Abstract
Protein–protein interactions are essential biologic processes that occur at inter- and intracellular levels. To gain insight into the various complex cellular functions of these interactions, it is necessary to assess them under physiologic conditions. Recent advances in various proteomic technologies allow to investigate protein–protein interaction networks in living cells. The combination of proximity-dependent labelling and chemical cross-linking will greatly enhance our understanding of multi-protein complexes that are difficult to prepare, such as organelle-bound membrane proteins. In this review, we describe our current understanding of mass spectrometry-based proteomics mapping methods for elucidating organelle-bound membrane protein complexes in living cells, with a focus on protein–protein interactions in mitochondrial subcellular compartments.
Keywords: BioID, mass spectrometry, mitochondria, proteome, XL-MS
Co-immunoprecipitation, yeast two-hybrid assay, or surface plasmon resonance approaches are commonly used to investigate the interactions between proteins of interest in the fields of cell biology and biochemistry. These approaches have various advantages and disadvantages, and care must be taken to ensure that the selected methods are optimal for addressing the specific experimental aims of the study. For analysis of the interactions between organelle-bound membrane protein complexes, such as mitochondrial proteins, it is particularly important to prepare the protein components such that the structures, topologies and functions remain intact. Various kinds of living cells, including yeasts and tissue culture, are used for studying interactomes among membrane-associated materials.
Rapid progress in the field of bio-imaging has led to the wide use of fluorescent or bioluminescent proteins such as green fluorescent protein (GFP) and luciferase as reporters for monitoring intermolecular interactions, such as by fluorescence resonance energy transfer (FRET) (1, 2) and bioluminescence resonance energy transfer (BRET) (3). Although the behaviour of fluorophore fusion proteins used in resonance energy transfer (RET) analysis may not precisely reflect that of the endogenous proteins (validation is needed), these systems are advantageous for monitoring interaction events of interest in living cells without considering the preparation issues mentioned above. Several issues regarding these systems, however, must be considered. For example, FRET requires an external light source to excite the fluorescent proteins, and the laser pulse often causes photobleaching of the cell materials. The BRET system eliminates this problem because the light is emitted by a donor moiety and occurs as a result of a chemical reaction (i.e. a natural resonance energy transfer process that occurs as a result of enzymatic activity via luciferase), but the overall BRET signal can be extremely weak relative to that of FRET. Another minor issue for FRET is that the photobleaching causes some intracellular proteins to autofluorescence, thereby increasing the background noise, which decreases the signal-to-noise ratio. Finally, another limitation of RET techniques is that the stoichiometry of the components is less than a few molecules even when using a split protomer (e.g. split GFP) (4).
Other approaches are therefore needed to resolve these issues for the analysis of protein–protein interactions in multi-protein complexes. Here, we describe several mass spectrometry (MS)-based proteomics methods that are useful for elucidating organelle-bound membrane protein complexes in living cells, with a focus on protein–protein interactions in mitochondrial subcellular compartments.
Proximity-dependent biotin identification
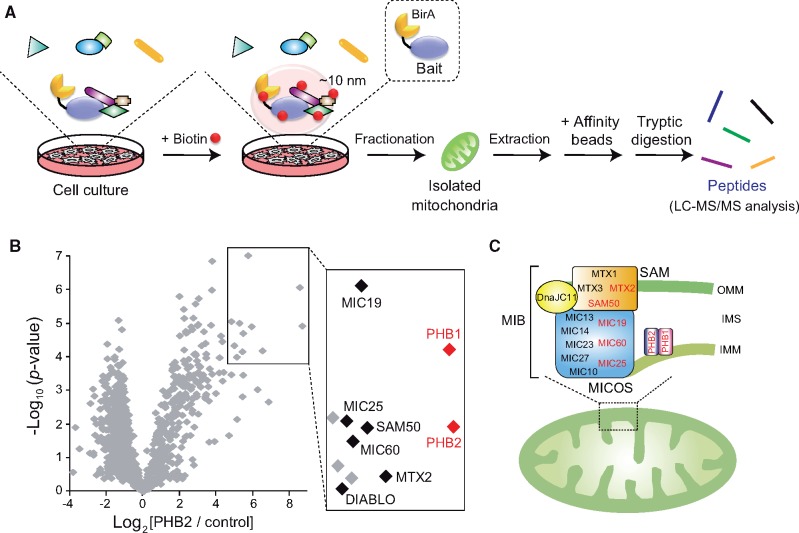
Proximity-dependent labelling methods in living cells were recently developed to address several of the concerns and limitations mentioned above (5). The techniques are powerful and ideal tools for evaluating a network of protein–protein interactions under physiologic conditions, especially for detecting low-affinity and/or transient associations, such as interactomes involved in signal transduction. A proximity-based biotin labelling method, proximity-dependent biotin identification (BioID), was established by Roux et al. (6). The basic principal of the BioID system is that a protein of interest (termed bait) that harbours a promiscuous biotin ligase (BirA R118G mutant, from Escherichia coli) catalyses the biotinylation of nearby endogenous proteins with lysine side chains (within ∼10 nm) after the addition of a biotin supplement to the tissue culture medium (5, 6) (Fig. 1A, specialized protocol for mitochondrial interactomes). Because biotinylation of the targets is a covalent modification process, these biotin-labelled proteins are resistant to stringent cell lysis treatment with the protein extraction and washing process, and affinity purification (such as streptavidin beads). As a result, BioID coupled with MS analysis reveals biotin-labelled proteins and allows for mapping of protein–protein interaction networks with high spatial resolution in living cells (6). Further, targeting of the nearby proteins can be enhanced by using a smaller type of biotin ligase (from Aquifex aeolicus; BioID2) to fuse the bait protein, which also reduces amount of biotin supplementation that is required (7).
Fig. 1.
Applying the BioID assay to elucidate protein interaction in mitochondrial complexes. (A) Schematic view of the BioID method in mitochondrial proteins. Cultured cells expressing proteins of interest (Bait) fused with the promiscuous BirA are incubated with biotin for 18 h. The mitochondrial fraction isolated from the cells is then lysed, and the extracted biotinylated proteins are affinity-purified followed by tryptic digestion and identification by LC-MS/MS. (B) Volcano plot showing PHB2 versus control (non-bait) plotted against the P-value of quadruplicate results. Box in the figure shows significantly increased proteins (abundance ration > 5, P < 0.0001), and the most enriched proteins are labelled in the right enlarged view. This figure is reproduced from a previous paper (8). (C) Model of PHB complexes in mitochondria. PHB complexes localized in IMM seem to assemble with MIB complex that subassemblies with MICOS subcomplex and SAM. The candidates identified by the BioID assay in (B) are coloured in red. Note that the scheme does not show information on the stoichiometry.
The BioID2 assay was applied to elucidate a mitochondrial macromolecular complex in mammalian cells; BirA from A. aeolicus was fused to the C-terminal of prohibitin 2 (PHB2), an inner mitochondrial membrane (IMM)-bound protein (8). It is important to consider the effects of the BirA fusion tag at either the N- or C-termini on the biologic activity and the organelle localization in the target protein, particularly mitochondrial proteins (due to the potential masking of the mitochondrial targeting sequence). BirA fused to PHB2 behaved in an almost identical manner to the intact protein and biotinylated a large number of mitochondrial proteins (8). MS proteomic analysis mapping revealed significant enrichment of a select group of mitochondrial proteins, including its PHB1 isoform, mitochondrial contact site and cristae organizing system (MICOS) subcomplex (9), and sorting and assembly machinery (SAM) (10) in the PHB2-BirA fraction (Fig. 1B), showing the assembly of PHB2 with the mitochondrial intermembrane space (IMS) bridging (MIB) complex (11, 12) (Fig. 1C).
BioID methods have been successfully applied to reveal associations between membrane proteins that exhibit a wide range of subcellular localizations, not only in the mitochondria but also in the nuclear lamina (6, 13), cell–cell junctions (14) and endoplasmic reticulum (15). Although the BioID approach is potentially useful for elucidating subcellular protein–protein interactions in living cells, the system has some inherent limitations. If the topology of the bait and target face the opposite direction across the membrane, the catalytic activity from the BirA may not reach the target side. Further, if the surface of the target molecule proximate to the bait lacks lysine residues, the approach may fail.
Cross-linking coupled with MS
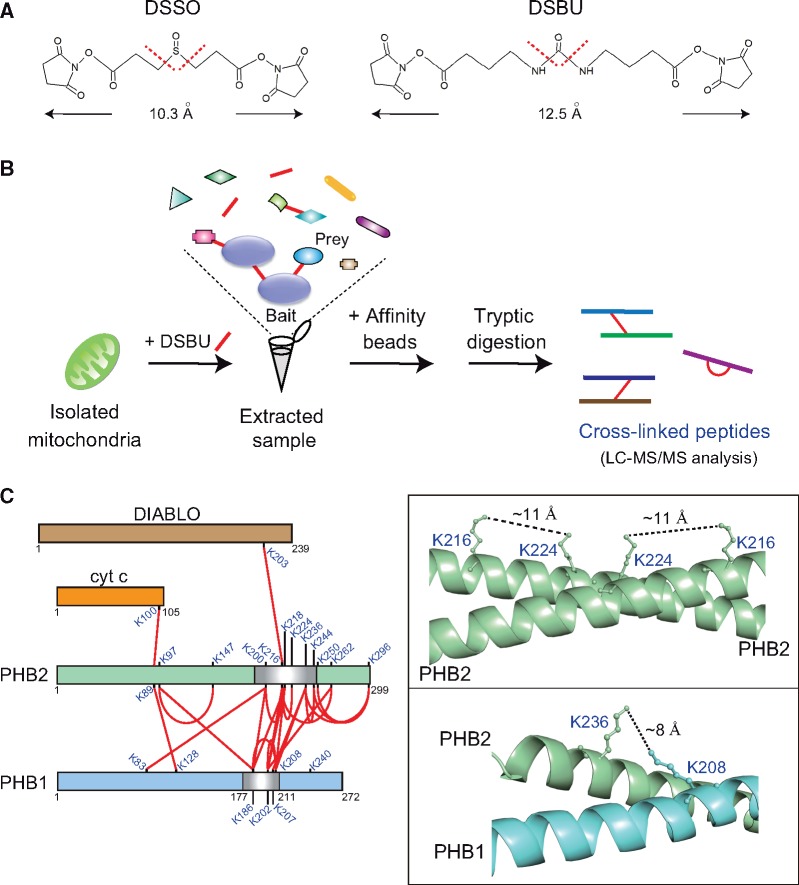
Chemical cross-linking is a classical approach used to freeze protein complexes, especially for capturing transient protein–protein interactions. The use of membrane-permeable cross-linkers allows the reagents to permeate into the cellular compartments, and in combination with MS (cross-linking coupled with MS: XL-MS) (16, 17) provides vital insight into the spatial arrangement of protein complexes in organelles at the subunit level. XL-MS is challenging, however, because of the complex fragmentation pattern of cross-linked peptides, which frequently prevents unambiguous identification of the cross-linked peptides. MS-cleavable cross-linkers, such as disuccinimidyl sulfoxide (DSSO; containing cleavable C − S bonds) or disuccinimidyl dibutyric urea (DSBU; containing cleavable C − N bonds) (Fig. 2A), are recent innovations (18). The advantages of these MS-cleavable cross-linkers is that their cleavage (in the gas phase during MS) generates distinguishable ion doublets that enable the cleaved products to be identified with a database search (16, 19) (Fig. 2B). A number of dynamic interactions of the ribosome have been successfully captured using this method in HeLa cells (16).
Fig. 2.
Overview of the XL-MS analysis. (A) Chemical structures and spacer lengths of the MS-cleavable cross-linking reagents DSSO and DSBU. Collision-induced dissociation-associated cleavage sites in the cross-linkers are indicated by red dotted lines. (B) Schematic view of the XL-MS method in mitochondrial proteins. Mitochondria are chemically cross-linked with DSBU, followed by extraction of the protein lysates. The extracted cross-linked proteins are affinity-purified for subsequent analysis and identification by LC-MS/MS. (C) Cross-link (red lines) map showing all observed Lys-Lys cross-linked pairs between PHB2 and other mitochondrial proteins. Cross-links within PHB1 or PHB2 between monomers or within one monomer cannot be distinguished in this analysis. The box in the right two panels shows the crystal structure of the PHB2 homo-dimer (PDB 6IQE; top) and predicted PHB1-PHB2 hetero-dimer model in which observed cross-link pairs are labelled. The cross-link map on the left was reproduced from a previous paper (8). The structures in the figure were depicted using PyMOL.
Aiming to elucidate at mitochondrial interactomes, Bruce et al. (20, 21) expanded the XL-MS method in mitochondria isolated from several murine tissues and revealed a large-scale mitochondrial proteome, including MICOS and oxidative phosphorylation (OXPHOS) complexes. Heck et al. (22) also identified multiple stoichiometries and conformations of OXPHOS supercomplexes in intact mitochondria from murine heart. XL-MS has further provided important insight into the spatial arrangement of the PHB complexes in mitochondria (mentioned in the above section). XL-MS of mitochondria isolated from human embryonic kidney (HEK) 293 cells with DSBU revealed a number of unique Lys-Lys cross-links in the PHB complexes (8). The majority of identified peptide pairs were homotypic (PHB2-PHB2) and heterotypic (PHB1-PHB2), and substantial interactions were observed between PHB2 and DIABLO, or between PHB2 and cytochrome c, both of which are involved in apoptosis (Fig. 2C, left). Some of the observed cross-links within PHB complexes show good agreement with the high-resolution three-dimensional PHB2 crystal structure (8). The linkage distances between PHB2(Lys216) and PHB2(Lys224) or between PHB1(Lys208) and PHB2(Lys236) are well within the empirically derived maximum linkable length (12.5 Å) of DSBU (Fig. 2C, right).
Although the XL-MS method has provided insight into many protein complexes for mapping three-dimensional structures at low resolution, the cross-linking process may cause artefactual protein–protein contact. A minor limitation of the XL-MS method is that cross-links within the target cannot be distinguished as intramolecular or intermolecular, as in the case of PHB complexes.
Isobaric chemical labelling for multiplex quantitative proteomics
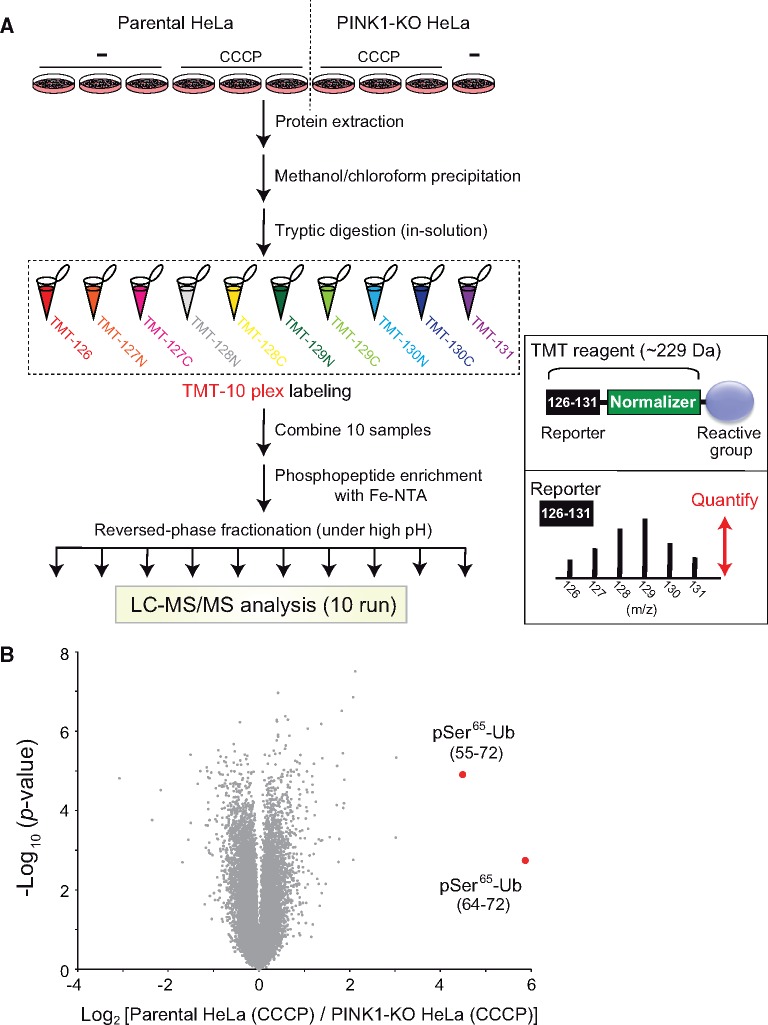
Finally, we briefly describe another type of protein–protein interaction (kinase-substrate interaction for phosphorylation) in sub-mitochondrial compartments. Loss-of-function mutations in PTEN-induced kinase 1 (PINK1), a mitochondrial serine/threonine-protein kinase, cause early-onset autosomal recessive Parkinson’s disease (23). Upon the loss of the mitochondrial membrane potential, such as in damaged mitochondria, PINK1 switches the import pathway from the IMM to the outer mitochondrial membrane (OMM), where interactions with the translocase of the outer membrane complex stabilize PINK1, resulting in an increased the PINK1 concentration at the OMM (24). PINK1 then forms a large multi-protein complex on the OMM and is activated through intermolecular autophosphorylation (25). The activated PINK1 catalyses the phosphorylation of both ubiquitin (Ser65) and the ubiquitin-like domain of Parkin (Ser65), leading to mitochondrial translocation and activation of Parkin (26). Ubiquitination of various mitochondrial proteins by the activated Parkin ultimately leads to selective elimination of damaged mitochondria via the autophagy pathway. Experimentally, the phosphorylation of ubiquitin by PINK1 was substantially confirmed by isobaric tandem mass tag (TMT) labelling combined with phosphopeptide enrichment and liquid chromatography-tandem mass spectroscopy (LC-MS/MS). The TMT method (27) allows for simultaneous identification and quantification of chemically labelled peptides from up to 11 samples labelled at the peptide level (Fig. 3A). Phosphoproteomic analysis of parental and PINK1-deficient HeLa cells quantified 31,286 unique phosphopeptides and revealed a PINK1-dependent increase in the ubiquitin peptide containing phosphorylated Ser65 (Fig. 3B). Thus, global large-scale quantitative phosphoproteomic analysis is a useful strategy for analysing kinase-substrate interactions (29, 30).
Fig. 3.
Example of multiplex quantitative proteomics. (A) Workflow of the TMT/Fe(III)-nitrilotriacetate (Fe-NTA)-based quantitative phosphoproteomic analysis. Parental or PINK-knockout HeLa cells (28) were treated with or without carbonyl cyanide m-chlorophenylhydrazone (CCCP). Cellular proteins were purified by methanol/chloroform precipitation and digested with trypsin. The digested peptides for each sample were labelled with TMT-10plex reagents, and all TMT-labelled samples were pooled and subjected to phosphopeptide enrichment using Fe-NTA complexes. After reversed-phase fractionation under high-pH conditions, each fraction was analysed by LC-MS/MS. Right bottom insert shows a schematic of the TMT reagent structure (top) with its functional regions (reporter and mass-normalizer) and peptide quantitation is accomplished by comparing the intensities of the reporter ions in the MS/MS spectra (bottom). (B) A volcano plot showing changes in the phosphopeptides of CCCP-treated parental HeLa cells and CCCP-treated PINK1-knockout HeLa cells. Plots of the two ubiquitin-derived peptides (corresponding to amino acids 55–72 and 64–72) containing phosphorylated Ser65 are shown in red.
Future perspectives
MS-based proteomic methods are ideal for increasing the number of molecules analysed in protein–protein interactions when elucidating multi-protein complexes. A novel technique, using peroxidase-based proximity labelling was also developed and one approach, known as engineered ascorbate peroxidase, generates a snapshot of proximate proteins with a rapid labelling time (within a few minutes) (31, 32).
This review provides several examples of the application of proteomic analysis to examine intermolecular interactions that have been advanced mainly in mitochondrial proteins. Applications to other organelle-bound molecules or cytoplasmic proteins as well as for studies of complexes in higher organisms (e.g. mice) are highly anticipated.
Acknowledgements
We are grateful to our lab members for critically reading the manuscript and providing valuable comments. We are also grateful to Taro Tamada (National Institutes for Quantum and Radiological Science and Technology) for assistance with the figures. We acknowledge many researchers who have contributed to the field of proteomic analysis and apologize that space limitations prevent us from citing all of their other articles.
Funding
This study was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (JSPS KAKENHI Grants No. 17H03667, 17K19561 and 18H04863 to T.K., and 17K08635, 18KK0229 and 19H04966 to H.K.) and Joint Usage and Joint Research Programs, the Institute of Advanced Medical Sciences, Tokushima University to T.K.
Author contribution
T.K. and H.K. contributed to review the field of proteomic analysis and wrote the manuscript.
Conflict of Interest
None declared.
Glossary
Abbreviations
- BioID
proximity-dependent biotin identification
- BirA
Bifunctional ligase/repressor BirA
- BRET
bioluminescence resonance energy transfer
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- DSBU
disuccinimidyl dibutyric urea
- DSSO
disuccinimidyl sulfoxide
- Fe-NTA
Fe(III)-nitrilotriacetate
- FRET
fluorescence resonance energy transfer
- GFP
green fluorescent protein
- HEK293
human embryonic kidney 293
- IMM
inner mitochondrial membrane
- IMS
intermembrane space
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- MIB
mitochondrial intermembrane space bridging
- MICOS
mitochondrial contact site and cristae organizing system
- OMM
outer mitochondrial membrane
- OXPHOS
oxidative phosphorylation
- PHB
prohibitin
- PINK1
PTEN-induced kinase 1
- RET
resonance energy transfer
- SAM
sorting and assembly machinery
- TMT
tandem mass tag
- XL-MS
cross-linking coupled with mass spectrometry
References
- 1. Jares-Erijman E.A., Jovin T.M. (2003) FRET imaging. Nat. Biotechnol. 21, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 2. Miyawaki A. (2011) Development of probes for cellular functions using fluorescent proteins and fluorescence resonance energy transfer. Annu. Rev. Biochem. 80, 357–373 [DOI] [PubMed] [Google Scholar]
- 3. Pfleger K.D., Eidne K.A. (2006) Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nat. Methods 3, 165–174 [DOI] [PubMed] [Google Scholar]
- 4. Sasaki O., Yoshizumi T., Kuboyama M., Ishihara T., Suzuki E., Kawabata S., Koshiba T. (2013) A structural perspective of the MAVS-regulatory mechanism on the mitochondrial outer membrane using bioluminescence resonance energy transfer. Biochim. Biophys. Acta 1833, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 5. Kim D.I., Roux K.J. (2016) Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol. 26, 804–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roux K.J., Kim D.I., Raida M., Burke B. (2012) A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim D.I., Jensen S.C., Noble K.A., KC B., Roux K.H., Motamedchaboki K., Roux K.J. (2016) An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 27, 1188–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshinaka T., Kosako H., Yoshizumi T., Furukawa R., Hirano Y., Kuge O., Tamada T., Koshiba T. (2019) Structural basis of mitochondrial scaffolds by prohibitin complexes: insight into a role of the coiled-coil region. iScience 19, 1065–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rampelt H., Zerbes R.M., van der Laan M., Pfanner N. (2017) Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta 1864, 737–746 [DOI] [PubMed] [Google Scholar]
- 10. Becker T., Vögtle F.N., Stojanovski D., Meisinger C. (2008) Sorting and assembly of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1777, 557–563 [DOI] [PubMed] [Google Scholar]
- 11. Ott C., Ross K., Straub S., Thiede B., Götz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T., Kozjak-Pavlovic V. (2012) Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell Biol. 32, 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huynen M.A., Mühlmeister M., Gotthardt K., Guerrero-Castillo S., Brandt U. (2016) Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim. Biophys. Acta 1863, 91–101 [DOI] [PubMed] [Google Scholar]
- 13. Chojnowski A., Sobota R.M., Ong P.F., Xie W., Wong X., Dreesen O., Burke B., Stewart C.L. (2018) 2C-BioID: an advanced two component BioID system for precision mapping of protein interactomes. iScience 10, 40–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo Z., Neilson L.J., Zhong H., Murray P.S., Zanivan S., Zaidel-Bar R. (2014) E-cadherin interactome complexity and robustness resolved by quantitative proteomics. Sci. Signal 7, rs7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Vliet A.R., Giordano F., Gerlo S., Segura I., Van Eygen S., Molenberghs G., Rocha S., Houcine A., Derua R., Verfaillie T., Vangindertael J., De Keersmaecker H., Waelkens E., Tavernier J., Hofkens J., Annaert W., Carmeliet P., Samali A., Mizuno H., Agostinis P. (2017) The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with Filamin-A and F-Actin remodeling. Mol. Cell 65, 885–899 [DOI] [PubMed] [Google Scholar]
- 16. Liu F., Rijkers D.T., Post H., Heck A.J.R. (2015) Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 [DOI] [PubMed] [Google Scholar]
- 17. Leitner A., Faini M., Stengel F., Aebersold R. (2016) Crosslinking and mass spectrometry: an integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 [DOI] [PubMed] [Google Scholar]
- 18. Yu C., Huang L. (2018) Cross-linking mass spectrometry: an emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu F., Lössl P., Scheltema R., Viner R., Heck A.J.R. (2017) Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 8, 15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schweppe D.K., Chavez J.D., Lee C.F., Caudal A., Kruse S.E., Stuppard R., Marcinek D.J., Shadel G.S., Tian R., Bruce J.E. (2017) Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. USA. 114, 1732–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chavez J.D., Lee C.F., Caudal A., Keller A., Tian R., Bruce J.E. (2018) Chemical crosslinking mass spectrometry analysis of protein conformations and supercomplexes in heart tissue. Cell Syst. 6, 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu F., Lössl P., Rabbitts B.M., Balaban R.S., Heck A.J.R. (2018) The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell. Proteomics 17, 216–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W.P., Latchman D.S., Harvey R.J., Dallapiccola B., Auburger G., Wood N.W. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 24. Lazarou M., Jin S.M., Kane L.A., Youle R.J. (2012) Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M., Shiba-Fukushima K., Sato S., Shimizu H., Fukunaga Y., Taniguchi H., Komatsu M., Hattori N., Mihara K., Tanaka K., Matsuda N. (2012) PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 3, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamano K., Matsuda N., Tanaka K. (2016) The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 17, 300–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L., Elias J.E. (2017) Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol. Biol. 1550, 185–198 [DOI] [PubMed] [Google Scholar]
- 28. Okatsu K., Koyano F., Kimura M., Kosako H., Saeki Y., Tanaka K., Matsuda N. (2015) Phosphorylated ubiquitin chain is the genuine Parkin receptor. J. Cell Biol. 209, 111–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raaijmakers L.M., Giansanti P., Possik P.A., Mueller J., Peeper D.S., Heck A.J., Altelaar A.F. (2015) PhosphoPath: visualization of phosphosite-centric dynamics in temporal molecular networks. J. Proteome Res. 14, 4332–4341 [DOI] [PubMed] [Google Scholar]
- 30. Narushima Y., Kozuka-Hata H., Tsumoto K., Inoue J., Oyama M. (2016) Quantitative phosphoproteomics-based molecular network description for high-resolution kinase-substrate interactome analysis. Bioinformatics 32, 2083–2088 [DOI] [PubMed] [Google Scholar]
- 31. Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., Ting A.Y. (2015) Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hung V., Udeshi N.D., Lam S.S., Loh K.H., Cox K.J., Pedram K., Carr S.A., Ting A.Y. (2016) Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 [DOI] [PMC free article] [PubMed] [Google Scholar]