Abstract
Background
Circular RNAs (circRNAs) are key regulators that take part in the carcinogenesis and development of breast cancer. The current study aimed to identify the expression of and explored the function of circRNA-0001283 in breast cancer.
Material/Methods
Breast cancer tissue samples were tested using high-throughput sequencing to identify the levels of relative genes; and proteins were addressed by using quantitative real-time polymerase chain reaction (qRT-PCR) and western-blot. Cell ability and cell apoptosis were investigated by Cell Counting Kit-8 (CCK-8) and flow cytometry. Invasion was detected by Transwell invasion assay. The identification of target genes was analyzed by dual-luciferase reporter assay.
Result
Downregulation of circRNA-0001283 expression was observed in breast cancer tissue samples. Ectopic expression of circRNA-0001283 remarkably suppressed cell viability and invasion, and induced apoptosis in breast cancer cells. Furthermore, circRNA-0001283 bound to miR-187 and decreased the expression of miR-187, which resulted in inhibition in cell growth and invasion. Finally, we showed that circRNA-0001283 positively regulated HIPK3 expression by sponging miR-187.
Conclusions
The results reveal a new functional circRNA-0001283 in breast cancer and may provide targets for developing novel therapeutic strategies for breast cancer.
MeSH Keywords: Cell Proliferation; DNA, Circular; Neoplasm Invasiveness
Background
Breast cancer is a common malignant tumor [1]. Although the cure rate has increased, the mortality of this disease remains high in women with breast cancer [2]. Therefore, gaining more insights into the molecular mechanisms that promote breast cancer progression, and developing novel therapies, are still urgently needed.
Breast carcinogenesis is a complex process involving a number of modulators and pathways. Non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are important regulators implicated in the formation and development of breast cancer [3]. Circular RNAs (circRNAs) are non-coding RNAs discovered recently, which form a continuous cycle of covalent closures [4]. CircRNAs play critical functions in a number of biological processes [5]. Emerging evidence shows that circRNAs take part in various diseases, including cardiovascular disease, neurological disorders, diabetes mellitus, and osteoarthritis [6–9]. Recently, the critical role of circRNA in cancer has been also revealed [10]. Aberrant expression of circRNAs was related to prevention, prognosis, and drug resistance of cancers [11–13]. But, the working mechanism of circRNAs is not fully understood in breast cancer.
In the present study, high-throughput sequencing was utilized to investigate the expression profile of circRNA in breast cancer. A significantly downregulated circRNA, circRNA-0001283 was discovered. Overexpression of circRNA-0001283 suppressed cell proliferation and invasion, and enhanced apoptosis in breast cancer cells. Moreover, our data demonstrated that circRNA-0001283 functions via miR-187/HIPK3 axis. Our findings might represent novel targets for the treatment of breast cancer.
Material and Methods
Clinical specimens
Breast cancer samples were obtained between 2016 and 2017 from the First Affiliated Hospital of Gannan, Medical University. Informed consents from all patients were obtained.
Cell culture
MCF-7, MDA-MB-231, MDA-MB-468 MDA-MB-453, and MCF-10A was purchased from the American Type Culture Collection (ATCC, USA). Cells were incubated in Dulbecco’s Modified Eagle’s Medium (Invitrogen, USA) or RPMI-1640 medium (Gibco, USA) with addition of 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin and streptomycin. All cells were cultured and incubated in a 5% CO2 atmosphere at 37°C.
CircRNA analysis
Total RNA was isolated from 10 pairs of breast cancer tissues using TRIzol reagent (Invitrogen, USA). CircRNA was enriched and analyzed by high-throughput sequencing.
Quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA was purified from the tissue and cells by TRIzol reagent (Invitrogen, USA). RNA was retrieved into cDNA (complementary DNA) by PrimeScript RT reagent kit (Takara, Shiga, Japan) as the template of following amplification experiment by using the SYBR Premix ExTaqII (TliRNaseHPlus) kit (Takara). Samples were normalized to the internal control. The values were calculated using 2−ΔΔCT method.
Luciferase reporter gene assay
The circRNA target genes were predicted by starBase, wild type and mutant circRNA-0001283 (with mutated miR-187 binding sites) were constructed using luciferase reporter plasmids. HEK293T cells were co-transfected with plasmids and miR-187 mimics using Lipofectamine 2000. Similarly, wild type and mutant HIPK3 3′UTR (with mutated miR-187 binding sites) were constructed using dual luciferase reporter plasmids. HEK293T cells were co-transfected with plasmids and miR-187 mimics using Lipofectamine 2000. After incubated for 48 hours, luciferase activity was measured and analyzed by a multifunctional fluorescent enzyme marker (Tecan, Switzerland).
Cell proliferation and apoptosis analysis
Cell proliferation was detected by a Cell Counting Kit-8 (CCK-8) according to the manufacturer’s instructions (Beyotime, China). A FITC Annexin V apoptosis detection kit (BD Biosciences, USA) was used to detect the apoptosis: 1×106 cells were stained with Annexin V/propidium iodide (PI) at 4°C for 15 minutes. Cells were then washed by precooled phosphate-buffered saline (PBS) and suspended in buffer for the next analysis.
Cell invasion assay
Treated cells were cultured in a Transwell chamber with Matrigel-coated membrane (BD Biosciences, Bedford, MA, USA). After culture for 24 hours, cells were fixed with 4% paraformaldehyde. After that, cells were incubated with crystal violet. Five visual fields were selected for counting under the microscope.
Statistical analysis
Data were expressed as mean±standard deviation (SD). Significance was analyzed using SPSS 18.0. Student’s t-test and one-way ANOVA were used for analysis. Significance was considered as the P value <0.05. All experiments were conducted in 3 times.
Results
CircRNA-0001283 is downregulated in breast cancer tissues
To screen dysregulated circRNAs, microarray analysis was performed with 10 pairs of cancer and adjacent non-cancer tissues. A heat map showed the 23 most downregulated circRNAs (Figure 1A). RNA fluorescence in situ hybridization (FISH) assay revealed that high localization of circRNA-0001283 was observed in the cytoplasm (Figure 1B). Next, the expression of circRNA-0001283 in breast cancer tissues and cell lines were also detected. The expression of circRNA-0001283 was markedly lower in cancer tissues than in normal tissues (Figure 1C). Downregulation of circRNA-0001283 was also found in human breast cancer cells compared to normal epithelial breast cells MCF-10A (Figure 1D).
Figure 1.
CircRNA-0001283 is downregulated in breast cancer samples and cell lines. (A) A heatmap shows most 23 downregulated circRNAs in 3 paired samples of tumor issues (Tumor) and corresponding normal tissues (Normal) by microarray analysis. (B) The subcellular localization of circRNA-0001283 was determined by RNA fluorescence in situ hybridization. (C) Expression levels of circRNA-0001283 in 10 paired samples of breast cancer were determined by quantitative real-time polymerase chain reaction (qRT-PCR). (D) Expression levels of circRNA-0001283 in cell lines were examined by qRT-PCR. * P<0.05, ** P<0.01.
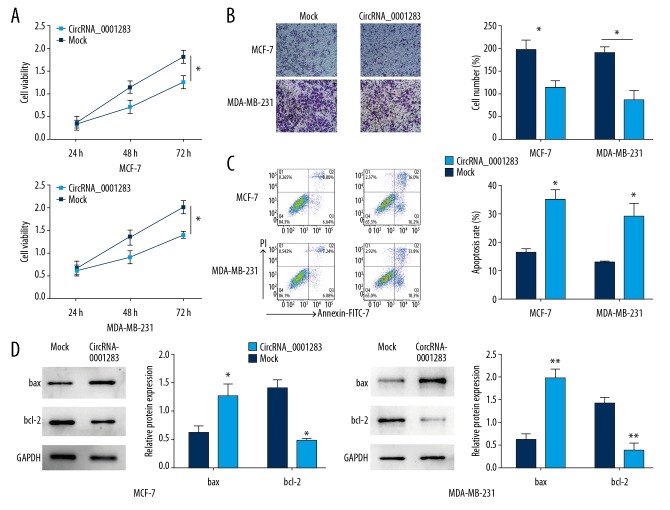
CircRNA-0001283 represses cell growth and invasion and enhances apoptosis
To further explored the effects of circRNA-0001283 on cancer cells, we carried out CCK-8 assay, and invasion and apoptosis analysis. Overexpression of circRNA-0001283 highly inhibited the growth of MCF7 and MDA-MB-231 cells (Figure 2A). Invasion assay revealed that ectopic expression of circRNA-0001283 remarkably inhibited the invasion abilities of MCF7 and MDA-MB-231 cells (Figure 2B). Moreover, overexpression of circRNA-0001283 markedly enhanced the apoptotic rate of breast cancer cells (Figure 2C). Consistent with these results, overexpression of circRNA-0001283 augmented the level of Bax and downregulated anti-apoptotic protein Bcl2 (Figure 2D). Together, these results suggest that circRNA-0001283 has a tumor suppressive function in breast cancer.
Figure 2.
CircRNA-0001283 represses cell growth and migration and enhances apoptosis in breast cancer cells. (A) The effect of overexpression of circRNA-0001283 on breast cancer cell proliferation was determined by Cell Counting Kit-8 assay. (B) The effect of overexpression of circRNA-0001283 on breast cancer cell invasion was determined by Transwell invasion assay. (C) The effect of overexpression of circRNA-0001283 on breast cancer cell apoptosis was accessed by flow cytometry. (D) Effects of overexpression of circRNA-0001283 on the expression of proteins related to apoptosis were detected. * P<0.05, ** P<0.01.
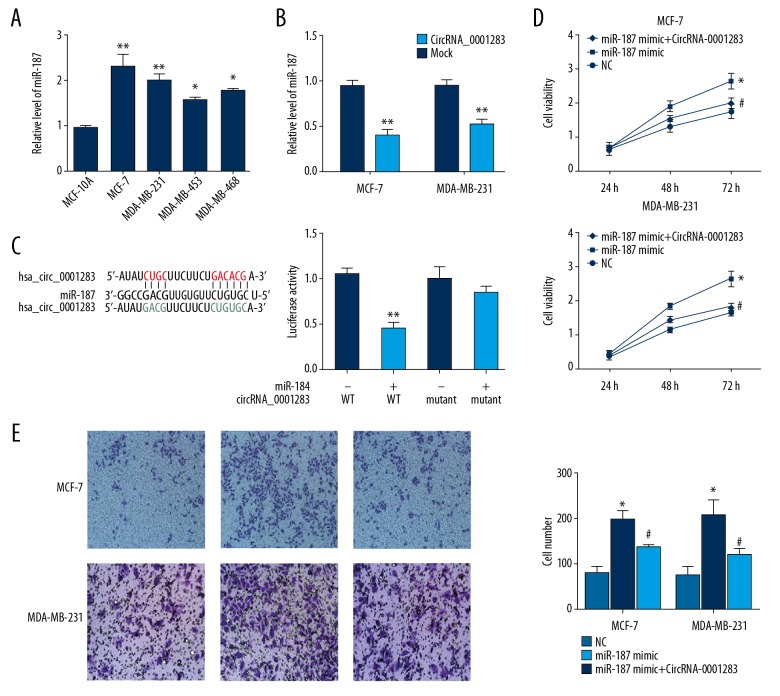
CircRNA-0001283 sponges miR-187
CircRNAs normally act as sponges to exert their function. Bioinformatics prediction suggested miR-187 to be a candidate target of circRNA-0001283. MiR-187 was shown to be upregulated in breast cancer cells compared with normal epithelial breast cells MCF-10A (Figure 3A). Overexpression of circRNA-0001283 markedly decreased the expression miR-187 (Figure 3B). To determine the direct interaction between circRNA-0001283 and miR-187, we performed luciferase reporter assay. The results showed that miR-187 remarkable inhibited the activity of wild type circRNA-0001283, however, it showed no obvious effect on the circRNA-0001283 mutant (Figure 3C). This result indicated that there is a direct interaction between circRNA-0001283 and miR-187. Moreover, ectopic expression of miR-187 remarkably increased the viability of MCF-7/MDA-MB-231 cells, and this effect of miR-187 was abolished by overexpression of circRNA-0001283 (Figure 3D). Similarly, transfection of circRNA-0001283 blocked the promotion of breast cancer cell migration induced by miR-187 (Figure 3E). Together, these results suggest that miR-187 contributes to circRNA-0001283-mediated inhibition in the growth and invasion of breast cancer cells.
Figure 3.
CircRNA-0001283 targets miR-187. (A) MiR-187 expression in breast cancer cells were determined by quantitative real-time polymerase chain reaction. (B) MiR-187 expression was decreased following circRNA-0001283 overexpression. (C) Luciferase reporter gene assay in MCF-7 cells overexpressing circRNA-0001283 (wild type/mutant) and miR-187 (NC/mimic). (D) Overexpression of circRNA-0001283 decreased breast cancer cell proliferation induced by miR-187. (E) Overexpression of circRNA-0001283 inhibited breast cancer cell invasion induced by miR-187. * P<0.05, ** P<0.01 versus NC. # P<0.05 versus miR-187 mimic.
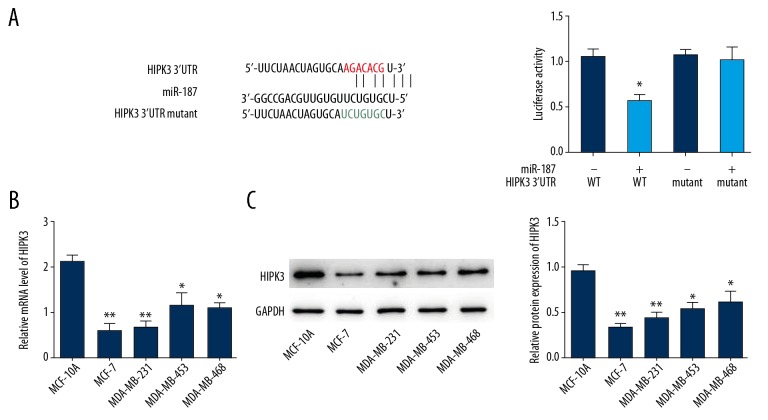
MiR-187 targets HIPK3
MiRNAs mediate their roles via regulating target genes. Targetscan and microRNA.org were used to explore the possible downstream targets of miR-187, and HIPK3 was predicted to be a potential target (Figure 4A). To confirm the direct association between miR-187 and HIPK3, we carried out the luciferase reporter gene assay. As expect, the luciferase activity of the wild type HIPK3 3′UTR was markedly reduced upon transfection with miR-187, while no notable changes shown in the HIPK3 3′UTR mutant (Figure 4A). Furthermore, a remarkable decrease of the mRNA and protein expression of HIPK3 were detected in breast cancer cells (Figure 4B, 4C). Then, we determined the influence of miR-187 on circRNA-001283-induced HIPK3 expression. MiR-187 mimics abolished the enhanced effect of circRNA-001283 on HIPK3 expression (Supplementary Figure 1B). Moreover, the functional activity of HIPK3 was detected in MCF-7 cells. The result showed that overexpression of HIPK3 significantly inhibited the migration of MCF-7 cells. Moreover, ectopic expression of HIPK3 reduced cell migration increased by miR-187 (Supplementary Figure 1A).
Figure 4.
HIPK3 is a target of miR-187. (A) Luciferase reporter assay in MCF-7 cells transfected with HIPK3 3′UTR (wild type/mutant) and miR-187 (NC/mimic). (B) HIPK3 mRNA level in breast cancer cells was detected by quantitative real-time polymerase chain reaction. (C) HIPK3 protein level in breast cancer cells was examined by western blot. * P<0.05, ** P<0.01.
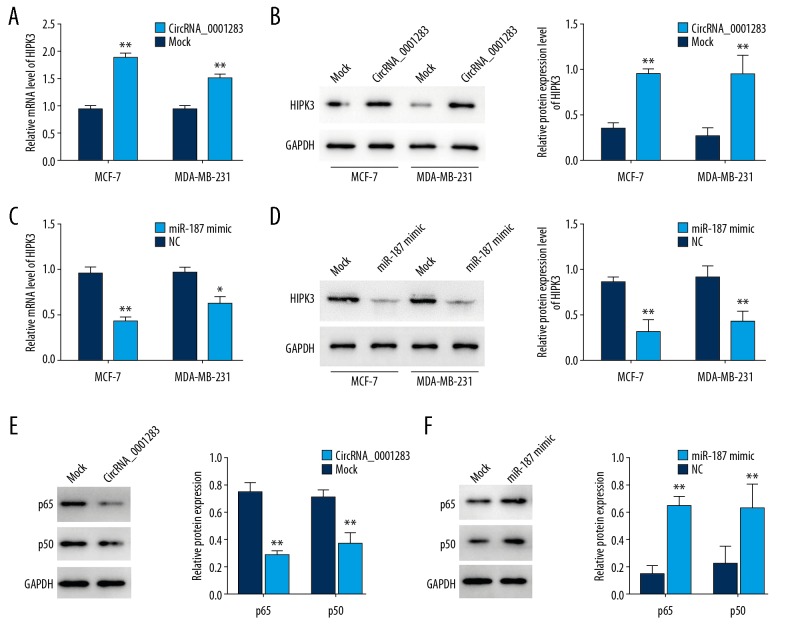
CircRNA-0001283 regulates NF-κB signaling via miR-187/HIPK3 axis
The mRNA and protein levels of HIPK3 were markedly enhanced in cells with circRNA-0001283 overexpression (Figure 5A, 5B). On the contrary, miR-187 mimic obviously inhibited HIPK3 expression (Figure 5C, 5D). Moreover, ectopic expression with circRNA-0001283 reduced the levels of p65 and p50 (Figure 5E), while transfection of miR-187 highly increased the levels of p65 and p50 (Figure 5F). Collectively, these results suggest that circRNA-0001283 may regulate NF-κB signaling via miR-187/HIPK3 cascade.
Figure 5.
CircRNA-0001283 regulates HIPK3 expression via miR-187. (A, B) Overexpression of circRNA-0001283 increased HIPK3 mRNA and protein levels. (C, D) Transfection of miR-187 decreased HIPK3 mRNA and protein levels. (E) Overexpression of circRNA-0001283 decreased the levels of p65 and p50. (F) Transfection of miR-187 increased the levels of p65 and p50.
Discussion
Exploration of circRNA expression prolife is an effective way for identifying novel regulators in breast cancer. Here, we showed that circRNA-0001283 was remarkably decreased in breast cancer tissues and cells. Our results demonstrated that circRNA-0001283 was related to cell growth, invasion, and apoptosis. CircRNA-0001283 exerted its function as a ceRNA (competing endogenous RNA) that bound to miR-187 and blocked the suppressive effect of miR-187 on HIPK3 expression.
Previous studies reported that circRNAs can act as either oncogenic or suppressive factors in breast cancer. For example, upregulated circ-UBE2D2 is associated with poor prognosis and enhances the progression of breast cancer [14]. Zhang et al. showed that in triple-negative breast cancer, circRNA_069718 promotes cell viability and invasion [15]. Yang et al. reported that circ_0103552 facilitates breast cancer cell proliferation and migration and decreases apoptotic cells [16]. CircRNA_0025202 inhibits colony formation and migration, and it promotes cell apoptosis in breast cancer [17]. CircTADA2As is demonstrated to be a negative regulator for breast cancer progression and metastasis [18]. Our data showed that circRNA-0001283 was decreased in the tissues of breast cancer. Overexpression of circRNA-0001283 inhibited cell proliferation and invasion and induced apoptosis, indicating a suppressive role of circRNA-0001283 in breast cancer.
Numbers of studies have revealed that circRNAs function in cancer via diverse mechanisms, such as acting as scaffolds of protein complexes, regulating protein subcellular localization, controlling gene expression, sequestering RNA-binding proteins, and functioning as competing endogenous RNA (ceRNA) [19,20]. MiRNAs are supposed to be important modulators in cancer [21,22]. Multiple circRNAs have been revealed to exert their function via sponging miRNAs [23–28]. It has been reported that miR-187 enhances cell growth and migration and represses the apoptosis of bladder cancer cells. High miR-187 expression is related to the advanced oral carcinoma [29]. In breast cancer, miR-187 is suggested to be a prognostic factor and facilitates aggressive invasion [30]. Here, we demonstrated that circRNA-0001283 suppresses breast cancer cell proliferation and invasion via regulating miR-187 expression [31].
In our further exploration of the mechanism of circRNA-0001283-mediated inhibition in breast cancer, we identified that HIPK3 was a downstream of circRNA-0001283. HIPK3 is a member of HIPK family [32]. The role of HIPK3 in cancer has been revealed previously. For instance, Curtin et al. reported that JNK modulates HIPK3 expression to promote resistance to Fas-mediated apoptosis in prostate cancer cells [33]. HIPK3 level is decreased in non-small cell lung cancer (NSCLC). Lower HIPK3 expression was associated with poorer survival in patients with NSCLC [34]. Here, our data showed that circRNA-0001283 increased the level of HIPK3 via downregulation of miR-187 expression.
Our data revealed that circRNA-0001283 was downregulated in breast cancer tissues. Overexpression of circRNA-0001283 repressed cell proliferation and invasion, and elevated apoptosis in breast cancer cells. Mechanically, we found that circRNA_069718 downregulated HIPK3 expression by sponging miR-187. These findings may support circRNA-0001283/miR-187/HIKP3 as potential targets for the treatment of breast cancer.
Conclusions
The results of our study revealed a new functional circRNA-0001283 in breast cancer progression and may provide novel targets for the treatment of breast cancer.
Supplementary Data
The influence of miR-187 on circRNA-001283-induced HIPK3 expression. (A) The effect of overexpression of miR-187 mimics on breast cancer cell invasion was determined by Transwell invasion assay. (B) MiR-187 mimics abolished the enhanced effect of circRNA-001283 on HIPK3 expression.
Abbreviation
- circRNA
circular RNA
- qRT-PCR
quantitative real-time reverse transcription PCR
- ceRNA
competing endogenous RNA
- NSCLC
non-small cell lung cancer
- miRNA
microRNA
- lncRNA
long non-coding RNA
Footnotes
Source of support: This research was supported by Jiangxi Youth Science Foundation (grant no. 2015BAB215021) and National Natural Science Foundation of China (grant no. 81601349)
Conflict of interest
None.
Availability of data and materials
Data used to support our findings in this study are available from the corresponding author upon request.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: Comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12:477–88. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagini S. Breast cancer: Current molecular therapeutic targets and new players. Anticancer Agents Med Chem. 2017;17:152–63. doi: 10.2174/1871520616666160502122724. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–41. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao J, Zhang T, Jiao X, et al. Hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J Cell Physiol. 2020;235(2):1287–95. doi: 10.1002/jcp.29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Shen B, Zeng Y. The therapeutic potential and role of miRNA, lncRNA, and circRNA in osteoarthritis. Curr Gene Ther. 2019;19(4):255–63. doi: 10.2174/1566523219666190716092203. [DOI] [PubMed] [Google Scholar]
- 7.Wu F, Han B, Wu S, et al. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. 2019;39(37):7369–93. doi: 10.1523/JNEUROSCI.0299-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16:503–14. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, She G, Zhou W, et al. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr J. 2019;66:431–41. doi: 10.1507/endocrj.EJ18-0291. [DOI] [PubMed] [Google Scholar]
- 10.Santer L, Bar C, Thum T. Circular RNAs: A novel class of functional RNA molecules with a therapeutic perspective. Mol Ther. 2019;27(8):P1350–63. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Deng X, Xiao G, et al. Circ_0001730 promotes proliferation and invasion via the miR-326/Wnt7B axis in glioma cells. Epigenomics. 2019;11:1335–52. doi: 10.2217/epi-2019-0121. [DOI] [PubMed] [Google Scholar]
- 12.Shang J, Chen WM, Wang ZH, et al. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp Hematol. 2019;70:42–54e3. doi: 10.1016/j.exphem.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Abu N, Hon KW, Jeyaraman S, et al. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11:875–84. doi: 10.2217/epi-2019-0042. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Li J, Du C, et al. Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl Oncol. 2019;12:1305–13. doi: 10.1016/j.tranon.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Xu HD, Xing XJ, et al. CircRNA_069718 promotes cell proliferation and invasion in triple-negative breast cancer by activating Wnt/beta-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23:5315–22. doi: 10.26355/eurrev_201906_18198. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Song C, Chen Y, et al. Circular RNA circ_0103552 forecasts dismal prognosis and promotes breast cancer cell proliferation and invasion by sponging miR-1236. J Cell Biochem. 2019;120:15553–60. doi: 10.1002/jcb.28822. [DOI] [PubMed] [Google Scholar]
- 17.Sang Y, Chen B, Song X, et al. CircRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–52. doi: 10.1016/j.ymthe.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu JZ, Shao CC, Wang XJ, et al. CircTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–88. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Pan LZ, Hu M, Ma JY. Molecular network-based identification of circular RNA-associated ceRNA network in papillary thyroid cancer. Pathol Oncol Res. 2019 doi: 10.1007/s12253-019-00697-y. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garofalo M, Leva GD, Croce CM. MicroRNAs as anti-cancer therapy. Curr Pharm Des. 2014;20:5328–35. doi: 10.2174/1381612820666140128211346. [DOI] [PubMed] [Google Scholar]
- 23.Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–61. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Xiao L, Ding B, Xu S, et al. CircRNA_0058097 promotes tension-induced degeneration of endplate chondrocytes by regulating HDAC4 expression through sponge adsorption of miR-365a-5p. J Cell Biochem. 2020;121(1):418–29. doi: 10.1002/jcb.29202. [DOI] [PubMed] [Google Scholar]
- 25.Xie B, Zhao Z, Liu Q, et al. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253–61. doi: 10.1016/j.gene.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 26.Yang R, Xing L, Zheng X, et al. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18:4. doi: 10.1186/s12943-018-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Yu L, Liu Y. CircRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516:546–50. doi: 10.1016/j.bbrc.2019.06.087. [DOI] [PubMed] [Google Scholar]
- 28.Zhao F, Chen CW, Yang WW, et al. Hsa_circRNA_0059655 plays a role in salivary adenoid cystic carcinoma by functioning as a sponge of miR-338-3p. Cell Mol Biol (Noisy-le-grand) 2018;64:100–6. [PubMed] [Google Scholar]
- 29.Lin SC, Kao SY, Chang JC, et al. Up-regulation of miR-187 modulates the advances of oral carcinoma by targeting BARX2 tumor suppressor. Oncotarget. 2016;7:61355–65. doi: 10.18632/oncotarget.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulrane L, Madden SF, Brennan DJ, et al. MiR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res. 2012;18:6702–13. doi: 10.1158/1078-0432.CCR-12-1420. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Lin C, Zhao L, et al. Oncogene miR-187-5p is associated with cellular proliferation, migration, invasion, apoptosis and an increased risk of recurrence in bladder cancer. Biomed Pharmacother. 2018;105:461–69. doi: 10.1016/j.biopha.2018.05.122. [DOI] [PubMed] [Google Scholar]
- 32.Kim YH, Choi CY, Lee SJ, et al. Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem. 1998;273:25875–79. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 33.Curtin JF, Cotter TG. JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU 145 prostate carcinoma cells. J Biol Chem. 2004;279:17090–100. doi: 10.1074/jbc.M307629200. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Qian L, Yang J, et al. The expression level and prognostic value of HIPK3 among non-small-cell lung cancer patients in China. Onco Targets Ther. 2018;11:7459–69. doi: 10.2147/OTT.S166878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The influence of miR-187 on circRNA-001283-induced HIPK3 expression. (A) The effect of overexpression of miR-187 mimics on breast cancer cell invasion was determined by Transwell invasion assay. (B) MiR-187 mimics abolished the enhanced effect of circRNA-001283 on HIPK3 expression.