Abstract
Background
Osteoporosis is a metabolic osteopathy characterized by abnormal bone mass and microstructure that has become a public health problem worldwide. Cuscutae semen (CS) is a traditional Chinese medicine (TCM) that has a positive effect on the prevention and treatment of osteoporosis. However, the mechanism of CS is unclear. Therefore, this study aimed to reveal the possible molecular mechanism involved in the effects of CS on osteoporosis based on a network pharmacology approach.
Material/Methods
The inactive and active ingredients of CS were identified by searching the pharmacology analysis platform of the Chinese medicine system (TCMSP), and the targets of osteoporosis were screened in the relevant databases, such as GeneCards, PubMed, and the Comparative Toxicogenomics Database (CTD). The network of “medicine-ingredients-disease-targets (M-I-D-T)” was established by means of network pharmacology, and the key targets and core pathways were determined by R analysis. Molecular docking methods were used to evaluate the binding activity between the target and the active ingredients of CS.
Results
Eleven active ingredients were identified in CS, and 175 potential targets of the active ingredients were also identified from the TCMSP. Moreover, we revealed 22 539 targets related to osteoporosis in the 3 well-established databases, and we determined the intersection of the disease targets and the potential targets of the active ingredients; 107 common targets were identified and used in further analysis. Additionally, biological processes and signaling pathways involved in target action, such as fluid shear stress, atherosclerosis, cancer pathways, and the TNF signaling pathway, were determined. Finally, we chose the top 5 common targets, CCND1, EGFR, IL6, MAPK8, and VEGFA, for molecular docking with the 11 active ingredients of CS.
Conclusions
This study suggested that CS has multiple ingredients and multiple targets relevant to the treatment of osteoporosis. We determined that the active ingredient, sesamin, may be the most crucial ingredient of CS for the treatment of osteoporosis. Additionally, the network pharmacology method provided a novel research approach to analyze the function of complex ingredients.
MeSH Keywords: Medicine, Chinese Traditional; Osteoporosis; Pharmacology
Background
Osteoporosis (OP) is a systemic disease characterized by reduced bone strength and damaged bone microstructure, resulting in increased bone fragility and proneness to fracture [1]. According to the World Health Organization (WHO) statistics, 200 million people suffer from OP, and with the aging of the global population, this figure will continue to increase [2]. As a result of this phenomenon, the soaring expense of medical treatment and nursing has become an indisputable fact. It is predicted that in China, expenses due to OP-related fracture treatment will reach 25.43 billion dollars in 2050 [3]. At present, anti-osteoporosis drugs mainly include anti-catabolic agents, anabolic agents and supplements that utilize other mechanisms, such as calcium, vitamin K2 and strontium [4].
Because traditional Chinese medicine (TCM) can play a positive role in the treatment of OP [5], an increased number of researchers have begun to explore Chinese herbal medicines. Cuscutae semen (CS), which is derived from the dry and mature seeds of Cuscuta australis R. Br. or Cuscuta chinensis Lam., is a widely used Chinese medicine with a long history of use [6,7]. Previous studies have shown that CS can nourish the kidneys and liver, prevent miscarriages and protect the eyesight [8]. Moreover, it also plays a certain role in the treatment of impotence and the seminal inhibition of the growth of tumor cells [9,10]. In addition to these biological functions, Yao et al. showed that CS can promote the proliferation of bone marrow mesenchymal stem cells and osteoblasts and inhibit osteoclast activity in rat bone cells. Moreover, Yang et al. demonstrated that CS can induce osteogenic activity in human osteoblast-like MG-63 cells [11]. Although some ingredients of CS have been extracted and verified [12–14], the identities of numerous other components and how they relieve OP by influencing bone metabolism are still largely unknown.
Recently, the concept of network pharmacology has been proposed as a new method to predict the mechanisms of the effects of drug therapy on disease at the whole organismal level [15]. With the aid of molecular biology and related database information, network pharmacology has shifted from the traditional “one drug, one target” strategy to the “drug-target-pathway-disease” strategy to provide a more comprehensive understanding of TCM mechanisms [16]. Therefore, this study adopted a network pharmacology approach to analyze and construct an “ingredient-target-pathway” network of the effects of CS on the treatment of OP. The multitarget and multiple pathway network of CS was also revealed from a holistic point of view, providing a reference for further exploring the mechanism underlying its treatment effects on OP.
Material and Methods
Acquisition of chemical ingredients and screening of active ingredients
“Cuscutae semen” was used as the key word to search for all the chemical information about CS in the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (http://lsp.nwsuaf.edu.cn/tcmsp.php) [17] and in the literature review. By referring to multiple standards in the literature, an oral availability (OB) greater than 30% and a pharmacokinetic value (DL) greater than 0.18 were used as the limiting conditions to screen the active ingredients in the database [18]. As a result, 11 chemical ingredients were identified, and PubChem (http://pubchem.ncbi.nlm.nih.gov/) was used to retrieve the active ingredients and obtain their 3D structures in mol2 format for further analysis.
Target prediction of the active ingredients
The targets of the active ingredients in CS were queried by using the TCMSP target module, and the target protein name was transformed into a gene name by using Perl (http://www.perl.org/) and the UniProt database (http://www.UniProt.org/).
Target prediction of disease
To reveal the genes possibly related to disease, “osteoporosis” was used as the keyword, and the GeneCards database (https://www.genecards.org/), PubMed website (https://www.ncbi.nlm.nih.gov) and CTD database (https://ctdbase.org/tools/batchQuery.go) were utilized. All of these online tools are continuously updated with information about human genes and genetic diseases, providing a relatively comprehensive overview of research results. We removed duplicate targets from the search results.
Intersection of active ingredients and disease targets
We first downloaded the R package (https://www.rproject.org/) and entered the command code to install the toolkit for drawing a Venn diagram in R. Then, by using the previously prepared files that contained the active ingredients of CS and the disease targets, a specific command code was entered in R, which generated the Venn diagram and a list describing the specific outcomes of the analysis. This “ingredients to disease” list was used in the following steps.
Network construction and analysis
The “ingredients to disease” list was imported into Cytoscape 3.6.1 software (https://cytoscape.org/); then, the CS ingredient names and OP names were also introduced into Cytoscape to construct the model of the medicine-ingredients-disease-targets (M-I-D-T) network. In the network construction, nodes were used to represent molecules or target proteins, and edges were used to represent the relationships among ingredients, disease and targets.
Construction of the protein interaction network
The “ingredients to disease” list was imported into the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string-db.org), which is a protein–protein interaction (PPI) database that can search for known proteins and predict PPIs [19]. In the operating interface, we limited the species to “Homo sapiens” and set the minimum interaction threshold to 0.7 to determine the relationships between potential targets of CS in the treatment of OP. Next, we utilized the R package to screen the hub proteins. The basic principle was to determine the number of junction nodes between all proteins and the top 30 proteins.
Gene ontology and pathway enrichment analysis
Bioconductor (http://www.bioconductor.org/) provides tools for the analysis and interpretation of high-throughput genomic data. It uses the programming software R, which is an open source and open development software [20]. With the help of the R package, we successfully installed this useful analysis tool and then ran the code. The enrichment analysis of Gene Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were carried out on the genes in the ingredient-OP target network, and the results were obtained (P-adjusted value <0.01). By using the count score, we selected the top 20 for presentation.
Molecular docking
Five targets with a maximum count score in the target interaction network obtained from the PPI analysis were selected. They were mitogen-activated protein kinase 8 (MAPK8), epidermal growth factor receptor (EGFR), cyclin D1 (CCND1), interleukin-6 (IL-6), and vascular endothelial growth factor A (VEGFA). All of these were searched in the PDB database (http://www.rcsb.org/pdb/home/home.do), which is the most important database containing atomic-level 3D structural data for biological macromolecules (proteins, nucleic acids and sugars) [21], and the conformations were screened according to the following conditions: 1) protein structure was obtained by an x-ray diffraction method; 2) protein structure had a resolution less than 3 Å; 3) protein was typed; and 4) protein structures reported in previous docking studies were preferred. Then, Autotools was used to remove the excess protein chains and ligands, and hydrogenation was performed to remove the water molecules. AutoGrid was used to calculate the energy lattice, to set the grid box coordinates, and to set the distance of each small grid point to 0.1 nm. Finally, Autodock Vina 4.2 (http://vina.scripps.edu/), which is an open source program used for molecular docking that was designed and implemented by Dr. Oleg Trott at the Molecular Graphics Lab at The Scripps Research Institute [22], was used for batch docking of potential active ingredients in CS with the 5 proteins, and the results returned 9 conformations. The predominant conformation was analyzed and plotted with Free Maestro by Schrodinger (https://www.schrodinger.com/freemaestro).
Results
Active ingredients of CS
Seventy-six chemical ingredients of CS were identified in the TCMSP database. After setting the filtering criteria mentioned above, 11 active ingredients of CS were determined, which are shown in Table 1.
Table 1.
Active components of Cuscutae semen (CS).
| Mol ID | Mol name | OB (%) | DL |
|---|---|---|---|
| MOL001558 | Sesamin | 56.55 | 0.83 |
| MOL000184 | NSC63551 | 39.25 | 0.76 |
| MOL000354 | Isorhamnetin | 49.6 | 0.31 |
| MOL000358 | beta-Sitosterol | 36.91 | 0.75 |
| MOL000422 | kaempferol | 41.88 | 0.24 |
| MOL005043 | Campest-5-en-3beta-ol | 37.58 | 0.71 |
| MOL005440 | Isofucosterol | 43.78 | 0.76 |
| MOL005944 | Matrine | 63.77 | 0.25 |
| MOL006649 | Sophranol | 55.42 | 0.28 |
| MOL000953 | CLR | 37.87 | 0.68 |
| MOL000098 | Quercetin | 46.43 | 0.28 |
Potential targets of the active ingredients
Through searching the TCMSP target module, 175 potential targets of CS active ingredients and their corresponding symbols were collected (Supplementary Table 1).
Potential targets of OP
In this study, 3 internationally recognized databases of disease genes were searched, and 22 539 potential targets were retrieved after removing the duplicate targets. These targets were closely related to the occurrence and development of OP (Supplementary Table 2).
Ingredient and disease targets intersection
After inputting the potential targets of the ingredients and the disease targets into the R platform, the intersection of the 2 types of targets was determined. The Venn diagram showed that 107 potential targets had relationships with active ingredients and OP (Figure 1). The common gene names of the 107 potential targets are shown in Supplementary Table 3.
Figure 1.
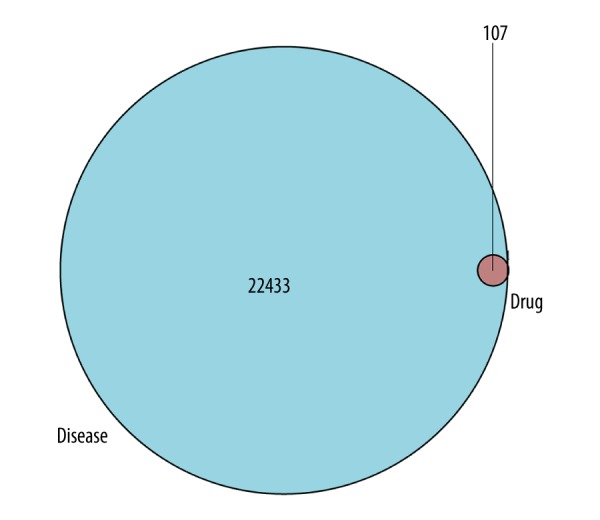
Venn diagram of 107 potential common targets.
M-I-T-D network
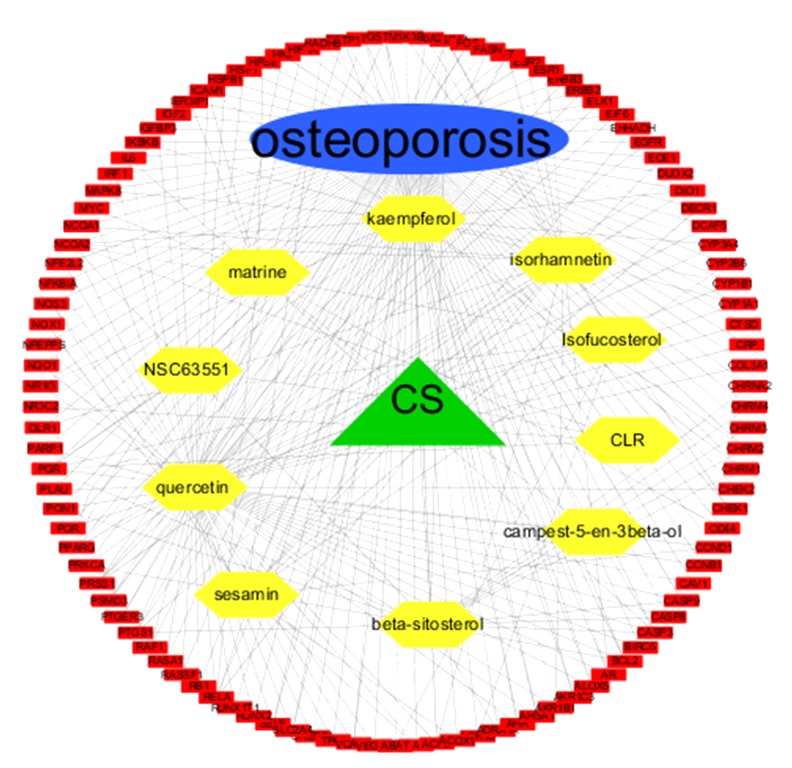
As shown in Figure 2, a medicine-ingredients-targets-disease network was generated and indicated that these 4 components had close relationships with each other.
Figure 2.
A medicine-ingredients-targets-disease network of 4 parts. CS: Cuscutae Semen; yellow: active ingredients of CS; red: 107 potential common targets.
PPI network
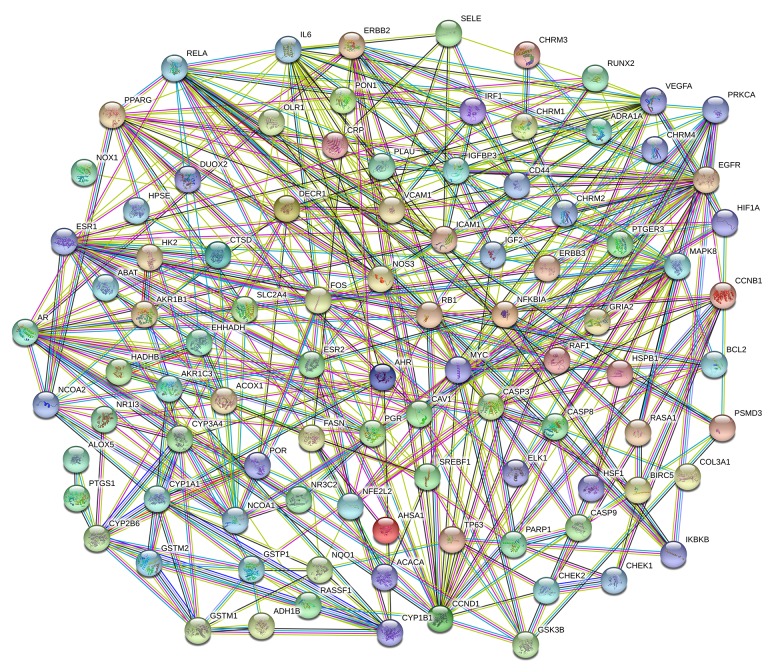
In the “ingredients and disease intersection” part of the PPI network, OP targets and active ingredient potential targets showed 107 duplicate genes. These genes may become targets for the treatment of OP. To study the interaction of the targets in vivo and search for the hub genes, PPI network analysis of the potential target groups was carried out (Figure 3). The linked tables with different colors represent the different meanings of the biological information. Moreover, the top 30 genes that had a close relationship with other genes were represented via a bar plot that clearly described these 30 gene targets in terms of their key positions in the PPI network. These genes were MAPK8, EGFR, CCND1, IL-6, VEGFA, ESR1, AR, MYC, CASP3, PPARG, RELA, DECR1, NCOA1, ERBB2, FOS, ICAM1, NOS3, CYP1A1, PRKCA, CYP2B6, PGR, CASP8, CAV1, CYP3A4, GSTP1, NCOA2, RB1, VCAM1, AHR, and CD44 (Figure 4).
Figure 3.
Protein–protein interaction (PPI) network analysis of 107 potential target: cyan line: from curated databases; purple line: experimentally determined; green line: gene neighborhood; red line: gene fusions; blue line: gene co-occurrence; yellow line: text mining; black line: co-expression; baby blue line: protein homology.
Figure 4.
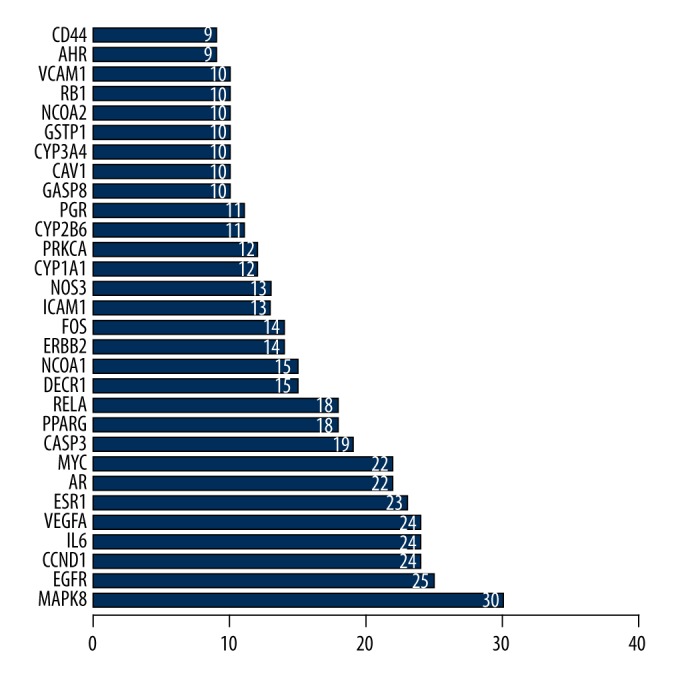
Top 30 targets from protein–protein interaction (PPI) network.
GO and KEGG pathway enrichment analysis
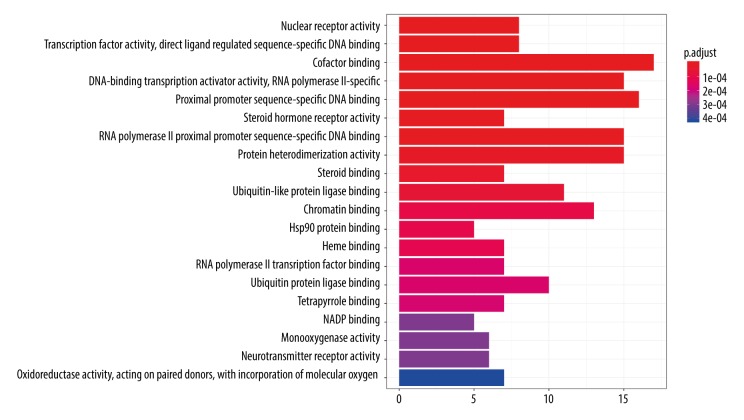
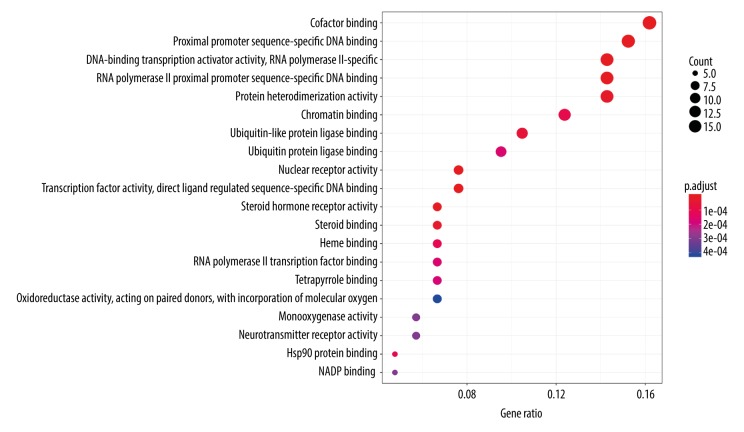
To illustrate the mechanism underlying the effects of CS active ingredients on OP more comprehensively and concretely, we performed GO enrichment analysis of the 107 common targets in the ingredient-disease target network. As a result, the top 20 enriched GO terms were identified, which are shown in a bar plot (P-adjusted value <0.01, Figure 5). For example, in biological processes, the targets of CS were enriched in cofactor binding (GO: 0048037), proximal promoter sequence-specific DNA binding (GO: 0000987), DNA-binding transcription activator activity, RNA polymerase II proximal promoter sequence-specific binding (GO: 0001228), RNA polymerase II proximal promoter sequence-specific DNA binding (GO: 0000978), protein heterodimerization activity (GO: 0046982), chromatin binding (GO: 0003682), ubiquitin-like protein ligase binding (GO: 0044389), enzyme activator activity (GO: 0008047), and other processes. Meanwhile, a dot plot indicated the gene ratio of the number of target genes involved in one biological process to the number of all annotated genes. The higher the ratio, the higher the level of enrichment is. The size of the dot reflects the number of target genes in the analysis, and the different colors of the dots indicate the different P-adjusted value ranges (Figure 6).
Figure 5.
Top 20 enriched Gene Ontology (GO) terms selected from 107 common targets. (P-adjust value <0.01).
Figure 6.
A dot plot to describe P-adjust value range of top 20 targets.
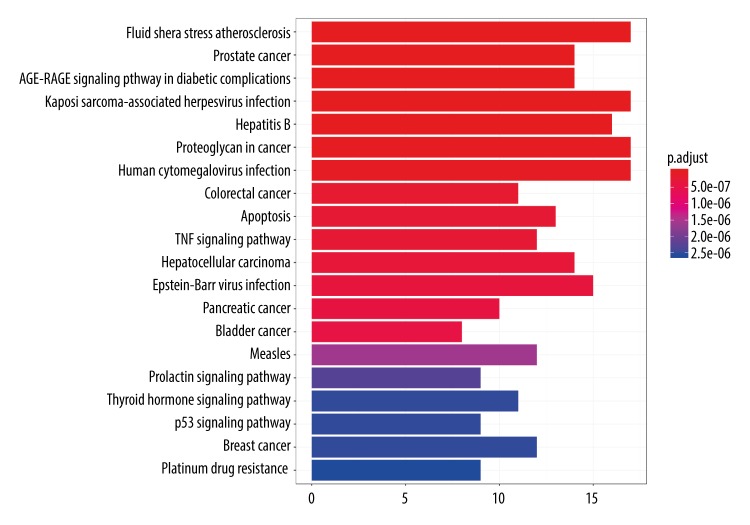
To elucidate the critical pathways among the 107 potential targets in terms of OP therapy, the top 20 pathways were filtered according to a P-adjusted value < 0.01 (Figure 7) and included pathways involved in fluid shear stress and atherosclerosis (hsa05418), Kaposi sarcoma-associated herpesvirus infection (hsa05167), proteoglycans in cancer (hsa05205), human cytomegalovirus infection (hsa05163), hepatitis B infection (hsa05161), Epstein-Barr virus infection (hsa05169), prostate cancer (hsa05215), AGE-RAGE signaling in diabetic complications (hsa04933), hepatocellular carcinoma (hsa05225), apoptosis (hsa04210), TNF signaling (hsa04668), measles infection (hsa05162), breast cancer, (hsa05224), colorectal cancer (hsa05210), thyroid hormone signaling (hsa04919), pancreatic cancer (hsa05212), p53 signaling (hsa04115), bladder cancer (hsa05219), prolactin signaling (hsa04917), and platinum drug resistance (hsa01524).
Figure 7.
Top 20 pathways from Kyoto Encyclopedia of Genes and Genomes (KEGG). (P-adjust value <0.01).
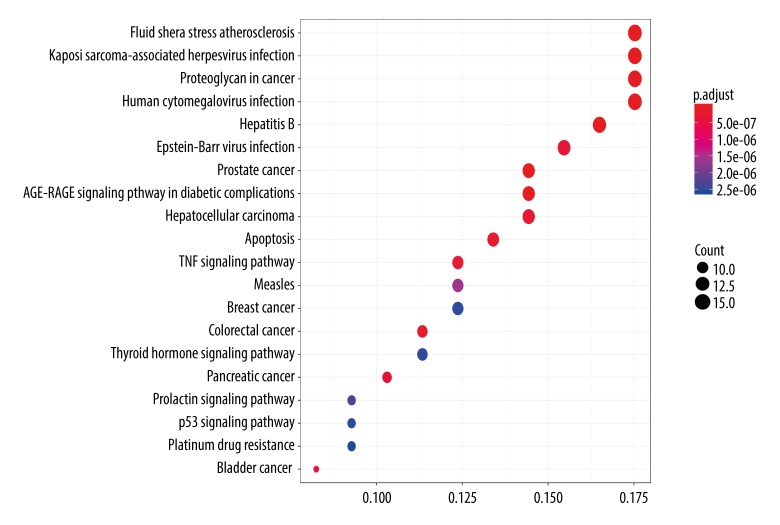
By applying an identical analytical method as that used for the GO analysis, a dot plot showing the relevant pathways was obtained that showed the same data as the GO dot plot (Figure 8).
Figure 8.
A dot plot to describe P-adjust value range of top 20 pathways.
Active ingredient-main target molecular docking
Because 107 potential targets were obtained, the top 5 targets (CCND1, EGFR, IL-6, MAPK8, and VEGFA), which had higher scores, were selected for molecular docking with 11 active ingredients of CS. The ranking of the binding free energy of the 11 small molecule compounds was achieved, as shown in Table 2. This table indicates that the active ingredient sesamin bound to all 5 proteins better than the other ingredients.
Table 2.
Binding free energy of 11 small molecules.
| Affinity (kcal/mol) | CCND1 | EGFR | IL6 | MAPK8 | VEGFA |
|---|---|---|---|---|---|
| Sesamin | −7.1 | −9.3 | −7.3 | −8.7 | −6.9 |
| Isorhamnetin | −6.6 | −8.4 | −6.2 | −7.6 | −6.0 |
| CLR | −6.2 | −8.2 | −6.9 | −8.0 | −5.6 |
| Sophranol | −6.6 | −8 | −5.7 | −7.9 | −5.4 |
| Matrine | −6.5 | −7.8 | −6.1 | −7.5 | −5.4 |
| Kaempferol | −6.7 | −8.2 | −6.2 | −7.6 | −6.2 |
| Isofucosterol | −6.3 | −7.4 | −6.6 | −8.8 | −6.0 |
| Campest-5-en-3beta-ol | −6.5 | −7.8 | −6.2 | −8.3 | −5.6 |
| NSC63551 | −6.6 | −8.0 | −6.4 | −7.9 | −6.3 |
| Quercetin | −7.1 | −8.0 | −6.0 | −7.0 | −6.9 |
| beta-Sitosterol | −6.2 | −7.1 | −6.2 | −8.1 | −5.8 |
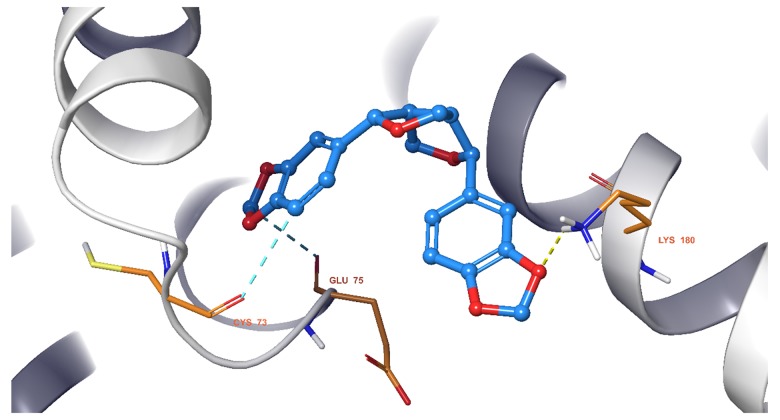
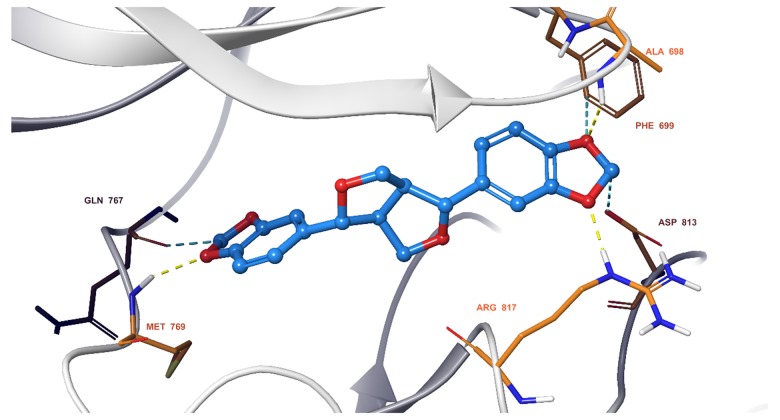
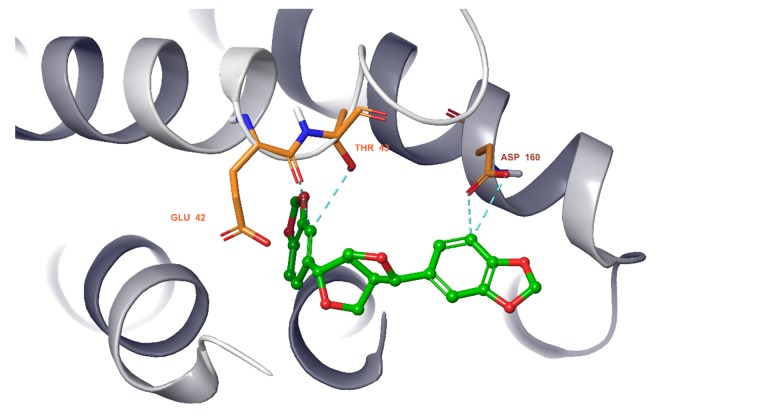
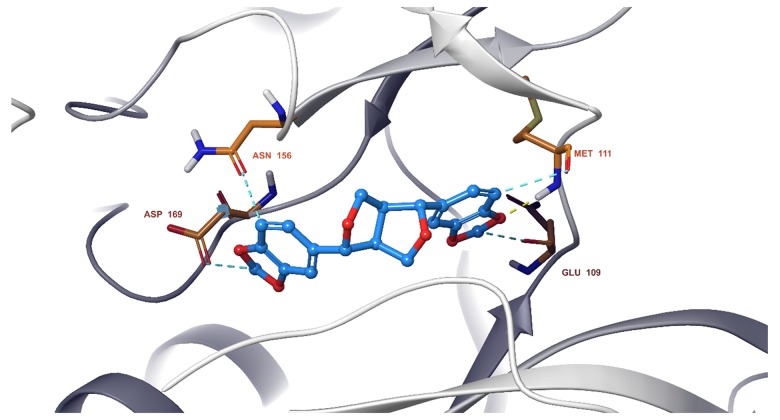
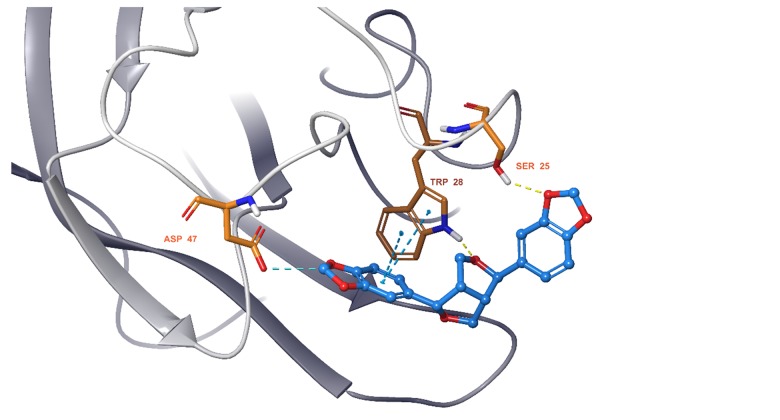
We observed that all 11 ingredients entered the active pocket of the enzyme. For example, the analysis of sesamin indicated that sesamin bound to the active pocket of CCND1 and interacted with the H-bond of the amino group on LYS180 and weakly interacted via an aromatic hydrogen bond with CYS73 and GLU75 (Figure 9). In Figure 10, sesamin bound to the active pockets of EGFR and interacted with MET769, ALA698, and ARG817 to form hydrogen bonds and with GLN767, ASP813, and PHE699 to form weakly aromatic hydrogen bonds. Sesamin combined with the active pocket of IL-6 and interacted with and formed weakly aromatic hydrogen bonds with GLU42, THR43, and ASP 160, as shown in Figure 11. Additionally, sesamin was linked to the active pocket of MAPK8 and interacted with MET111 to form hydrogen bonds. It also interacted with ASN156, ASP169, GLU109, and MET109 to form weakly aromatic hydrogen bonds (Figure 12). Finally, sesamin bound to the active pocket of VEGFA and interacted with SER25 and TRP28 to form hydrogen bonds. The aromatic ring of RP28 interacted equally with the Pi-Pi bond and weakly with ASP47 to form aromatic hydrogen bonds (Figure 13).
Figure 9.
Sesamin bound to the active pocket of cyclin D1 (CCND1).
Figure 10.
Sesamin bound to the active pocket of epidermal growth factor receptor (EGFR).
Figure 11.
Sesamin bound to the active pocket of interleukin-6 (IL6).
Figure 12.
Sesamin bound to the active pocket of mitogen-activated protein kinase 8 (MAPK8).
Figure 13.
Sesamin bound to the active pocket of vascular endothelial growth factor A (VEGFA).
Discussion
OP is characterized by low bone mass and a higher risk of fracture due to bone fragility. CS is a kind of bioactive ingredient extracted from plants. In recent years, it has been shown to play a vital role in bone metabolism through antioxidative and antiapoptotic effects [23]. Because of the variety of components in CS, the mechanism underlying its influence on OP is still unclear. Therefore, this study used a network pharmacology method to uncover the relevant relationships from multiple angles.
For example, certain studies on the relationship between sesamin, quercetin, and kaempferol and OP have been reported. Sesamin, also known as flax and flax oil, originated in the western regions of ancient China and belongs to the flax seed family [24]. It is mainly distributed in tropics worldwide. A previous study reported the protective effect of sesame oil against bone loss in an ovariectomized (OVX) rat model [25]. Orawan et al. indicated that after treatment with sesamin of human fetal osteoblasts and human adipose-derived stem cells, osteoblast differentiation could be activated through the p38 and ERK/MAPK pathways [26]. Moreover, Ma et al. reported that sesamin had the ability to promote the osteoblastic differentiation of BMSCs by regulating the Wnt/β-catenin pathway [27]. Tsuji et al. verified the inhibitory effect of quercetin on bone loss in an ovariectomized mouse model [28]. In vivo experiments also indicated that quercetin could improve BMSC activity and osteogenic differentiation ability [29]. A study by Adhikary et al. showed that supplementation with kaempferol could increase the speed of the healing of fractures caused by glucocorticoids and minimize bone loss in rats [30].
In this study, 107 common targets were analyzed via GO and KEGG enrichment analyses, and certain biological functions and signal pathways that had relationships with the ingredients and OP were determined. Based on the results, we speculated that the main biological functions involved DNA binding and protein or cofactor binding. Through these fundamental processes, CS may act on the relevant targets and thus affect the associated signaling pathways to exert drug effects. These pathways mainly play a role in certain cancers, TNF signaling pathways and stress signaling pathways. In a previous study, fluid shear stress (FSS) played a vital role in facilitating the proliferation and differentiation of osteoblasts [31] and reducing apoptosis [32]. As indicated by the bioinformatic findings of this study, the top 5 potential targets that were selected all had connections with OP.
MAPK8 performs the specific phosphorylation of the transcription factor c-Jun in the nucleus and is the kinase of c-Jun. Thus, MAPK8 is also called c-Jun amino-terminal kinase (JNK) [33]. JNK is one of most critical pathways related to osteoclastogenesis [34]. Some studies have confirmed this finding. Lee et al. used methylglyoxal to treat RAW264.7 macrophages and discovered that JNK was the most likely factor involved in activating osteoclasts [35]. Additionally, in another study, after treatment with a JNK inhibitor, researchers found that both the expression of mature osteoblast markers and mineralization of osteoblasts declined; however, upon the overexpression of JNK, osteoblast differentiation was enhanced [36]. Cyclin D1 (CCND1) plays an important role in regulating cell proliferation. A study indicated that chlorogenic acid exerted positive effects on BMSCs by activating cyclin D1 [37]. Other studies have also shown that enhanced cyclin D1 levels are associated with bone anabolism and anti-apoptosis [38–40]. Sato et al. focused on glucocorticoid-induced OP and demonstrated that when cyclin D1 was downregulated, bone formation was inhibited [41]. Interleukin (IL)-6 is a potent bone resorption factor that induces osteocyte differentiation and promotes osteoclast formation [42,43]. A previous study showed that IL-6 was associated with increased activity of osteoclasts during postmenopausal OP [44]. In addition, animal experiments also revealed that knockout of the mouse IL-6 gene could prevent bone loss after ovariectomy (OVX) [45]. Therefore, IL-6 plays an important role in the pathogenesis of OP [46]. Vascular endothelial growth factor A (VEGFA) is a homologue of the VEGF family. It has been confirmed that there are functional VEGF receptors in primary osteoblasts, which allow VEGF to play a role in promoting osteoblast proliferation and bone remodeling [47]. A recent study also indicated that in mesenchymal stem cells (MSCs), VEGFA overexpression could enhance cell vitality and proliferation, and the expression levels of type I and type II collagen were evidently upregulated [48]. Meanwhile, a study by Min et al. showed that VEGFA also functions in osteoclast differentiation [49]. Epidermal growth factor receptor (EGFR) is the receptor of EGF, which affects osteoprogenitor maintenance and new bone formation [50]. At the same time, a study by Liu et al. revealed an interesting phenomenon in which the expression of p-EGFR on the endosteal surface of cortical bone was decreased in 15-month-old mice compared with that in 3-month-old mice [51]. This finding indicated that EGFR had a relationship with age in bone metabolism. Moreover, knockdown of EGFR in osteoblastic cells led to bone loss due to a decreased number of bone marrow mesenchymal progenitors [52]. In addition, it has also been reported that the mechanism of the regulation by EGFR of bone development involves the negative regulation of mTOR signaling during the process of osteoblastic differentiation [53]. As Table 2 indicates, the lower the docking score, the higher the affinity of the docking molecule and the target was. Therefore, according to all the docking results, sesamin bound well to all 5 proteins. At the same time, based on the docking structure, we presume that the binding can be further improved by increasing the number of hydrogen bond interactions to enhance activity, which may allow sesamin to become a crucial agent for treating OP.
Conclusions
By using a novel analysis approach, we tested the hypothesis that the Chinese herbal medicine, Cuscutae semen (CS), had a positive influence on the treatment and prevention of OP. Accordingly, this study further revealed the CS pharmacodynamic basis and mechanism of action involved in the treatment of OP at the systemic level. The most active ingredient in CS that produces its effect was predicted.
However, because this study depended on database and statistical code analysis to make predictions about the effectiveness of drugs, some limitations should be considered. Hence, our future research will focus on experimental studies to verify these hypotheses.
Supplementary Data
Potential targets and corresponding symbols of Cuscutae semen (CS) active components.
| Mol Id | Mol name | Target | Symbol |
|---|---|---|---|
| MOL001558 | Sesamin | G1/S-specific cyclin-D1 | CCND1 |
| MOL001558 | Sesamin | Fatty acid synthase | FASN |
| MOL001558 | Sesamin | Acetyl-CoA carboxylase 1 | ACACA |
| MOL001558 | Sesamin | Nitric oxide synthase, endothelial | NOS3 |
| MOL001558 | Sesamin | Endothelin-converting enzyme 1 | ECE1 |
| MOL001558 | Sesamin | Cytochrome P450 2B6 | CYP2B6 |
| MOL001558 | Sesamin | Sterol regulatory element-binding protein 1 | SREBF1 |
| MOL001558 | Sesamin | NADPH oxidase 1 | NOX1 |
| MOL001558 | Sesamin | Peroxisomal acyl-coenzyme A oxidase 1 | ACOX1 |
| MOL001558 | Sesamin | Peroxisomal bifunctional enzyme | EHHADH |
| MOL001558 | Sesamin | Trifunctional enzyme subunit beta, mitochondrial | HADHB |
| MOL001558 | Sesamin | 2,4-dienoyl-CoA reductase, mitochondrial | DECR1 |
| MOL000184 | NSC63551 | Progesterone receptor | PGR |
| MOL000354 | Isorhamnetin | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000354 | Isorhamnetin | Estrogen receptor | ESR1 |
| MOL000354 | Isorhamnetin | Androgen receptor | AR |
| MOL000354 | Isorhamnetin | Peroxisome proliferator activated receptor gamma | PPARG |
| MOL000354 | Isorhamnetin | Estrogen receptor beta | ESR2 |
| MOL000354 | Isorhamnetin | Glycogen synthase kinase-3 beta | GSK3B |
| MOL000354 | Isorhamnetin | Trypsin-1 | PRSS1 |
| MOL000354 | Isorhamnetin | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000354 | Isorhamnetin | Serine/threonine-protein kinase Chk1 | CHEK1 |
| MOL000354 | Isorhamnetin | Aldose reductase | AKR1B1 |
| MOL000354 | Isorhamnetin | Nuclear receptor coactivator 1 | NCOA1 |
| MOL000354 | Isorhamnetin | Coagulation factor VII | F7 |
| MOL000354 | Isorhamnetin | Acetylcholinesterase | ACHE |
| MOL000354 | Isorhamnetin | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000354 | Isorhamnetin | Glutamate receptor 2 | GRIA2 |
| MOL000354 | Isorhamnetin | Transcription factor p65 | RELA |
| MOL000354 | Isorhamnetin | Oxidized low-density lipoprotein receptor 1 | OLR1 |
| MOL000358 | beta-Sitosterol | Progesterone receptor | PGR |
| MOL000358 | beta-Sitosterol | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000358 | beta-Sitosterol | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M3 | CHRM3 |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M1 | CHRM1 |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M4 | CHRM4 |
| MOL000358 | beta-Sitosterol | Alpha-1A adrenergic receptor | ADRA1A |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M2 | CHRM2 |
| MOL000358 | beta-Sitosterol | Neuronal acetylcholine receptor subunit alpha-2 | CHRNA2 |
| MOL000358 | beta-Sitosterol | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000358 | beta-Sitosterol | Apoptosis regulator Bcl-2 | BCL2 |
| MOL000358 | beta-Sitosterol | Caspase-9 | CASP9 |
| MOL000358 | beta-Sitosterol | Caspase-3 | CASP3 |
| MOL000358 | beta-Sitosterol | Caspase-8 | CASP8 |
| MOL000358 | beta-Sitosterol | Protein kinase C alpha type | PRKCA |
| MOL000358 | beta-Sitosterol | Serum paraoxonase/arylesterase 1 | PON1 |
| MOL000422 | Kaempferol | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000422 | Kaempferol | Androgen receptor | AR |
| MOL000422 | Kaempferol | Peroxisome proliferator activated receptor gamma | PPARG |
| MOL000422 | Kaempferol | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000422 | Kaempferol | Trypsin-1 | PRSS1 |
| MOL000422 | Kaempferol | Progesterone receptor | PGR |
| MOL000422 | Kaempferol | Muscarinic acetylcholine receptor M1 | CHRM1 |
| MOL000422 | Kaempferol | Acetylcholinesterase | ACHE |
| MOL000422 | Kaempferol | Muscarinic acetylcholine receptor M2 | CHRM2 |
| MOL000422 | Kaempferol | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000422 | Kaempferol | Coagulation factor VII | F7 |
| MOL000422 | Kaempferol | Transcription factor p65 | RELA |
| MOL000422 | Kaempferol | Inhibitor of nuclear factor kappa-B kinase subunit beta | IKBKB |
| MOL000422 | Kaempferol | Apoptosis regulator Bcl-2 | BCL2 |
| MOL000422 | Kaempferol | Activator of 90 kDa heat shock protein ATPase homolog 1 | AHSA1 |
| MOL000422 | Kaempferol | Caspase-3 | CASP3 |
| MOL000422 | Kaempferol | Mitogen-activated protein kinase 8 | MAPK8 |
| MOL000422 | Kaempferol | Peroxisome proliferator-activated receptor gamma | PPARG |
| MOL000422 | Kaempferol | Cytochrome P450 3A4 | CYP3A4 |
| MOL000422 | Kaempferol | Cytochrome P450 1A1 | CYP1A1 |
| MOL000422 | Kaempferol | Intercellular adhesion molecule 1 | ICAM1 |
| MOL000422 | Kaempferol | E-selectin | SELE |
| MOL000422 | Kaempferol | Vascular cell adhesion protein 1 | VCAM1 |
| MOL000422 | Kaempferol | Cytochrome P450 1B1 | CYP1B1 |
| MOL000422 | Kaempferol | Arachidonate 5-lipoxygenase | ALOX5 |
| MOL000422 | Kaempferol | Glutathione S-transferase P | GSTP1 |
| MOL000422 | Kaempferol | Aryl hydrocarbon receptor | AHR |
| MOL000422 | Kaempferol | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 |
| MOL000422 | Kaempferol | Solute carrier family 2, facilitated glucose transporter member 4 | SLC2A4 |
| MOL000422 | Kaempferol | Nuclear receptor subfamily 1 group I member 3 | NR1I3 |
| MOL000422 | Kaempferol | Type I iodothyronine deiodinase | DIO1 |
| MOL000422 | Kaempferol | Glutathione S-transferase Mu 1 | GSTM1 |
| MOL000422 | Kaempferol | Glutathione S-transferase Mu 2 | GSTM2 |
| MOL000422 | Kaempferol | Aldo-keto reductase family 1 member C3 | AKR1C3 |
| MOL005043 | Campest-5-en-3beta-ol | Progesterone receptor | PGR |
| MOL005440 | Isofucosterol | Progesterone receptor | PGR |
| MOL005440 | Isofucosterol | Mineralocorticoid receptor | NR3C2 |
| MOL005440 | Isofucosterol | 4-aminobutyrate aminotransferase, mitochondrial | ABAT |
| MOL005440 | Isofucosterol | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL005440 | Isofucosterol | Alcohol dehydrogenase 1B | ADH1B |
| MOL005440 | Isofucosterol | Nuclear receptor coactivator 2 | NCOA2 |
| MOL005944 | Matrine | Transcription factor p65 | RELA |
| MOL005944 | Matrine | Interleukin-6 | IL6 |
| MOL005944 | matrine | Caspase-3 | CASP3 |
| MOL005944 | Matrine | Myc proto-oncogene protein | MYC |
| MOL005944 | Matrine | Intercellular adhesion molecule 1 | ICAM1 |
| MOL005944 | Matrine | Heparanase | HPSE |
| MOL005944 | Matrine | Immediate early response 3-interacting protein 1 | IER3IP1 |
| MOL005944 | Matrine | CD44 antigen | CD44 |
| MOL000953 | CLR | Progesterone receptor | PGR |
| MOL000953 | CLR | Mineralocorticoid receptor | NR3C2 |
| MOL000953 | CLR | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000098 | Quercetin | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000098 | Quercetin | Androgen receptor | AR |
| MOL000098 | Quercetin | Peroxisome proliferator activated receptor gamma | PPARG |
| MOL000098 | Quercetin | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000098 | Quercetin | Aldose reductase | AKR1B1 |
| MOL000098 | Quercetin | Trypsin-1 | PRSS1 |
| MOL000098 | Quercetin | Coagulation factor VII | F7 |
| MOL000098 | Quercetin | Acetylcholinesterase | ACHE |
| MOL000098 | Quercetin | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000098 | Quercetin | Transcription factor p65 | RELA |
| MOL000098 | Quercetin | Epidermal growth factor receptor | EGFR |
| MOL000098 | Quercetin | Vascular endothelial growth factor A | VEGFA |
| MOL000098 | Quercetin | G1/S-specific cyclin-D1 | CCND1 |
| MOL000098 | Quercetin | Apoptosis regulator Bcl-2 | BCL2 |
| MOL000098 | Quercetin | Proto-oncogene c-Fos | FOS |
| MOL000098 | Quercetin | Eukaryotic translation initiation factor 6 | EIF6 |
| MOL000098 | Quercetin | Caspase-9 | CASP9 |
| MOL000098 | Quercetin | Urokinase-type plasminogen activator | PLAU |
| MOL000098 | Quercetin | Retinoblastoma-associated protein | RB1 |
| MOL000098 | Quercetin | Interleukin-6 | IL6 |
| MOL000098 | Quercetin | Activator of 90 kDa heat shock protein ATPase homolog 1 | AHSA1 |
| MOL000098 | Quercetin | Caspase-3 | CASP3 |
| MOL000098 | Quercetin | Cellular tumor antigen p53 | TP63 |
| MOL000098 | Quercetin | ETS domain-containing protein Elk-1 | ELK1 |
| MOL000098 | Quercetin | NF-kappa-B inhibitor alpha | NFKBIA |
| MOL000098 | Quercetin | NADPH--cytochrome P450 reductase | POR |
| MOL000098 | Quercetin | Caspase-8 | CASP8 |
| MOL000098 | Quercetin | RAF proto-oncogene serine/threonine-protein kinase | RAF1 |
| MOL000098 | Quercetin | Protein kinase C alpha type | PRKCA |
| MOL000098 | Quercetin | Hypoxia-inducible factor 1-alpha | HIF1A |
| MOL000098 | Quercetin | Protein CBFA2T1 | RUNX1T1 |
| MOL000098 | Quercetin | Receptor tyrosine-protein kinase erbB-2 | ERBB2 |
| MOL000098 | Quercetin | Peroxisome proliferator-activated receptor gamma | PPARG |
| MOL000098 | Quercetin | Acetyl-CoA carboxylase 1 | ACACA |
| MOL000098 | Quercetin | Cytochrome P450 3A4 | CYP3A4 |
| MOL000098 | Quercetin | Caveolin-1 | CAV1 |
| MOL000098 | Quercetin | Myc proto-oncogene protein | MYC |
| MOL000098 | Quercetin | Cytochrome P450 1A1 | CYP1A1 |
| MOL000098 | Quercetin | Intercellular adhesion molecule 1 | ICAM1 |
| MOL000098 | Quercetin | E-selectin | SELE |
| MOL000098 | Quercetin | Vascular cell adhesion protein 1 | VCAM1 |
| MOL000098 | Quercetin | Prostaglandin E2 receptor EP3 subtype | PTGER3 |
| MOL000098 | Quercetin | Baculoviral IAP repeat-containing protein 5 | BIRC5 |
| MOL000098 | Quercetin | Dual oxidase 2 | DUOX2 |
| MOL000098 | Quercetin | Nitric oxide synthase, endothelial | NOS3 |
| MOL000098 | Quercetin | Heat shock protein beta-1 | HSPB1 |
| MOL000098 | Quercetin | Maltase-glucoamylase, intestinal | MGAM |
| MOL000098 | Quercetin | Cytochrome P450 1B1 | CYP1B1 |
| MOL000098 | Quercetin | G2/mitotic-specific cyclin-B1 | CCNB1 |
| MOL000098 | Quercetin | Arachidonate 5-lipoxygenase | ALOX5 |
| MOL000098 | Quercetin | Glutathione S-transferase P | GSTP1 |
| MOL000098 | Quercetin | Nuclear factor erythroid 2-related factor 2 | NFE2L2 |
| MOL000098 | Quercetin | NAD(P)H dehydrogenase [quinone] 1 | NQO1 |
| MOL000098 | Quercetin | Poly [ADP-ribose] polymerase 1 | PARP1 |
| MOL000098 | Quercetin | Aryl hydrocarbon receptor | AHR |
| MOL000098 | Quercetin | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 |
| MOL000098 | Quercetin | Solute carrier family 2, facilitated glucose transporter member 4 | SLC2A4 |
| MOL000098 | Quercetin | Collagen alpha-1(III) chain | COL3A1 |
| MOL000098 | Quercetin | DDB1- and CUL4-associated factor 5 | DCAF5 |
| MOL000098 | Quercetin | Nuclear receptor subfamily 1 group I member 3 | NR1I3 |
| MOL000098 | Quercetin | Serine/threonine-protein kinase Chk2 | CHEK2 |
| MOL000098 | Quercetin | Heat shock factor protein 1 | HSF1 |
| MOL000098 | Quercetin | C-reactive protein | CRP |
| MOL000098 | Quercetin | Runt-related transcription factor 2 | RUNX2 |
| MOL000098 | Quercetin | Ras association domain-containing protein 1 | RASSF1 |
| MOL000098 | Quercetin | Cathepsin D | CTSD |
| MOL000098 | Quercetin | Insulin-like growth factor-binding protein 3 | IGFBP3 |
| MOL000098 | Quercetin | Insulin-like growth factor II | IGF2 |
| MOL000098 | Quercetin | Interferon regulatory factor 1 | IRF1 |
| MOL000098 | Quercetin | Receptor tyrosine-protein kinase erbB-3 | ERBB3 |
| MOL000098 | Quercetin | Serum paraoxonase/arylesterase 1 | PON1 |
| MOL000098 | Quercetin | Type I iodothyronine deiodinase | DIO1 |
| MOL000098 | Quercetin | Puromycin-sensitive aminopeptidase | NPEPPS |
| MOL000098 | Quercetin | Hexokinase-2 | HK2 |
| MOL000098 | Quercetin | Ras GTPase-activating protein 1 | RASA1 |
| MOL000098 | Quercetin | Glutathione S-transferase Mu 1 | GSTM1 |
| MOL000098 | Quercetin | Glutathione S-transferase Mu 2 | GSTM2 |
107 common genes of ingredients and osteoporosis.
| CCND1 | NCOA2 |
| FASN | CHEK1 |
| ACACA | AKR1B1 |
| NOS3 | NCOA1 |
| ECE1 | F7 |
| CYP2B6 | ACHE |
| SREBF1 | GABRA1 |
| NOX1 | GRIA2 |
| ACOX1 | RELA |
| EHHADH | OLR1 |
| HADHB | CHRM3 |
| DECR1 | CHRM1 |
| PGR | CHRM4 |
| PTGS1 | ADRA1A |
| ESR1 | CHRM2 |
| AR | CHRNA2 |
| PPARG | BCL2 |
| ESR2 | CASP9 |
| GSK3B | CASP3 |
| PRSS1 | CASP8 |
| PRKCA | AKR1C3 |
| PON1 | NR3C2 |
| IKBKB | ABAT |
| AHSA1 | ADH1B |
| MAPK8 | IL6 |
| CYP3A4 | MYC |
| CYP1A1 | HPSE |
| ICAM1 | IER3IP1 |
| SELE | CD44 |
| VCAM1 | EGFR |
| CYP1B1 | VEGFA |
| ALOX5 | FOS |
| GSTP1 | EIF6 |
| AHR | PLAU |
| PSMD3 | RB1 |
| SLC2A4 | TP63 |
| NR1I3 | ELK1 |
| DIO1 | NFKBIA |
| GSTM1 | POR |
| GSTM2 | RAF1 |
| HIF1A | HSPB1 |
| RUNX1T1 | CCNB1 |
| ERBB2 | NFE2L2 |
| CAV1 | NQO1 |
| PTGER3 | PARP1 |
| BIRC5 | COL3A1 |
| DUOX2 | DCAF5 |
| CHEK2 | IGF2 |
| HSF1 | IRF1 |
| CRP | ERBB3 |
| RUNX2 | NPEPPS |
| RASSF1 | HK2 |
| CTSD | RASA1 |
| IGFBP3 |
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (No.81660054)
Conflicts of Interest
None.
Supplementary Table 2. Potential targets of osteoporosis.
Supplementary Table 2 available from the corresponding author on request.
References
- 1.Marwick C. Consensus panel considers osteoporosis. JAMA. 2000;283:2093–95. [PubMed] [Google Scholar]
- 2.Kanis JA. WHO technical report. University of Sheffield; UK: 2007. p. 66. [Google Scholar]
- 3.Si L, Winzenberg TM, Jiang Q, et al. Projection of osteoporosis-related fractures and costs in China: 2010–2050. Osteoporos Int. 2015;26:1929–37. doi: 10.1007/s00198-015-3093-2. [DOI] [PubMed] [Google Scholar]
- 4.Comas-Calonge A, Figueiredo R, Gay-Escoda C. Surgical treatment vs. conservative treatment in intravenous bisphosphonate-related osteonecrosis of the jaws. Systematic review. J Clin Exp Dent. 2017;9:e302–7. doi: 10.4317/jced.53504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei X, Xu A, Shen H, et al. Qianggu capsule for the treatment of primary osteoporosis: Evidence from a Chinese patent medicine. BMC Complement Altern Med. 2017;17:108. doi: 10.1186/s12906-017-1617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajimehdipoor H, Kondori BM, Amin GR, et al. Development of a validated HPLC method for the simultaneous determination of compounds in Cuscuta chinensis Lam. By ultra-violet detection. DARU J Fac Pharm. 2012;20:57. doi: 10.1186/2008-2231-20-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng HZ, Dong ZH, She J. Modern study of traditional Chinese medicine. Beijing Xue Yuan Press of the People’s Republic of China; Beijing, China: 1998. pp. 4110–20. [Google Scholar]
- 8.Yang S, Xu X, Xu H, et al. Purification, characterization and biological effect of reversing the kidney-yang deficiency of polysaccharides from semen cuscutae. Carbohydr Polym. 2017;175:249–56. doi: 10.1016/j.carbpol.2017.07.077. [DOI] [PubMed] [Google Scholar]
- 9.Lin MK, Yu YL, Chen KC, et al. Kaempferol from Semen cuscutae attenuates the immune function of dendritic cells. Immunobiology. 2011;216:1103–9. doi: 10.1016/j.imbio.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh TC, Lu X, Guo J, et al. Effects of herbal preparation Equiguard on hormone-responsive and hormone-refractory prostate carcinoma cells: mechanistic studies. Int J Oncol. 2002;20:681–89. [PubMed] [Google Scholar]
- 11.Yang HM, Shin HK, Kang YH, et al. Cuscuta chinensis extract promotes osteoblast differentiation and mineralization in human osteoblast-like MG-63 cells. J Med Food. 2009;12:85–92. doi: 10.1089/jmf.2007.0665. [DOI] [PubMed] [Google Scholar]
- 12.Du XM, Kohinata K, Kawasaki T, et al. Components of the ether-insoluble resin glycoside-like fraction from Cuscuta chinensis. Phytochemistry. 1998;48:843–50. doi: 10.1016/s0031-9422(97)00990-4. [DOI] [PubMed] [Google Scholar]
- 13.Bao X, Wang Z, Fang J, et al. Structural features of an immunostimulating and antioxidant acidic polysaccharide from the seeds of Cuscuta chinensis. Planta Med. 2002;68:237–43. doi: 10.1055/s-2002-23133. [DOI] [PubMed] [Google Scholar]
- 14.He XH, Yang WZ, Meng AH, et al. Two new lignan glycosides from the seeds of Cuscuta chinensis. J Asian Nat Prod Res. 2010;12:934–39. doi: 10.1080/10286020.2010.506434. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25:1110–11. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 16.Poornima P, Kumar JD, Zhao Q, et al. Network pharmacology of cancer: from understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacol Res. 2016;111:290–302. doi: 10.1016/j.phrs.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Xue R, Fang Z, Zhang M, et al. TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013;41:D1089–95. doi: 10.1093/nar/gks1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi XQ, Yue SJ, Tang YP, et al. A network pharmacology approach to investigate the blood enriching mechanism of Danggui buxue Decoction. J Ethnopharmacol. 2019;235:227–42. doi: 10.1016/j.jep.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–68. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanstrup J, Broeckling CD, Helmus R, et al. The metaRbolomics toolbox in bioconductor and beyond. Metabolites. 2019;9(10) doi: 10.3390/metabo9100200. pii: E200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose PW, Prlić A, Altunkaya A, et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017;45:D271–81. doi: 10.1093/nar/gkw1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnapee S, Li J, Yang X, et al. Cuscuta chinensis Lam.: A systematic review on ethnopharmacology, phytochemistry and pharmacology of an important traditional herbal medicine. J Ethnopharmacol. 2014;157:292. doi: 10.1016/j.jep.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 24.Kamal-Eldin A, Moazzami A, Washi S. Sesame seed lignans: potent physiological modulators and possible ingredients in functional foods & nutraceuticals. Recent Pat Food Nutr Agric. 2011;3:17–29. doi: 10.2174/2212798411103010017. [DOI] [PubMed] [Google Scholar]
- 25.Boulbaroud S, Mesfioui A, Arfaoui A, et al. Preventive effects of flaxseed and sesame oil on bone loss in ovariectomized rats. Pak J Biol Sci. 2008;11(13):1696–701. doi: 10.3923/pjbs.2008.1696.1701. [DOI] [PubMed] [Google Scholar]
- 26.Wanachewin O, Boonmaleerat K, Pothacharoen P, et al. Sesamin stimulates osteoblast differentiation through p38 and ERK1/2 MAPK signaling pathways. BMC Complement Altern Med. 2012;12:71. doi: 10.1186/1472-6882-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma ZP, Zhang ZF, Yang YF, et al. Sesamin promotes osteoblastic differentiation and protects rats from osteoporosis. Med Sci Monit. 2019;25:5312–20. doi: 10.12659/MSM.915529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuji M, Yamamoto H, Sato T, et al. Dietary quercetin inhibits bone loss without effect on the uterus in ovariectomized mice. J Bone Miner Metab. 2009;27:673–81. doi: 10.1007/s00774-009-0088-0. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z, Min J, Zhao Y, et al. Quercetin rescued TNF-alpha-induced impairments in bone marrow-derived mesenchymal stem cell osteogenesis and improved osteoporosis in rats. Am J Transl Res. 2018;10:4313–21. [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikary S, Choudhary D, Ahmad N, et al. Dietary flavonoid kaempferol inhibits glucocorticoid-induced bone loss by promoting osteoblast survival. Nutrition. 2018;53:64–76. doi: 10.1016/j.nut.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Arnsdorf EJ, Tummala P, Kwon RY, et al. Mechanically induced osteogenic differentiation – the role of RhoA, ROCKII and cytoskeletal dynamics. J Cell Sci. 2009;122:546–53. doi: 10.1242/jcs.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan SD, Kuijpers-Jagtman AM, Semeins CM, et al. Fluid shear stress inhibits TNFalpha-induced osteocyte apoptosis. J Dent Res. 2006;85:905–9. doi: 10.1177/154405910608501006. [DOI] [PubMed] [Google Scholar]
- 33.Khodayari A, Ghaderian SMH, Jafarian M, et al. Evaluation of PKM2 and MAPK8IP2 polymorphism in ameloblastic carcinoma: A retrospective quantitative study. Int J Mol Cell Med. 2012;1:203–9. [PMC free article] [PubMed] [Google Scholar]
- 34.Roy B. Biomolecular basis of the role of diabetes mellitus in osteoporosis and bone fractures. World J Diabetes. 2013;4:101–13. doi: 10.4239/wjd.v4.i4.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KM, Lee CY, Zhang G, et al. Methylglyoxal activates osteoclasts through JNK pathway leading to osteoporosis. Chem Biol Interact. 2019;308:147–54. doi: 10.1016/j.cbi.2019.05.026. [DOI] [PubMed] [Google Scholar]
- 36.Matsuguchi T, Chiba N, Bandow K, et al. JNK activity is essential for Atf4 expression and late-stage osteoblast differentiation. J Bone Miner Res. 2009;24:398–410. doi: 10.1359/jbmr.081107. [DOI] [PubMed] [Google Scholar]
- 37.Zhou RP, Lin SJ, Wan WB, et al. Chlorogenic acid prevents osteoporosis by Shp2/PI3K/Akt pathway in ovariectomized rats. PLoS One. 2016;11:e0166751. doi: 10.1371/journal.pone.0166751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhee Y, Allen MR, Condon K, et al. PTH receptor signaling in osteocytes governs periosteal bone formation and intracortical remodeling. J Bone Miner Res. 2011;26(5):1035–46. doi: 10.1002/jbmr.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer S, Han L, Bartell SM, et al. Sirtuin1 (Sirt1) promotes cortical bone formation by preventing β-catenin sequestration by FoxO transcription factors in osteoblast progenitors. J Biol Chem. 2014;289(35):24069–78. doi: 10.1074/jbc.M114.561803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou W, Yang S, Zhang T, et al. Hypoxia enhances glucocorticoid-induced apoptosis and cell cycle arrest via the PI3K/Akt signaling pathway in osteoblastic cells. J Bone Miner Metab. 2015;33(6):615–24. doi: 10.1007/s00774-014-0627-1. [DOI] [PubMed] [Google Scholar]
- 41.Sato AY, Cregor M, Delgado-Calle J, et al. Protection from glucocorticoid-induced osteoporosis by anti-catabolic signaling in the absence of Sost/Sclerostin. J Bone Miner Res. 2016;31(10):1791–802. doi: 10.1002/jbmr.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clowes JA, Riggs BL, Khosla S. The role of the immune system in the pathophysiology of osteoporosis. Immunol Res. 2005;208:207–27. doi: 10.1111/j.0105-2896.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- 43.Caetano-Lopes J, Canhão H, Fonseca JE. Osteoimmunology: The hidden immune regulation of bone. Autoimmun Rev. 2009;8:250–55. doi: 10.1016/j.autrev.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 44.Manolagas SC, Bellido T, Jilka RL. New insights into the cellular, biochemical, and molecular basis of postmenopausal and senile osteoporosis: Roles of IL-6 and gp130. Int J Immunopharmacol. 1995;17(2):109–16. doi: 10.1016/0192-0561(94)00089-7. [DOI] [PubMed] [Google Scholar]
- 45.Zhu S, He H, Gao C, et al. Ovariectomy induced bone loss in TNFα and IL6 knockout mice is regulated by different mechanisms. J Mol Endocrinol. 2018;60(3):185–98. doi: 10.1530/JME-17-0218. [DOI] [PubMed] [Google Scholar]
- 46.Karsenty G, Olson E. Bone and muscle endocrine functions: Unexpected paradigms of inter-organ communication. Cell. 2016;164(6):1248–56. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Street J, Lenehan B. Vascular endothelial growth factor regulates osteoblast survival – evidence for an autocrine feedback mechanism. J Orthop Surg Res. 2009;4:19. doi: 10.1186/1749-799X-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Q, Sun W-X, Zhang Z-F. High expression of VEGFA in MSCs promotes tendon-bone healing of rotator cuff tear via microRNA-205-5p. Eur Rev Med Pharmacol Sci. 2019;23:4081–88. doi: 10.26355/eurrev_201905_17909. [DOI] [PubMed] [Google Scholar]
- 49.Min JK, Kim YM, Kim Y-M, et al. Vascular endothelial growth factor up-regulates expression of receptor activator of NF-kappa B (RANK) in endothelial cells. Concomitant increase of angiogenic responses to RANK ligand. J Biol Chem. 2003;278:39548–57. doi: 10.1074/jbc.M300539200. [DOI] [PubMed] [Google Scholar]
- 50.Chandra A, Lan S, Zhu J, et al. Epidermal growth factor receptor (EGFR) signaling promotes proliferation and survival in osteoprogenitors by increasing early growth response 2 (EGR2) expression. J Biol Chem. 2013;288:20488–98. doi: 10.1074/jbc.M112.447250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu G, Xie Y, Su J, et al. The role of EGFR signaling in age-related osteoporosis in mice cortical bone. FASEB J. 2019;33(10):11137–47. doi: 10.1096/fj.201900436RR. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Tamasi J, Lu X, et al. Epidermal growth factor receptor plays an anabolic role in bone metabolism in vivo. J Bone Miner Res. 2011;26:1022–34. doi: 10.1002/jbmr.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linder M, Hecking M, Glitzner E, et al. EGFR controls bone development by negatively regulating mTOR-signaling during osteoblast differentiation. Cell Death Differ. 2018;25:1094–106. doi: 10.1038/s41418-017-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Potential targets and corresponding symbols of Cuscutae semen (CS) active components.
| Mol Id | Mol name | Target | Symbol |
|---|---|---|---|
| MOL001558 | Sesamin | G1/S-specific cyclin-D1 | CCND1 |
| MOL001558 | Sesamin | Fatty acid synthase | FASN |
| MOL001558 | Sesamin | Acetyl-CoA carboxylase 1 | ACACA |
| MOL001558 | Sesamin | Nitric oxide synthase, endothelial | NOS3 |
| MOL001558 | Sesamin | Endothelin-converting enzyme 1 | ECE1 |
| MOL001558 | Sesamin | Cytochrome P450 2B6 | CYP2B6 |
| MOL001558 | Sesamin | Sterol regulatory element-binding protein 1 | SREBF1 |
| MOL001558 | Sesamin | NADPH oxidase 1 | NOX1 |
| MOL001558 | Sesamin | Peroxisomal acyl-coenzyme A oxidase 1 | ACOX1 |
| MOL001558 | Sesamin | Peroxisomal bifunctional enzyme | EHHADH |
| MOL001558 | Sesamin | Trifunctional enzyme subunit beta, mitochondrial | HADHB |
| MOL001558 | Sesamin | 2,4-dienoyl-CoA reductase, mitochondrial | DECR1 |
| MOL000184 | NSC63551 | Progesterone receptor | PGR |
| MOL000354 | Isorhamnetin | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000354 | Isorhamnetin | Estrogen receptor | ESR1 |
| MOL000354 | Isorhamnetin | Androgen receptor | AR |
| MOL000354 | Isorhamnetin | Peroxisome proliferator activated receptor gamma | PPARG |
| MOL000354 | Isorhamnetin | Estrogen receptor beta | ESR2 |
| MOL000354 | Isorhamnetin | Glycogen synthase kinase-3 beta | GSK3B |
| MOL000354 | Isorhamnetin | Trypsin-1 | PRSS1 |
| MOL000354 | Isorhamnetin | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000354 | Isorhamnetin | Serine/threonine-protein kinase Chk1 | CHEK1 |
| MOL000354 | Isorhamnetin | Aldose reductase | AKR1B1 |
| MOL000354 | Isorhamnetin | Nuclear receptor coactivator 1 | NCOA1 |
| MOL000354 | Isorhamnetin | Coagulation factor VII | F7 |
| MOL000354 | Isorhamnetin | Acetylcholinesterase | ACHE |
| MOL000354 | Isorhamnetin | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000354 | Isorhamnetin | Glutamate receptor 2 | GRIA2 |
| MOL000354 | Isorhamnetin | Transcription factor p65 | RELA |
| MOL000354 | Isorhamnetin | Oxidized low-density lipoprotein receptor 1 | OLR1 |
| MOL000358 | beta-Sitosterol | Progesterone receptor | PGR |
| MOL000358 | beta-Sitosterol | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000358 | beta-Sitosterol | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M3 | CHRM3 |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M1 | CHRM1 |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M4 | CHRM4 |
| MOL000358 | beta-Sitosterol | Alpha-1A adrenergic receptor | ADRA1A |
| MOL000358 | beta-Sitosterol | Muscarinic acetylcholine receptor M2 | CHRM2 |
| MOL000358 | beta-Sitosterol | Neuronal acetylcholine receptor subunit alpha-2 | CHRNA2 |
| MOL000358 | beta-Sitosterol | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000358 | beta-Sitosterol | Apoptosis regulator Bcl-2 | BCL2 |
| MOL000358 | beta-Sitosterol | Caspase-9 | CASP9 |
| MOL000358 | beta-Sitosterol | Caspase-3 | CASP3 |
| MOL000358 | beta-Sitosterol | Caspase-8 | CASP8 |
| MOL000358 | beta-Sitosterol | Protein kinase C alpha type | PRKCA |
| MOL000358 | beta-Sitosterol | Serum paraoxonase/arylesterase 1 | PON1 |
| MOL000422 | Kaempferol | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000422 | Kaempferol | Androgen receptor | AR |
| MOL000422 | Kaempferol | Peroxisome proliferator activated receptor gamma | PPARG |
| MOL000422 | Kaempferol | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000422 | Kaempferol | Trypsin-1 | PRSS1 |
| MOL000422 | Kaempferol | Progesterone receptor | PGR |
| MOL000422 | Kaempferol | Muscarinic acetylcholine receptor M1 | CHRM1 |
| MOL000422 | Kaempferol | Acetylcholinesterase | ACHE |
| MOL000422 | Kaempferol | Muscarinic acetylcholine receptor M2 | CHRM2 |
| MOL000422 | Kaempferol | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000422 | Kaempferol | Coagulation factor VII | F7 |
| MOL000422 | Kaempferol | Transcription factor p65 | RELA |
| MOL000422 | Kaempferol | Inhibitor of nuclear factor kappa-B kinase subunit beta | IKBKB |
| MOL000422 | Kaempferol | Apoptosis regulator Bcl-2 | BCL2 |
| MOL000422 | Kaempferol | Activator of 90 kDa heat shock protein ATPase homolog 1 | AHSA1 |
| MOL000422 | Kaempferol | Caspase-3 | CASP3 |
| MOL000422 | Kaempferol | Mitogen-activated protein kinase 8 | MAPK8 |
| MOL000422 | Kaempferol | Peroxisome proliferator-activated receptor gamma | PPARG |
| MOL000422 | Kaempferol | Cytochrome P450 3A4 | CYP3A4 |
| MOL000422 | Kaempferol | Cytochrome P450 1A1 | CYP1A1 |
| MOL000422 | Kaempferol | Intercellular adhesion molecule 1 | ICAM1 |
| MOL000422 | Kaempferol | E-selectin | SELE |
| MOL000422 | Kaempferol | Vascular cell adhesion protein 1 | VCAM1 |
| MOL000422 | Kaempferol | Cytochrome P450 1B1 | CYP1B1 |
| MOL000422 | Kaempferol | Arachidonate 5-lipoxygenase | ALOX5 |
| MOL000422 | Kaempferol | Glutathione S-transferase P | GSTP1 |
| MOL000422 | Kaempferol | Aryl hydrocarbon receptor | AHR |
| MOL000422 | Kaempferol | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 |
| MOL000422 | Kaempferol | Solute carrier family 2, facilitated glucose transporter member 4 | SLC2A4 |
| MOL000422 | Kaempferol | Nuclear receptor subfamily 1 group I member 3 | NR1I3 |
| MOL000422 | Kaempferol | Type I iodothyronine deiodinase | DIO1 |
| MOL000422 | Kaempferol | Glutathione S-transferase Mu 1 | GSTM1 |
| MOL000422 | Kaempferol | Glutathione S-transferase Mu 2 | GSTM2 |
| MOL000422 | Kaempferol | Aldo-keto reductase family 1 member C3 | AKR1C3 |
| MOL005043 | Campest-5-en-3beta-ol | Progesterone receptor | PGR |
| MOL005440 | Isofucosterol | Progesterone receptor | PGR |
| MOL005440 | Isofucosterol | Mineralocorticoid receptor | NR3C2 |
| MOL005440 | Isofucosterol | 4-aminobutyrate aminotransferase, mitochondrial | ABAT |
| MOL005440 | Isofucosterol | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL005440 | Isofucosterol | Alcohol dehydrogenase 1B | ADH1B |
| MOL005440 | Isofucosterol | Nuclear receptor coactivator 2 | NCOA2 |
| MOL005944 | Matrine | Transcription factor p65 | RELA |
| MOL005944 | Matrine | Interleukin-6 | IL6 |
| MOL005944 | matrine | Caspase-3 | CASP3 |
| MOL005944 | Matrine | Myc proto-oncogene protein | MYC |
| MOL005944 | Matrine | Intercellular adhesion molecule 1 | ICAM1 |
| MOL005944 | Matrine | Heparanase | HPSE |
| MOL005944 | Matrine | Immediate early response 3-interacting protein 1 | IER3IP1 |
| MOL005944 | Matrine | CD44 antigen | CD44 |
| MOL000953 | CLR | Progesterone receptor | PGR |
| MOL000953 | CLR | Mineralocorticoid receptor | NR3C2 |
| MOL000953 | CLR | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000098 | Quercetin | Prostaglandin G/H synthase 1 | PTGS1 |
| MOL000098 | Quercetin | Androgen receptor | AR |
| MOL000098 | Quercetin | Peroxisome proliferator activated receptor gamma | PPARG |
| MOL000098 | Quercetin | Nuclear receptor coactivator 2 | NCOA2 |
| MOL000098 | Quercetin | Aldose reductase | AKR1B1 |
| MOL000098 | Quercetin | Trypsin-1 | PRSS1 |
| MOL000098 | Quercetin | Coagulation factor VII | F7 |
| MOL000098 | Quercetin | Acetylcholinesterase | ACHE |
| MOL000098 | Quercetin | Gamma-aminobutyric acid receptor subunit alpha-1 | GABRA1 |
| MOL000098 | Quercetin | Transcription factor p65 | RELA |
| MOL000098 | Quercetin | Epidermal growth factor receptor | EGFR |
| MOL000098 | Quercetin | Vascular endothelial growth factor A | VEGFA |
| MOL000098 | Quercetin | G1/S-specific cyclin-D1 | CCND1 |
| MOL000098 | Quercetin | Apoptosis regulator Bcl-2 | BCL2 |
| MOL000098 | Quercetin | Proto-oncogene c-Fos | FOS |
| MOL000098 | Quercetin | Eukaryotic translation initiation factor 6 | EIF6 |
| MOL000098 | Quercetin | Caspase-9 | CASP9 |
| MOL000098 | Quercetin | Urokinase-type plasminogen activator | PLAU |
| MOL000098 | Quercetin | Retinoblastoma-associated protein | RB1 |
| MOL000098 | Quercetin | Interleukin-6 | IL6 |
| MOL000098 | Quercetin | Activator of 90 kDa heat shock protein ATPase homolog 1 | AHSA1 |
| MOL000098 | Quercetin | Caspase-3 | CASP3 |
| MOL000098 | Quercetin | Cellular tumor antigen p53 | TP63 |
| MOL000098 | Quercetin | ETS domain-containing protein Elk-1 | ELK1 |
| MOL000098 | Quercetin | NF-kappa-B inhibitor alpha | NFKBIA |
| MOL000098 | Quercetin | NADPH--cytochrome P450 reductase | POR |
| MOL000098 | Quercetin | Caspase-8 | CASP8 |
| MOL000098 | Quercetin | RAF proto-oncogene serine/threonine-protein kinase | RAF1 |
| MOL000098 | Quercetin | Protein kinase C alpha type | PRKCA |
| MOL000098 | Quercetin | Hypoxia-inducible factor 1-alpha | HIF1A |
| MOL000098 | Quercetin | Protein CBFA2T1 | RUNX1T1 |
| MOL000098 | Quercetin | Receptor tyrosine-protein kinase erbB-2 | ERBB2 |
| MOL000098 | Quercetin | Peroxisome proliferator-activated receptor gamma | PPARG |
| MOL000098 | Quercetin | Acetyl-CoA carboxylase 1 | ACACA |
| MOL000098 | Quercetin | Cytochrome P450 3A4 | CYP3A4 |
| MOL000098 | Quercetin | Caveolin-1 | CAV1 |
| MOL000098 | Quercetin | Myc proto-oncogene protein | MYC |
| MOL000098 | Quercetin | Cytochrome P450 1A1 | CYP1A1 |
| MOL000098 | Quercetin | Intercellular adhesion molecule 1 | ICAM1 |
| MOL000098 | Quercetin | E-selectin | SELE |
| MOL000098 | Quercetin | Vascular cell adhesion protein 1 | VCAM1 |
| MOL000098 | Quercetin | Prostaglandin E2 receptor EP3 subtype | PTGER3 |
| MOL000098 | Quercetin | Baculoviral IAP repeat-containing protein 5 | BIRC5 |
| MOL000098 | Quercetin | Dual oxidase 2 | DUOX2 |
| MOL000098 | Quercetin | Nitric oxide synthase, endothelial | NOS3 |
| MOL000098 | Quercetin | Heat shock protein beta-1 | HSPB1 |
| MOL000098 | Quercetin | Maltase-glucoamylase, intestinal | MGAM |
| MOL000098 | Quercetin | Cytochrome P450 1B1 | CYP1B1 |
| MOL000098 | Quercetin | G2/mitotic-specific cyclin-B1 | CCNB1 |
| MOL000098 | Quercetin | Arachidonate 5-lipoxygenase | ALOX5 |
| MOL000098 | Quercetin | Glutathione S-transferase P | GSTP1 |
| MOL000098 | Quercetin | Nuclear factor erythroid 2-related factor 2 | NFE2L2 |
| MOL000098 | Quercetin | NAD(P)H dehydrogenase [quinone] 1 | NQO1 |
| MOL000098 | Quercetin | Poly [ADP-ribose] polymerase 1 | PARP1 |
| MOL000098 | Quercetin | Aryl hydrocarbon receptor | AHR |
| MOL000098 | Quercetin | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 |
| MOL000098 | Quercetin | Solute carrier family 2, facilitated glucose transporter member 4 | SLC2A4 |
| MOL000098 | Quercetin | Collagen alpha-1(III) chain | COL3A1 |
| MOL000098 | Quercetin | DDB1- and CUL4-associated factor 5 | DCAF5 |
| MOL000098 | Quercetin | Nuclear receptor subfamily 1 group I member 3 | NR1I3 |
| MOL000098 | Quercetin | Serine/threonine-protein kinase Chk2 | CHEK2 |
| MOL000098 | Quercetin | Heat shock factor protein 1 | HSF1 |
| MOL000098 | Quercetin | C-reactive protein | CRP |
| MOL000098 | Quercetin | Runt-related transcription factor 2 | RUNX2 |
| MOL000098 | Quercetin | Ras association domain-containing protein 1 | RASSF1 |
| MOL000098 | Quercetin | Cathepsin D | CTSD |
| MOL000098 | Quercetin | Insulin-like growth factor-binding protein 3 | IGFBP3 |
| MOL000098 | Quercetin | Insulin-like growth factor II | IGF2 |
| MOL000098 | Quercetin | Interferon regulatory factor 1 | IRF1 |
| MOL000098 | Quercetin | Receptor tyrosine-protein kinase erbB-3 | ERBB3 |
| MOL000098 | Quercetin | Serum paraoxonase/arylesterase 1 | PON1 |
| MOL000098 | Quercetin | Type I iodothyronine deiodinase | DIO1 |
| MOL000098 | Quercetin | Puromycin-sensitive aminopeptidase | NPEPPS |
| MOL000098 | Quercetin | Hexokinase-2 | HK2 |
| MOL000098 | Quercetin | Ras GTPase-activating protein 1 | RASA1 |
| MOL000098 | Quercetin | Glutathione S-transferase Mu 1 | GSTM1 |
| MOL000098 | Quercetin | Glutathione S-transferase Mu 2 | GSTM2 |
107 common genes of ingredients and osteoporosis.
| CCND1 | NCOA2 |
| FASN | CHEK1 |
| ACACA | AKR1B1 |
| NOS3 | NCOA1 |
| ECE1 | F7 |
| CYP2B6 | ACHE |
| SREBF1 | GABRA1 |
| NOX1 | GRIA2 |
| ACOX1 | RELA |
| EHHADH | OLR1 |
| HADHB | CHRM3 |
| DECR1 | CHRM1 |
| PGR | CHRM4 |
| PTGS1 | ADRA1A |
| ESR1 | CHRM2 |
| AR | CHRNA2 |
| PPARG | BCL2 |
| ESR2 | CASP9 |
| GSK3B | CASP3 |
| PRSS1 | CASP8 |
| PRKCA | AKR1C3 |
| PON1 | NR3C2 |
| IKBKB | ABAT |
| AHSA1 | ADH1B |
| MAPK8 | IL6 |
| CYP3A4 | MYC |
| CYP1A1 | HPSE |
| ICAM1 | IER3IP1 |
| SELE | CD44 |
| VCAM1 | EGFR |
| CYP1B1 | VEGFA |
| ALOX5 | FOS |
| GSTP1 | EIF6 |
| AHR | PLAU |
| PSMD3 | RB1 |
| SLC2A4 | TP63 |
| NR1I3 | ELK1 |
| DIO1 | NFKBIA |
| GSTM1 | POR |
| GSTM2 | RAF1 |
| HIF1A | HSPB1 |
| RUNX1T1 | CCNB1 |
| ERBB2 | NFE2L2 |
| CAV1 | NQO1 |
| PTGER3 | PARP1 |
| BIRC5 | COL3A1 |
| DUOX2 | DCAF5 |
| CHEK2 | IGF2 |
| HSF1 | IRF1 |
| CRP | ERBB3 |
| RUNX2 | NPEPPS |
| RASSF1 | HK2 |
| CTSD | RASA1 |
| IGFBP3 |