Abstract
Antiviral-resistant influenza viruses in the clinical environment, especially type B, are reported rarely. A stem cell transplant recipient remained influenza B positive for 2 months, despite repeated antiviral treatments. Laboratory tests demonstrated the evolution and persistence of neuraminidase inhibitor-resistant influenza B virus with a substitution at codon 119.
Keywords: antiviral resistance, influenza B virus, neuraminidase inhibitors, neuraminidase mutation, stem cell transplant
Introduction
Seasonal influenza is a major public health and clinical problem, causing an average of more than 200 000 hospitalizations and 23 000 deaths each year from influenza-associated complications in the United States alone [1, 2]. Influenza infection is associated with increased morbidity and mortality in immunocompromised hosts, where the clinical course often is prolonged, with higher rates of viral pneumonia and protracted viral shedding than in other individuals [3, 4].
Neuraminidase inhibitors (NAIs), including oseltamivir and zanamivir, constitute the main class of antivirals used for clinical treatment. Their antiviral activity is mediated by binding to the catalytic site of the influenza neuraminidase enzyme (NA), preventing the release of newly formed viral particles from the surface of infected respiratory epithelial cells. Although the NAs of influenza A and B viruses have only approximately 30% amino acid homology [5], the 19 amino acids of the catalytic site targeted by NAIs are highly conserved [5], so that NAIs are active against both influenza A and B types. And although most currently circulating influenza A and B strains are susceptible to NAIs, the possible emergence of NAI-resistant influenza viruses is a serious clinical concern. In immunocompromised patients, the combination of sustained influenza virus replication, decreased host antiviral responses, and selective pressure from antiviral therapy promote the emergence of resistant strains at higher frequencies than in other patient populations. The laboratory characterization of NAI-resistant influenza viruses recovered from patients during antiviral treatment, and correlation with clinical parameters, are important for understanding the generation and transmissibility of resistant variants. Clinical cases of influenza A virus (IAV) with NA sequence variants manifesting as NAI-resistant phenotypes in immunocompromised individuals have been reported, and the impact of these NA changes on viral fitness have been well studied [6]. However, NAI-resistant influenza B viruses (IBVs) with characterized NA variants, while reported during some surveillance studies and in a few clinical cases [7–18], have been reported less often. Consequently, IBV infections not responding to NAI therapy do not tend to be investigated for resistance as promptly as those with IAV.
Here, we describe the emergence of an NA glutamic acid to valine substitution at position 119 (E119V) with concurrent reduced inhibition by oseltamivir and peramivir and summarize virus characteristics, antiviral regimens, and clinical outcome during IBV infection in a stem cell transplant (SCT) recipient. This substitution has been identified previously and characterized in clinical isolates and recombinant IAVs [16, 18, 19] in association with reduced susceptibility to NAI. Substitutions at position 119 of IBV have been reported in cell culture isolates [20, 21]. We report here a case of IBV carrying the E119V NA variant isolated clinically during IBV human infection.
Case Report
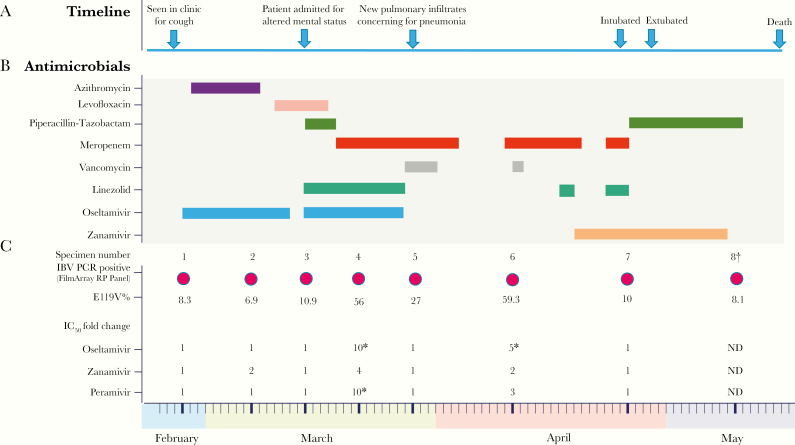
A 58-year-old man with myelodysplastic syndrome diagnosed in August 2007 underwent a mismatched unrelated donor haplo-cord SCT on December 2017. The SCT was complicated by graft-versus-host-disease (GVHD) of the skin and gut (January 2018), requiring immunosuppressive treatment with antithymocyte globulin and sirolimus. In February 2018, he developed persistent cough and loose stools in absence of fever. A nasopharyngeal (NP) swab was tested using a multiplex respiratory pathogen polymerase chain reaction (PCR) assay (FilmArray Respiratory Pathogen [RP] panel [BioFire Diagnostics, LLC, Salt Lake City, UT]) and was positive for IBV. He had not received the seasonal influenza vaccine. A 10-day course of oseltamivir (75 mg twice daily, orally) was started immediately 1 day after symptom onset. Chest computed tomography (CT) scan demonstrated ground glass opacities compatible with pneumonia; thus, azithromycin was also initiated. The patient’s symptoms initially improved, but he was admitted for altered mental status, diarrhea, and hypotension in early March 2018. A repeat FilmArray RP panel was still positive for IBV, so oseltamivir therapy (75 mg twice daily, orally) was extended and broad-spectrum antibiotics were started for possible superimposed pneumonia. All blood and respiratory cultures were negative for other pathogens. Despite antiviral and antibiotic therapy, the patient deteriorated and became hypoxic, requiring intubation and transfer to the intensive care unit. Chest CT scan was notable for new multifocal pulmonary infiltrates. All cultures remained negative, while IBV was detected in both NP swab and bronchoalveolar lavage (BAL) specimens. After completing several cycles of broad-spectrum antibiotics, his respiratory status improved, he was extubated, and oseltamivir was discontinued at day 28 of therapy. The patient remained hospitalized for management of his GVHD and intermittent fever. In April 2018, the FilmArray RP panel was repeated because of his intermittent low-grade fevers and persistent cough and he was still IBV positive.
Given the prolonged influenza shedding and oseltamivir treatment, viral sequencing and phenotypic resistance testing of IBV isolated from the NP specimens was performed. The BAL specimen was processed by an outside laboratory and residual sample was not available for further analysis. Sequencing of the IBV NA gene demonstrated the presence of an E119V substitution (IAV N2 numbering). Zanamivir treatment (10 mg inhaled every 12 hours) was initiated, but the patient’s clinical status continued to decline with worsening of his GVHD and a gastrointestinal bleed. He ultimately passed away in May 2018. A timeline of the events, treatments, and evolution of the NA mutation is shown in Figure 1.
Figure 1.
Clinical Course and Emergence of the E119V Influenza B Virus Variant. The figure summarizes (A) the clinical history, (B) timeline of antimicrobial treatment, and (C) timeline of FilmArray RP Panel testing, the percentage of the E119V variant in the total IBV population as determined by sequencing on primary clinical specimens, and IC50-fold changes compared to baseline for the approved NAIs: oseltamivir, zanamivir, and peramivir in cultured viral isolates. Decreased susceptibility to the tested NAI was observed in the samples with an asterisk. The IC50-fold change cutoff for reduced NAI susceptibility is ≥5.. IBV indicates influenza B virus; PCR, polymerase chain reaction; IC50, half maximal inhibitory concentration; E119V%, percentage of E119V mutation in the IBV population; and ND, not done. † this isolate did not grow to a high titer and its NA activity was too low for NAI susceptibility testing.
METHODS
Specimens
NP swabs were collected as part of routine clinical care using standard procedures.
Molecular Testing
Respiratory pathogen testing was performed with the FilmArray RP panel. Total nucleic acid was isolated from NP swab specimens using an automated platform (NucliSens easyMAG, bioMérieux, Inc., Durham, NC) and QIAamp kit (Qiagen, Germantown, MD) in accordance with the manufacturers’ protocols. Nucleic acids extracted using the easyMAG platform were tested by real-time reverse transcriptase-PCR to confirm the presence of IBV and identify the lineage, and nucleic acids extracted using QIAamp kits were sequenced using high-throughput DNA sequencing methods [22, 23]. Sequences were analyzed with the IRMA bioinformatics pipeline [24] and further assessed for minor variant percentages using Geneious Pro (version 9.1.5) (Geneious Pro, Newark, NJ).
Virus Culture
Influenza-positive specimens were inoculated into Madin-Darby Canine Kidney cells (MDCK) (Atlanta line, originally provided by the U.S. Centers for Disease Control and Prevention (CDC), maintained and provided by the Media and Cell Core Facility, Wadsworth Center) and cultured according to World Health Organization (WHO) protocols [25]. Viruses were harvested at 50% or more cytopathic effect.
Phenotypic Testing
Cultured viruses were tested in a NA-Fluor™ phenotypic assay for antiviral susceptibility [26] against the NAIs oseltamivir, peramivir, and zanamivir (Carbosynth, San Diego, CA).
RESULTS
From late February to early May 2018, while the patient was symptomatic, a total of 8 NP swab specimens were collected, all of which tested positive for IBV with the FilmArray RP panel. These samples were processed for genotypic and phenotypic susceptibility analyses. All 8 samples contained sufficient IBV to enable robust sequence analysis (>90% full-length whole-genome coverage, >600 × depth, at NA codon 119). Influenza B virus was cultured successfully from 7 of these samples. One of the isolates did not grow to high titer and its NA activity was too low for NAI susceptibility testing.
An E119V NA substitution was detected in <10% of the IBV virus (Yamagata lineage) population in the first sample collected at time of diagnosis. The percentage of the NAI-resistant variant rose to 56% after initiating oseltamivir treatment, and it reached its highest level of 59.3% of the total population after prolonged therapy (Figure 1). Upon discontinuation of oseltamivir, the virus population became dominated by wild-type virus, with E119V comprising ≤10% of the virus population in the last 2 specimens of the series. No other NA sequence changes were observed in the Next-Generation Sequence (NGS) data. Two nonsynonymous changes were seen in the NGS sequence data of the hemagglutinin gene: G140R in samples 4, 5, 7, 8, and G526R in sample 8. Neither of these substitutions have been reported to be of significance.
Among the 7 cultured isolates suitable for phenotypic analysis, 5 were determined to be susceptible to all tested NAI antivirals. However, the cultured isolate from a specimen collected in March 2018 (sample 4), which contained a predominance of the E119V IBV variant, showed reduced oseltamivir and peramivir susceptibility. A timeline of the patient’s clinical course, treatment, and emergence of the IBV-resistant variant is summarized in Figure 1.
Discussion
The extremely rapid emergence and world-wide spread of oseltamivir-resistant H1N1 in 2008 demonstrated the importance of vigilant monitoring of patient populations for the evolution of antiviral-resistant influenza viruses. The susceptibility of immunocompromised patients to prolonged influenza infection, necessitating multiple courses of treatment, have been demonstrated repeatedly to cause an increased incidence of NAI-resistant IAV. Reports of NAI-resistant IBV infection, even in immunocompromised patients undergoing antiviral therapy, are rare.
In this report, we detail the investigation of an IBV Yamagata infection in a SCT recipient with prolonged clinical course despite prolonged antiviral treatment. Genotypic and phenotypic analyses demonstrated the presence of an E119V substitution that is associated with decreased NAI susceptibility in IAV [19, 27]. Previous studies suggest that sequence changes conferring NAI resistance do not adversely affect IBV fitness as they do for IAV [28]. In our patient, a mixed population with a minor component of the E119V variant was present at the time of diagnosis, and it became dominant during NAI therapy but diminished upon cessation of NAI therapy. This E119V variant only outgrew the wild-type virus under the selective pressure of the NAI, suggesting a fitness disadvantage associated with this variant in the absence of antiviral drug. This is in agreement with in vitro data, which found that recombinant IBV bearing the E116V substitution was attenuated up to 10-fold in comparison to wild-type virus (likely as a consequence of an overactive NA protein), adversely affecting the balance between the HA and NA activities of the virus that are required for efficient viral replication [18, 21]. This may differ from influenza A/H3N2 virus, in which the E119V oseltamivir-resistant virus could be stably maintained for prolonged periods even in the absence of antiviral-selective pressure [29].
The NAI’s half maximal inhibitory concentration (IC50) values suggest that at the time of diagnosis the IBV isolated from the patient was susceptible to all NAIs. However, the patient did not respond to treatment; this is likely because the NAI-resistant E119V variant continued to replicate and became the predominant strain during NAI therapy. This is supported by the finding that the IBV isolate recovered in culture from the specimen 4 collected in March 2018 contained a predominance of the E119V IBV variant and showed reduced oseltamivir and peramivir susceptibility, but full susceptibility to zanamivir.
Suboptimal clinical efficacy of oseltamivir for the treatment of IBV has been reported [30]. Although an oseltamivir-resistant variant emerged in our patient after a prolonged course of oseltamivir treatment, possibly suggesting a suboptimal therapeutic efficacy, we cannot exclude that other factors, including the patient’s immunocompromised state posttransplant and increased viral replication, may have contributed to the lack of initial response to oseltamivir in our patient.
This case highlights the limited options for influenza treatment. At the time the case presented, all United States Food and Drug Administration-approved agents with activity against circulating influenza strains were NAIs. Although it was not possible to assess whether treatment with baloxavir, a cap-dependent endonuclease inhibitor, would have been helpful, sequence analysis of the IBV recovered from this patient did not reveal evidence of known baloxavir-resistant mutations [31]. To ensure the full range of IAV and IBV antiviral resistance variants are documented, we recommend that viruses from all influenza-infected patients that do not respond to treatment be characterized comprehensively using genotypic methods, including sequencing of viruses directly from clinical specimens and phenotypic methods.
Acknowledgments
We acknowledge Dr. Catherine Small and our colleagues on the transplant infectious disease and bone marrow transplant teams for their contribution to patient care. We also acknowledge the services of the Applied Genomics Technology Core staff, who performed the sequencing reactions.
Financial support. None reported.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Reed C, Chaves SS, Daily Kirley P, et al. Estimating influenza disease burden from population-based surveillance data in the United States. PLOS ONE 2015; 10:e0118369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rolfes MA, Millman AJ, Talley P, et al. Influenza-associated parotitis during the 2014-2015 influenza season in the United States. Clin Infect Dis 2018; 67:485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar D, Ferreira VH, Blumberg E, et al. A 5-year prospective multicenter evaluation of influenza infection in transplant recipients. Clin Infect Dis 2018; 67:1322–9. [DOI] [PubMed] [Google Scholar]
- 4. Boivin G, Goyette N, Bernatchez H. Prolonged excretion of amantadine-resistant influenza a virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin Infect Dis 2002; 34:E23–5. [DOI] [PubMed] [Google Scholar]
- 5. Colman PM, Hoyne PA, Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol 1993; 67:2972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen HT, Fry AM, Gubareva LV. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther 2012; 17:159–73. [DOI] [PubMed] [Google Scholar]
- 7. Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis 1998; 178:1257–62. [DOI] [PubMed] [Google Scholar]
- 8. Hurt AC, Iannello P, Jachno K, et al. Neuraminidase inhibitor-resistant and -sensitive influenza B viruses isolated from an untreated human patient. Antimicrob Agents Chemother 2006; 50:1872–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ison MG, Gubareva LV, Atmar RL, Treanor J, Hayden FG. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J Infect Dis 2006; 193:760–4. [DOI] [PubMed] [Google Scholar]
- 10. Hatakeyama S, Sugaya N, Ito M, et al. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA 2007; 297:1435–42. [DOI] [PubMed] [Google Scholar]
- 11. Bastien N, Gubbay JB, Richardson D, Sleeman K, Gubareva L, Li Y. Detection of an influenza B virus strain with reduced susceptibility to neuraminidase inhibitor drugs. J Clin Microbiol 2011; 49:4020–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sleeman K, Sheu TG, Moore Z, et al. Influenza B viruses with mutation in the neuraminidase active site, North Carolina, USA, 2010-11. Emerg Infect Dis 2011; 17:2043–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fage C, Abed Y, Checkmahomed L, Venable MC, Boivin G. In vitro properties and virulence of contemporary recombinant influenza B viruses harboring mutations of cross-resistance to neuraminidase inhibitors. Viruses 2019; 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abed Y, Fage C, Lagüe P, et al. Reduced susceptibility to neuraminidase inhibitors in influenza B isolate, Canada. Emerg Infect Dis 2019; 25:838–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Escuret V, Collins PJ, Casalegno JS, et al. A novel I221L substitution in neuraminidase confers high-level resistance to oseltamivir in influenza B viruses. J Infect Dis 2014; 210:1260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escuret V, Frobert E, Bouscambert-Duchamp M, et al. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors. J Clin Virol 2008; 41:25–8. [DOI] [PubMed] [Google Scholar]
- 17. van der Vries E, Ip DK, Cowling BJ, et al. Outcomes and susceptibility to neuraminidase inhibitors in individuals infected with different influenza b lineages: the influenza resistance information study. J Infect Dis 2016; 213:183–90. [DOI] [PubMed] [Google Scholar]
- 18. Jackson D, Barclay W, Zürcher T. Characterization of recombinant influenza B viruses with key neuraminidase inhibitor resistance mutations. J Antimicrob Chemother 2005; 55:162–9. [DOI] [PubMed] [Google Scholar]
- 19. Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis 2011; 203:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Staschke KA, Colacino JM, Baxter AJ, et al. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology 1995; 214:642–6. [DOI] [PubMed] [Google Scholar]
- 21. Burnham AJ, Baranovich T, Marathe BM, Armstrong J, Webster RG, Govorkova EA. Fitness costs for Influenza B viruses carrying neuraminidase inhibitor-resistant substitutions: underscoring the importance of E119A and H274Y. Antimicrob Agents Chemother 2014; 58:2718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou B, Lin X, Wang W, et al. Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics. J Clin Microbiol 2014; 52:1330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McGinnis J, Laplante J, Shudt M, George KS. Next generation sequencing for whole genome analysis and surveillance of influenza A viruses. J Clin Virol 2016; 79:44–50. [DOI] [PubMed] [Google Scholar]
- 24. Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 2016; 17:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. World Health Organization. Manual on Animal Influenza Diagnosis and Surveillance. https://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf. Published May 2002. Accessed November 17, 2019. [Google Scholar]
- 26. Hurt AC, Okomo-Adhiambo M, Gubareva LV. The fluorescence neuraminidase inhibition assay: a functional method for detection of influenza virus resistance to the neuraminidase inhibitors. Methods Mol Biol 2012; 865:115–25. [DOI] [PubMed] [Google Scholar]
- 27. Abed Y, Baz M, Boivin G. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir Ther 2006; 11:971–6. [PubMed] [Google Scholar]
- 28. Farrukee R, Leang SK, Butler J, et al. Influenza viruses with B/Yamagata- and B/Victoria-like neuraminidases are differentially affected by mutations that alter antiviral susceptibility. J Antimicrob Chemother 2015; 70:2004–12. [DOI] [PubMed] [Google Scholar]
- 29. Hurt AC, Leang SK, Tiedemann K, et al. Progressive emergence of an oseltamivir-resistant A(H3N2) virus over two courses of oseltamivir treatment in an immunocompromised paediatric patient. Influenza Other Respir Viruses 2013; 7:904–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawai N, Ikematsu H, Iwaki N, et al. A comparison of the effectiveness of oseltamivir for the treatment of influenza A and influenza B: a Japanese multicenter study of the 2003-2004 and 2004-2005 influenza seasons. Clin Infect Dis 2006; 43:439–44. [DOI] [PubMed] [Google Scholar]
- 31. Hayden FG, Sugaya N, Hirotsu N, et al. ; Baloxavir Marboxil Investigators Group Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N Engl J Med 2018; 379:913–23. [DOI] [PubMed] [Google Scholar]