ABSTRACT
Several studies have revealed the functional importance of autophagy in both adipogenesis and carcinogenesis. Here, we investigated autophagy as a link between tumorigenesis and adipogenesis using 3T3-L1 cells, which have been shown to closely mimic the in vivo differentiation process. The relative levels of LC3-II/I showed that autophagy was the highest after 4–6 days of initiation of differentiation and it diminished thereafter. Furthermore, chloroquine (CQ), a late autophagy inhibitor, effectively inhibited adipogenic differentiation of 3T3-L1 cells, suggesting that autophagy may have a positive impact on adipogenic differentiation. Notably, real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis showed that CQ completely blocked the mRNA expression of three adipokines (adiponectin, leptin, and peroxisome proliferator-activated receptor-γ (PPARγ)), which increased proportionally to adipocyte differentiation. Using adipokine antibody arrays, we also found that among 38 adipokines examined, 6 adipokines were significantly differentially regulated in mature adipocytes compared to those in preadipocytes. A comparative analysis of adipokine production revealed that CQ-treated adipocytes displayed a profile similar to that of preadipocytes. Subsequently, CQ treatment significantly inhibited the migration capacity of v-Ha-ras-transformed cells in both 3T3-L1 adipocyte-conditioned medium and co-culture with 3T3-L1 using a transwell plate. Taken together, our results suggest that autophagy inhibition blocks the production of mediators relevant to the adipogenic process and may significantly contribute to reducing obesity-related cancer risk.
KEYWORDS: Adipocytes, adipokines, autophagy, chloroquine, tumors
Introduction
Obesity is increasing worldwide and is a well-known risk factor for several cancers (De Pergola and Silvestris 2013). However, its underlying mechanism is not yet well understood. Adipose-secreted adipokines might be one of the underlying factors that have been suggested to explain the association between obesity and increased risk of certain cancers (Gui et al. 2017). Moreover, a previous study demonstrated that adipokines from mature adipocytes exhibited the ability to promote cell growth, motility, and invasion of breast cancer cells in vitro (Iyengar et al. 2003).
It has been reported that autophagy is critical for lipid accumulation and adipocyte differentiation factors (Jansen et al. 2012). Genetic disruption of autophagy-related genes significantly impedes adipogenesis in cultured preadipocytes and in mice (Baerga et al. 2009). On the other hand, there is a contrasting report that autophagy is downregulated in the adipose tissue derived from high fat-fed mice and in hypertrophic 3T3-L1 adipocytes (Yoshizaki et al. 2012). Thus, the link between autophagy and cellular program of adipogenesis remains to be further defined.
Besides its contrasting role in adipocyte differentiation, autophagy also has opposing, context-dependent roles in cancer (Levy et al. 2017). Autophagy has been known to function as a tumor suppressor by removing damaged organelles/proteins and limiting cell growth (Mathew et al. 2009). In contrast, other evidence indicates that the predominant role of autophagy in cancer cells is to maintain tumor cell survival (Degenhardt et al. 2006). Our previous studies also showed that autophagy could play a dual role in either pro-cell survival or pro-cell death in response to anticancer drugs (Hwang et al. 2018).
In this study, we investigated autophagy as a link between tumorigenicity and adipocyte differentiation. The 3T3-L1 cell line, which is used extensively as an in vitro model to generate adipocyte-like cells, was used as the adipocyte differentiation model. Our study revealed that autophagy is specifically required for the initial stage of adipogenic differentiation. Furthermore, our results suggested that autophagy might regulate adipogenesis and carcinogenesis via crosstalk with adipokines secreted by adipocytes. Thus, the results of our report could contribute to the development of auxiliary therapy against obesity-related cancer risk.
Materials and methods
Reagents and antibodies
Proteome Profiler Mouse Adipokine Array Kit (Catalog # ARY013) was purchased from R&D Systems (Minneapolis, MN, USA). Dulbecco’s modified Eagle’s medium (DMEM), and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific (Carlsbad, CA, USA). Insulin, 3-isobutyl-1-methylxanthine (IBMX), dexamethasone (DEX), Oil Red O dye, and chloroquine (CQ) were purchased from Sigma–Aldrich (St. Louis, MO, USA).
Cell lines and culture conditions
Murine preadipocyte (3T3-L1) cells were purchased from the American Type Culture Collection (Manassas, VA, USA). 3T3-L1 preadipocytes were cultured in DMEM-F12 supplemented with 10% FBS and kept at 37°C in a 5% CO2 incubator. The 3T3-L1 cells used for all following investigations were of low passage number and were routinely subdivided at <70% confluence. The v-Ha-ras-transformed NIH3T3 (Ras-NIH3T3) cells were maintained at 37°C in DMEM supplemented with 10% FBS.
Differentiation of 3T3-L1 cells and oil red O staining
For adipocyte differentiation, 3T3-L1 preadipocytes were seeded in 48-well culture plates (1.5 × 104 cells/well) and grown to confluence. Differentiation of 2-day post-confluent preadipocytes (designated as day 0) was initiated with medium containing MDI (10 μg/mL insulin, 1 μM DEX, and 0.5 mM IBMX) in DMEM-F12 with 10% FBS. Three days later, the medium was replaced with DMEM-F12 containing 10% FBS and 10 μg/mL insulin. Another 2 days later, the medium was changed to DMEM-F12 + 10% FBS. Differentiated cells were used for functional assays on days 8–9 after differentiation was initiated. In vitro, differentiated cells were fixed for 20 min in buffered formalin and stained with Oil Red O for 3 h. For the quantification of lipid droplets, stained cells were dried completely and extracted with isopropanol (0.5 mL/well of a 24-well plate). The optical density of the extracted dye was measured at 510 nm using a SpectraMax 190 Microplate Reader (Molecular Devices, Sunnyvale, CA, USA).
Autophagy monitoring assay by conversion of LC3
Autophagy was measured by the conversion of LC3-I to LC3-II by immunoblot analysis as previously (Hwang et al. 2018). Detection was achieved using the Bio-Rad ChemiDoc XRS+ instrument (Hercules, CA, USA), and the data were visualized using the Bio-Rad Image Lab software version 5.2.1 (Bio-Rad Laboratories).
Cell viability assay
The cells were seeded in quadruplicate in 96-well microtiter plates (Costar, Cambridge, MA, USA) at a density of 5 × 103 cells/well and then incubated at 37°C in a humidified 5% CO2/95% air incubator. On the indicated days, the cells were incubated with WST-1 reagent at 37°C for 3 h. The absorbance of the samples against a background control (medium alone), which served as a blank, was measured at 450 nm using a SpectraMax 190 microplate reader.
Real-time quantitative reverse transcription-PCR (qPCR) analysis
Total RNA isolation, reverse transcription, and PCR reaction were performed as described previously (Kim et al. 2017). All primers were synthesized by Bioneer (Daejeon, Korea). The primer sequences used for the qPCR analysis are listed in Table 1. The real-time PCR data were normalized for differences in β-actin levels by analysis with the 2−ΔΔCt method.
Table 1. Primer sequence for real-time quantitative PCR analysis.
| Genes | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Adiponectin | GTGCAGGTTGGATGGCAGGCA | CAGTGACGCGGGTCTCCAGC |
| Leptin | CCCTGTGGAGGTGAGCGGGA | CCAGCCACCACGAGCCTTCG |
| PPARγ | GCCTTCGCTGATGCACTGCC | CAGCAACCATTGGGTCAGCTCT |
Proteome Profiler™ adipokine array analysis
Proteome Profiler Mouse Adipokine Array Kits were used to simultaneously detect the relative expression levels of 38 adipokines using protein lysates from 3T3-L1 adipocytes. The array was performed according to the manufacturer’s instructions. Briefly, protein (250 μg) was incubated overnight with the adipokine antibody cocktail. The antigen–antibody reaction was detected by chemiluminescent detection substrate (Thermo Fisher Scientific, Rockford, IL, USA) and images were captured by a Molecular imager (Bio-Rad). Densitometric analysis of immunoreactions on the captured image was performed by Image Lab software version 5.2.1 (Bio-Rad).
Culture of Ras-NIH3T3 cells in adipocyte-conditioned medium
When 3T3-L1 adipocytes were differentiated to day 12 in the absence and presence of CQ, the medium was replaced with 1% FBS/DMEM for 24 h. The supernatants were collected and used as an adipocyte-conditioned medium (Ad-CM) for subsequent in vitro studies. To test the effect of Ad-CM on the growth of Ras-NIH3T3 cells, different concentrations of Ad-CM were prepared in a fresh 1% FBS/DMEM medium.
Co-culture of Ras-NIH 3T3cells with 3T3-L1 adipocytes
To mimic the physiological environment of the obesity-related tumor, Ras-NIH3T3 cells and 3T3-L1 adipocytes were co-cultured using a transwell culture system (0.4 mm pore size). Firstly, 3T3-L1 cells were seeded in 6-well plates and differentiated into adipocytes in the absence and presence of CQ until day 12. Ras-NIH3T3 cells were seeded in the upper chamber of a 6-well transwell culture system at a density of 4.25 × 104 cells/well and co-cultivated with or without mature adipocytes in the lower chamber. Ras-NIH3T3 cells cultivated alone in similar conditions served as controls.
In vitro cell migration assay
An in vitro cell migration assay was carried out using a 24-well transwell culture system as described previously (Hwang et al. 2018). For quantitation, the crystal violet dye retained on the filters was acid extracted, and cell migration was measured by reading the absorbance at 550 nm. Each experiment was performed in triplicate.
Results
The role of autophagy in 3T3-L1 adipocyte differentiation
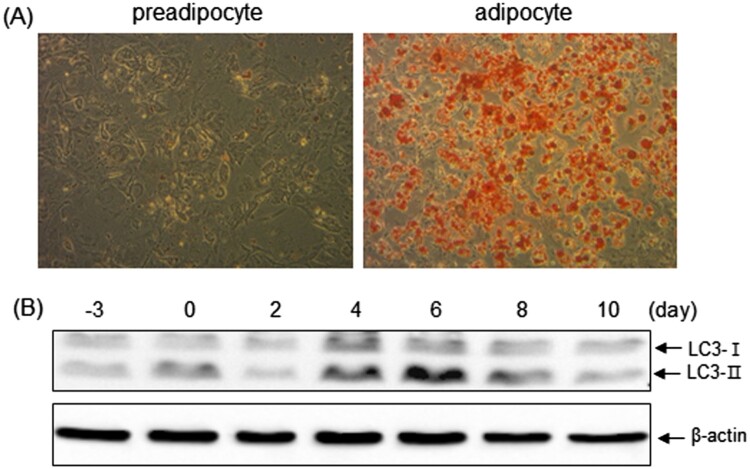
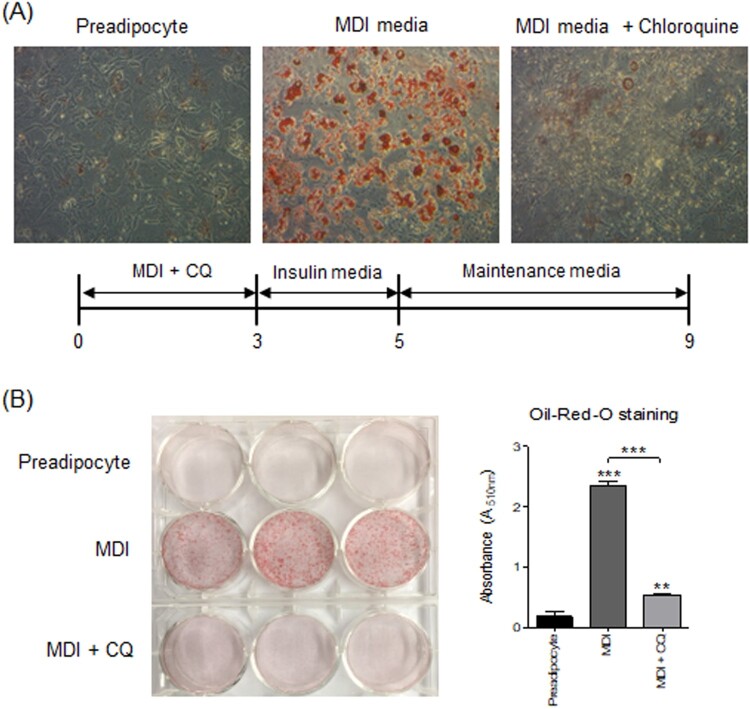
Differentiation of 3T3-L1 preadipocytes into adipocytes was monitored by staining with the neutral lipid-specific dye Oil Red O, which reveals the formation of visible lipid droplets. As shown in Figure 1A, 3T3-L1 cells treated with MDI readily differentiated into morphologically distinct fat-laden adipocytes. To reveal the role of autophagy during differentiation of 3T3-L1 preadipocytes, we measured LC3-I to LC3-II conversion, which is a good indicator of autophagy. The relative levels of LC3-II/I showed that autophagy reached a maximum after 4–6 days of initiation of differentiation and diminished thereafter (Figure 1B). In addition, treatment with CQ, which is known to inhibit the fusion of autophagosome with lysosome (Kimura et al. 2013), impaired differentiation of 3T3-L1 cells (Figure 2A and B). These data collectively suggest that autophagy may have a positive impact on adipogenic differentiation.
Figure 1.
Autophagy induction during adipocyte differentiation of 3T3-L1 preadipocytes. (A) Morphological evidence of adipocyte differentiation at day 9 of the conversion process was monitored by the appearance of fat droplets utilizing light microscopy after oil red O staining. (B) At −3, 0, 2, 4, 6, 8 and 10 days after initiation of differentiation, the change in the electrophoretic mobility of LC3 from a non-autophagic (LC3-I) form to an autophagic membrane-recruited (LC3-II) form was determined by immunoblotting. β-Actin expression was assessed as protein loading control. The presented results are representative of at least three independent experiments.
Figure 2.
The effect of the late-stage autophagy inhibitor chloroquine on 3T3-L1 adipocyte differentiation. The 3T3-L1 preadipocytes were treated with 10 μM CQ in differentiation medium for 3 days before being switched to insulin medium. (A) A representative image of Oil Red O staining of cells at day 9. (B) Left, A representative image of a 6-well plates stained with Oil Red O at day 9; Right, Quantification of Oil Red O staining (n = 3/treatment).
Late-stage autophagy inhibitor chloroquine affects the production of adipokines
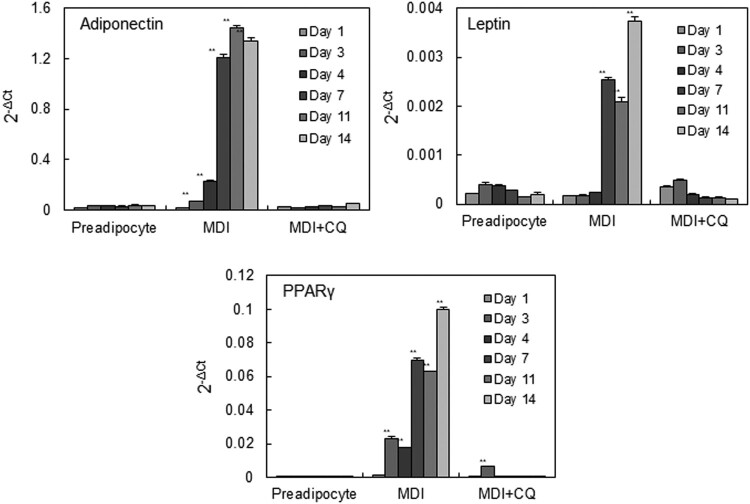
Adipokines secreted by the adipose tissue play an important role in adipocyte differentiation (Hauner 2005). Thus, we investigated whether autophagy is also closely linked to adipokine production. Among the adipokines, leptin, adiponectin, and PPARγ are currently the most prominent due to their functions in the regulation of adipogenesis (Deng and Scherer 2010). The qPCR assay revealed that the expressions of three adipokine mRNA were first detected on day 3 of initiation of differentiation and then increased progressively as the cells acquired a terminally differentiated phenotype (Figure 3). Notably, CQ completely blocked the increase of mRNA expression of these adipokines in 3T3-L1 cells during MDI-induced adipocyte differentiation.
Figure 3.
The effect of chloroquine on adipokine mRNA expression during 3T3-L1 adipocyte differentiation. Total RNA was isolated from 3T3-L1 cells at 1, 3, 4, 7, 11, and 14 days after initiation of differentiation in the presence or absence of 10 μM CQ. The mRNA levels of three adipokines (adiponectin, leptin, and PPARγ) were measured by real-time qRT-PCR. For quantitative analysis of gene expression, the comparative threshold cycle (Ct) method for relative quantification (2−ΔΔCt) was used. The expression of the target genes was normalized to β-actin expression. Values represent the mean ± standard deviation (SD) of quadruplicate determinants from one of three representative experiments. **p < .01 compared to preadipocytes, as determined by Dunnett’s test.
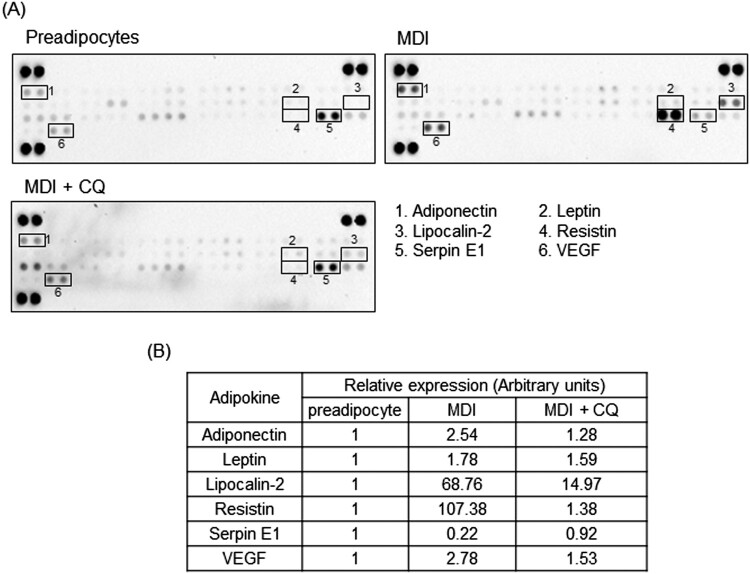
In addition, Proteome Profiler™ mouse adipokine antibody arrays were performed to identify the differentially expressed adipokines in differentiated adipocytes. Figure 4A depicts images of chemiluminescent reaction spots that represent the expression of 38 adipokines in 3T3-L1 cells. Among 38 adipokines examined, we found 6 adipokines (adiponectin, lipocalin-2, resistin, VEGF, leptin, and serpin E1/PAI-1) that were significantly differentially regulated in mature adipocytes compared to those in preadipocytes (Figure 4B). CQ-treated adipocytes displayed a profile similar to that of preadipocytes, with a significant reduction of adiponectin, lipocalin-2, resistin, and VEGF expression, but only a minor effect on leptin. Interestingly, serpin E1/PAI-1 was downregulated in differentiated cells but recovered to its normal level after CQ treatment. These observations suggest that autophagy modulates several key factors of adipogenic differentiation.
Figure 4.
Chloroquine modulates adipokine production in 3T3-L1 adipocytes. 3T3-L1 preadipocytes were differentiated to day 7 into mature adipocytes in the presence or absence of 10 μM CQ. The cell lysates from preadipocytes and mature adipocytes were assessed for adipokine expression using an antibody adipokine array. In (A), images of chemiluminescent reaction spots on adipokine profiler membranes representing the expression of various adipokines in 3T3-L1 cells differentiated in the presence and absence of 30 μM of CQ. In (B), a comparative expression of adipokines (in arbitrary units) obtained from densitometric analysis of the chemiluminescent reaction spots is tabulated.
Chloroquine breaks the crosstalk between Ras-NIH3T3 cells and adipocytes
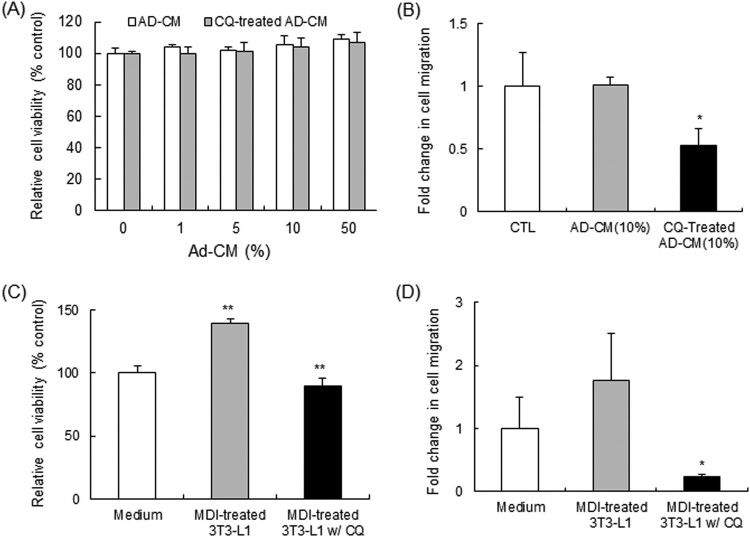
Adipokines secreted from mature adipocytes might be one of the factors that have been suggested to explain the association between obesity and increased risk of certain cancers (Gui et al. 2017). Thus, we evaluated the effect of Ad-CM on Ras-NIH3T3 cells, which clearly show morphologically transformed foci of cells. Ras-NIH3T3 cells were cultured for 48 h in 1%, 5%, 10%, and 50% Ad-CM in fresh complete medium. Ad-CM exerted no effect on the viability of Ras-NIH3T3 cells, regardless of CQ treatment, as compared to cells cultured in normal media (Figure 5A). However, cell migration assay showed that CQ-treated Ad-CM greatly abrogated the migration capacity of Ras-NIH3T3 cells (Figure 5B). Next, to mimic the physiological environment of obesity-related cancer, Ras-NIH3T3 cells were co-cultured using a transwell culture system with differentiated 3T3-L1 cells. After co-cultivation with 3T3-L1 cells, the viability of Ras-NIH3T3 cells was significantly increased by 39% (p < 0.01) (Figure 5C). The migration capacity was somewhat greater in Ras-NIH3T3 cells co-cultivated with mature adipocytes, although it was statistically insignificant (Figure 5D). However, the migration capacity of Ras-NIH3T3 cells was greatly suppressed when co-cultivated with 3T3-L1 cells treated with CQ. These results suggest that impaired autophagy deregulates the production of adipokines relevant to the adipogenic process, and may significantly contribute to the migration capacity of tumor cells.
Figure 5.
Chloroquine breaks the crosstalk between Ras-NIH 3T3 cells and adipocytes. (A and B) Ad-CM was collected from 3T3-L1 adipocytes that were differentiated to day 12 in the absence and presence of CQ. In (A), Ras-NIH 3T3 cells were cultured in the presence of increasing concentrations of Ad-CM for 48 h. Cell viability was then evaluated using WST-1 reagent. The values represent mean ± SD of quadruplicates from one of three representative experiments. In (B), Ras-NIH 3T3 cells were incubated for 4 h in 10% Ad-CM, and then migration was measured in vitro using transwell system. One representative data were presented. n = 3; *p < .05 compared to cells cultured in normal Ad-CM. (C and D) Ras-NIH 3T3 cells were co-cultured with adipocytes that were differentiated in the presence or absence of CQ for 48 h using a transwell system. In (C), Ras-NIH 3T3 cells were trypsinized and cell viability was evaluated using WST-1 reagent. The values represent mean ± SD of quadruplicates from one of three representative experiments. In (D), the migration capacity of Ras-NIH 3T3 cells was measured in vitro using transwell system. One representative data were presented. n = 3; **p < .01 and *p < .05 compared to cells grown alone.
Discussion
Autophagy may play a crucial role in modulating adipogenic differentiation (Jansen et al. 2012), provoking a considerable interest in the possibility of targeting autophagy for the prevention and treatment of obesity-related diseases. In this study, we found that the relative levels of LC3-II/I were increased most significantly on days 4–6 after initiation of adipogenic differentiation and then returned to normal levels thereafter. Consistent with our results, it has been reported that autophagy is essential and most active during the initial stage of adipocyte differentiation, but it is dispensable during the later stages (Skop et al. 2014). We used CQ, which is currently the most widely used autophagy inhibitor, to block autophagy in 3T3-L1 preadipocytes. In particular, CQ is the only clinically available autophagy inhibitors approved by the US Food and Drug Administration (Mauthe et al. 2018). Although the type III PI3K inhibitor 3-MA was the first identified inhibitor of autophagy, a recent study presents a finding that 3-MA could promote autophagy under nutrient-rich conditions for a prolonged period (Wu et al. 2010). Our results revealed that, when treated with CQ, 3T3-L1 cells exhibited drastically reduced the efficiency of adipocyte differentiation. These data collectively suggest that autophagy may have a positive impact on adipogenic differentiation. However, the precise mechanisms by which autophagy induces adipogenic differentiation are poorly understood. An intriguing hypothesis is that autophagy increases the stability of PPARγ2, the key regulator of adipogenesis (Zhang et al. 2013). In addition, the full-length isoform of CCAAT enhancer binding protein β (C/EBPβ-LAP) has been reported to promote early adipogenic differentiation by directly activating expression of PPARγ2 (Lechner et al. 2013). Several studies have used RNA-Seq analysis to investigate the genes related to adipogenesis, some of which have been associated with tumorigenesis. The RNA-Seq study by Mangiola et al. (2018) suggests that periprostatic adipose tissue from patients with high-risk prostate cancer has a distinct transcriptional signature, which was confirmed by qPCR in an expanded cohort. In particular, Leptin and Wnt have been demonstrated as the top two transcripts elevated in mouse high fat diet colon shared with tumors, using next generation RNA sequencing and pathway analysis (Penrose et al. 2017).
Adipokines secreted from mature adipocytes might be one of the factors that have been suggested to explain the association between obesity and increased risk of certain cancers (Gui et al. 2017). Here, we evaluated the role of autophagy in the expression of three adipokines (adiponectin, leptin, and PPARγ), which are currently the most prominent due to their functions in the regulation of adipogenesis (Deng and Scherer 2010). The real-time qRT-PCR analysis showed that the expression of these three adipokines increased proportionally to adipocyte differentiation. In addition, adipokine antibody array showed that among 38 adipokines examined, 5 adipokines were upregulated during differentiation of 3T3-L1 preadipocytes into adipocytes, with a time-course comparable to that of other adipocyte markers. These five adipokines included adiponectin, lipocalin-2, resistin, leptin, and VEGF. In particular, lipocalin-2 and resistin levels were most dramatically increased in parallel with adipocyte differentiation. Recently, it has been reported that resistin is upregulated in patients with breast cancer and promotes breast cancer cell growth, invasion, and migration (Lee et al. 2016). Lipocalin-2 (NGAL) was found to attenuate autophagy to exacerbate cardiac apoptosis induced by myocardial ischemia (Sung et al. 2017). Unexpectedly, we found that adiponectin expression was significantly increased in 3T3-L1 adipocytes, although it has been reported that adiponectin characteristically differs from most adipokines as it is negatively correlated with obesity (Shehzad et al. 2012). However, unlike the results obtained in qRT-PCR analysis, no significant increase in leptin level was observed in adipokine antibody arrays. Norman et al. (2003) reported that differentiated 3T3-L1 adipocytes express only small amounts of leptin. Interestingly, we observed that serpin E1/PAI, which has well-documented pro-tumorigenic functions (Fang et al. 2012), was downregulated in differentiated 3T3-L1 adipocytes. Our findings are consistent with those of Liang et al. (2006), in which serpin E1/PAI-1 deficiency promoted adipocyte differentiation. As expected, CQ-treated cells displayed an adipokine profile similar to that of preadipocytes. Five upregulated adipokines returned to their original levels when treated with CQ. The serpin E1/PAI-1 also returned to its original level when treated with CQ.
Subsequently, the culture of Ras-NIH3T3 cells in 3T3-L1 Ad-CM and co-culture of 3T3-L1 and Ras-NIH3T3 cells using a transwell plate were performed to clarify the relationship between adipocytes and tumor cells. Ad-CM exerted no effect on both the proliferative ability and migration capacity of Ras-NIH3T3 cells. However, our co-culture system showed that Ras-NIH3T3 cells co-cultivated with mature 3T3-L1 adipocytes exhibited enhanced migration, suggesting that mature adipocytes are able to stimulate the migration capacities of cancer cell lines. Co-culture with adipocytes also has an enhancing effect on Ras-NIH3T3 cell proliferation. Conversely, CQ treatment significantly inhibited the migration capacity of Ras-NIH3T3 cells in both the Ad-CM model and the co-culture system. In particular, these results suggest that the deregulation of adipokine levels in adipocytes after CQ treatment might contribute to the suppressed migration capacity of tumor cells. This may probably be due to the difference of expression ratio relative to each of the other adipokines, rather than of the absolute expression level of each adipokine between control and CQ-treated adipocytes.
Taken together, our results revealed that autophagy is essential and most active during the initial stage of adipocyte differentiation, but it is dispensable during the later stages. More importantly, CQ treatment blocked the production of mediators relevant to the adipogenic process in the co-culture system, and might significantly contribute to the suppression of obesity-related cancer development. These results suggest that CQ broke the crosstalk between these two cell lines, possibly contributing to its chemopreventive properties in cancer. Thus, in future studies on the effects of obesity and subsequent alterations in adipokine expression in tumor cells, it is important to verify the role of autophagy as a potential therapeutic target.
Funding Statement
This research was supported by the Incheon National University Research Grant in 2018.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Baerga R, Zhang Y, Chen PH, Goldman S, Jin S.. 2009. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 5:1118–1130. doi: 10.4161/auto.5.8.9991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 10:51–64. doi: 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Scherer PE.. 2010. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Ann N Y Acad Sci. 1212:E1–E19. doi: 10.1111/j.1749-6632.2010.05875.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pergola G, Silvestris F.. 2013. Obesity as a major risk factor for cancer. J Obes. 2013. 291546. doi: 10.1155/2013/291546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Placencio VR, DeClerck YA.. 2012. Protumorigenic activity of plasminogen activator inhibitor-1 through an antiapoptotic function. J Natl Cancer Inst. 104:1470–1484. doi: 10.1093/jnci/djs377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Pan Q, Chen X, Xu S, Luo X, Chen L.. 2017. The association between obesity related adipokines and risk of breast cancer: a meta-analysis. Oncotarget. 8:75389–75399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauner H. 2005. Secretory factors from human adipose tissue and their functional role. Proc Nutr Soc. 64:163–169. doi: 10.1079/PNS2005428 [DOI] [PubMed] [Google Scholar]
- Hwang S-H, Han B-I, Lee M.. 2018. Knockout of ATG5 leads to malignant cell transformation and resistance to Src family kinase inhibitor PP2. J Cell Physiol. 233:506–515. doi: 10.1002/jcp.25912 [DOI] [PubMed] [Google Scholar]
- Iyengar P, Combs TP, Shah SJ, Gouon-Evans V, Pollard JW, Albanese C, Flanagan L, Tenniswood MP, Guha C, Lisanti MP, et al. 2003. Adipocyte-secreted factors synergistically promote mammary tumorigenesis through induction of anti-apoptotic transcriptional programs and proto-oncogene stabilization. Oncogene. 22:6408–6423. doi: 10.1038/sj.onc.1206737 [DOI] [PubMed] [Google Scholar]
- Jansen HJ, van Essen P, Koenen T, Joosten LA, Netea MG, Tack CJ, Stienstra R.. 2012. Autophagy activity is up-regulated in adipose tissue of obese individuals and modulates proinflammatory cytokine expression. Endocrinology. 153:5866–5874. doi: 10.1210/en.2012-1625 [DOI] [PubMed] [Google Scholar]
- Kim JH, Ahn JH, Lee M.. 2017. Upregulation of MicroRNA-1246 is associated with BRAF inhibitor resistance in Melanoma cells with Mutant BRAF. Cancer Res Treat. 49:947–959. doi: 10.4143/crt.2016.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A, Isaka Y.. 2013. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 73:3–7. doi: 10.1158/0008-5472.CAN-12-2464 [DOI] [PubMed] [Google Scholar]
- Lechner S, Mitterberger MC, Mattesich M, Zwerschke W.. 2013. Role of C/EBPβ-LAP and C/EBPβ-LIP in early adipogenic differentiation of human white adipose-derived progenitors and at later stages in immature adipocytes. Differentiation. 85:20–31. doi: 10.1016/j.diff.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Lee JO, Kim N, Lee HJ, Lee YW, Kim SJ, Park SH, Kim HS.. 2016. Resistin, a fat-derived secretory factor, promotes metastasis of MDA-MB-231 human breast cancer cells through ERM activation. Sci Rep. 6:18923. doi: 10.1038/srep18923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JMM, Towers CG, Thorburn A.. 2017. Targeting autophagy in cancer. Nat Rev Cancer. 17:528–542. doi: 10.1038/nrc.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Kanjanabuch T, Mao SL, Hao CM, Tang YW, Declerck PJ, Hasty AH, Wasserman DH, Fogo AB, Ma LJ.. 2006. Plasminogen activator inhibitor-1 modulates adipocyte differentiation. Am J Physiol Endocrinol Metab. 290:E103–E113. doi: 10.1152/ajpendo.00605.2004 [DOI] [PubMed] [Google Scholar]
- Mangiola S, Stuchbery R, Macintyre G, Clarkson MJ, Peters JS, Costello AJ, Hovens CM, Corcoran NM.. 2018. Periprostatic fat tissue transcriptome reveals a signature diagnostic for high-risk prostate cancer. Endocr Relat Cancer. 25:569–581. doi: 10.1530/ERC-18-0058 [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 137:1062–1075. doi: 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F.. 2018. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 14:1435–1455. doi: 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman D, Isidori AM, Frajese V, Caprio M, Chew SL, Grossman AB, Clark AJ, Michael Besser G, Fabbri A.. 2003. ACTH and α-MSH inhibit leptin expression and secretion in 3T3-L1 adipocytes: model for a central–peripheral melanocortin-leptin pathway. Mol Cell Endocrinol. 200:99–109. doi: 10.1016/S0303-7207(02)00410-0 [DOI] [PubMed] [Google Scholar]
- Penrose HM, Heller S, Cable C, Nakhoul H, Baddoo M, Flemington E, Crawford SE, Savkovic SD.. 2017. High-fat diet induced leptin and Wnt expression: RNA-sequencing and pathway analysis of mouse colonic tissue and tumors. Carcinogenesis. 38:302–311. doi: 10.1093/carcin/bgx001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad A, Iqbal W, Shehzad O, Lee YS.. 2012. Adiponectin: regulation of its production and its role in human diseases. Hormones. 11:8–20. doi: 10.1007/BF03401534 [DOI] [PubMed] [Google Scholar]
- Skop V, Cahova M, Dankova H, Papackova Z, Palenickova E, Svoboda P, Zidkova J, Kazdova L.. 2014. Autophagy inhibition in early but not in later stages prevents 3T3-L1 differentiation: effect on mitochondrial remodeling. Differentiation. 87:220–229. doi: 10.1016/j.diff.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Sung HK, Chan YK, Han M, Jahng JWS, Song E, Danielson E, Berger T, Mak TW, Sweeney G.. 2017. Lipocalin-2 (NGAL) attenuates autophagy to exacerbate cardiac apoptosis induced by myocardial ischemia. J Cell Physiol. 232:2125–2134. doi: 10.1002/jcp.25672 [DOI] [PubMed] [Google Scholar]
- Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM.. 2010. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 285:10850–10861. doi: 10.1074/jbc.M109.080796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki T, Kusunoki C, Kondo M, Yasuda M, Kume S, Morino K, Sekine O, Ugi S, Uzu T, Nishio Y, et al. 2012. Autophagy regulates inflammation in adipocytes. Biochem Biophys Res Commun. 417:352–357. doi: 10.1016/j.bbrc.2011.11.114 [DOI] [PubMed] [Google Scholar]
- Zhang C, He Y, Okutsu M, Ong LC, Jin Y, Zheng L, Chow P, Yu S, Zhang M, Yan Z.. 2013. Autophagy is involved in adipogenic differentiation by repressesing proteasome-dependent PPARγ2 degradation. Am J Physiol Endocrinol Metab. 305:E530–E539. doi: 10.1152/ajpendo.00640.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]