Clinical microbiology is experiencing revolutionary advances in the deployment of molecular, genome sequencing-based, and mass spectrometry-driven detection, identification, and characterization assays. Laboratory automation and the linkage of information systems for big(ger) data management, including artificial intelligence (AI) approaches, also are being introduced. The initial optimism associated with these developments has now entered a more reality-driven phase of reflection on the significant challenges, complexities, and health care benefits posed by these innovations.
KEYWORDS: artificial intelligence, automation, biobanking, clinical microbiology, consolidation, surveillance
SUMMARY
Clinical microbiology is experiencing revolutionary advances in the deployment of molecular, genome sequencing-based, and mass spectrometry-driven detection, identification, and characterization assays. Laboratory automation and the linkage of information systems for big(ger) data management, including artificial intelligence (AI) approaches, also are being introduced. The initial optimism associated with these developments has now entered a more reality-driven phase of reflection on the significant challenges, complexities, and health care benefits posed by these innovations. With this in mind, the ongoing process of clinical laboratory consolidation, covering large geographical regions, represents an opportunity for the efficient and cost-effective introduction of new laboratory technologies and improvements in translational research and development. This will further define and generate the mandatory infrastructure used in validation and implementation of newer high-throughput diagnostic approaches. Effective, structured access to large numbers of well-documented biobanked biological materials from networked laboratories will release countless opportunities for clinical and scientific infectious disease research and will generate positive health care impacts. We describe why consolidation of clinical microbiology laboratories will generate quality benefits for many, if not most, aspects of the services separate institutions already provided individually. We also define the important role of innovative and large-scale diagnostic platforms. Such platforms lend themselves particularly well to computational (AI)-driven genomics and bioinformatics applications. These and other diagnostic innovations will allow for better infectious disease detection, surveillance, and prevention with novel translational research and optimized (diagnostic) product and service development opportunities as key results.
INTRODUCTION
The health care market is subject to pressure caused by a variety of disruptive factors. These include population density, population dynamics and composition, disease prevalence and severity, economic status, cost of tests, and others (1). Further pressure originates from changes in the nature of health care delivery itself, government and payer initiatives, the attitude of insurance organizations, consumer education and expectation, and rapid changes in technology. These diverse pressures have prompted a push to consolidate biomedical laboratory analyses, where resources and services are centralized and serve a large(r) population for purposes of enhanced efficiency, increased standardization, and potentially earlier time to results. Initially, consolidation efforts were mostly driven by business considerations, including diagnostic costs, privatization, and scarcities of appropriately qualified personnel. More recently, secondary benefits, including integrated databases and reporting systems as well as more easily managed biorepositories, delivered additional value to consolidation. Furthermore, emerging technologies and platforms are more easily assimilated in bigger laboratories, leaving the smaller ones at risk of being left behind. These new and usually quite complex technologies used in consolidated laboratories already require (multiple) accreditation levels to comply with European Conformité Européenne (CE) or American Food and Drug Administration (FDA) guidelines. In addition, these regulatory validations are increasingly required to address the roll-out of innovative diagnostics to deal with rising rates of health care-associated infections and antimicrobial resistance (AMR) (for more detail, see, for instance, https://www.fda.gov/news-events/speeches-fda-officials/fdas-strategic-approach-combating-antimicrobial-resistance-09142018). Note that throughout the text the term “validation” will be used when referring to the process in which a proof-of-principle test will be transformed into a clinically relevant, reliable, and reproducible laboratory test. Improvement in diagnostic capacity of the clinical microbiology laboratory is still much needed. Beyond technological development, capacity also involves the ability to share data repositories, samples, and strain collections, to closely collaborate with public and private stakeholders (including regulatory agencies), and ultimately to embed rapid academic access to new diagnostic tools (2) while maintaining or improving clinical utility (3–5).
We wish to illustrate, in part from personal experiences, both the advantages, challenges, and, most importantly, the consequences of such wholesale consolidation and its effects upon the adequate introduction of new technologies. If consolidation is to realize maximal benefit, it will require careful planning and subsequent reassessment against the proposed original model at local, national, and international levels. Although health economic arguments are the often-quoted drivers behind laboratory consolidation efforts, the benefits of consolidation should never compromise the quality, rapid availability, and value of the diagnostics. We shall also discuss secondary benefits relating to the relative ease with which new technologies can be introduced into bigger consolidated laboratories.
In 2015, Sautter and Thomson critically reviewed laboratory consolidation in a point-counterpoint discussion (6). The authors concluded, in agreement, that financial pressure was one of the biggest, if not the biggest, parameter driving consolidation. They also agreed that reductions in turnaround time (TAT) and technological developments and improvements were important secondary drivers toward consolidation. At that stage in 2015, there was a lack of clarity on the true costs and savings of consolidation and the medical value for patient care needed to be substantiated. The effects of longer courier routes, maintenance of technical expertise among personnel, diagnostic menus, and test result communication associated with consolidation needed to be studied. Here, we provide an update to that discussion by also considering the transformative technologies that have recently become available to clinical microbiologists.
(Part of this work was presented during an oral presentation at the 13th International Conference on Genomics in Shenzhen, China [2018].)
OVERVIEW OF THE DIFFERENT CONSOLIDATED LABORATORY MODELS
There are many examples of the consolidation of routine microbiology laboratories worldwide with different practice and business models. Each of these models has distinct advantages and constraints from economic and patient care perspectives, which cannot be generalized across the different models. Of note, there are very few public documents where the situation before and after consolidation is clearly quantified. There are certainly no peer-reviewed publications on the financial details and the proper maintenance of diagnostic excellence by the consolidated laboratories or good evidence supporting a particular model. The lack of guidelines describing what best practice looks like for all steps of the diagnostic process also makes it difficult to compare the different models. Four models have emerged from consolidation efforts, each with specific potential clinical/economic advantages and constraints:
Model 1: large private industrial laboratory practices servicing many healthcare jurisdictions within a country or even countries.
The U.S. laboratories were at the forefront of such a consolidation model, with Quest Diagnostics formed in 1967 and LabCorp in 1971. In Europe, laboratory amalgamation started later and involved the consolidation of small, medium, and larger individual laboratories. In a country such as Belgium, laboratory consolidation has led to a nearly linear decrease in the number of independent laboratories, from 496 in 1996 to 148 in 2017 (7). In this model, cost reduction and/or profit-making is not necessarily directly linked to patient care.
Model 2: large consolidated health maintenance organization (HMO) systems servicing several hospitals where a core high-volume centralized facility supports a larger tiered laboratory network.
Three representative U.S. examples of such private hospital laboratory consolidation are BJC Healthcare (resulting from the merging of Barnes–Jewish, Inc., with Christian Health Services), providing service for 15 hospitals in the Midwest region, Geisinger Health System, serving 10 mid-Atlantic region hospitals, and Northwell Health Laboratories, serving 23 hospitals primarily in the New York region. Labor Berlin GmbH, among the biggest diagnostic laboratory consortia in Europe, was founded in 2011 and consolidated the in vitro diagnostics departments of the two major hospital networks of the State of Berlin, Charité–Universitäts Medizin Berlin and Vivantes Network of Health. Labor Berlin’s mission is to offer state-of-the-art diagnostic services to Charité and Vivantes hospitals and to be a major hub for refining and pushing boundaries in research and development for new in vitro diagnostic services, methodology, and hardware. Its sister company, Labor Berlin Services, provides services to outside medical entities. Together, they serve over 24,000 hospital beds and perform more than 8 million bacteriological analyses per year. Labor Berlin, as the diagnostic laboratory of the largest University Hospital in Europe (Charité), has built a strong research and development platform and has set up a diagnostic clinical scientist program to attract promising young microbiologists, virologists, and laboratory medicine specialists. The latter must be considered the main advantage of consolidated laboratories; having all expertise under a single roof provides huge teaching and education advantages.
Model 3: consolidation through private-public partnerships.
Located in Central London, Health Services Laboratories (HSL) is the result of a 2016 public-private partnership (PPP) between The Doctors Laboratory (the largest independent provider of clinical pathology services in the UK), the Royal Free London NHS Foundation Trust (the Royal Free London), and the University College London Hospitals NHS Foundation Trust (UCLH). HSL is the largest pathology provider in the UK, and both NHS Foundation Trusts are investors in the business. HSL is directed by a Board of Managers and Clinicians from all 3 parties. It performs over 36 million tests a year in a ca. 10,000-m2 facility for approximately 3 million hospital and community patients. Like HSL, the Calgary Laboratory Services (CLS) model in Alberta, Canada, which was formed in 1996, represents a valuable example (8, 9).
Model 4: consolidation due to healthcare reforms to create a large tiered laboratory network of urban/rural practice.
In China, the population dynamics are such that tertiary health care units of the four Tier-One megalopolises (Shanghai, Beijing, Shenzhen, and Guangzhou, as classified by the Chinese Tier system) and the 30 Tier-Two cities (which are provincial capitals hosting 3 to 15 million inhabitants) had to adapt routine clinical microbiology services to high-throughput practices since the late 1990s. This move was part of an overall health care reformation, which included a significant change in the urban as well as rural health care systems, beginning in 2003 (10) and leading to the implementation of a near-universal Chinese health care system (11). Four infectious disease outbreaks of major economic impact (avian influenza, swine influenza, severe acute respiratory syndrome, and now coronavirus disease 2019 (COVID-19)), together with national surveillance programs (e.g., for prevention of methicillin-resistant Staphylococcus aureus [MRSA] colonization and infection), allowed tertiary health care units and, in particular, clinical microbiology laboratories to enjoy 2 decades of continuous governmental support and investment (12). Although the development and sales of quality diagnostic national products has expanded in parallel with the opening of the diagnostic market to foreign diagnostic companies, the market forces seem not to be the main factors driving the consolidation of clinical microbiology laboratories in China, which is quite different from other geographic areas. Consolidation of clinical microbiology laboratories relates mostly to the reorganization of central governmental institutions (e.g., academic hospitals, public health institutions, and/or the army hospitals) (13). For example, the 301 Hospital or People’s Liberation Army General Hospital (PLAGH) is a G3A military hospital located in Beijing that currently has 4,000 patient beds and consists of a setup servicing anything east of the Great Wall and north of the Yangtze River. This structure, however, represents the Chinese exception rather than the rule. In terms of private services, again consolidation depends on local (provincial-level) permissions and allowances. The largest Chinese private diagnostic companies in the infectious diseases domain (e.g., Dian Diagnostics in Hangzhou) still conform to publicly funded schemes and do not have a direct consumer link. Some exceptions can be noted (e.g., in Guangzhou and Shanghai), where local regulations allow for more flexibility. Otherwise, a very tight, top-down-regulated picture emerges.
The publication of the UK Public NHS Improvement operational productivity and performance plan, aiming to establish new pathology networks across England by providing more responsive, top-quality, and efficient services by 2021, represents another example of consolidation upon health care reforms (https://improvement.nhs.uk/documents/6113/Pathology_networking_state_of_the_nation.pdf). This plan was developed based on the following premise:
Consolidating pathology services allows for the most consistent, clinically appropriate turnaround times ensuring the right test is available at the right time. It makes better use of our highly skilled workforce to deliver improved, earlier diagnostic services supporting better patient outcomes. Taking a hub and spoke approach to this consolidation can ensure an appropriate critical mass to support specialist diagnostics, so that patients have equal access to key tests and services are sustainable.
In conclusion, laboratory consolidation certainly is not new, and many other examples of amalgamation can be easily provided. Despite the existence of different business models, consolidating clinical microbiology laboratories to large-scale network proportions will offer early access to advanced, often expensive technologies with high diagnostic value for very complex clinical cases. This is because with networks’ high-test volumes comes buying power, which is out of reach to individual small hospitals. For example, high-volume tests (e.g., screening tests for sexually transmitted diseases, hospital-acquired infections, or tuberculosis) can be performed at significant cost savings on high-throughput platforms compared to mid-volume or low-volume platforms (14). Partnering of local forces with institutes such as the Foundation for Innovative New Diagnostics (FIND) is key to success. Likewise, it is impractical for individual small laboratories to perform highly specialized tests when the technical expertise and testing platforms may only be available in consolidated laboratories. In addition, promoting the rational ordering and use of diagnostic tests can be helpful in optimizing patient care (15). They also represent opportunities for test and equipment manufacturers wishing to take forward emerging diagnostics stuck in the academic space through to validation, EU and FDA accreditation, and, ultimately, introduction into the global marketplace. In other words, consolidated laboratories leverage academic progress through their volume, and this is where bigger is surely better.
CONSOLIDATED LABORATORY DESIGNS
Despite the four different business models presented above, most consolidated laboratories share a design that is described below. The overall layout of consolidated laboratories is depicted in Fig. 1.
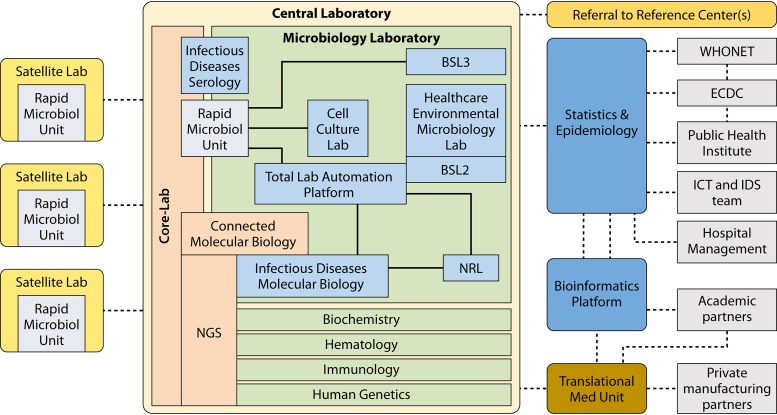
FIG 1.
Global layout scheme for a consolidated clinical microbiology laboratory organization. The central laboratory comprises, among others, a clinical microbiology unit harboring all key technological facilities, including high-tech facilities for molecular testing, MALDI-TOF MS, and NGS, as well as cell culture laboratory for virus and intracellular bacteria detection and one or more National Reference Laboratories (NRL). At the management level (right part), there are opportunities for reaching out to external specialized facilities (WHONET, ECDC, etc.) as well as all the required IT and informatics facilities. On the left, the satellite laboratories are depicted. These can vary in number and complexity but should be within relatively close proximity and focused on rapid response technology only. (Adapted from reference 18.)
Central Laboratory Structure
In general, the central clinical laboratory combines different types of expertise: biochemistry, hematology, immunology, human genetics, microbiology, and others. The microbiological expertise includes all classical subspecialty areas (e.g., bacteriology, mycobacteriology, mycology, parasitology, and virology) and technologies (e.g., microscopy, culture, microbial identification, antimicrobial susceptibility testing [AST], immunoassays/serology, and molecular diagnostics). These, in turn, may be supported by several local and delocalized rapid response laboratories within and outside the same institute (16). The latter may generate logistical issues because of the need for rapid and secure specimen transport. As previously demonstrated in Switzerland and more recently in WakeMed Health & Hospitals (Raleigh, North Carolina), drones represent useful vehicles for such transport (17).
The main medical-managerial goals of such amalgamated laboratory structures are to consolidate the screening of large volumes of samples or highly specialized diagnostic testing and to effectively separate the urgent from the less urgent activities while retaining access to more niche-specific testing and, of course, excellent overall quality levels (18).
To reduce the turnaround times (TATs) and improve the laboratory’s efficiency, implementation of 24-h/7-day (24/7) diagnostic activities for intensive care units (ICUs) and emergency care departments (ECs) must be prioritized. Still, on a global scale for the health care system as a whole, 24/7 availability must become the de facto norm.
Test TATs in centralized facilities are challenged by the potential delays introduced by specimen transit time and information technology (IT) barriers to seamless postanalytical validation and reporting of results. Therefore, the consolidation of clinical microbiology laboratories involves an array of platforms linked to a central laboratory information system (LIS), with the importance of middleware solutions becoming increasingly evident. Middleware is defined as software linking instruments directly to the LIS without intervention by technicians or data operators. Links between hospital and laboratory information systems (HIS and LIS) are commonly designed and customized for adequate data quality control and dissemination. Such IT systems are essential to verify and track orders and to appropriately direct to completion all testing on a given sample (19). For example, splitting bronchoalveolar lavage (BAL) specimens for routine microbiology-virology, mycobacteriology, mycology, and fungal antigen testing may be challenging, especially in instances where a specimen can only be partially handled via automated processes. Decisions related to interfacing each component instrument, such as parallel identification and antimicrobial susceptibility testing (ID/AST) and blood culture instruments, etc., will be required, and the physical logistics of specimen management are complex. The bigger the laboratory, the larger the number of samples and the more complex the logistics will be. Middleware, however, may provide solutions for gathering all data on a culture or sample together for consideration and action by laboratory technologists before submission to the LIS for final patient report generation. Additionally, middleware can offer the potential for real-time tracking and improving of all work flowing through the centralized laboratory to anticipate technical needs. What seems clear is that early consideration, development, and deployment of an integrated and seamless LIS as “big gets bigger” is mandatory.
Satellite Laboratory Structure
Despite the consolidation of the vast majority of analytical tests, there nevertheless remains a clear need for maintaining satellite laboratories for testing when and where transit delays are unacceptable; examples here include, among others, point-of-care screening and satellite blood culture facilities. Additionally, some nonautomated, manual specimen processing can be performed in satellite facilities (e.g., limited numbers of Gram stains and cultures) and then interpreted in the central laboratory by “telemicrobiology,” where the microbiologist validates a result without having the physical evidence, such as culture plates, directly at hand (20). This can also happen within the satellite laboratory, where a certain part of the laboratory can be prepared for the performance of urgent, important testing for which the need for speed is very high. Even if most samples are analyzed in one central laboratory, validation of results from the satellite laboratories via a telemicrobiology system makes it possible to maintain the virtual or even physical presence of a clinical microbiologist in the satellite laboratory. Maintaining clinical microbiologist presence on site allows, on the one hand, provision of high-quality laboratory services and, on the other, strong professional interaction between microbiologists and patient-facing clinicians. It is likely to also drive better implementation of infection control and antibiotic stewardship. Circumstantial evidence obtained in a rural laboratory supports this hypothesis: adding just a single staff microbiologist on site resulted in better use of antibiotics and lowered costs (21).
CONSOLIDATION THROUGH LABORATORY AUTOMATION
The most difficult practical challenges faced by a big consolidated laboratory include neglected but critical sample logistics and the subsequent sample processing (e.g., inoculation on culture media, plate reading, microbial identification, AST, and the increasingly diverse downstream methods for proteomic and genomic analysis) (22, 23). Total laboratory automation (TLA) increasingly addresses such shortcomings and provides solutions to some of the issues (24). Over the coming years, with the further maturation of TLA systems, the number of problems solved is likely to increase steadily.
However, the “when” and “how” of moving from more classical methodologies to partial or full automation in a way that preserves the accuracy and “intelligence” of the prior established technologies while preserving trust in those who will act on its results is a question far from easily resolved. This is especially true if microorganisms of interest can be relatively easily cultured and differentiated through the use of highly specific screening media, such as selective and/or chromogenic agars or broths (25). Automation is most valuable when it reduces manual activity that is slow, repetitive, and does not require human analysis and interpretation. The initial stages of traditional microbiology culture include plate or broth inoculation and incubation, followed by inspection for colony formation. As already mentioned above, this still represents a substantial portion of the labor used currently in laboratories and is estimated to apply to approximately 70% to 80% of all current activities (26), making automation of these processes of potential benefit even to those laboratories with a modest workload. Furthermore, where does the balance between reduction in hands-on time (and, therefore, labor costs), TAT, and quality lie? This question must be addressed and resolved before taking the first steps toward a laboratory merger.
One of the key aspects, the relevance of which is often underestimated, is the complexity of specimens and the diagnostic tests performed. In clinical microbiology laboratories, numerous different types of clinical materials are submitted for analysis on a daily basis. This represents one of the fundamental differences between clinical microbiology and clinical chemistry and explains why clinical chemistry laboratories have already been largely and successfully automated. The first decisions to be taken upon receipt of a clinical specimen are whether the quality and quantity of the sample suffice and whether additional preanalytic manipulations are required. Apparently simple decisions, such as whether the sample containers need to be opened manually or not or whether the sample needs to be split for several analyses, have to be included in the design of the workflow between the arrival of a specimen and its inclusion in a certain test. This process can be at least partly automated but (in our experience) requires continuous monitoring and significant residual manual input from appropriately qualified and experienced laboratory technologists. Although comparison of manual and automated urine cultures showed that automation increased the number of positive samples and generated greater microbial diversity among positive samples, there was no evidence for better time to result, and the different positivity rates were not significantly different (27). Additional health economics studies describing the financial consequences of TLA are still much needed.
When direct-from-specimen tests (enzyme-linked immunosorbent assay [ELISA] from serum, antigen [diffusion] tests for respiratory specimens, direct cell sorting for urine samples, etc.) are available, they need to be performed before more time-consuming and complex assays are attempted, including direct inoculation on culture media and sample processing for non-culture-driven testing (e.g., PCR or antibody-mediated tests). PCR tests, especially those that are multiplexed, are likely to remain dominant for several decades (28). To optimize workflow and minimize TAT, direct-from-specimen tests (e.g., immunoassays and multiplex PCR) should be performed in a rapid microbiology unit rather than an infectious disease immunoassay or molecular biology laboratory (Fig. 1). More complex or time-consuming tests will likely require further interpretation before result release; some, but not all, of these processes currently can be fully automated. Finally, follow-up analyses may need to be performed, and ID normally drives the selection of an appropriate AST panel. Again, different technologies can be incorporated and a variety of semiautomated methods are currently available, although one has to realize that the actual workflow codefines the timing of the testing protocol (29).
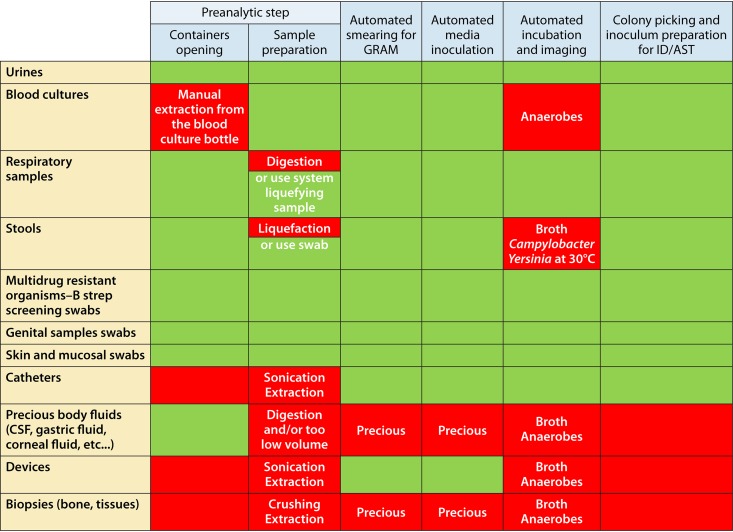
Such a rapidly emerging variety of technology solutions and their combinations for downstream reflex testing makes the design of optimal microbiology diagnostic pathways exceedingly complex. This precedes the subsequent composite interpretation of results that are sometimes couched in analytical uncertainty. We have attempted to distill a simplified scheme assessing a broad overview of all of these activities in Fig. 2.
FIG 2.
Two-dimensional workflow gap analysis for guiding different clinical specimens through the diagnostic microbiology pipeline. Sample types (column on the left) are evaluated versus some of the diagnostic processes. Green fields identify appropriate compatibility, and red fields identify a need for optimization.
The details described above emphasize the fact that even in a semiautomated laboratory, human intervention and expertise remain very important. The ability to visually read a positive culture can have a major impact on the interpretation. There is also risk in triggering downstream tests on the basis of a preliminary and perhaps incorrect presumptive microbial ID for reasons of clinical urgency. Accordingly, there continues to be everyday tension between the drive to properly reduce TAT and the (more time-consuming) need for a confirmed rather than presumptive result. We consider these issues important, if not fundamental, to the overall quality of the routine clinical microbiology laboratory. Consolidated laboratories must consider how to incorporate these essential, “automation unfriendly,” and labor-intensive realities for improvement of clinical quality while still allowing relatively high-throughput result reporting. Finally, the current instruments for automation might replace human manipulation more than provide real innovation. As long as the culture plate remains at the center of the microbiology laboratory, paradigm-shifting innovation will not be easily designed and implemented.
Is Total Laboratory Automation Already Practical and Mature?
Although automation has been present in diagnostic laboratories for over 30 years, TLA had not entered clinical microbiology laboratories until recently. No single breakthrough technology has been proven to be cost-effective or versatile enough to replace culture-based diagnostics. Still, diagnostic alternatives, such as multiplex PCR, have primarily replaced immunoassays, viral cultures, and bacterial cultures for slow-growing or difficult-to-grow bacteria. By streamlining the workflow process and reducing the TAT, extended automation from the front end up to the postanalytical phase will bring significant benefits to the laboratory through its flexibility, scalability, and interchangeability. Such automation encompasses instruments that perform specimen processing and tracks systems transporting plates to and from incubators, digital cameras capturing plate images, automated incubators associated with digital reading stations, organism identification, AST, and software managing these processes (Fig. 2 provides a methodological review).
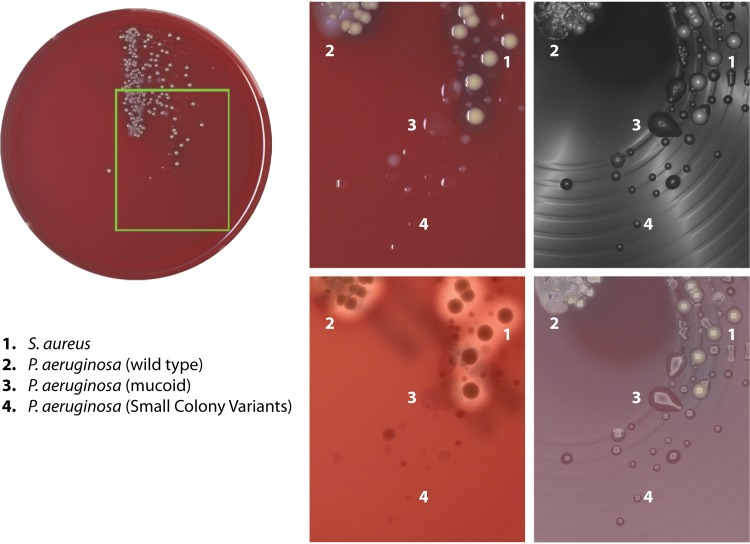
Classical culture-based microbiology provides important subject matter as to how TLA improves processes at various levels. Automated plate and broth inoculation saves labor by eliminating repetitive and manual tasks while adding the benefit of producing more isolated bacterial colonies (27, 30, 31). Due to the variety of incoming specimens, a preanalytical step may be required. Samples range from biopsy specimens (including hard-to-manage bone tissue, for instance) or devices, semisolids (stools), and samples of various viscosities, such as urine, sputum, and other body fluids (aspirates, cerebrospinal fluids, etc.). Moreover, such samples may be infectious, requiring sometimes strict and cumbersome laboratory infection prevention measures. In addition to this range of different sample consistencies, a considerable proportion of microbiology specimens is precious and/or could only be collected by invasive (surgical) procedures (cerebrospinal fluids, bones, biopsy specimens, etc.). Although liquid specimens generally are handled well with automated specimen processing, other specimen types may be processed in a semiautomated mode or in a fully manual mode (32). Some containers are incompatible with streaking instrumentation (e.g., caps that cannot be gripped by the instrument); therefore, one of the key challenges is container standardization. Automated systems are able to prepare slides for staining, but stain imaging and reading are still conducted separately and manually. With continuous automated incubation of plated media, the laboratory can achieve a smooth(er) workflow. However, it is still limited, as it usually does not include anaerobic or microaerophilic conditions. The next critical step is plate imaging during automated incubation. The system has to ensure the production of high-quality images for the detection of bacterial growth and to differentiate colony morphotypes at the earliest stages of incubation (33). Imaging technology has the ability to detect bacterial growth earlier, particularly for slow-growing bacteria (24, 34, 35). It is also a powerful tool to differentiate and track bacterial morphotypes, such as those for P. aeruginosa mucoid or small-colony variants in cystic fibrosis patients’ sputum (Fig. 3). Since plate images are stored digitally, the review of growth over time and examination of a patient’s culture history can be done independently of staffing levels at any given hour. Some systems enable taking images of plates not incubated in the device due to their unsupported atmosphere requirements in order to ensure production of high-quality images and to enforce traceability. Systematic reviews on the precise technological needs and requirements for the imaging equipment are currently lacking. It is clear that huge progress has been made in this field but that successful implementation requires maintenance of human expertise needed to complement the automated processes. Clear data on the economic gains of implementing full laboratory automation are scarce, although the first papers describing reduced TAT using automated systems also prompt the issue of the profitability of such approaches (36).
FIG 3.
Detection of bacterial morphotypes on culture plates inoculated with sputum samples from cystic fibrosis patients using high-resolution imaging technology. The actual plate is shown on the left, and the four panels on the right show the same region of the plate captured using different illumination technologies. Different colony morphologies for P. aeruginosa and Staphylococcus aureus have been highlighted numerically.
In parallel with the development of automation, significant advances have been recorded for the improvement of culture-based diagnostics. Collectively called culturomics, the development of new media, alternative atmospheres, intelligent culture containers, and molecular or metabolic follow-up has identified thousands of new bacterial species inhabiting the various niches in humans and animals (37–41). The discovery and isolation of new organisms by such novel, alternative culture techniques is likely to lead to new paradigms of bacterial infection and pathogenesis. How laboratory automation will rise to this challenge of flexibility and the need for rapid implementation of new tests based on these emerging research findings remains unanswered to date.
Recent advances in biome research have demonstrated that the microbiota composition prior to, during, and after an episode of sepsis may be associated with increased susceptibility and worse outcome (42). From this perspective, the insertion of specific biome diagnostics (and, just as importantly, their evidence-based interpretation, not necessarily by microbiologists) and/or the identification of new markers into the laboratory workflow represent one of most interesting challenges for the coming years (43).
Automation and Artificial Intelligence
Digital approaches have been central in the development of laboratory consolidation and automation, since the associated increase in specimen pathway complexity requires intelligent instrumentation and integrated communication between systems and equipment (44). Communication needs to be integrated via the LIS rather than simply connecting single instruments and surrounding information systems (45). There is also an important role to play for middleware that can facilitate the communication between different instruments and the LIS. Beyond data communication, artificial intelligence (AI) represents a new direction for laboratory data management. AI is usually defined as the theory and development of computer systems that perform tasks normally requiring human intelligence (such as visual perception, speech recognition, and decision-making, among others) (46). Predicting trends in infectious diseases while using clinical, diagnostic, and epidemiological data is one of the exciting applications of AI and deep learning. As an example, the application of AI for the detection of negative urine screening cultures was shown to significantly reduce the diagnostic workload (47). The workload could be reduced more than 40% by different algorithms that included not only actual diagnostic data but also demographics and prior culture data for the patients involved. As another example, predicting outbreaks of viral infection can be improved by over 20% in accuracy when surveillance data are analyzed using recent developments in the field of neural networks and deep-learning algorithms (48). Similar approaches, when applied to the continuous monitoring and detailed analysis of patients’ tympanic temperature, allows for the distinction between patients with infectious versus noninfectious fever (49). Even the analysis of basic and patient-specific electronic medical records (EMR) on admission notes, vital signs, and requested diagnostic tests can stratify patients with infections with reasonable accuracy (50).
Developments in the field of AI are in their infancy, and mature solutions seem to be restricted to the public health environment (51); such approaches are ripe for application to clinical microbiology and infected patients. AI-driven programs for effectively reducing antibiotic usage have already been introduced (52), and significantly more specific tools to manage patient care through better use of clinical and epidemiological big data will become available in the decade to come. The first studies describing the role of bioinformatics in translating fundamental microbiome research into prophylactic and therapeutic interventions have been published (53). Although these initial assessments are descriptive for the most part, it is obvious that the application of AI in new fields of fundamental microbiological research will accelerate the development of real diagnostic and clinical applications.
Aspects of AI, such as machine learning (ML), are becoming central in various domains of clinical microbiology laboratory automation (54). ML is an application of AI that provides systems with the capability to learn and improve from experience without being formally programmed to do so. ML focuses on computer programs that can access and use data during the actual learning process. ML algorithms search for and identify patterns in (huge) data sets in order to make better decisions based on their recognition. The primary aim is to allow the computer to learn automatically without human intervention and to constantly adjust and fine-tune the accuracy of conclusions in these decision spaces. ML processes are highly relevant and applicable to the expansion and extension of the clinical impact of automated diagnostic systems (55, 56). It is likely that ML applied to this field will release innovation and improvement in all categories of diagnostics. ML will be able to distill novel diagnostic algorithms from preexisting data and refine the accuracy of such algorithms after the inclusion of new high-quality data (57). The practice of ML and its generic use in clinical microbiology was recently reviewed by Qu et al. (58). As a clinically relevant example, ML can be used to predict the development of complicated Clostridioides difficile infection (e.g., leading to ICU admission, development of toxic megacolon, need for colectomy, or death) based on cumulative data extracted from patients’ individual electronic health records (59). ML can also be used for the automated classification of microbial species (60), but a potentially interesting use of ML is in the development of tools that generate AST results entirely based on genomic data (61). This type of investigation queries huge databases that combine existing genome sequences and their associated, high-quality predefined in vitro AST data with newly defined genome sequences in order to develop in silico antibiograms (62, 63). One successful manner in which ML facilitates this type of correlation is through the cataloging of short oligonucleotides of a certain length. The number and nature of such short sequence motifs, known as k-mers, correlates with MICs for the various microbial strains included and has predictive value for new sequences (containing or not containing the k-mers) (64). Such approaches have been successfully tested for Mycobacterium tuberculosis and Staphylococcus aureus in proof-of-concept studies. For species with a less clonal population structure, these so-called geno-to-pheno (G2P) analyses currently are defined as being much more cumbersome to perform. Studies performed for Pseudomonas aeruginosa showed useful performance for certain antibiotics, whereas for others results were significantly less robust (62).
Many laboratories are developing tools that correlate genome sequences with antibiotic susceptibility levels. This development occurs at the interface of microbiology, data science, and ML development (65). Of note, such applications also can be used to search for new molecular markers for antibiotic resistance (66). The overall strategy was to leverage years’ worth of microbiological data by applying it to ML applications to correlate the most common bacterial species with their AST pattern. Most of these systems are currently in evaluation for use in routine, high-throughput diagnostics and show promise (67 and references therein). While ML algorithms will of course not completely replace doctors and medical professionals in validating microbial test results, they have the potential to make validation much more time-efficient and may help to further minimize (human) errors and highlight inconsistencies. In addition, there are ethical and legal considerations that still need to be dealt with. Institutes such as the U.S. FDA are performing their initial explorations of the quality and reproducibility of the G2P systems (68). Nevertheless, ML will ultimately help to bridge the gap between a shortage of qualified personnel and the increasing volume, multidisciplinary character, and complexity of microbial test results.
It should be noted that universal automation in microbiology for all sample and specimen type workflows remains a considerable challenge. Variable and often limited types and volumes (cerebrospinal fluid, vitreous fluid, etc.), as well as preparation submethodology (extraction, sonication, crushing, etc.) and prioritizing test protocols for precious surgical specimens, add another layer of complexity possibly not fully compatible with AI requirements (22). As such, a balance needs to be reached between universal sample processing approaches and open systems for the orchestration of specimen workflow by middleware. Openness remains a clear challenge for manufacturers due to aspects of competition, instrument design, and regulatory requirements, among others. First steps here must preserve the microbiologist’s full control and oversight of the entire processing chain for final decisions based on personal knowledge and expertise; the “smartness” of the ML-driven automated solution then could be calibrated against this human reference and reiterated until ready for application.
AI is obviously also central in various aspects of clinical microbiology laboratory automation (54). A key example, for instance, is scalable decisional algorithms (plate-reading software) through which image analysis will facilitate extraction of additional information from a simple two-dimensional picture (growth/no growth, localization of isolated colonies, colony counting, the appearance of the colony of the growth medium [e.g., chromogenic culture media or plates containing blood, allowing for the observation of hemolysis], presumptive identification, and colony picking recommendation). The same capabilities must be provided to the microbiologist when reading images instead of reading physical culture plates to complete tasks in a faster, safer, and more reproducible manner. Once images become digital objects, new capabilities and performance will follow through better image analysis and decisional algorithms. Plate-reading software should include smart displays and ergonomic capabilities, which will lead to a huge improvement in logistic efficiency thanks to image sorting based on multiple criteria (specimen type, patient characteristics, time of incubation, etc.) instead of manual plate sorting and dealing only with plates of interest. Expert software systems allowing intelligent and automated result interpretation are in much need as well. Another clear issue is the global need to use the same diagnostic “languages,” of which there are many options. This, however, is still a domain of intense (strategic, political, and scientific) discussions where final choices have not been made yet (69). In conclusion, the question of the successful application of AI- and ML-driven diagnostics support is not one of if but when. We consider that the speed with which this happens will relate to ownership of the strategy and its implementation. By this we mean that several stakeholders are involved, namely, (i) the laboratory itself, (ii) the instrument manufacturers, (iii) the owners of the laboratory, and (iv) the academics and IT specialists responsible for developing the algorithmic solutions themselves. It is currently far from clear how these essential parties will work best together to (for example) agree on how best to share intellectual property (IP) and make this happen. On a final note, AI needs data, and most data of interest are essentially mobile. Thus, mobile devices will significantly increase in importance in this field of AI-accessible health care (70).
Do Microbiology Laboratories Benefit from Automation?
Most microbiology laboratories can benefit from automation, particularly those that process large numbers of relatively uncomplicated specimen types (e.g., urine specimens). It should be recognized, however, that growth of bacteria per se now is no longer an essential step, as nucleic acid amplification testing (NAAT) and hypersensitive antigen/antibody assays (enzyme immunoassay, ELISA, and single molecular array [SiMoA]) continue to be developed to play an ever-increasing role in infectious disease diagnostics (28, 71, 72). Indeed, the same sample may be subjected to multiple testing scenarios (Fig. 2). To this end, full automation should also accommodate the workflow for those specimens needing culture, nucleic acid extraction, direct immunochemistry assays, or a combination of each. Clearly, it is not always the size and throughput of the laboratory that is the single defining factor for the introduction of automation. For some laboratories, the number and types of special specimens received define the value of automation. Equally important may be the fact that for some of the special specimens, classical technologies have to be maintained. Still, the opportunities in the automation field are considered revolutionary by many (36, 73, 74). For these reasons, modularity and scalability (e.g., integration of multiple incubation/imaging systems, free-standing inoculators, incubators, culture plate processing systems, and integrated workstations) become important design considerations for manufacturers, such that a laboratory of any size would have access to components of the complete system. In addition, automation in laboratories within geographic regions where competition for skilled labor is high or acute shortages of qualified microbiology personnel occur would likely be advantageous (75). In this age of telemedicine, sharing digital images of bacterial growth on agar plates as well as stained smears in real time across the Internet for consultation (20), and even directed workup of cultures, represents a milestone in improving the quality of microbiology testing offered in distant (rural) hospital laboratories. Such digital technologies also have particular relevance to the potential transformation of services in low-resource settings (76). Additionally, sharing images with physicians can be educational and, optimally, inform clinical decisions.
Microbiology often requires unique judgment, objective thinking, and rapid decision-making. Automation will not replace these personal skills but may release and “enrich” the time for key decision-making individuals at the critical stages of culture evaluation by eliminating repetitive preanalytical and analytical tasks, including specimen sorting and plating (77). This is a key aspect where those with skills need to be properly aligned with skillful decision-making. In fact, there would be considerable labor savings if incubators were simply capable of cataloging the location of individual cultures for on-demand retrieval and review. Those components downstream of culture review would need to interface with systems selected for microbial ID/AST as well as (next-generation) sequence-based analysis.
Can Automation Improve Clinical Outcomes?
Automation in microbiology holds the promise of specimen processing on a real-time basis that could reduce TAT, improve patient outcomes, and limit unnecessary antibiotic use. Many examples of such advantages of automation to reduce patient identification error rate exist in clinical chemistry (78, 79). However, a significant reduction in TAT in microbiology remains elusive or even poorly defined. In manual processing laboratories, TAT is determined by operational hours, the delay and length of culture incubation (which can vary from 12 to 24 h depending upon when during the day-evening-night specimens are received and culture plates incubated), and workflow practices. Specimen transport delays from patient to laboratory must also be considered for TAT. Automation generally provides better isolation of colonies, thereby reducing the need for subcultures. However, workflow changes will be required to realize these advantages in the timeliness and quality of the results.
Automation, however, does not imply that a system runs by itself. Additional around-the-clock shifts could require additional people that, in combination with depreciation of the equipment, could lead to increasing per-test costs. It remains to be seen if AI algorithms would permit accurate interpretation and processing of positive culture results without any human intervention. If pertinent patient results are generated and posted on the laboratory information system overnight but not acted upon by medical personnel, then the opportunity is lost and the additional expense of 24-h operation is not realized in terms of patient outcome (80). There are increasing data that demonstrate the true impact and benefit of clinical microbiology results, such as rapid microbial identification (RMI), on patient management and outcomes (81, 82). As an example, most microbiology result-driven changes resulted in treatment escalation in the general patient population and treatment deescalation in the oncological patient population (83). Martiny et al. also demonstrated that in the pediatric population, RMI is particularly helpful in confirming contamination by cutaneous bacteria but never led to deescalation of treatment, most likely because there is a somewhat paradoxical reluctance to stop treatment in a patient who is improving. The delay in modifying the treatment was high (>4 h in about 50% of cases), suggesting that communication needs to be improved further. These and other observations shed some thoughtful light on the significant impact of RMI (or lack of impact) in antimicrobial stewardship initiatives and emphasize the need for matrix-assisted laser desorption ionization (MALDI) ID processes to be combined with an antimicrobial stewardship program (84–86) and, by extension, the role of clinical pharmacists who need to be included in, and contribute to, better coordination of care (87).
SECONDARY BENEFITS OF CONSOLIDATING LABORATORY SERVICES
Beyond the patient-centered improvements described above, the ability to develop a sustainable consolidated clinical microbiology laboratory service represents an opportunity for other health fields, from public health surveillance to translational medicine.
Consolidation and Real-Time Microbiological Surveillance
The detection of abnormal events or the sudden increase or emergence of certain types of infection can be adequately examined in consolidated laboratories with or without the support of satellite facilities (88). The use of a bacterial real-time laboratory-based surveillance system (BALYSES) for monitoring patients infected with certain bacterial species and the Marseille Antibiotic Resistance Surveillance System (MARSS), which surveys β-lactam resistance phenotypes for 15 species, demonstrated this (88). BALYSES and MARSS enabled the detection of 52 abnormal events for 24 bacterial species, leading to 19 official reports in 1 year. This shows that in big laboratory settings, the integrated use of historic and actual data can improve the level of clinical insight. Long-term longitudinal analysis using similar tools also showed its usefulness (89). During an 11-year surveillance period, hundreds of events were surveyed weekly, including clinical samples, diagnostic tests, and antibacterial resistance patterns. This detailed analysis revealed a mean number of 0.5 alerts/week for abnormal microbiological events. The recent development of FilmArray Trend, a commercial cloud epidemiology system based on the integration of different laboratories’ exported data from FilmArray respiratory panel (RP) tests, represents another way to easily investigate geographical dynamics of respiratory diseases on a large scale without any home-made development (www.syndromictrends.com). This example of “virtual laboratory consolidation” will play an important role in data collection and comparison. In addition, consolidated clinical microbiology laboratories can actively support ongoing surveillance, e.g., on AMR, by connecting some or all of the produced data (under appropriate management and regulatory structures) to national public health surveillance systems or international networks, such as EARS-Net (European Union), CAESAR (Central Asia and Eastern Europe), ReLAVRA (Latin America), or the Global Antimicrobial Resistance Surveillance System (GLASS), thanks to WHONET software (90, 91). The Infection Response Through Virus Genomics–ICONIC consortium in London showed similar sensitivity and specificity for the monitoring of viral infections over extended periods of time (92). The clinical workflow implemented by the members of ICONIC covered sample handling (also at long distance), sequencing of viral genomes, interpretation of the genomic data, and ultimately clinical reporting (93). The consortium delivered refined maps showing influenza A virus hospital transmission chains and the frequent nosocomial introductions from the community. The emergence of novel subclades was defined and their limited spread in the hospital taken as being representative of adequate infection control. This was true both within the specific London/UK context as well as in Brussels/Belgium for comparisons of the detection sensitivity of influenza A subclade identification over a given season (7). Of note, genomic surveillance is becoming more and more common in many European countries (94). Recently a more complete review on the use of overall, well-connected public health surveillance was published from within the ECDC (95). Activities include harmonization of laboratory diagnostics, antimicrobial susceptibility testing and molecular typing methods, a multicenter method validation, technical capacity mapping, training of staff, and quality assessment of testing. Again, this is an important example of politically and medically driven virtual laboratory consolidation. Key priorities included optimization and broader use of rapid diagnostics, further integration of whole-genome sequencing (WGS), and electronic linkage of laboratory and public health systems. Obviously, this task is expected to become increasingly simple with the consolidation of physical laboratories into larger units.
Consolidation and Implementation of New(er) Technologies
Translational research using new techniques and more holistic approaches in data management support the overall feasibility of consolidated clinical microbiology, with or without the physical integration of laboratories. A number of high-profile translational research initiatives are based on the availability of big infrastructural units (100,000 Genomes Project in the UK and the Precision Medicine Initiative in the United States) that would support the eventual technological transfer of high-throughput analytical approaches from the research into the (consolidated) clinical laboratory. The interaction of consolidated clinical laboratory networks with basic academic health scientists (e.g., health economists, biostatisticians, bioinformaticians, molecular biologists, physicists, etc.) perhaps is inevitable under the umbrella of such large consortia. As a result, the speed at which promising diagnostic tests move through the academic pipeline into clinical application(s) may increase while at the same time maintaining the breadth of creative approaches.
Simple text messaging and the transfer of pictures using smartphone technology has already been shown to be important in outbreak settings (96). More integrated phone-based diagnostic platforms have been developed as well and already quietly find their way into laboratories (97). In addition, early-stage microbiological research investigating the potential for micromanipulation and microfluidics are emerging (98, 99). Identification using MALDI–time of flight mass spectrometry (MALDI-TOF MS) and sequence-based analysis are becoming routinely available in the bigger laboratories (100). Indeed, the expanded use of new(er) techniques continues to lessen the level of microbiology expertise required of bench technologists to rapidly, accurately, and consistently identify microorganisms. However, a deeper understanding and appreciation of these different “-omics” processes and an easy solution to provide universally accessible downstream abilities to query databases would be highly desirable. The need for more deeply developed bioinformatics approaches is essential (101). There are already significant efforts to move next-generation sequencing (NGS) to the heart of the microbiology laboratory as a universal tool for pathogen detection and identification (102), antimicrobial resistance prediction, and molecular epidemiology (103, 104), where it has been scientifically demonstrated to be promising (105–107). However, further studies are needed to better identify the benefits of the integration of NGS into current testing algorithms (108–110).
In addition, it is likely that novel resistance mechanisms first will be identified by growth-based methods or subsequent to highlighted treatment failures and then confirmed by comparative sequence analysis, so clinical microbiologists will have to be familiar with both technologies (111). Sequence-based resistance detection will remain easier to detect than full susceptibility, primarily since it will be challenging to adequately predict full antibiotic susceptibility. Ultimately, testing gene expression differences using RNA approaches may be more likely to generate adequate diagnostic or therapeutic profiles (112). More details can be found in two excellent recent reviews (113, 114). Consolidated laboratories are unlikely to be the first routine laboratories that start implementing these technologies, but this should not absolve them of a watchful and engaged eye on the rapidity with which such approaches will find their way into routine clinical microbiology. Alternatively, consolidated laboratories might provide the platform for introduction of such new technologies if there were sufficient throughput, quality, and financial incentives to do so.
Consolidated Biobanking
Collecting clinical specimens for retrospective use and combining these with extensive clinical, diagnostic, geographic, and demographic data in biobanks is essential for research purposes and the development of improved diagnostic tools and by contributing to patient care on the basis of retrospective tracing of specific pathogens/conditions. The NIH Human Microbiome Project, which served as a catalyst for human microbiome research, highlights the added value of large consolidated biobanking (115, 116). The International Agency for Research on Cancer (IARC) Biobank, the UK Biobank, and the China Kadoorie Biobank are three other major examples. While the latter two reflect large national population cohorts, the IARC Biobank is centered on a certain pathology across a large number of geographical areas (117). Consolidated microbiology laboratories and the sheer number of specimens they process will directly affect biobanking by increasing their operational capacities and flexibility through greater automation and connectivity/integration to health care workflows and/or middleware in order to ensure their effectiveness and sustainability (118). Sharing samples, patients’ data, and methods through networks is now more important than ever and often requires interaction with less traditional collaborating specialties, such as engineering, law, and computer science, to foster translational medicine (119).
As a result, biobanks are expected to be repositioned from Mertonian functionalism (for the good of society as a whole) to agency-based frameworks that focus on performance-related processes (120). Biobanking will become increasingly embedded in health care systems that use longitudinal samples from individuals as part of their personal care and will favor translational research, aiming to bring products and therapies quickly to market (121, 122). This is anticipated by the creation of new standards (ISO/DIS 20387 Biotechnology–Biobanking, 2018) and alignment with existing best practices, supporting the deeper clinical integration of such infrastructure facilities (123).
CONCLUDING REMARKS
Many challenges exist for the development of the microbiology laboratory of the future. These include the expanding need for automation, increasingly voluminous data and their management and automated interpretation, the governance of data privacy, the introduction of new high-throughput and data-intense technologies in routine practice, biobanking, local entrepreneurship, and others (124). Providing solutions that generate optimal and affordable health care services need to be at the core of all of these activities. For patient management, consolidated laboratories need to offer the same quality of patient care by supporting clinical decision-making despite delocalization of sample analysis. Determining what tests will remain on satellite sites and what tests will be sent to the consolidated laboratory must be determined by the various stakeholders, including care providers at the referring sites.
The consolidation of microbiological laboratories is often linked to the purchase of expensive laboratory-dedicated infrastructure and equipment, including, for example, MALDI-TOF MS, next-generation sequencing, full laboratory automation, and automated molecular diagnostics or AI-driven solutions. Consolidation also allows for expansion of laboratory testing to a 24/7 model, something that may not be available or even possible in smaller health care settings. Around-the-clock testing has the potential for achieving reduced TATs, in turn potentially resulting in shortened lengths of hospital stay and improved patient outcomes as long as the health care provider also rises to the 24-h challenge. New developments in communication technologies need to underpin consultation between laboratories and/or clinicians and even a rethink of workflows.
For disease surveillance, consolidated laboratories should provide integrated real-time epidemiology for a large percentage of clinically relevant pathogens, covering viruses, bacteria, fungi, and parasites. To implement such structures, there is a clear need for the extensive integration of existing data streams, e.g., genomics and electronic health records (eHR) (including laboratory records), as well as the development of new solutions, most likely a wider array of middleware solutions, preferably taken up in partnerships with other stakeholder groups. For biobanking and translational medical research, stronger integration into clinical workflow is needed to provide ongoing support for discovery science and precision medicine.
It has to be noted that the true costs or profits of consolidation largely remain to be determined. Obviously, there are costs incurred with prolonged opening times, offset by labor expenditures and operational costs. On the other hand, 24/7 service delivery allows optimal and lean use of laboratory space and resources and, therefore, allows for an increase of volume of analysis without additional investments. Outcome studies that assess these issues are needed. Furthermore, the logistics of moving samples across different sites toward a centralized location remains a substantial technical challenge that is only too often underestimated. Strategies that investigate different possibilities, such as direct, local molecular testing or even automation in smaller laboratory settings, also need to be taken into account.
The maintenance of microbiological competency at the highest level among technologists working at satellite sites, distant from the consolidated microbiological laboratories, needs to be supported by technological innovation, aiding clinician-specialist interactions. In our current view, there are at least two major options: consolidation and automation should jointly focus on improvement of patient outcome and/or heightened laboratory income. In our opinion, both can be simultaneously realized with sufficient strategic thought and planning concerning the points we raise here.
ACKNOWLEDGMENTS
We gratefully acknowledge the help of all those involved in the development of the LHUB–ULB and the Groupement Hospitalier Universitaire de Bruxelles (GHUB). Jean-François Gorse (bioMérieux, France) is gratefully acknowledged for his insightful input in the automation sections of the manuscript. We are grateful to Jost Landgrebe (Cognotekt, Cologne) for critically reading and commenting on the manuscript.
V.G. thanks the UCLH Biomedical Research Centre’s support of his academic activities.
O.V., M.H., A.D., V.G., and Z.K. have no personal or financial interests to declare. A.V.B. and G.D. are employees of bioMérieux and P.M. is an employee of BD Diagnostic Systems, companies designing, developing, and selling diagnostic tests for infectious diseases. bioMérieux and BD Diagnostic Systems had no part in the design and writing of this work.
Where authors are identified as personnel of the International Agency for Research on Cancer/WHO, the authors alone are responsible for the views expressed in this article and do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/WHO.
Biographies

Olivier Vandenberg was trained as a clinical microbiologist (2001) and completed his Ph.D. in Biomedical Sciences (2006) at Université Libre de Bruxelles (ULB), Brussels, Belgium. He is currently Head of the Innovation and Business Development Unit of the University Laboratory of Brussels (LHUB-ULB) and Honorary Senior Lecturer of the Division of Infection & Immunity, University College London (UCL). In addition, he is presently Professor of Microbiology in the School of Public Health and in the Faculty of Medicine of the Université Libre de Bruxelles (ULB). His research focuses on the clinical impact of new diagnostic tools in both industrialized and low-resource settings. Since 2000, he has supervised the consolidation of several microbiology laboratories, allowing the implementation of infectious disease surveillance programs in industrialized but also in low- and middle-income countries. Besides this, he collaborates with different manufacturers in the development of new approaches for the diagnosis and control of infectious diseases.

Géraldine Durand joined bioMérieux in 2007, and her current position is Europe R&D Microbiology Director in charge of microbiology product development for culture, identification, and antimicrobial susceptibility testing. She received her Pharm D from the University of Lyon, France. After an internship in medical biology and obtaining an MSc at the French National Reference Center for Staphylococci, she completed her residency in clinical biology at the Hospices Civils de Lyon, where she earned her Ph.D. She conducted her research work at the French National Reference Center for Staphylococci. Her research was focused on methicillin-resistant Staphylococcus aureus (MRSA). She described a new emerging MRSA clone (Geraldine clone). Her current research is directed at rapid microbiology, mass spectrometry, next-generation sequencing, and laboratory automation, mainly driven by the medical value brought by imaging technologies for bacterial culture.

Marie Hallin completed her M.D. in 2000, followed by residency training in the Laboratory Medicine at the Université Libre de Bruxelles, Brussels, Belgium. She subsequently completed a Ph.D. thesis on the comparative genomics of methicillin-resistant Staphylococcus aureus epidemic clones. She is currently Head of the Department of Microbiology of the University Laboratory of Brussels (LHUB-ULB). Dr. Hallin’s primary research focus is the diagnosis and understanding of the epidemiology of multidrug-resistant pathogens. She also studies the clinical impact of user‐friendly field‐adapted diagnostics for nonexpert users.

Andreas Diefenbach is Professor and Chair of Microbiology at Charité–Universitätsmedizin Berlin and Director of Microbiology at Labor Berlin–Charité and Vivantes GmbH. His laboratory studies the development and function of the innate immune system. Andreas studied Medicine at the University of Erlangen and graduated in Microbiology and Immunology (1998). He obtained postdoctoral training at the Department of Molecular and Cellular Biology, University of California Berkeley (1999 to 2002). Prior to joining Charité and Labor Berlin, he was an Assistant Professor at the Skirball Institute of Biomolecular Medicine, New York University (2003 to 2006), a Full Professor at the University of Freiburg (2006 to 2013), and Professor and Chair of Medical Microbiology at the Johannes-Gutenberg-University Mainz (2013 to 2016). As a Physician Scientist, Andreas has been supported by grants from the Howard Hughes Medical Institute, the European Research Council, and the Deutsche Forschungsgemeinschaft (DFG). He is the coordinator of the DFG Priority Program 1937 (“Innate Lymphoid Cells”). He was awarded the Main Scientific Prize of the German Society of Hygiene and Microbiology and was a Kavli Fellow of the National Academy of Sciences USA and Alexander-von-Humboldt-Stiftung. Since 2017, he is recognized as one of the Highly Cited Researchers (Clarivate Analytics).

Vanya Gant is an academic microbiology and infectious disease clinician with a Ph.D. in cellular immunology. He contributed to the design and cocreation of the Health Services Laboratories, a consolidated public/private partnership in London, delivering over 35 million tests a year. He coauthored a book on artificial neural networks in clinical medicine and is Co-Principal Investigator for an NIHR program grant on antimicrobial resistance. He has a keen interest in industry pullthrough from academic innovation and implementation science.

Patrick Murray directed clinical laboratories in academia (Washington University in Saint Louis, University of Maryland) for 25 years and in the government (National Institutes of Health) for 10 years. In July 2011 he retired from the NIH and accepted his current position at BD Life Sciences as Vice President, Worldwide Scientific Affairs. He is a fellow of the American Academy of Microbiology and the Infectious Disease Society of America and has authored more than 275 research articles and 20 books.

Zisis Kozlakidis completed his Ph.D. in microbiology (2005) at Imperial College London, UK. He is currently Head of Laboratory Services and Biobanking at the International Agency for Cancer Research, World Health Organization (WHO) and Visiting Professor at the School of Medical Sciences, Central South University, Changsha, China, and he holds a visiting faculty post at Cass Business School, University of London. His research focuses on the clinical impact of new diagnostic and precision medicine tools, particularly in low-resource settings. Since 2009, he has supervised the creation and operation of several microbiology biobanks globally, supporting integrated healthcare projects and infectious disease surveillance programs. He was named UK Healthcare Innovator of the year in 2018 by the Institute of Engineering and Technology, UK. He has authored more than 60 peer-reviewed papers and is a Turnberg Fellow of the UK Academy of Medical Sciences.

Alex van Belkum, fellow of the American Academy of Microbiology, is a research microbiologist who, after a 15-year academic career, moved from university to industry about ten years ago. He has a global interest in translational aspects of fundamental microbiological research and the development of innovative diagnostics for microbial infectious diseases. Alex van Belkum published over 550 peer-reviewed papers and has an H factor slowly approaching 100. Importantly, he is the father to three and grandfather to his four best friends.
REFERENCES
- 1.Thomson S, Figueras J, Evetovits T. 2014. Economic crisis, health systems and health in Europe: impact and implications for policy. European Observatory on Health System and Policies, Copenhagen WHO, Copenhagen, Denmark. [PubMed] [Google Scholar]
- 2.Peeling RW, Murtagh M, Olliaro PL. 2019. Epidemic preparedness: why is there a need to accelerate the development of diagnostics? Lancet Infect Dis 19:e172–e178. doi: 10.1016/S1473-3099(18)30594-2. [DOI] [PubMed] [Google Scholar]
- 3.Miller MB, Atrzadeh F, Burnham CA, Cavalieri S, Dunn J, Jones S, Mathews C, McNult P, Meduri J, Newhouse C, Newton D, Oberholzer M, Osiecki J, Pedersen D, Sweeney N, Whitfield N, Campos J, ASM Clinical and Public Health Microbiology Committee and the ASM Corporate Council . 2019. Clinical utility of advanced microbiology testing tools. J Clin Microbiol 57:e00495-19. doi: 10.1128/JCM.00495-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naugler C, Church DL. 2019. Automation and artificial intelligence in the clinical laboratory. Crit Rev Clin Lab Sci 56:98–110. doi: 10.1080/10408363.2018.1561640. [DOI] [PubMed] [Google Scholar]
- 5.Plebani M. 2016. Harmonization in laboratory medicine: requests, samples, measurements and reports. Crit Rev Clin Lab Sci 53:184–196. doi: 10.3109/10408363.2015.1116851. [DOI] [PubMed] [Google Scholar]
- 6.Sautter RL, Thomson RB Jr. 2015. Consolidated clinical microbiology laboratories. J Clin Microbiol 53:1467–1472. doi: 10.1128/JCM.02569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Wijngaert S, Bossuyt N, Ferns B, Busson L, Serrano G, Wautier M, Thomas I, Byott M, Dupont Y, Nastouli E, Hallin M, Kozlakidis Z, Vandenberg O. 2019. Bigger and better? Representativeness of the Influenza A surveillance using one consolidated clinical microbiology laboratory data set as compared to the Belgian Sentinel Network of Laboratories. Front Public Health 7:150. doi: 10.3389/fpubh.2019.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church DL, Hall P. 1999. Centralization of a regional clinical microbiology service: the Calgary experience. Can J Infect Dis 10:393–402. doi: 10.1155/1999/372382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright JR., Jr 2015. Calgary laboratory services: a unique Canadian model for an academic department of pathology and laboratory medicine succeeding in the face of provincial integration of public, private, and academic laboratories. Acad Pathol 2:2374289515619944. doi: 10.1177/2374289515619944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagstaff A, Yip W, Lindelöw M, Hsiao WC. 2009. China’s health system and its reform: a review of recent studies. Health Econ 18:S7–S23. doi: 10.1002/hec.1518. [DOI] [PubMed] [Google Scholar]
- 11.Blumenthal D, Hsiao W. 2015. Lessons from the East–China’s rapidly evolving health care system. N Engl J Med 372:1281–1285. doi: 10.1056/NEJMp1410425. [DOI] [PubMed] [Google Scholar]
- 12.Burns LR, Liu GG. 2017. China’s healthcare system and reform (part II: healthcare reform). Cambridge University Press, Cambridge, UK. [Google Scholar]
- 13.Zhang L, Wang H, Li Q, Zhao MH, Zhan QM. 2018. Big data and medical research in China. BMJ 360:j5910. doi: 10.1136/bmj.j5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paramasivan CN, Lee E, Kao K, Mareka M, Kubendiran G, Kumar TA, Keshavjee S, Satti H, Alabi G, Raviglione M, Roscigno G. 2010. Experience establishing tuberculosis laboratory capacity in a developing country setting. Int J Tuber Lung Dis 14:59–64. [PubMed] [Google Scholar]
- 15.Wertheim BM, Aguirre AJ, Bhattacharyya RP, Chorba J, Jadhav AP, Kerry VB, Macklin EA, Motyckova G, Raju S, Lewandrowski K, Hunt DP, Wright DE. 2017. An educational and administrative intervention to promote rational laboratory test ordering on an academic general medicine service. Am J Med 130:47–53. doi: 10.1016/j.amjmed.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drancourt M, Michel-Lepage A, Boyer S, Raoult D. 2016. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev 29:429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poljak M, Šterbenc A. 28 September 2019. Use of drones in clinical microbiology and infectious diseases: current status, challenges and barriers. Clin Microbiol Infect doi: 10.1016/j.cmi.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Vandenberg O, Kozlakidis Z, Schrenzel J, Struelens MJ, Breuer J. 2018. Control of infectious diseases in the era of European clinical microbiology laboratory consolidation: new challenges and opportunities for the patient and for public health surveillance. Front Med (Lausanne) 5:15. doi: 10.3389/fmed.2018.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhoads DD, Sintchenko V, Rauch CA, Pantanowitz L. 2014. Clinical microbiology informatics. Clin Microbiol Rev 27:1025–1047. doi: 10.1128/CMR.00049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mascart G, Martiny D, Miendje Y, Tummers G, Mahadeb B, Vandenberg O, Hallin M. 2018. The future of automation in bacteriology. Ann Biol Clin (Paris) 76:365–372. doi: 10.1684/abc.2018.1366. [DOI] [PubMed] [Google Scholar]
- 21.Libertin CR, Watson SH, Tillett WL, Peterson JH. 2017. Dramatic effects of a new antimicrobial stewardship program in a rural community hospital. Am J Infect Control 45:979–982. doi: 10.1016/j.ajic.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Ledeboer NA, Dallas SD. 2014. The automated clinical microbiology laboratory: fact or fantasy? J Clin Microbiol 52:3140–3146. doi: 10.1128/JCM.00686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina G, Greub G. 2016. Automation in bacteriology: a changing way to perform clinical diagnosis in infectious diseases. Clin Microbiol Infect 22:215–216. doi: 10.1016/j.cmi.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 24.Bailey AL, Burnham CD. 2019. Reducing the time between inoculation and first-read of urine cultures using total lab automation significantly reduces turn-around-time of positive culture results with minimal loss of first-read sensitivity. Eur J Clin Microbiol Infect Dis 38:1135–1141. doi: 10.1007/s10096-019-03512-3. [DOI] [PubMed] [Google Scholar]
- 25.Perry JD. 2017. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin Microbiol Rev 30:449–479. doi: 10.1128/CMR.00097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dauwalder O, Landrieve L, Laurent F, de Montclos M, Vandenesch F, Lina G. 2016. Does bacteriology laboratory automation reduce time to results and increase quality management? Clin Microbiol Infect 22:236–243. doi: 10.1016/j.cmi.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 27.Quiblier C, Jetter M, Rominski M, Mouttet F, Böttger EC, Keller PM, Hombach M. 2016. Performance of Copan WASP for routine urine microbiology. J Clin Microbiol 54:585–592. doi: 10.1128/JCM.02577-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AK, Houpt ER. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 29.Broyer P, Perrot N, Rostaing H, Blaze J, Pinston F, Gervasi G, Charles MH, Dachaud F, Dachaud J, Moulin F, Cordier S, Dauwalder O, Meugnier H, Vandenesch F. 2018. An automated sample preparation instrument to accelerate positive blood cultures microbial identification by MALDI-TOF mass spectrometry (Vitek MS). Front Microbiol 9:911. doi: 10.3389/fmicb.2018.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croxatto A, Dijkstra K, Prod'hom G, Greub G. 2015. Comparison of inoculation with InoqulA and WASP automated systems with manual inoculation. J Clin Microbiol 53:2298–2307. doi: 10.1128/JCM.03076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iversen J, Stendal G, Gerdes CM, Meyer CH, Andersen CØ, Frimodt-Møller N. 2016. Comparative evaluation of inoculation of urine samples with the Copan WASP and BD Kiestra InoqulA instruments. J Clin Microbiol 54:328–332. doi: 10.1128/JCM.01718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croxatto A, Prod'hom G, Faverjon F, Rochais Y, Greub G. 2016. Laboratory automation in clinical bacteriology: what system to choose? Clin Microbiol Infect 22:217–235. doi: 10.1016/j.cmi.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Croxatto A, Marcelpoil R, Orny C, Morel D, Prod'hom G, Greub G. 2017. Towards automated detection, semi-quantification and identification of microbial growth in clinical bacteriology: a proof of concept. Biomed J 40:317–328. doi: 10.1016/j.bj.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein S, Nurjadi D, Horner S, Heeg K, Zimmermann S, Burckhardt I. 2018. Significant increase in cultivation of Gardnerella vaginalis, Alloscardovia omnicoles, Actinotignum schaalii, and Actinomyces spp. in urine samples with total laboratory automation. Eur J Clin Microbiol Infect Dis 37:1305–1311. doi: 10.1007/s10096-018-3250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lainhart W, Burnham C. 2018. Enhanced recovery of fastidious organisms from urine culture in the setting of total laboratory automation. J Clin Microbiol 56:e00546-18. doi: 10.1128/JCM.00546-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey A, Ledeboer N, Burnham CD. 2019. Clinical microbiology is growing up: the total laboratory automation revolution. Clin Chem 65:634–643. doi: 10.1373/clinchem.2017.274522. [DOI] [PubMed] [Google Scholar]
- 37.Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D. 2013. The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur J Clin Microbiol Infect Dis 32:637–645. doi: 10.1007/s10096-012-1787-3. [DOI] [PubMed] [Google Scholar]
- 38.Dubourg G, Lagier JC, Armougom F, Robert C, Hamad I, Brouqui P, Raoult D. 2013. The proof of concept that culturomics can be superior to metagenomics to study atypical stool samples. Eur J Clin Microbiol Infect Dis 32:1099. doi: 10.1007/s10096-013-1843-7. [DOI] [PubMed] [Google Scholar]
- 39.Greub G. 2012. Culturomics: a new approach to study the human microbiome. Clin Microbiol Infect 18:1157–1159. doi: 10.1111/1469-0691.12032. [DOI] [PubMed] [Google Scholar]
- 40.Lagier JC, Bilen M, Cadoret F, Drancourt M, Fournier PE, La Scola B, Raoult D. 2018. Naming microorganisms: the contribution of the IHU Méditerranée Infection, Marseille, France. New Microbes New Infect 26:S89–S95. doi: 10.1016/j.nmni.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mailhe M, Ricaboni D, Vitton V, Gonzalez JM, Bachar D, Dubourg G, Cadoret F, Robert C, Delerce J, Levasseur A, Fournier PE, Angelakis E, Lagier JC, Raoult D. 2018. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol 18:157. doi: 10.1186/s12866-018-1304-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Haak BW, Prescott HC, Wiersinga WJ. 2018. Therapeutic potential of the gut microbiota in the prevention and treatment of sepsis. Front Immunol 9:2042. doi: 10.3389/fimmu.2018.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamburini FB, Andermann TM, Tkachenko E, Senchyna F, Banaei N, Bhatt AS. 2018. Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat Med 24:1809–1814. doi: 10.1038/s41591-018-0202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burnham CA, Dunne WM Jr, Greub G, Novak SM, Patel R. 2013. Automation in the clinical microbiology laboratory. Clin Chem 59:1696–1702. doi: 10.1373/clinchem.2012.201038. [DOI] [PubMed] [Google Scholar]
- 45.Bourbeau PP, Ledeboer NA. 2013. Automation in clinical microbiology. J Clin Microbiol 51:1658–1665. doi: 10.1128/JCM.00301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beam AL, Kohane IS. 2016. Translating artificial intelligence into clinical care. JAMA 316:2368–2369. doi: 10.1001/jama.2016.17217. [DOI] [PubMed] [Google Scholar]
- 47.Burton RJ, Albur M, Eberl M, Cuff SM. 2019. Using artificial intelligence to reduce diagnostic workload without compromising detection of urinary tract infections. BMC Med Inform Decis Mak 19:171. doi: 10.1186/s12911-019-0878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chae S, Kwon S, Lee D. 2018. Predicting infectious disease using deep learning and big data. IJERPH 15:1596. doi: 10.3390/ijerph15081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dakappa PH, Prasad K, Rao SB, Bolumbu G, Bhat GK, Mahabala C. 2018. Classification of infectious and noninfectious diseases using artificial neural networks from 24-hour continuous tympanic temperature data of patients with undifferentiated fever. Crit Rev Biomed Eng 46:173–183. doi: 10.1615/CritRevBiomedEng.2018025917. [DOI] [PubMed] [Google Scholar]
- 50.Tou H, Yao L, Wei Z, Zhuang X, Zhang B. 2018. Automatic infection detection based on electronic medical records. BMC Bioinformatics 19:117. doi: 10.1186/s12859-018-2101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong ZSY, Zhou J, Zhang Q. 2019. Artificial intelligence for infectious disease big data analytics. Infect Dis Health 24:44–48. doi: 10.1016/j.idh.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Nault V, Pepin J, Beaudoin M, Perron J, Moutquin JM, Valiquette L. 2017. Sustained impact of a computer-assisted antimicrobial stewardship intervention on antimicrobial use and length of stay. J Antimicrob Chemother 72:933–940. doi: 10.1093/jac/dkw468. [DOI] [PubMed] [Google Scholar]
- 53.van den Bogert B, Boekhorst J, Pirovano W, May A. 2019. On the role of bioinformatics and data science in industrial microbiome applications. Front Genet 10:721. doi: 10.3389/fgene.2019.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shapshak P, Somboonwit C, Sinnott JT. 2017. Artificial intelligence and virology–quo vadis. Bioinformation 13:410–411. doi: 10.6026/97320630013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith KP, Kang AD, Kirby JE. 2017. Automated interpretation of blood culture Gram stains by use of a deep convolutional neural network. J Clin Microbiol 56:e01521-17. doi: 10.1128/JCM.01521-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreini P, Bonechi S, Bianchini M, Garzelli A, Mecocci A. 2016. Automatic image classification for the urinoculture screening. Comput Biol Med 70:12–22. doi: 10.1016/j.compbiomed.2015.12.025. [DOI] [PubMed] [Google Scholar]
- 57.Rajkomar A, Dean J, Kohane I. 2019. Machine learning in medicine. N Engl J Med 380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 58.Qu K, Guo F, Liu X, Lin Y, Zou Q. 2019. Application of machine learning in microbiology. Front Microbiol 10:827. doi: 10.3389/fmicb.2019.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li BY, Oh J, Young VB, Rao K, Wiens J. 2019. Using machine learning and the electronic health record to predict complicated Clostridium difficile infection. Open Forum Infect Dis 6:ofz186. doi: 10.1093/ofid/ofz186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murali A, Bhargava A, Wright ES. 2018. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6:140. doi: 10.1186/s40168-018-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macesic N, Polubriaginof F, Tatonetti NP. 2017. Machine learning: novel bioinformatics approaches for combating antimicrobial resistance. Curr Opin Infect Dis 30:511–517. doi: 10.1097/QCO.0000000000000406. [DOI] [PubMed] [Google Scholar]
- 62.Jaillard M, van Belkum A, Cady KC, Creely D, Shortridge D, Blanc B, Barbu EM, Dunne WM Jr, Zambardi G, Enright M, Mugnier N, Le Priol C, Schicklin S, Guigon G, Veyrieras JB. 2017. Correlation between phenotypic antibiotic susceptibility and the resistome in Pseudomonas aeruginosa. Int J Antimicrob Agents 50:210–218. doi: 10.1016/j.ijantimicag.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 63.Tanmoy AM, Westeel E, De Bruyne K, Goris J, Rajoharison A, Sajib MSI, van Belkum A, Saha SK, Komurian-Pradel F, Endtz HP. 2018. Salmonella enterica serovar Typhi in Bangladesh: exploration of genomic diversity and antimicrobial resistance. mBio 9:e02112-18. doi: 10.1128/mBio.02112-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahé P, Tournoud M. 2018. Predicting bacterial resistance from whole-genome sequences using k-mers and stability selection. BMC Bioinformatics 19:383. doi: 10.1186/s12859-018-2403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Landgrebe J, Smith B. 8 April 2019. Making AI meaningful again. Synthese doi: 10.1007/s11229-019-02192-y. [DOI] [Google Scholar]
- 66.Jaillard M, Lima L, Tournoud M, Mahé P, van Belkum A, Lacroix V, Jacob L. 2018. A fast and agnostic method for bacterial genome-wide association studies: bridging the gap between k-mers and genetic events. PLoS Genet 14:e1007758. doi: 10.1371/journal.pgen.1007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su M, Satola SW, Read TD. 2018. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57:e01405-18. doi: 10.1128/JCM.01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.U.S. FDA. Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD). Discussion paper and request for feedback. U.S. Food and Drug Administration, Washington, DC. [Google Scholar]
- 69.Gansel X, Mary M, van Belkum A. 2019. Semantic data interoperability, digital medicine, and e-health in infectious disease management: a review. Eur J Clin Microbiol Infect Dis 38:1023–1034. doi: 10.1007/s10096-019-03501-6. [DOI] [PubMed] [Google Scholar]
- 70.Sim I. 2019. Mobile devices and health. N Engl J Med 381:956–968. doi: 10.1056/NEJMra1806949. [DOI] [PubMed] [Google Scholar]
- 71.Banz A, Lantz A, Riou B, Foussadier A, Miller M, Davies K, Wilcox M. 2018. Sensitivity of single-molecule array assays for detection of Clostridium difficile toxins in comparison to conventional laboratory testing algorithms. J Clin Microbiol 56:e00452-18. doi: 10.1128/JCM.00452-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pollock NR, Banz A, Chen X, Williams D, Xu H, Cuddemi CA, Cui AX, Perrotta M, Alhassan E, Riou B, Lantz A, Miller MA, Kelly CP. 2019. Comparison of Clostridioides difficile stool toxin concentrations in adults with symptomatic infection and asymptomatic carriage using an ultrasensitive quantitative immunoassay. Clin Infect Dis 68:78–86. doi: 10.1093/cid/ciy415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yarbrough ML, Lainhart W, McMullen AR, Anderson NW, Burnham CD. 2018. Impact of total laboratory automation on workflow and specimen processing time for culture of urine specimens. Eur J Clin Microbiol Infect Dis 37:2405–2411. doi: 10.1007/s10096-018-3391-7. [DOI] [PubMed] [Google Scholar]
- 74.De Socio GV, Di Donato F, Paggi R, Gabrielli C, Belati A, Rizza G, Savoia M, Repetto A, Cenci E, Mencacci A. 2018. Laboratory automation reduces time to report of positive blood cultures and improves management of patients with bloodstream infection. Eur J Clin Microbiol Infect Dis 37:2313–2322. doi: 10.1007/s10096-018-3377-5. [DOI] [PubMed] [Google Scholar]
- 75.Humphreys H, Nagy E, Kahlmeter G, Ruijs GJHM. 2010. The need for European professional standards and the challenges facing clinical microbiology. Eur J Clin Microbiol Infect Dis 29:617–621. doi: 10.1007/s10096-010-0906-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lan NPH, Van Dung N, Visser C, Anh TTN, Bay PVB, Hong TTK, Brinke P, Hendriks W, Osinga T, van der Waals F, Botma J, Hien TT, Farrar JJ, van Doorn HR, Chau NVV, de Jong MD. 2014. Network building and knowledge exchange with telemicrobiology. Lancet Glob Health 2:e78. doi: 10.1016/S2214-109X(13)70112-8. [DOI] [PubMed] [Google Scholar]
- 77.Glasson J, Hill R, Summerford M, Olden D, Papadopoulos F, Young S, Giglio S. 2017. Multicenter evaluation of an image analysis device (APAS): comparison between digital image and traditional plate reading using urine cultures. Ann Lab Med 37:499–504. doi: 10.3343/alm.2017.37.6.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angeletti S, De Cesaris M, Hart JG, Urbano M, Vitali MA, Fragliasso F, Dicuonzo G. 2015. Laboratory automation and intra-laboratory turnaround time: experience at the University Hospital Campus Bio-Medico of Rome. J Lab Autom 20:652–658. doi: 10.1177/2211068214566458. [DOI] [PubMed] [Google Scholar]
- 79.Markin RS, Whalen SA. 2000. Laboratory automation: trajectory, technology, and tactics. Clin Chem 46:764–771. doi: 10.1093/clinchem/46.5.764. [DOI] [PubMed] [Google Scholar]
- 80.Brecher SM. 2017. Waltzing around sacred cows on the way to the future. J Clin Microbiol 56:e01779-17. doi: 10.1128/JCM.01779-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vlek AL, Bonten MJ, Boel CH. 2012. Direct matrix-assisted laser desorption ionization time-of-flight mass spectrometry improves appropriateness of antibiotic treatment of bacteremia. PLoS One 7:e32589. doi: 10.1371/journal.pone.0032589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Delport JA, Strikwerda A, Armstrong A, Schaus D, John M. 2017. MALDI-ToF short incubation identification from blood cultures is associated with reduced length of hospitalization and a decrease in bacteremia associated mortality. Eur J Clin Microbiol Infect Dis 36:1181–1186. doi: 10.1007/s10096-017-2906-y. [DOI] [PubMed] [Google Scholar]
- 83.Martiny D, Debaugnies F, Gateff D, Gérard M, Aoun M, Martin C, Konopnicki D, Loizidou A, Georgala A, Hainaut M, Chantrenne M, Dediste A, Vandenberg O, Van Praet S. 2013. Impact of rapid microbial identification directly from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry on patient management. Clin Microbiol Infect 19:E568–E581. doi: 10.1111/1469-0691.12282. [DOI] [PubMed] [Google Scholar]
- 84.Perez KK, Olsen RJ, Musick WL, Cernoch PL, Davis JR, Peterson LE, Musser JM. 2014. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J Infect 69:216–225. doi: 10.1016/j.jinf.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 85.Patel TS, Kaakeh R, Nagel JL, Newton DW, Stevenson JG. 2017. Cost analysis of implementing matrix-assisted laser desorption ionization-time of flight mass spectrometry plus real-time antimicrobial stewardship intervention for bloodstream infections. J Clin Microbiol 55:60–67. doi: 10.1128/JCM.01452-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beganovic M, Costello M, Wieczorkiewicz SM. 2017. Effect of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) alone versus MALDI-TOF MS combined with real-time antimicrobial stewardship interventions on time to optimal antimicrobial therapy in patients with positive blood cultures. J Clin Microbiol 55:1437–1445. doi: 10.1128/JCM.02245-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Toklu HZ, Hussain A. 2013. The changing face of pharmacy practice and the need for a new model of pharmacy education. J Young Pharm 5:38–40. doi: 10.1016/j.jyp.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abat C, Chaudet H, Colson P, Rolain JM, Raoult D. 2015. Real-time microbiology laboratory surveillance system to detect abnormal events and emerging infections, Marseille, France. Emerg Infect Dis 21:1302–1310. doi: 10.3201/eid2108.141419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colson P, Rolain JM, Abat C, Charrel R, Fournier PE, Raoult D. 2015. EPIMIC: a simple homemade computer program for real-time epidemiological surveillance and alert based on microbiological data. PLoS One 10:e0144178. doi: 10.1371/journal.pone.0144178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O'Brien TF, Clark A, Peters R, Stelling J. 2019. Why surveillance of antimicrobial resistance needs to be automated and comprehensive. J Glob Antimicrob Resist 17:8–15. doi: 10.1016/j.jgar.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 91.Tornimbene B, Eremin S, Escher M, Griskeviciene J, Manglani S, Pessoa-Silva CL. 2018. WHO global antimicrobial resistance surveillance system early implementation 2016–17. Lancet Infect Dis 18:241–242. doi: 10.1016/S1473-3099(18)30060-4. [DOI] [PubMed] [Google Scholar]
- 92.Harvala H, Frampton D, Grant P, Raffle J, Ferns RB, Kozlakidis Z, Kellam P, Pillay D, Hayward A, Nastouli E, ICONIC Consortium . 2017. Emergence of a novel subclade of influenza A(H3N2) virus in London, December 2016 to January 2017. Euro Surveill 22:30466. doi: 10.2807/1560-7917.ES.2017.22.8.30466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Houlihan CF, Frampton D, Ferns RB, Raffle J, Grant P, Reidy M, Hail L, Thomson K, Mattes F, Kozlakidis Z, Pillay D, Hayward A, Nastouli E. 2018. Use of whole-genome sequencing in the investigation of a nosocomial influenza virus outbreak. J Infect Dis 218:1485–1489. doi: 10.1093/infdis/jiy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Revez J, Espinosa L, Albiger B, Leitmeyer KC, Struelens MJ, ECDC National Microbiology Focal Points and Experts Group . 2017. Survey on the use of whole-genome sequencing for infectious diseases surveillance: rapid expansion of European national capacities, 2015–2016. Front Public Health 5:347. doi: 10.3389/fpubh.2017.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Albiger B, Revez J, Leitmeyer KC, Struelens MJ. 2018. Networking of public health microbiology laboratories bolsters Europe’s defenses against infectious diseases. Front Public Health 6:46. doi: 10.3389/fpubh.2018.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bastawrous A, Armstrong MJ. 2013. Mobile health use in low- and high-income countries: an overview of the peer-reviewed literature. J R Soc Med 106:130–142. doi: 10.1177/0141076812472620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rateni G, Dario P, Cavallo F. 2017. Smartphone-based food diagnostic technologies: a review. Sensors (Basel) 17:1453. doi: 10.3390/s17061453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang WH, Wang CH, Yang SY, Lin YC, Wu JJ, Lee MS, Lee GB. 2014. Rapid isolation and diagnosis of live bacteria from human joint fluids by using an integrated microfluidic system. Lab Chip 14:3376–3384. doi: 10.1039/c4lc00471j. [DOI] [PubMed] [Google Scholar]
- 99.Sackmann EK, Fulton AL, Beebe DJ. 2014. The present and future role of microfluidics in biomedical research. Nature 507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 100.Dubourg G, Raoult D. 2016. Emerging methodologies for pathogen identification in positive blood culture testing. Expert Rev Mol Diagn 16:97–111. doi: 10.1586/14737159.2016.1112274. [DOI] [PubMed] [Google Scholar]
- 101.Kao RR, Haydon DT, Lycett SJ, Murcia PR. 2014. Supersize me: how whole-genome sequencing and big data are transforming epidemiology. Trends Microbiol 22:282–291. doi: 10.1016/j.tim.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parize P, Muth E, Richaud C, Gratigny M, Pilmis B, Lamamy A, Mainardi JL, Cheval J, de Visser L, Jagorel F, Ben Yahia L, Bamba G, Dubois M, Join-Lambert O, Leruez-Ville M, Nassif X, Lefort A, Lanternier F, Suarez F, Lortholary O, Lecuit M, Eloit M. 2017. Untargeted next-generation sequencing-based first-line diagnosis of infection in immunocompromised adults: a multicentre, blinded, prospective study. Clin Microbiol Infect 23:574.e1–574.e6. doi: 10.1016/j.cmi.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 103.Reuter S, Ellington MJ, Cartwright EJ, Köser CU, Török ME, Gouliouris T, Harris SR, Brown NM, Holden MT, Quail M, Parkhill J, Smith GP, Bentley SD, Peacock SJ. 2013. Rapid bacterial whole-genome sequencing to enhance diagnostic and public health microbiology. JAMA Intern Med 173:1397–1404. doi: 10.1001/jamainternmed.2013.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 107.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goggin KP, Gonzalez-Pena V, Inaba Y, Allison KJ, Hong DK, Ahmed AA, Hollemon D, Natarajan S, Mahmud O, Kuenzinger W, Youssef S, Brenner A, Maron G, Choi J, Rubnitz JE, Sun Y, Tang L, Wolf J, Gawad C. 19 December 2019. Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol doi: 10.1001/jamaoncol.2019.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Allcock RJN, Jennison AV, Warrilow D. 2017. Towards a universal molecular microbiological test. J Clin Microbiol 55:3175–3182. doi: 10.1128/JCM.01155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petersen LM, Martin IW, Moschetti WE, Kershaw CM, Tsongalis GJ. 2019. Third-generation sequencing in the clinical laboratory: exploring the advantages and challenges of nanopore sequencing. J Clin Microbiol 58:e01315-19. doi: 10.1128/JCM.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, Wollam A, Reyes J, Panesso D, Rojas NL, Shamoo Y, Murray BE, Weinstock GM, Arias CA. 2014. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother 58:4527–4534. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barczak AK, Gomez JE, Kaufmann BB, Hinson ER, Cosimi L, Borowsky ML, Onderdonk AB, Stanley SA, Kaur D, Bryant KF, Knipe DM, Sloutsky A, Hung DT. 2012. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc Natl Acad Sci U S A 109:6217–6222. doi: 10.1073/pnas.1119540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E. 2018. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24:335–341. doi: 10.1016/j.cmi.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chiu CY, Miller SA. 2019. Clinical metagenomics. Nat Rev Genet 20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.NIH Human Microbiome Portfolio Analysis Team. 2019. A review of 10 years of human microbiome research activities at the US National Institutes of Health, fiscal years 2007–2016. Microbiome 7:31. doi: 10.1186/s40168-019-0620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mendy M, Caboux E, Sylla BS, Dillner J, Chinquee J, Wild C, BCNet Survey Participants . 2014. Infrastructure and facilities for human biobanking in low- and middle-income countries: a situation analysis. Pathobiology 81:252–260. doi: 10.1159/000362093. [DOI] [PubMed] [Google Scholar]
- 118.Simeon-Dubach D, Henderson MK. 2014. Sustainability in biobanking. Biopreserv Biobank 12:287–291. doi: 10.1089/bio.2014.1251. [DOI] [PubMed] [Google Scholar]
- 119.Hulsen T, Jamuar SS, Moody AR, Karnes JH, Varga O, Hedensted S, Spreafico R, Hafler DA, McKinney EF. 2019. From big data to precision medicine. Front Med (Lausanne) 6:34. doi: 10.3389/fmed.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Doucet M, Yuille M, Georghiou L, Dagher G. 2017. Biobank sustainability: current status and future prospects. BSAM 5:1–7. doi: 10.2147/BSAM.S100899. [DOI] [Google Scholar]
- 121.Caulfield T, Burningham S, Joly Y, Master Z, Shabani M, Borry P, Becker A, Burgess M, Calder K, Critchley C, Edwards K, Fullerton SM, Gottweis H, Hyde-Lay R, Illes J, Isasi R, Kato K, Kaye J, Knoppers B, Lynch J, McGuire A, Meslin E, Nicol D, O'Doherty K, Ogbogu U, Otlowski M, Pullman D, Ries N, Scott C, Sears M, Wallace H, Zawati MH. 2014. A review of the key issues associated with the commercialization of biobanks. J Law Biosci 1:94–110. doi: 10.1093/jlb/lst004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kinkorová J, Topolčan O. 2018. Biobanks in Horizon 2020: sustainability and attractive perspectives. EPMA J 9:345–353. doi: 10.1007/s13167-018-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simeon-Dubach D, Kozlakidis Z. 2018. New standards and updated best practices will give modern biobanking a boost in professionalism. Biopreserv Biobank 16:1–2. doi: 10.1089/bio.2017.0126. [DOI] [PubMed] [Google Scholar]
- 124.Burnham CA, McAdam AJ. 2019. Total laboratory automation: a micro-comic strip. J Clin Microbiol 57:e01036-19. doi: 10.1128/JCM.01036-19. [DOI] [PMC free article] [PubMed] [Google Scholar]