Abstract
Background
The presentation of clinical leptospirosis has been historically associated with animal workers, slaughterhouse workers and medical veterinarians. This association has shifted to be related to flooding events and outdoor activities; few cases are related to high-risk factors found in immunosuppressed patients. Scarcely a handful of cases have serological evidence of immune response against Leptospira serovar Bratislava representing serogroup Australis, a serovar associated with poor reproductive performance in swine and horses, and recently with cats.
Case presentation
Herein, we describe a rare clinical presentation of disseminated Leptospira infection in an immunosuppressed 65-year-old woman. She was admitted to the emergency room with fever, bacteraemia, bilateral uveitis and pulmonary involvement. The patient denied outdoor activities; she only had wide exposure to faeces and urine from cats living in her home. Her medical history included idiopathic thrombocytopenic purpura (ITP) diagnosed at the age of 18. She did not respond to medical treatment, and a splenectomy was performed. At age 60, she was diagnosed with Chronic Myeloid Leukemia (CML), and was treated with a tyrosine kinase inhibitor (TKI) –Imatinib. The patient voluntarily discontinued the treatment for the last 6 months. After extensive workup, no microorganisms were identified by the commonly used stains in microbiology. The diagnosis was performed through dark-field microscopy, microagglutination test (MAT), Leptospira genus-specific PCR, the IS1500 PCR for identification of pathogenic species, and 16S based sequencing for the genus identification.
Conclusion
Immunosuppressed patients may acquire uncommon infections from ubiquitous microorganisms. In this case, serology evidence of exposure to Leptospira serovar Bratislava by MAT and the presence of the Leptospira genus were identified. It should be on mind for the diagnosis in otherwise healthy patients, and thoroughly search on splenectomised patients exposed to animals. Additionally, this report highlights the usefulness of PCR for diagnosis of this potentially life-threatening illness.
Keywords: Leptospirosis, Leptospira, Weil’s disease, Serovar Bratislava, Idiopathic thrombocytopenic purpura, Immunosuppression, Chronic myeloid leukemia, Splenectomy
Background
Leptospirosis is a worldwide zoonosis caused by pathogenic Leptospira. The global incidence of severe leptospirosis is estimated to be more than 1 million per year, with a fatality rate ranging around 10%. In the past, the disease was related to professionals of rural areas and the flooding season. Due to climate change conditions, leptospirosis is an increasing public health issue in many developing countries [1, 2]. Currently, its presentation in urbanised areas is related to outdoor activities and animal contact. Mexico is considered a country of moderate incidence [3]; it has geographic regions with the environmental conditions of temperature and precipitation favourable for Leptospira infection all along the year such as the states of Tabasco and Quintana Roo [4, 5].
The clinical presentation of acute leptospirosis is typically a rapid onset, febrile, systemic disease. The signs are mild and self-limiting; the leptospiremic (initial) phase includes high spiking fever, headache, conjunctivitis, and myalgias, which last 4 to 9 days. There is no pathognomonic clinical feature; however, acute onset and high spike fever indicate the clinician to consider leptospirosis among the differential diagnosis and the medical history that points out a probable exposure to Leptospira. The severe presentation of leptospirosis, in addition to the above symptoms, also includes jaundice, haemorrhage, and acute renal failure. Unusual manifestations include pulmonary, cardiovascular, neurological, gastrointestinal, ocular and other systemic symptoms [6, 7].
Leptospirosis in an asplenic patient is rare and has not been reported previously. Herein we describe a leptospirosis case due to a cats and swine related Leptospira serovar in an asplenic patient, who had a history of non-responsive IPT and under chronic phase of CML.
Case presentation
A 65-year-old female was admitted to the emergency room. The patient had a medical history of idiopathic thrombocytopenic purpura (ITP) diagnosed at the age of 18, refractory to steroid-based treatment. A splenectomy was performed, and she remained with normal platelet counts. She was diagnosed at 60 years-old with chronic myeloid leukaemia (CML), and received treatment with tyrosine kinase inhibitor (TKI)-Imatinib. The diagnosis was confirmed by bone marrow aspiration and fluorescent in situ hybridisation (FISH), in which 22% were negative cells and 78% positive cells for the Philadelphia chromosome. The qPCR for BCR-ABL/ABL was 40.22% (15,420.24 ABL copies, and 6202.4 BCR-ABL copies). On admission, she admitted to voluntarily had stopped the Imatinib treatment 6 months before due to a major depression, had fever and worsening eyesight (later diagnosed as acute uveitis) (Fig. 1). She had no history of travelling to leptospirosis endemic areas, nor alcohol, drug or tobacco abuse, neither other medications intake. Nevertheless, she had close contact with her pet cat. She was admitted to the hospital with acute respiratory failure, bacteraemia and uveitis.
Fig. 1.
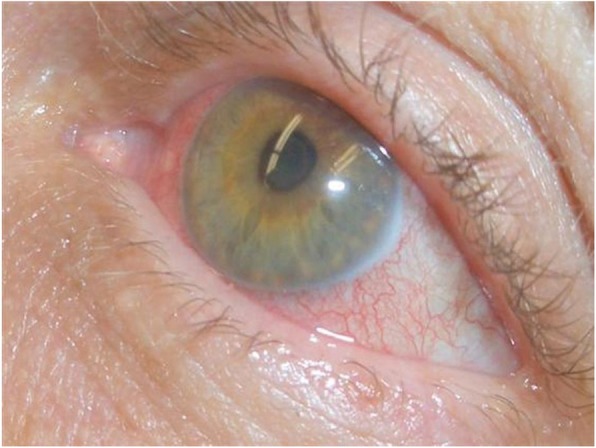
Clinical view of the inflammatory process in the patient’s left eye
Laboratory tests showed leukocyte count of 144.6 × 103/ml, hemoglobin 12.4 g/dl, platelets 763 × 103/ml, glucose level of 116 mg/dl, and albumin level of 3.0 mg/dl. Liver function test showed malnourishment and low protein level. Renal function test showed high creatinine level indicating an acute kidney injury mainly associated with the inflammatory response, and electrolytes were within normal limits. Additionally, the levels of C-reactive protein and procalcitonin were moderately high (Supplementary Table 1). Due to respiratory deterioration, a sputum culture was performed, and E. coli was isolated. In México, at the time the patient was treated, there was an outbreak of influenza, reason why a diagnostic qPCR for the influenza virus was performed; the result was negative. The chest X-Ray revealed multiple pneumonia foci (Fig. 2). She was started on empirical broad antibiotic regimen. Blood and urine cultures collected on admission showed no microorganisms grown from conventional medium or seen on Gram, Ziehl-Nielsen, and Giemsa stains. The blood cultures in the BacT/ALERT® FA medium (bioMérieux, Durham NC), after 2 days of incubation at 34 °C, were positive. Unstained spirochaete-like microorganisms were observed by dark-field microscopy. Due to clinical and microbiological finding, the antimicrobial treatment was changed to penicillin 20 million UI/ IV, plus doxycycline 100 mg twice a day. Subsequently, two blood samples were tested by Microagglutination test (MAT) separated from each other by 10 days (Supplementary Methodology Document). Differential diagnosis were performed, and negative results were reported. After the successful treatment, the CML treatment was resumed with a second-generation TKI (Dasatinib). A year later, the FISH analysis showed 35 negative cells and 65 positive cells for the Philadelphia chromosome. The aspiration of bone marrow showed an adequate maturation, compatible with CML in chronic phase.
Fig. 2.
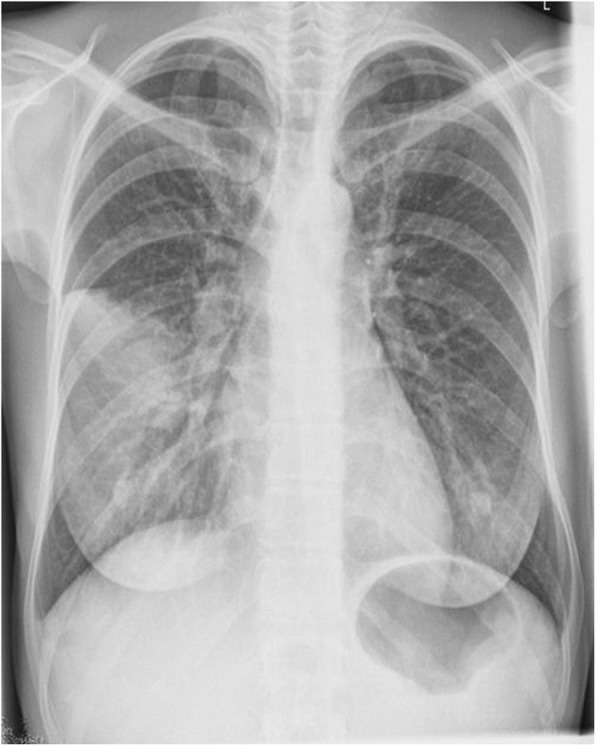
In the chest x-ray, the lung had alveolar occupation of the right middle lobe due to an atypical pneumonia
Microagglutination test (MAT)
Microagglutination test (MAT) was performed as previously described by the Pan American Health Organization [8]. The patient’s sera were diluted to 1/50 for a screening test and 50 μl of live cultures of 4 to 7 days in EMJH medium of twelve L. interrogans serovars were used as antigens (Autumnalis, Bataviae, Bratislava, Canicola, Celledoni, Grippotyphosa, Hardjoprajitno, Icterohaemorrhagiae, Pomona, Pyrogenes, Tarassovi, Wolffi and Mini) (Supplementary Table 2). Each serovar was added into a column of a 96-well flat end microtitre plate (Nunc, Maryland, USA); a negative control was included for each serovar. The plate was gently stirred and incubated 1 hour at room temperature. Reading was performed in dark-field microscopy (Carl Zeiss, Germany). The patient’s sera were serially diluted from 1/25 to 1/1600, and the microagglutination was observed for each serovar. The final titre represents the maximum dilution in which the agglutination grade two was noted.
Leptospira isolation
From the patient’s blood sample, three drops were seeded into EMJH liquid medium and maintained at 30 °C. Cultures were periodically observed in dark-field microscopy to spot spirochaetal forms and maintained for 6 months before being considered negative cultures. In parallel, the patient’s blood sample was inoculated to a Syrian golden hamster (Mesocricetus auratus) for bacterial isolation [9] (Supplementary Methodology Document and Supplementary Table 3).
DNA extraction
DNA was extracted from the patient’s blood samples and the hamster’s tissues with the DNeasy Blood &Tissue Kit (QIAGEN, Cal, USA), according to the manufacturer’s instructions and suspended in 50 μL of nuclease-free water. DNA was quantified using an Epoch microplate spectrophotometer (Biotech) and stored at 4 °C.
PCR
The Leptospira genus-specific PCR based on the 23S rDNA was used to amplify a 482 bp fragment [10]. A second PCR was performed to identify only pathogenic strains based on the amplification of the insertion sequence IS1500 of L. interrogans (sensu lato) [11] (Supplementary Methodology Document). The electrophoresis of the amplified products were stained with ethidium bromide on 1.6% agarose gels and visualised. Further genus identification was performed based on sequencing of the 16S rRNA gene, using the primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGCYTACCTTGTTACGACTT-3′) with DNA extracted from the first blood sample as template [12]. A PCR product of 1432 bp was amplified, purified, and DNA was sequenced in both directions. Nucleotide sequence was determined with Taq FS Dye Terminator Cycle Sequencing Fluorescence-Based Sequencing, and analysed on an Applied Biosystems 3730 DNA sequencing system (Foster City, CA, USA). The sequence was registered in Genbank under the accession number MN545905. It has 100% homology with Leptospira interrogans strains (CP011410.1, AE016823.1, AE010300.2, and CP018146.1) and variable homology with reference sequences (Supplementary Methodology Document and Supplementary Table 4).
In summary, the results support the diagnosis of leptospirosis; abundant spirochaetal forms, at least 106 Leptospires/mL were observed in dark field microscopy, The MAT results (Supplementary Table 2) indicated the serovar Bratislava as the most probable causing serovar because the patient’s serum reacted with Bratislava (Jez Bratislava) in titres of 1/320. This titre was the highest in the first sample and stayed in a steady-state in the second sample. There was no other serovar with titres at this level. Titres to Serovars Bataviae (Van Tienen); Canicola (Hond Utech IV); Grippotyphosa (Moska V); Hardjo (Hardjoprajitno), and Pomona (Pomona) were lower than 1/80 in both samples, and those for Pyrogenes (Salinem) fell from 1/160 to 1/80 in the second sample. During the Leptospira isolation attempt, its survival was sustained for about a month in the EMJH medium, but it did not flourish. The PCRs based on the23S rDNA and the IS1500 identified the presence of the bacteria DNA in the patient’s blood samples, in the hamster tissues (Supplementary Table 3), and the Leptospira genus was identified by the 16S based sequencing.
Discussion and conclusions
Our patient was in the chronic phase of CML and voluntarily stopped TKI treatment losing the haematological response, which coincided with leptospirosis. The primary mechanism of immunosuppression related to the acquisition of the Leptospira infection was asplenic status. In theory, CML could also be associated with deregulation of the immune system. However, we are aware that there is no strong clinical evidence to support it.
Mild Leptospirosis cases escape diagnosis in immunosuppressed patients due to HIV (Human Immunodeficiency Virus) or other conditions [13]. To the best of our knowledge, there are no reports of leptospirosis in splenectomised patients. Splenectomy is a second-line treatment for ITP when previous therapeutic measures had failed. The procedure is not strictly “curative” because the immune mechanism persists, and the consequence is that the patient has a permanent immunosuppression condition [14]. Therefore, splenectomy is associated with an increase of overwhelming post-splenectomy infections (OPSI), defined as infections that require admission to intensive care unit as a late complication following splenectomy [15]. OPSI are caused by unusual encapsulated bacteria, including Streptococcal pneumonia, Haemophilus influenzae, Neisseria meningitidis, and other organisms such as Capnocytophaga canimorsus [16–18], and Cryptococcus neoformans [19]. The risk of OPSI is minimised by the administration of a broad vaccination scheme. In Mexico, the Leptospira preventive vaccination in humans has not been approved. Therefore, the patient’s vaccination scheme did not include it.
The asplenic status is a specific risk factor that increase up to 2.2% the risk to develop pneumonia [20] According to guidelines, the pathogen isolated from blood cultures is the definite cause of the pneumonia except for the sputum isolation of Legionella pneumophila, Mycobacterium tuberculosis, Pneumocystis carinii cysts or trophozoites [21]. Otherwise, the isolation from sputum are presumptive, but must be confirmed by more than one sample with heavy to moderate growth. The diagnosis of E. coli pneumonia is based on radiographic evidence of bronchopneumonia of the lower lobes coupled with positive sputum and positive blood cultures for E. coli [22, 23]. To uphold the diagnosis of pneumonia due to E. coli, at least two of these criteria should be met, and be supported by clinical suspicion and the patient’s history. In this case, not all the criteria to diagnose E. coli pneumonia were met, and the E. coli sputum culture was considered a contamination due to oropharyngeal secretions [22].
Individuals subjected to splenectomy may form antibodies quite normally to antigens given subcutaneously but not respond well to antigens administered intravenously [24]. The asplenic status of the patient is relevant in the development of leptospirosis because the spleen participates in the removal of Leptospira, and various bloodborne pathogens as it produces opsonins that promote phagocytosis [25]. In the hamster model, Leptospira circulates in blood in the leptospiremic phase of the disease, and the histopathological changes that are produced in the spleen include cellular necrosis in the splenic cord, dilated sinusoids, congested hemorrhagic areas, and infiltration of inflammatory cells in the splenic parenchyma and sinusoids [26].
During the acute phase of illness, conjunctiva congestion, panuveitis with or without hypopyon are common clinical findings [27]. Therefore, it was not possible to differentiate whether the uveitis was a consequence of previous treatment or due to leptospirosis. In this immunosuppressed patient, not all haematological manifestations were due to leptospirosis, although some abnormalities may be associated with it [28].
Three radiographic patterns have been described in patients with pulmonary involvement during leptospiral infection; small nodular densities, diffuse ground-glass densities and rarely, confluent areas of consolidation [29]. In this case, pneumonic foci were observed yet only serovar Grippotyphosa, Valbuzzi, and Australis have been associated with pulmonary manifestations [30–32].
Serogroup Icterohaemorrhagiae is the most frequent in human infections, followed by Canicola, Grippotyphosa, Pyrogenes, Pomona and Australis, which may vary in each geographic region [33]. This patient was exposed somehow to serovar Bratislava, a globally distributed serovar but with unknown epidemiology [34]. This serovar is mostly associated with swine and cattle reproductive failure [35, 36], and maintained by dogs and horses [34]. This serovar is currently related to rural and domestic cats. Cats can be infected without clinical signs or seem clinically unapparent [37–39], needing an extended incubation period to develop the disease [40]. Cats may shed as much Leptospira as dogs do [41, 42]. Therefore, cats can be a source of urban leptospirosis [38, 42–44], and in this case, the patient’s cat may have been a potential infection source.
The Bact/ALERT® Microbial Detection System detects the microbial growth via the colourimetric detection of changes in the CO2 concentration [45], and the Bact/ALERT® FA media can support viable Leptospira up to 9 days [46, 47]. In our laboratory experience, the negative results of the Gram, Ziehl-Nielsen, and Giemsa stains, coupled with the slight change in the CO2 level guided us to foresee the possibility of a slow-growing microorganism. The first blood sample was taken during the acute phase of leptospirosis, in which the patient was septicemic, reason why it was possible to visualise under dark-field microscopy.
Culture confirmation is the gold standard for clinical cases. We attempted Leptospira isolation during the leptospiremic phase in the hamster model, based on previous studies [9]. Nonetheless, Leptospira isolation and culture are difficult to be obtained; its persistence depends on the serovar [9, 48]. In particular, serovar Bratislava and close related strains are fastidious serovars [34]. Reason why our difficulties in achieving isolation are understood.
The MAT is a serogroup-specific test, its threshold titre is established according to the prevalence in each geographical region; for example, it is set at 1/100 for mainland France and 1/400 for endemic zones [33]. The official guidelines (NOM-029-SSA2–1999) establish the MAT threshold for humans at 1/80 in the first sample, confirmed by a second sample with the double or higher titres, or by the bacteria isolation or PCR. Titres under 1/80 are evidence of previous exposure to Leptospira, but not as the result of a current infection. The patient’s first sample showed a 1/320 titre to L. interrogans serovar Bratislava, and 1/160 to L. interrogans serovar Pyrogenes. The second sample showed titres as high as the previous sample to L. interrogans serovar Bratislava, low titres to L. interrogans serovar Hardjo and L. interrogans serovar Grippotyphosa. These MAT results are explained by two coexistence, the early and accurate treatment that halted leptospires quickly enough and the patient’s immunosuppression condition which are reflected as constant titres in the second sample, as observed by other researchers [49]. After treatment, and during the 10 months of follow up, MAT titres diminished, the uveitis improved, and she had no further leptospirosis signs.
Other diagnostic options include PCR amplification of bacterial DNA from blood during the first week after symptoms onset [33], qPCR applications [50–53], and sequence-based identification of Leptospira [54–56]. However, results should be validated by MAT because Leptospira is not always present in blood, as it could be removed if the treatment has begun. Additionally, MAT can be false negative considering that antibodies may rise until the second or third week of the disease [33]. In this case, the clinical judgment and suspicion was of paramount value to guide us to dark-field observation and the penicillin-based treatment [57], enhanced with doxycycline added along with intravenous fluids. Even though the unarguable conclusion of the infecting serovar cannot be drawn without isolation, the observations during blood culture and the MAT high titres to L. Bratislava gave us reasonable evidence of the Leptospira exposure.
In circumstances of atypical presentations of leptospirosis, diagnosis may be aided by PCR [58]. PCR detects the presence of nucleic acids of extremely low number of microorganisms; around two to ten cells [59, 60]. PCR cannot distinguish between viable and dead cells because all of them contribute to a positive signal. Even in culture-negative blood samples, PCR may be positive if the patient has received an effective antimicrobial drug but have not cleared nonviable organisms [61]. This condition happens in splenectomised patients, in which Leptospira may remain dead or alive for extended periods in comparison to immunocompetent patients, aside that they have inefficient antibody production. In this case, PCR confirmed the Leptospira genus as the aetiological agent and its circulation in the bloodstream, and ruled out a false positive diagnosis [62].
Finally, we want to emphasise the need of increased awareness in healthcare providers to consider leptospirosis in those cases in which infections may be reported as culture-negative severe infections [63], especially in asplenic patients, as this zoonosis may have an unusual presentation in immunosuppressed patients so that appropriate therapy can be initiated.
Supplementary information
Additional file 1: Supplementary. Supplementary Methodology document. Table 1. Laboratory tests summary. Supplementary Table 2. Results of the Microscopic agglutination test (MAT). Supplementary Table 3. Results summary. Supplementary Table 4.- Results of BLAST with Reference Sequences.
Acknowledgements
We are indebted to the Leptospira collection of the Microbiology and Immunology Department, Facultad de Medicina Veterinaria y Zootecnia, Universidad Nacional Autónoma de México. We thanks Dr. Aurora Loaeza del Castillo for her critical and careful review. We want to thank also the three anonymous reviewers for constructive criticism.
Abbreviations
- CML
Chronic Myeloid Leukemia
- EMJH
Ellinghausen-McCullough-Johnson-Harris
- FISH
Fluorescent In Situ Hybridisation
- ITP
Idiopathic Thrombocytopenic Purpura
- MAT
Microagglutination test
- OPSI
Overwhelming post-splenectomy infections
- PCR
Polymerase Chain Reaction
Authors’ contributions
JGM and ECC cared for the patient and collected data. DAL performed the dark-field microscopy and the microagglutination test. SAE and DMB did the clinical laboratory work. RHC, MLL and EMCC performed the Leptospira isolation assays in the hamster model, PCR and sequencing, carried out the literature search and drafted the manuscript. All authors made critical revisions and approved the final manuscript.
Funding
There are no conflicts of funding source.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information file.
Ethics approval and consent to participate
The experimental animal protocol was approved by the Comité Interno para el Cuidado y Uso de Los Animales de Laboratorio del Hospital General “Dr Manuel Gea González”.
We are exempt from ethical approval from Comité de Ética y Comité de investigación as it is not required in our hospital for clinical cases.
Consent for publication
The written informed consent was obtained from the patient’s kin for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Competing interests
“The authors declare that they have no competing interests.”
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
J. García-Méndez, Email: jogarciam@hotmail.com
E. Cervera-Ceballos, Email: ecerverac@incan.edu.mx
D. Atilano-López, Email: daniel.atilano@yahoo.com.mx
S. Arroyo-Escalante, Email: saraarroyoescalante@hotmail.com
D. Moncada-Barrón, Email: davidmoncadabarron@hotmail.com
M. Leyva-Leyva, Email: margaritaleyvaleyva@yahoo.com.mx
R. Hernández-Castro, Email: rigo37@gmail.com
E. M. Carrillo-Casas, Email: ekarri@gmail.com
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12879-020-4869-3.
References
- 1.Abela-Ridder B, Sikkema R, Hartskeerl RA. Estimating the burden of human leptospirosis. Int J Antimicrob Agents. 2010;36(Suppl 1):S5–S7. doi: 10.1016/j.ijantimicag.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Hartskeerl RA, Collares-Pereira M, Ellis WA. Emergence, control and re-emerging leptospirosis: dynamics of infection in the changing world. Clin Microbiol Infect. 2011;17(4):494–501. doi: 10.1111/j.1469-0691.2011.03474.x. [DOI] [PubMed] [Google Scholar]
- 3.Pappas G, Papadimitriou P, Siozopoulou V, Christou L, Akritidis N. The globalization of leptospirosis: worldwide incidence trends. Int J Infect Dis. 2008;12(4):351–357. doi: 10.1016/j.ijid.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Borbolla-Sala ME, García Vanegas L, Cadenas-Martínez MT, Hernandez-Hernandez R, De La Fuente Gutiérrez JC, Piña-Gutiérrez OE, Rodríguez León A. Leptospirosis durante la contingencia ambiental por inundación en Tabasco 2008. Salud en Tabasco. 2009;15(2, 3):860–7.
- 5.Sánchez-Montes S, Espinosa-Martínez DV, Ríos-Muñoz CA, Berzunza-Cruz M, Becker I. Leptospirosis in Mexico: epidemiology and potential distribution of human cases. PLoS One. 2015;10(7):e0133720. doi: 10.1371/journal.pone.0133720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bal AM. Unusual clinical manifestations of leptospirosis. J Postgrad Med. 2005;51(3):179–183. [PubMed] [Google Scholar]
- 7.Moschini J. Leptospirosis humana con compromiso del sistema nervioso central. Comunicación de dos casos y revisión de la literatura. Neurología Argentina. 2011;3(4):222–228. doi: 10.1016/j.neuarg.2011.06.009. [DOI] [Google Scholar]
- 8.Myers DM. Manual de Métodos para el diagnóstico de laboratorio de la leptospirosis. In: Nota Técnica No 30. Edited by PAHO/WHO; 1985.
- 9.Haake DA. Hamster model of leptospirosis. Curr Protoc Microbiol. 2006, Chapter 12:Unit 12E 12. [DOI] [PMC free article] [PubMed]
- 10.Woo TH, Patel BK, Smythe LD, Norris MA, Symonds ML, Dohnt MF. Identification of pathogenic Leptospira by TaqMan probe in a LightCycler. Anal Biochem. 1998;256(1):132–134. doi: 10.1006/abio.1997.2503. [DOI] [PubMed] [Google Scholar]
- 11.Zuerner R L, Bolin C A. Differentiation of Leptospira interrogans isolates by IS1500 hybridization and PCR assays. Journal of clinical microbiology. 1997;35(10):2612–2617. doi: 10.1128/JCM.35.10.2612-2617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lane DJ: 16S/23S rRNA sequencing In: Nucleic acid techniques in bacterial systematics. edn. Edited by Stackebrandt E, and Goodfellow, M. New York, NY: John Wiley and Sons; 1991: 115–175.
- 13.Jones S, Kim T. Fulminant leptospirosis in a patient with human immunodeficiency virus infection: case report and review of the literature. Clinical infectious diseases. 2001;33(5):E31–E33. doi: 10.1086/322645. [DOI] [PubMed] [Google Scholar]
- 14.Guía de Práctica clínica IMSS-143-08; Diagnóstico y tratamiento de Purpura Trombocitopénica inmunológica. In. México: Secretaría de Salud, Gobierno Federal; 2009.
- 15.Cullingford GL, Watkins DN, Watts AD, Mallon DF. Severe late postsplenectomy infection. Br J Surg. 1991;78(6):716–721. doi: 10.1002/bjs.1800780626. [DOI] [PubMed] [Google Scholar]
- 16.Di Sabatino A, Carsetti R, Corazza GR. Post-splenectomy and hyposplenic states. Lancet. 2011;378(9785):86–97. doi: 10.1016/S0140-6736(10)61493-6. [DOI] [PubMed] [Google Scholar]
- 17.King H, Shumacker HB., Jr splenic studies. I. Susceptibility to infection after splenectomy performed in infancy. Ann Surg. 1952;136(2):239–242. doi: 10.1097/00000658-195208000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubin LG, Schaffner W. clinical practice. Care of the asplenic patient. N Engl J Med. 2014;371(4):349–356. doi: 10.1056/NEJMcp1314291. [DOI] [PubMed] [Google Scholar]
- 19.Abbas H, Cifuentes-Kottkamp A, Abbas N, Cindrich R, Singh M. A rare presentation of cryptococcal meningitis and cerebellitis in an asplenic patient, seronegative for human immunodeficiency virus (HIV) American Journal of Case Reports. 2018;19(183–186):183. doi: 10.12659/AJCR.906869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai S-W, Lin C-L, Liao K-F. Risk of pneumonia among patients with splenectomy: a retrospective population-based cohort study. Ann Saudi Med. 2017;37(5):351–356. doi: 10.5144/0256-4947.2017.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrie TJ, Fine MJ, Obrosky DS, Coley C, Singer DE, Kapoor WN. Community-acquired pneumonia due to Escherichia coli. Clinical microbiology infection. 1998;4(12):717–723. doi: 10.1111/j.1469-0691.1998.tb00657.x. [DOI] [PubMed] [Google Scholar]
- 22.Tillotson JR, Lerner AM. Characteristics of pneumonias caused by Escherichia coli. N Engl J Med. 1967;277(3):115–122. doi: 10.1056/NEJM196707202770302. [DOI] [PubMed] [Google Scholar]
- 23.Jonas M, Cunha BA. Bacteremic Escherichia coli pneumonia. Arch Intern Med. 1982;142(12):2157–2159. doi: 10.1001/archinte.1982.00340250121017. [DOI] [PubMed] [Google Scholar]
- 24.Ellis EF, Smith RT. The role of the spleen in immunity. With special reference to the post-splenectomy problems in infants. Pediatrics. 1966;37(1):111–9. [PubMed]
- 25.Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol. 2005;5(8):606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 26.Muensoongnoen J, Phulsuksombati D, Parichatikanond P, Sangjan N, Pilakasiri C, Sripaoraya K, Roongruangchai J, Koedpuech K, Pilakasiri K. A histopathological study of hearts and spleens of hamsters (Mesocricetus auratus) infected with Leptospira interrogans, serovar Pyrogenes. Southeast Asian J Trop Med Public Health. 2006;37(4):720–728. [PubMed] [Google Scholar]
- 27.Rathinam SR. Ocular manifestations of leptospirosis. J Postgrad Med. 2005;51(3):189–194. [PubMed] [Google Scholar]
- 28.Laing RW, Teh C, Toh CH. Thrombotic thrombocytopenic purpura (TTP) complicating leptospirosis: a previously undescribed association. J Clin Pathol. 1990;43(11):961–962. doi: 10.1136/jcp.43.11.961-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Im JG, Yeon KM, Han MC, Kim CW, Webb WR, Lee JS, Han YC, Chang WH, Chi JG. Leptospirosis of the lung: radiographic findings in 58 patients. AJR Am J Roentgenol. 1989;152(5):955–959. doi: 10.2214/ajr.152.5.955. [DOI] [PubMed] [Google Scholar]
- 30.Watt G, Padre LP, Tuazon ML, Calubaquib C, Santiago E, Ranoa CP, Laughlin LW. Placebo-controlled trial of intravenous penicillin for severe and late leptospirosis. Lancet. 1988;1(8583):433–435. doi: 10.1016/S0140-6736(88)91230-5. [DOI] [PubMed] [Google Scholar]
- 31.Vijayachari P, Sehgal SC, Goris MG, Terpstra WJ, Hartskeerl RA. Leptospira interrogans serovar Valbuzzi: a cause of severe pulmonary haemorrhages in the Andaman Islands. J Med Microbiol. 2003;52(Pt 10):913–918. doi: 10.1099/jmm.0.05094-0. [DOI] [PubMed] [Google Scholar]
- 32.Yersin C, Bovet P, Merien F, Clement J, Laille M, Van Ranst M, Perolat P. Pulmonary haemorrhage as a predominant cause of death in leptospirosis in Seychelles. Trans R Soc Trop Med Hyg. 2000;94(1):71–76. doi: 10.1016/S0035-9203(00)90445-0. [DOI] [PubMed] [Google Scholar]
- 33.Picardeau M. Diagnosis and epidemiology of leptospirosis. Medecine et maladies infectieuses. 2013;43(1):1–9. doi: 10.1016/j.medmal.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Ellis William A. Current Topics in Microbiology and Immunology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. Animal Leptospirosis; pp. 99–137. [DOI] [PubMed] [Google Scholar]
- 35.Bolin CA, Cassells JA, Hill HT, Frantz JC, Nielsen JN. Reproductive failure associated with Leptospira interrogans serovar Bratislava infection of swine. J Vet Diagn Invest. 1991;3(2):152–154. doi: 10.1177/104063879100300209. [DOI] [PubMed] [Google Scholar]
- 36.Atxaerandio R, Aduriz G, Ziluaga I, Esteban JI, Maranda L, Mainar-Jaime RC. Serological evidence of Leptospira interrogans serovar Bratislava infection and its association with abortions in cattle in northern Spain. Veterinary record. 2005;156(12):376–380. doi: 10.1136/vr.156.12.376. [DOI] [PubMed] [Google Scholar]
- 37.Chan Kun-Wei, Hsu Yun-Hsiu, Hu Wei-Ling, Pan Ming-Jeng, Lai Jyh-Mirn, Huang Kwo-Ching, Chou Shih-Jen. Serological and PCR Detection of Feline Leptospira in Southern Taiwan. Vector-Borne and Zoonotic Diseases. 2014;14(2):118–123. doi: 10.1089/vbz.2013.1324. [DOI] [PubMed] [Google Scholar]
- 38.Agunloye CA, Nash AS. Investigation of possible leptospiral infection in cats in Scotland. J Small Anim Pract. 1996;37(3):126–129. doi: 10.1111/j.1748-5827.1996.tb02360.x. [DOI] [PubMed] [Google Scholar]
- 39.Larsson CE, Santa Rosa CA, Larsson MH, Birgel EH, Fernandes WR, Paim GV. Laboratory and clinical features of experimental feline leptospirosis. Int J zoonoses. 1985;12(2):111–119. [PubMed] [Google Scholar]
- 40.Arbour J, Blais MC, Carioto L, Sylvestre D. Clinical leptospirosis in three cats (2001-2009) J Am Anim Hosp Assoc. 2012;48(4):256–260. doi: 10.5326/JAAHA-MS-5748. [DOI] [PubMed] [Google Scholar]
- 41.Zaidi S, Bouam A, Bessas A, Hezil D, Ghaoui H, Ait-Oudhia K, Drancourt M, Bitam I. Urinary shedding of pathogenic Leptospira in stray dogs and cats, Algiers: a prospective study. PLoS One. 2018;13(5):e0197068. doi: 10.1371/journal.pone.0197068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez J, Blais MC, Lapointe C, Arsenault J, Carioto L, Harel J. Serologic and urinary PCR survey of leptospirosis in healthy cats and in cats with kidney disease. J Vet Intern Med. 2014;28(2):284–293. doi: 10.1111/jvim.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mylonakis ME, Bourtzi-Hatzopoulou E, Koutinas AF, Petridou E, Saridomichelakis MN, Leontides L, Siochu A. Leptospiral seroepidemiology in a feline hospital population in Greece. Veterinary record. 2005;156(19):615–616. doi: 10.1136/vr.156.19.615. [DOI] [PubMed] [Google Scholar]
- 44.Azócar-Aedo L, Monti G, Jara R. Leptospira spp. in domestic cats from different environments: prevalence of antibodies and risk factors associated with the Seropositivity. Animals. 2014;4(4):612–626. doi: 10.3390/ani4040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorpe TC, Wilson ML, Turner JE, DiGuiseppi JL, Willert M, Mirrett S, Reller LB. BacT/Alert: an automated colorimetric microbial detection system. J Clin Microbiol. 1990;28(7):1608–1612. doi: 10.1128/JCM.28.7.1608-1612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffith ME, Horvath LL, Mika WV, Hawley JS, Moon JE, Hospenthal DR, Murray CK. Viability of Leptospira in BacT/ALERT MB media. Diagn Microbiol Infect Dis. 2006;54(4):263–266. doi: 10.1016/j.diagmicrobio.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Girault D, Soupe-Gilbert ME, Geroult S, Colot J, Goarant C. Isolation of Leptospira from blood culture bottles. Diagn Microbiol Infect Dis. 2017;88(1):17–19. doi: 10.1016/j.diagmicrobio.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 48.Gomes-Solecki M, Santecchia I, Werts C. Animal models of leptospirosis: of mice and hamsters. Front Immunol. 2017;8:58. doi: 10.3389/fimmu.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faine S. Leptospira and leptospirosis: MediSci; 1999.
- 50.Ahmed Ahmed, Engelberts Mirjam F. M., Boer Kimberly R., Ahmed Niyaz, Hartskeerl Rudy A. Development and Validation of a Real-Time PCR for Detection of Pathogenic Leptospira Species in Clinical Materials. PLoS ONE. 2009;4(9):e7093. doi: 10.1371/journal.pone.0007093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villumsen S, Pedersen R, Borre MB, Ahrens P, Jensen JS, Krogfelt KA. Novel TaqMan(R) PCR for detection of Leptospira species in urine and blood: pit-falls of in silico validation. J Microbiol Methods. 2012;91(1):184–190. doi: 10.1016/j.mimet.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Thaipadungpanit J, Chierakul W, Wuthiekanun V, Limmathurotsakul D, Amornchai P, Boonslip S, Smythe LD, Limpaiboon R, Hoffmaster AR, Day NP, et al. Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipL32 genes for human leptospirosis in Thailand: a case-control study. PLoS One. 2011;6(1):e16236. doi: 10.1371/journal.pone.0016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slack A, Symonds M, Dohnt M, Harris C, Brookes D, Smythe L. Evaluation of a modified Taqman assay detecting pathogenic Leptospira spp. against culture and Leptospira-specific IgM enzyme-linked immunosorbent assay in a clinical environment. Diagn Microbiol Infect Dis. 2007;57(4):361–366. doi: 10.1016/j.diagmicrobio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Perez J, Goarant C. Rapid Leptospira identification by direct sequencing of the diagnostic PCR products in New Caledonia. BMC Microbiol. 2010;10:325. doi: 10.1186/1471-2180-10-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerqueira Gustavo M., McBride Alan J. A., Hartskeerl Rudy A., Ahmed Niyaz, Dellagostin Odir A., Eslabão Marcus R., Nascimento Ana L. T. O. Bioinformatics Describes Novel Loci for High Resolution Discrimination of Leptospira Isolates. PLoS ONE. 2010;5(10):e15335. doi: 10.1371/journal.pone.0015335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahmed SA, Sandai DA, Musa S, Hoe CH, Riadzi M, Lau KL, Tang TH. Rapid diagnosis of leptospirosis by multiplex PCR. Malays J Med Sci. 2012;19(3):9–16. [PMC free article] [PubMed] [Google Scholar]
- 57.Salud Sd: NORMA Oficial Mexicana NOM-029-SSA2–1999, Para la vigilancia epidemiológica, prevención y control de la leptospirosis en el humano. In: Diario Oficial de la Federación. Edited by Salud. EUM-Sd, vol. NOM-029-SSA2–1999. Mexico: Comité Consultivo Nacional de Normalización de Prevención y Control de Enfermedades; 2001:10.
- 58.Hall C, Lambourne J. The challenges of diagnosing leptospirosis. J Travel Medicine. 2014;21(2):139–140. doi: 10.1111/jtm.12095_1. [DOI] [PubMed] [Google Scholar]
- 59.Smythe LD, Smith IL, Smith GA, Dohnt MF, Symonds ML, Barnett LJ, McKay DB. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect Dis. 2002;2(1):13. doi: 10.1186/1471-2334-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Villumsen S, Pedersen R, Borre MB, Ahrens P, Jensen JS, Krogfelt KA. Novel TaqMan® PCR for detection of Leptospira species in urine and blood: pit-falls of in silico validation. J Microbiol Methods. 2012;91(1):184–190. doi: 10.1016/j.mimet.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Chierakul W, Limmathurotsakul D, Day NP, Peacock SJ. Molecular detection and speciation of pathogenic Leptospira spp. in blood from patients with culture-negative leptospirosis. BMC Infect Dis. 2011;11:338. doi: 10.1186/1471-2334-11-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agampodi SB, Matthias MA, Moreno AC, Vinetz JM. Utility of quantitative polymerase chain reaction in leptospirosis diagnosis: association of level of leptospiremia and clinical manifestations in Sri Lanka. Clinical infectious diseases. 2012;54(9):1249–1255. doi: 10.1093/cid/cis035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chong J, Jones P, Spelman D, Leder K, Cheng AC. Overwhelming post-splenectomy sepsis in patients with asplenia and hyposplenia: a retrospective cohort study. Epidemiology Infection. 2017;145(2):397–400. doi: 10.1017/S0950268816002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary. Supplementary Methodology document. Table 1. Laboratory tests summary. Supplementary Table 2. Results of the Microscopic agglutination test (MAT). Supplementary Table 3. Results summary. Supplementary Table 4.- Results of BLAST with Reference Sequences.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information file.


