Data were from a representative fed-batch fermentation repeated for three times.
, acetoin. The experiment was conducted in a 5-L bioreactor (BIOSTAT B, B. Braun Biotech International GmbH, Germany) with 4 L initial medium with an initial glucose concentration of 70 g/L approximately. Cultivation was carried out at 50°C and an initial pH of 7.0. The pH was maintained at 7.0 by automatic addition of 6 M acetic acid and 6 M NaOH. The agitation speed was 500 r/min and the aeration rate was 1 vvm. The fed-batch fermentation was conducted by feeding glucose solution when the residual glucose in the fermentation was lower than 10 g/L. Data were from a representative fed-batch fermentation repeated for three times.
Several recombinant Bacillus strains have been used to produce acetoin (Table 1). Based on a two-stage pH control strategy, the group of Li obtained a high acetoin concentration of 73.6 g/L with a productivity of 0.77 g/[L h] using metabolically engineered B. subtilis overexpressing 2,3-butanediol dehydrogenase (Zhang et al., 2014a). Fan et al. successfully improved the coproduction of uridine and acetoin by modification of acetoin metabolism in engineered B. subtilis. The recombinant strain can produce 60.48 g/L acetoin and 40.62 g/L uridine after 48 h of fed-batch fermentation (Fan et al., 2018). The group of Chen isolated a B. licheniformis strain WX-02 which can produce γ-poly-glutamic acid accompanied with 2,3-butanediol (Li X. et al., 2014). B. licheniformis WX-02ΔbudCΔacoR was constructed through deleting budC and acetoin degradation transcriptional activator encoding gene acoR. Fed-batch fermentation through a three-stage agitation strategy during which 2,3-butanediol was first accumulated and then converted to acetoin using B. licheniformis WX-02ΔbudCΔacoR can produce 78.79 g/L acetoin with a yield of 0.31 g/g and a productivity of 0.58 g/[L h] at 37°C (Li et al., 2017). B. licheniformis MW3 is a thermophilic strain which can produce 2,3-butanediol as its major fermentation product with higher yield and productivity than that of B. licheniformis WX-02 at 50°C, implying a latent capacity of strain MW3 for efficient acetoin production (Ge et al., 2016; Qiu et al., 2016). Thus, B. licheniformis MW3 (ΔbudCΔgdh) was constructed and the recombinant strain was able to produce 64.2 g/L acetoin with a relatively high productivity (2.378 g/[L h]) through a simple fermentation process. The values of productivity and yield of acetoin produced using B. licheniformis MW3 (ΔbudCΔgdh) were the highest ever obtained in acetoin production by recombinant Bacillus species. B. licheniformis MW3 (ΔbudCΔgdh) might be useful for the production of acetoin on a commercial scale.
TABLE 1.
Fermentation production of acetoin by recombinant Bacillus species.
| Strain | Engineering strategy | Concentration (g/L) | Yield (g/g) | Productivity (g/[L h]) | References |
| B. subtilis F126-2 | Disrupting 2,3-butanediol dehydrogenase gene bdhA and overexpression of alsSD operon based on a uridine producing strain | 60.48 | / | 1.26 | Fan et al., 2018 |
| B. subtilis BSA | Over-expression of 2,3-butanediol dehydrogenase in B. subtilis JNA 3-10, applying a two-stage pH control strategy | 73.6 | 0.408 | 0.77 | Zhang et al., 2014a |
| B. subtilis BMN | Moderate-expression of the water-forming NADH oxidase C in bdhA disrupted B. subtilis JNA 3-10 | 56.7 | 0.675 | 0.639 | Zhang et al., 2014b |
| B. subtilis 168/pMA5-zwf | Over-expression of glucose-6-phosphate dehydrogenase G6PDH in B. subtilis 168 | 43.3 | 0.33 | 0.36 | Bao et al., 2015 |
| B. subtilis BS168D | Blocking of 2,3-butanediol dehydrogenase gene bdhA in B. subtilis 168 | 24.6 | 0.246 | 0.342 | Zhang et al., 2017 |
| B. subtilis ZB02 | Over-expression of xylose transport protein AraE, xylose isomerase XylA, and xylulokinase XylB in B. subtilis 168ARSRCPΔacoAΔbdhA, using glucose, xylose, and arabinose as substrates | 62.2 | NRa | 0.864 | Zhang B. et al., 2016 |
| B. subtilis BSK814A4 | Deleting araR, bdhA, acoA and inserting a native xyl operon into genome-reduced B. subtilis strain BSK814 | 23.3 | 0.46 | 0.194 | Yan et al., 2018 |
| B. subtilis BSUW06 | Overexpressing alsSD operon and deleting bdhA, acoA, and pta in B. subtilis 168 | 19.8 | NR | 0.566 | Wang et al., 2012 |
| B. subtilis PAR | Moderate enhancement of ALsR expression using promoter PbdhA in B. subtilis 168 | 41.5 | 0.35 | 0.43 | Zhang X. et al., 2013 |
| B. licheniformis WX-02ΔbudCΔacoR | Deleting budC and acetoin degradation transcriptional activator encoding gene acoR in B. licheniformis WX-02 | 78.79 | 0.31 | 0.58 | Li et al., 2017 |
| B. licheniformis MW3 (ΔbudCΔgdh) | Deleting budC and gdh in B. licheniformis MW3 | 64.4 | 0.412 | 2.378 | This work |
aNR, not reported.
Conclusion
We constructed a recombinant strain B. licheniformis MW3 (ΔbudCΔgdh) for acetoin production. Acetoin at a high concentration (64.2 g/L) was produced with a productivity of 2.378 g/[L h] and a yield of 0.412 g/g through fed-batch fermentation by B. licheniformis MW3 (ΔbudCΔgdh). Because of its GRAS characteristic and ability to produce acetoin with high concentration, productivity, and yield, this strain should be a promising alternative for the practical production of acetoin.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
CL, YG, and MC performed experiments. CL, CG, and CM wrote the manuscript and conceived the study. CL, YG, MC, XG, and PL were involved in analysis and interpretation of experimental data. CM and PX coordinated the project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors greatly thank Prof. Dr. Friedhelm Meinhardt for kindly providing B. licheniformis MW3 and Prof. Dr. Armin Ehrenreich for plasmid pKVM1. The authors also thank Chengjia Zhang and Nannan Dong from Core Facilities for Life and Environmental Sciences (State Key Laboratory of Microbial Technology, Shandong University) for assistance in microbial fermentation.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31670041), the grants of National Key R&D Program of China (2019YFA0904900 and 2019YFA0904803), Shandong Provincial Funds for Distinguished Young Scientists (JQ 201806), Natural Science Foundation of Shandong Provincial (ZR2018PC008), Key R&D Program of Shandong Provincial (2019GSF107034 and 2019GSF107039), and Qilu Young Scholar of Shandong University. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00125/full#supplementary-material
References
- Bae S. J., Kim S., Hahn J. S. (2016). Efficient production of acetoin in Saccharomyces cerevisiae by disruption of 2,3-butanediol dehydrogenase and expression of NADH oxidase. Sci. Rep. 6:27667. 10.1038/srep27667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao T., Zhang X., Zhao X., Rao Z., Yang T., Yang S. (2015). Regulation of the NADH pool and NADH/NADPH ratio redistributes acetoin and 2,3-butanediol proportion in Bacillus subtilis. Biotechnol. J. 10 1298–1306. 10.1002/biot.201400577 [DOI] [PubMed] [Google Scholar]
- Dai J., Wang Z., Xiu Z. (2019). High production of optically pure (3R)-acetoin by a newly isolated marine strain of Bacillus subtilis CGMCC 13141. Bioprocess Biosyst. Eng. 42 475–483. 10.1007/s00449-018-2051-8 [DOI] [PubMed] [Google Scholar]
- Fan X., Wu H., Jia Z., Li G., Li Q., Chen N., et al. (2018). Metabolic engineering of Bacillus subtilis for the co-production of uridine and acetoin. Appl. Microbiol. Biotechnol. 102 8753–8762. 10.1007/s00253-018-9316-7 [DOI] [PubMed] [Google Scholar]
- Fu J., Huo G., Feng L., Mao Y., Wang Z., Ma H., et al. (2016). Metabolic engineering of Bacillus subtilis for chiral pure meso-2,3-butanediol production. Biotechnol. Biofuels. 9:90. 10.1186/s13068-016-0502-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Li K., Li L., Gao C., Zhang L., Ma C., et al. (2016). Contracted but effective: production of enantiopure 2,3-butanediol by thermophilic and GRAS Bacillus licheniformis. Green Chem. 18 4693–4703. 10.1039/C6GC01023G [DOI] [Google Scholar]
- Hartree E. F. (1972). Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 48 422–427. 10.1016/0003-2697(72)90094-2 [DOI] [PubMed] [Google Scholar]
- Ji X., Huang H., Ouyang P. (2011). Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol. Adv. 29 351–364. 10.1016/j.biotechadv.2011.01.007 [DOI] [PubMed] [Google Scholar]
- Jia X., Peng X., Liu Y., Han Y. (2017). Conversion of cellulose and hemicellulose of biomass simultaneously to acetoin by thermophilic simultaneous saccharification and fermentation. Biotechnol. Biofuels. 10:232. 10.1186/s13068-017-0924-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li K., Wang K., Chen C., Gao C., Ma C., et al. (2014). Efficient production of 2,3-butanediol from corn stover hydrolysate by using a thermophilic Bacillus licheniformis strain. Bioresour. Technol. 170 256–261. 10.1016/j.biortech.2014.07.101 [DOI] [PubMed] [Google Scholar]
- Li L., Wei X., Yu W., Wen Z., Chen S. (2017). Enhancement of acetoin production from Bacillus licheniformis by 2,3-butanediol conversion strategy: metabolic engineering and fermentation control. Process Biochem. 57 35–42. 10.1016/j.procbio.2017.03.027 [DOI] [Google Scholar]
- Li X., Gou X., Long D., Ji Z., Hu L., Xu D., et al. (2014). Physiological and metabolic analysis of nitrate reduction on poly-gamma-glutamic acid synthesis in Bacillus licheniformis WX-02. Arch. Microbiol. 196 791–799. 10.1007/s00203-014-1014-y [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu L., Li J., Du G., Chen J. (2019). Synthetic biology toolbox and chassis development in Bacillus subtilis. Trends Biotechnol. 37 548–562. 10.1016/j.tibtech.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Liu Z., Qin J., Gao C., Hua D., Ma C., Li L., et al. (2011). Production of (2S,3S)-2,3-butanediol and (3S)-acetoin from glucose using resting cells of Klebsiella pneumonia and Bacillus subtilis. Bioresour. Technol. 102 10741–10744. 10.1016/j.biortech.2011.08.110 [DOI] [PubMed] [Google Scholar]
- Qi G., Kang Y., Li L., Xiao A., Zhang S., Wen Z., et al. (2014). Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced D-2,3-butanediol production Bacillus licheniformis. Biotechnol. Biofuels. 7:16. 10.1186/1754-6834-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zhang J., Li L., Wen Z., Nomura C. T., Wu S., et al. (2016). Engineering Bacillus licheniformis for the production of meso-2,3-butanediol. Biotechnol. Biofuels. 9:117. 10.1186/s13068-016-0522-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachinger M., Bauch M., Strittmatter A., Bongaerts J., Evers S., Maurer K. H., et al. (2013). Size unlimited markerless deletions by a transconjugative plasmid-system in Bacillus licheniformis. J. Biotechnol. 167 365–369. 10.1016/j.jbiotec.2013.07.026 [DOI] [PubMed] [Google Scholar]
- Sun J., Zhang L., Rao B., Shen Y., Wei D. (2012). Enhanced acetoin production by Serratia marcescens H32 with expression of a water-forming NADH oxidase. Bioresour. Technol. 119 94–98. 10.1016/j.biortech.2012.05.108 [DOI] [PubMed] [Google Scholar]
- Wang M., Fu J., Zhang X., Chen T. (2012). Metabolic engineering of Bacillus subtilis for enhanced production of acetoin. Biotechnol. Lett. 34 1877–1885. 10.1007/s10529-012-0981-9 [DOI] [PubMed] [Google Scholar]
- Wang X., Lv M., Zhang L., Li K., Gao C., Ma C., et al. (2013). Efficient bioconversion of 2,3-butanediol into acetoin using Gluconobacter oxydans DSM 2003. Biotechnol. Biofuels 6:155. 10.1186/1754-6834-6-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tao F., Xin B., Liu H., Gao Y., Zhou N., et al. (2017). Switch of metabolic status: redirecting metabolic flux for acetoin production from glycerol by activating a silent glycerol catabolism pathway. Metab. Eng. 39 90–101. 10.1016/j.ymben.2016.10.020 [DOI] [PubMed] [Google Scholar]
- Waschkau B., Waldeck J., Wieland S., Eichstadt R., Meinhardt F. (2008). Generation of readily transformable Bacillus licheniformis mutants. Appl. Microbiol. Biotechnol. 78 181–188. 10.1007/s00253-007-1278-0 [DOI] [PubMed] [Google Scholar]
- Xiao Z., Lu J. (2014). Strategies for enhancing fermentative production of acetoin: a review. Biotechnol. Adv. 32 492–503. 10.1016/j.biotechadv.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Yan P., Wu Y., Yang L., Wang Z., Chen T. (2018). Engineering genome-reduced Bacillus subtilis for acetoin production from xylose. Biotechnol. Lett. 40 393–398. 10.1007/s10529-017-2481-4 [DOI] [PubMed] [Google Scholar]
- Yang T., Rao Z., Zhang X., Xu M., Xu Z., Yang S. (2017). Metabolic engineering strategies for acetoin and 2,3-butanediol production: advances and prospects. Crit. Rev. Biotechnol. 37 990–1005. 10.1080/07388551.2017.1299680 [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhang Z. (2019). Recent advances on production of 2, 3-butanediol using engineered microbes. Biotechnol. Adv. 37 569–578. 10.1016/j.biotechadv.2018.03.019 [DOI] [PubMed] [Google Scholar]
- Yuan H., Xu Y., Chen Y., Zhan Y., Wei X., Li L., et al. (2019). Metabolomics analysis reveals global acetoin stress response of Bacillus licheniformis. Metabolomics 15:25. 10.1007/s11306-019-1492-7 [DOI] [PubMed] [Google Scholar]
- Zhang B., Li X., Fu J., Li N., Wang Z., Tang Y., et al. (2016). Production of acetoin through simultaneous utilization of glucose, xylose, and arabinose by engineered Bacillus subtilis. PLoS One 11:e159298. 10.1371/journal.pone.0159298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhao X., Zhang J., Zhao C., Liu J., Tian Y., et al. (2017). Effect of deletion of 2,3-butanediol dehydrogenase gene (bdhA) on acetoin production of Bacillus subtilis. Prep. Biochem. Biotechnol. 47 761–767. 10.1080/10826068.2017.1320293 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu Q., Ge Y., Li L., Gao C., Xu P., et al. (2016). Biotechnological production of acetoin, a bio-based platform chemical, from a lignocellulosic resource by metabolically engineered Enterobacter cloacae. Green Chem. 18 1560–1570. 10.1039/C5GC01638J [DOI] [Google Scholar]
- Zhang X., Bao T., Rao Z., Yang T., Xu Z., Yang S., et al. (2014a). Two-Stage pH control strategy based on the pH preference of acetoin reductase regulates acetoin and 2,3-butanediol distribution in Bacillus subtilis. PLoS One 9:e91187. 10.1371/journal.pone.0091187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang R., Bao T., Rao Z., Yang T., Xu M., et al. (2014b). The rebalanced pathway significantly enhances acetoin production by disruption of acetoinreductase gene and moderate-expression of a new water-forming NADH oxidase in Bacillus subtilis. Metab. Eng. 23 34–41. 10.1016/j.ymben.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang R., Bao T., Yang T., Xu M., Li H., et al. (2013). Moderate expression of the transcriptional regulator AlsR enhances acetoin production by Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 40 1067–1076. 10.1007/s10295-013-1303-5 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li S., Liu L., Wu J. (2013). Acetoin production enhanced by manipulating carbon flux in a newly isolated Bacillus amyloliquefaciens. Bioresour. Technol. 130 256–260. 10.1016/j.biortech.2012.10.036 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.