Intestinal epithelial autophagy is crucial for host defense against invasive pathogens, and defects in this process occur frequently in patients with inflammatory bowel disease (IBD) and other mucosal disorders, but the exact mechanism that activates autophagy is poorly defined. Here, we investigated the role of RNA-binding protein HuR (human antigen R) in the posttranscriptional control of autophagy-related genes (ATGs) in the intestinal epithelium. We found that targeted deletion of HuR in intestinal epithelial cells (IECs) specifically decreased the levels of ATG16L1 in the intestinal mucosa.
KEYWORDS: IBD, mucosal defense, RNA-binding proteins, circular RNAs, epithelial homeostasis
ABSTRACT
Intestinal epithelial autophagy is crucial for host defense against invasive pathogens, and defects in this process occur frequently in patients with inflammatory bowel disease (IBD) and other mucosal disorders, but the exact mechanism that activates autophagy is poorly defined. Here, we investigated the role of RNA-binding protein HuR (human antigen R) in the posttranscriptional control of autophagy-related genes (ATGs) in the intestinal epithelium. We found that targeted deletion of HuR in intestinal epithelial cells (IECs) specifically decreased the levels of ATG16L1 in the intestinal mucosa. Intestinal mucosa from patients with IBD exhibited reduced levels of both HuR and ATG16L1. HuR directly interacted with Atg16l1 mRNA via its 3′ untranslated region and enhanced ATG16L1 translation, without affecting Atg16l1 mRNA stability. Circular RNA circPABPN1 blocked HuR binding to Atg16l1 mRNA and lowered ATG16L1 production. HuR silencing in cultured IECs also prevented rapamycin-induced autophagy, which was abolished by overexpressing ATG16L1. These findings indicate that HuR regulates autophagy by modulating ATG16L1 translation via interaction with circPABPN1 in the intestinal epithelium.
INTRODUCTION
The epithelium of the mammalian intestinal mucosa acts as a dynamic physical barrier and directly interfaces with a complex and diverse population of luminal bacteria (1–3). Although most members of this microbial community perform beneficial functions, certain pathogenic bacteria can evade this defense barrier and enter intestinal epithelial cells (IECs) (4). IEC-intrinsic innate immune responses limit bacterial invasion and maintain beneficial host-bacterium relationships (5, 6), but the cellular processes that control intestinal epithelial interactions with invasive pathogens remain largely unknown. Autophagy is an evolutionarily conserved process in which cytoplasmic pathogens and unwanted components are targeted to the lysosome for degradation (7–9). More than 30 autophagy-related genes (ATGs) are identified in mammals, and ATG16L1, the product of the Atg16l1 gene, plays an important role in the intestinal epithelium homeostasis, at least partially by interacting with A20 (10) and orchestrating interleukin-22 signaling (11). ATG16L1 also prevents necroptosis in the intestinal epithelium (12) and protects against tumor necrosis factor (TNF)-induced apoptosis during chronic colitis in mice (13). Genome-wide association studies have revealed the presence of polymorphisms and mutations in Atg16l1 and other Atg genes in patients with inflammatory bowel diseases (IBD) and other mucosal disorders associated with defective autophagy, Paneth cell dysfunction, and deregulation of mucosal defense (14–16).
HuR (encoded by the Elavl1 gene) is an extensively studied RNA-binding protein (RBP) and has emerged as a master posttranscriptional regulator of homeostasis in the intestinal epithelium (17–19). Posttranscriptional events, particular changes in mRNA stability and translation, are major mechanisms controlling gene expression in response to stressful environments (20). HuR typically interacts with U- or AU-rich elements located in the 3′ untranslated regions (3′-UTRs) and/or coding regions (CRs) of labile mRNAs, and this association enhances the stability and translation of target transcripts (21, 22). HuR also affects gene regulatory programs by interplaying with different noncoding RNAs (ncRNAs), including microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs). For example, HuR associates with lncRNA SPRY4-IT1 to synergistically increase the stability and translation of mRNAs encoding tight-junction proteins (23), and it also directly interacts with lncRNA H19 to prevent the processing of miRNA 675 (miR-675) from H19 (24). HuR competes with miR-195 to modulate Stim1 mRNA stability antagonistically (25) and also counteracts miR-330 to promote STAT3 translation (26). Targeted deletion of HuR in IECs inhibits regeneration of the intestinal mucosa (27), reduces tumor development (18), delays repair of damaged mucosa induced by mesenteric ischemia/reperfusion in the small intestine and by dextran sulfate sodium in the colon (28), impairs Paneth cell function (15), and alters Rac1 nucleocytoplasmic shuttling in the intestinal epithelium (29).
It has been also reported that HuR regulates expression of several ATGs in renal proximal tubular 2 (HK-2) cells (30) and hepatocellular carcinoma (HCC) cells (31), suggesting the involvement of HuR in the regulation of autophagy. However, all of these studies were conducted in cultured cells; therefore, the in vivo function of HuR in regulating ATG expression and autophagy activation in the intestinal epithelium remains to be fully investigated. Using mice with intestinal epithelium-specific ablation of HuR (IE-HuR−/− mice), we found that loss of HuR predominantly decreased the levels of ATG16L1 in the intestinal epithelium in vivo and further found that human intestinal mucosa with inflammation and injury/erosions from patients with IBD exhibited decreased levels of both HuR and ATG16L1. While HuR did not affect total Atg16l1 mRNA levels in vivo or in vitro, HuR promoted ATG16L1 translation by directly interacting with the 3′-UTR of Atg16l1 mRNA. Interestingly, circPABPN1 blocked HuR binding to Atg16l1 mRNA and prevented HuR-induced ATG16L1 translation. These findings indicate that HuR and circPABPN1 jointly regulate ATG16L1 expression in the intestinal epithelium and suggest that the HuR/circPABPN1/ATG16L1 axis is a promising therapeutic target for interventions to improve the intestinal mucosal defense in patients with compromised function of the intestinal barrier.
RESULTS
Targeted deletion of HuR in IECs inhibits ATG16L1 expression in vivo.
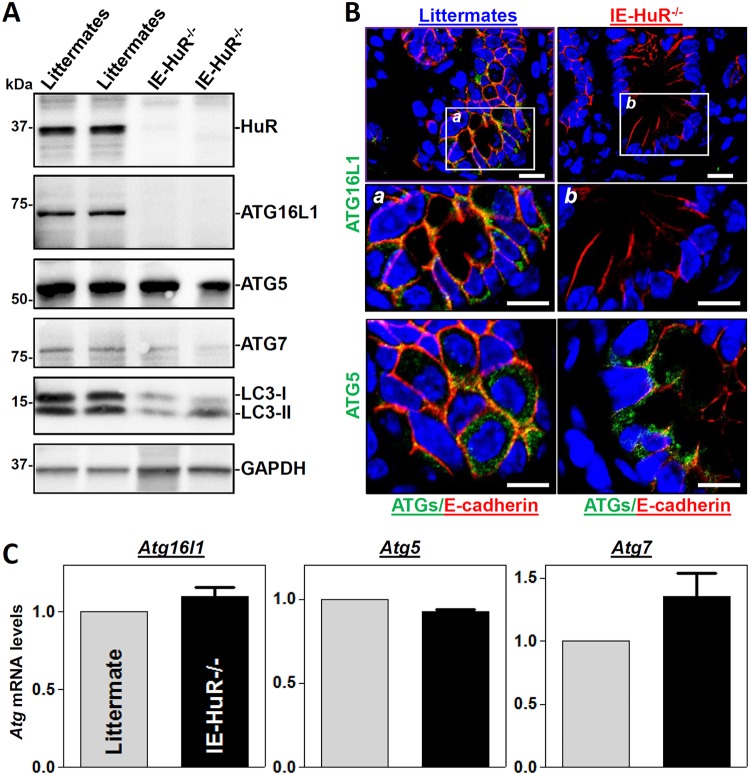
To investigate the in vivo function of HuR in the regulation of autophagy in the mammalian intestinal mucosa, IE-HuR−/− mice were generated by crossing HuRflox/flox (HuRfl/fl) mice with villin-Cre mice as described previously (27). HuR was undetectable in the small intestinal mucosa of IE-HuR−/− mice (Fig. 1A), but it was found at wild-type (WT) levels in other tissues and organs, such as gastric mucosa, lung, liver, and pancreas (data not shown), similar to our previous findings (27, 28). Targeted deletion of HuR in mice did not alter the overall morphology or structure of the small and large intestines (27). Interestingly, conditional HuR deletion in IECs markedly decreased the levels of ATG16L1 in the small intestinal mucosa, although it failed to alter tissue ATG5 abundance. In fact, ATG16L1 levels in the HuR-deficient intestinal epithelia from IE-HuR−/− mice were below the level of detection by Western blotting. Consistently, the levels of microtubule-associated protein light chain 3 I (LC3-I) and LC3-II were also decreased by ∼55% (n = 4; P < 0.05) in the intestinal mucosa of IE-HuR−/− mice compared with control littermates. On the other hand, the basal level of ATG7 in the intestinal mucosa was relatively low but was also reduced by ∼40% (n = 4; P < 0.05) in HuR-ablated mice. Western blot signals of ATG16L2 in the intestinal mucosa of both IE-HuR−/− and littermate mice were extremely low and difficult to detect. Immunohistochemical staining assays showed that immunoreactive signals of both ATG16L1 and ATG5 in the intestinal mucosa were normally located in the cytoplasm in control littermate mice, but ATG16L1 signals completely disappeared in the epithelia of IE-HuR−/− mice (Fig. 1B). In contrast, there were no differences in ATG5 immunoreactive signals in the intestinal mucosa between IE-HuR−/− mice and control littermates. In addition, targeted deletion of HuR in IECs did not alter transcription of the Atg genes, because the levels of all Atg16l1, Atg5, and Atg7 mRNAs in the intestinal mucosa of IE-HuR−/− mice were indistinguishable from those observed in control littermate mice (Fig. 1C). Together, these results indicate that HuR plays an important role in the regulation of autophagy in the intestinal epithelium primarily by modulating ATG16L1 expression.
FIG 1.
Targeted deletion of HuR in IECs decreases the levels of intestinal mucosal ATG16L1 in vivo. (A) Immunoblots of HuR and ATGs in the small intestinal mucosae obtained from control littermate and IE-HuR−/− mice. Total proteins were isolated from the intestinal mucosa and prepared for Western blot analysis. Equal loading was monitored by GAPDH. Three experiments were performed and showed similar results. (B) Immunohistochemical staining of ATG16L1 and ATG5 in the small intestinal mucosa. Green, ATG16L1 or ATG5; red, E-cadherin; blue, nuclei stained by DAPI (4′,6′-diamidino-2-phenylindole). Boxes define the area to be amplified. Scale bars, 25 μm. (C) Levels of mRNAs encoding ATG16L1, ATG5, and ATG7 in the small intestinal mucosa as measured by Q-PCR analysis. Values are the means ± SEM (n = 5).
Decreased HuR is associated with disrupted expression of ATG16L1 in patients with IBD.
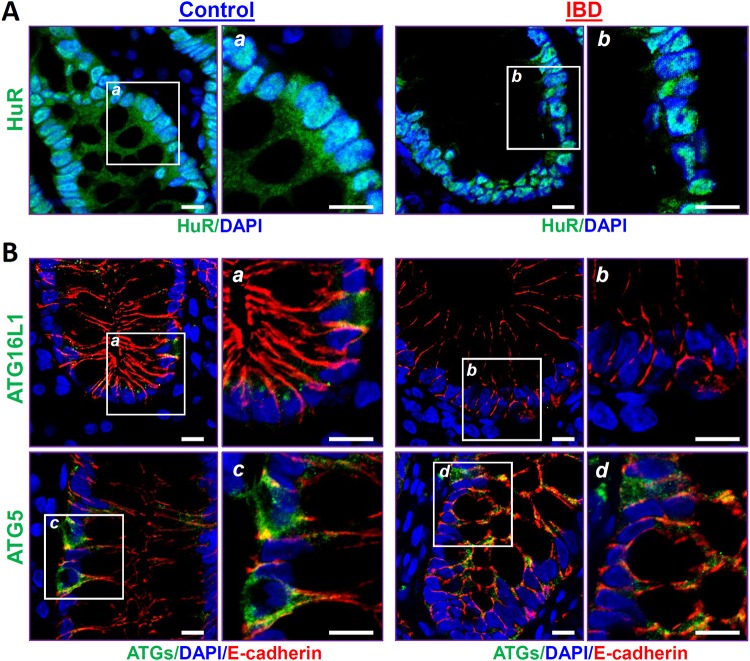
To determine the impact of HuR-regulated ATG16L1 on gut diseases in humans, we examined changes in the levels of HuR, ATG16L1, and ATG5 in the intestinal mucosae from patients with IBD by immunostaining assays. To keep the study focused, all tissue samples from IBD patients were collected from persons with ulcerative colitis (UC), rather than Crohn’s disease, for various measurements, whereas intestinal mucosae from patients without mucosal inflammation and injury/erosions served as controls. HuR was localized at both the cytoplasm and nucleus in the human intestinal epithelia of control individuals, as reported previously (15), but these HuR immunoreactive signals in the mucosal tissues from UC patients were decreased remarkably, particularly in the cytoplasm, compared with those observed in control patients (Fig. 2A). Importantly, the decreased abundance of intestinal mucosal HuR was accompanied by a specific reduction in the levels of ATG16L1 in patients with UC (Fig. 2B, top). In control individuals, both ATG16L1 and ATG5 were found predominantly in the cytoplasm of the intestinal mucosa, but ATG16L1 levels in the mucosal tissues from UC patients decreased dramatically compared with those in control patients. Notably, immunoreactive signals of ATG16L1 were almost undetectable in the intestinal samples obtained from patients with UC, although there were no changes in mucosal ATG5 levels (Fig. 2B, bottom). In addition, the decreased ATG16L1 levels following inhibition of HuR observed in UC patients were associated with delayed mucosal repair and gut barrier dysfunction, as evidenced by compromised epithelial regeneration and an inhibition of tight-junction expression (data not shown), as reported in our previous studies (3, 15). These results strongly suggest that disruption of HuR-regulated ATG16L1 expression in the intestinal mucosa contributes to the pathological process of IBD in humans.
FIG 2.
Association of decreased HuR with a reduction in ATG16L1 in the intestinal mucosae from patients with inflammatory bowel disease (IBD). (A) Immunostaining of HuR in the colonic mucosae from control individuals (without mucosal erosions/inflammation) and patients with IBD. Green, HuR; blue, nucleus stained by DAPI. Boxes define the area to be amplified. Scale bars, 25 μm. (B) Immunostaining staning of ATG16L1 (top) and ATG5 (bottom) in the colonic mucosa as described for panel A. Green, ATG16L1 or ATG5; red, E-cadherin (red); blue, nuclei stained by DAPI. All of these experiments were repeated in human tissue samples obtained from three control individuals or patients with IBD and showed similar results.
HuR silencing inhibits ATG16L1 translation in vitro.
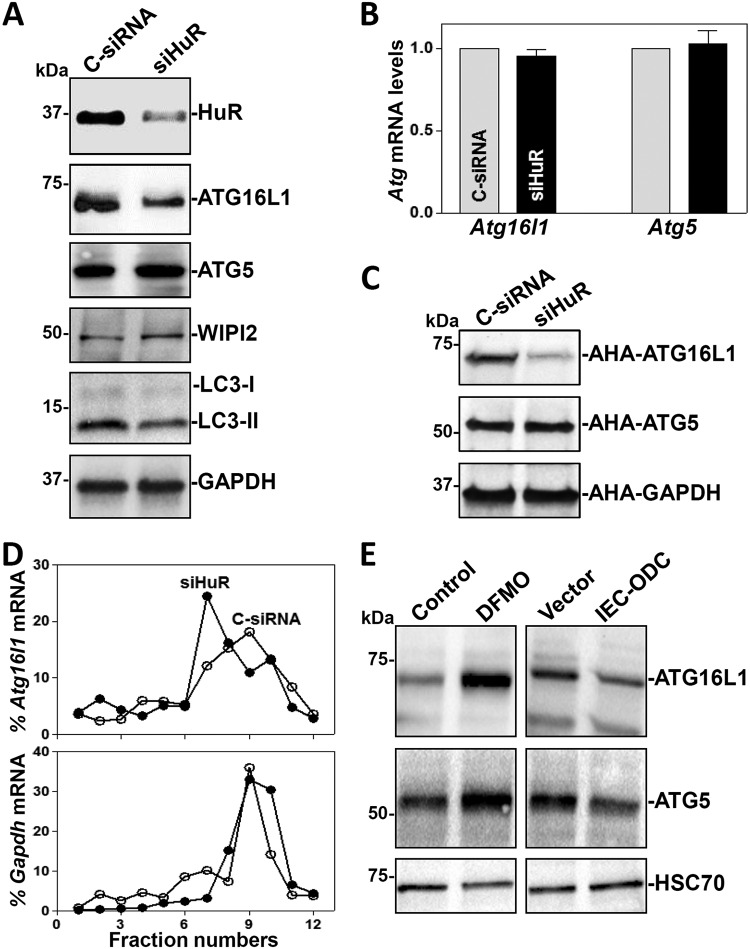
To gain a deeper understanding of the role of HuR in the regulation of ATG16L1 expression in the intestinal epithelium, we silenced HuR expression by use of small interfering RNA (siRNA) targeting the HuR mRNA (siHuR) in cultured IECs. The levels of HuR were decreased by ∼80% in cells at 48 h after transfection with siHuR (Fig. 3A); this reduction was specific, as the levels of other RBPs, such as CUGBP1 and TIAR, were not affected in HuR-silenced populations (data not shown). Consistent with the results in IE-HuR−/− mice, HuR silencing in cultured IECs also reduced ATG16L1 protein levels, but it failed to alter the levels of total Atg16l1 mRNA (Fig. 3B). HuR silencing also lowered the abundance of LC3-II proteins, without affecting the levels of ATG5 or WIPI2. To ascertain if the inhibition of ATG16L1 expression in HuR-silenced populations was due to a repression of ATG16L1 translation, we examined changes in the rate of new ATG16L1 protein synthesis after HuR silencing by assessing the de novo incorporation of labeled amino acids using the Click-iT technology (see Materials and Methods). As shown in Fig. 3C, newly synthesized ATG16L1 was markedly lower in HuR-silenced cells than in cells transfected with scramble control siRNA (C-siRNA). In contrast, there were no changes in nascent synthesis of ATG5 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) proteins after transfection with siHuR.
FIG 3.
HuR silencing inhibits ATG16L1 translation in cultured IECs. (A) Immunoblots of HuR, ATGs, and WIPI2 in cells transfected with siHuR or C-siRNA. Western blot analysis was carried out 48 h after transfection. (B) Levels of Atg mRNAs as measured by Q-PCR analysis in cells treated as described for panel A. Values are the means ± SEM (n = 3). (C) Newly synthesized ATG proteins as measured by l-azidohomoalanine (AHA) incorporation assays. (D) Distributions of mRNAs of Atg16l1 (top) and Gapdh (bottom) in each gradient fraction of polysomal profiles prepared from cells described for panel A. (E) Immunoblots of ATGs in cells exposed to DFMO for 6 days to deplete polyamines (left) and in cells with high levels of cellular polyamines by transfection with the Odc transgene (right). Three experiments were performed and showed similar results.
To further study the impact of HuR on ATG16L1 translation, we examined the relative distributions of Atg16l1 mRNA on individual fractions from polyribosome gradients prepared in cells in which HuR was silenced. Although HuR silencing did not affect global polysome profiles in IECs, as reported previously (32), the levels of Atg16l1 mRNA associated with actively translating fractions (fractions 9 and 10) decreased in HuR-silenced cells, with a marked shift of Atg16l1 mRNAs to low-translating fractions (fractions 7 and 8) (Fig. 3D, top). In contrast, Gapdh mRNA, encoding the housekeeping protein GAPDH, was distributed similarly in both groups (Fig. 3D, bottom). These results indicate that decreased HuR inhibits ATG16L1 expression primarily by lowering Atg16l1 mRNA translation in the intestinal epithelium.
Polyamines function as biological regulators of the intestinal epithelium homeostasis predominantly through control of HuR activity (33, 34). Depletion of polyamines by treating IECs with d,l-α-difluoromethylornithine (DFMO) for 6 days increased the levels of cytoplasmic HuR, whereas increasing the cellular polyamines via ectopic overexpression of the Odc gene (encoding a key enzyme for polyamine biosynthesis) reduced cytoplasmic HuR abundance and increased nuclear HuR levels (35). Interestingly, depletion of cellular polyamines by DFMO also increased the levels of cellular ATG16L1 and ATG5 proteins (Fig. 3E, left). In contrast, IECs overexpressing the Odc gene exhibited decreased ATG16L1 and ATG5 levels (Fig. 3E, right). Because there were no changes in ATG5 abundance in HuR-silenced cells and the mucosal tissues from IE-HuR−/− mice, these findings suggest that polyamines regulate ATG16L1 expression at least partially by altering HuR activity.
HuR interacts with Atg16l1 mRNA via its 3′-UTR.
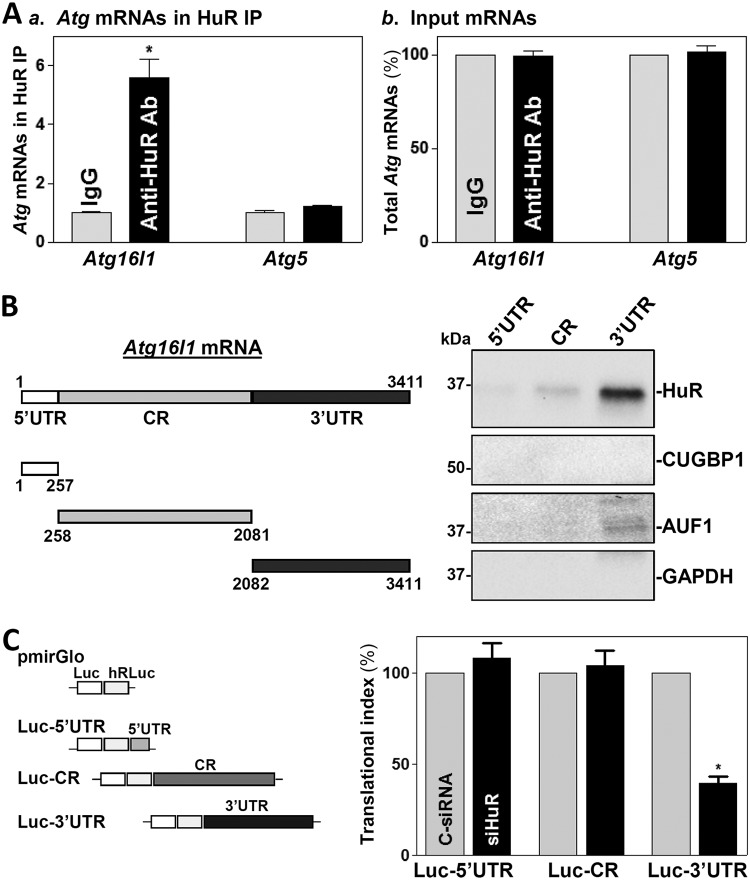
The Atg16l1 mRNA is a potential target of HuR, given that the human ATG16L1 mRNA shows extensive association with HuR (see Table S1 in the supplemental material) (http://starbase.sysu.edu.cn/). To determine whether HuR enhances ATG16L1 translation by directly interacting with the Atg16l1 mRNA, ribonucleoprotein (RNP) immunoprecipitation (RIP) assays were performed in IECs using anti-HuR antibody under conditions that preserved RNP integrity (36). Atg16l1 mRNA was highly enriched in HuR samples compared with control IgG samples, while HuR did not preferentially associate with Atg5 mRNA (Fig. 4A). The levels of Cdc42 mRNA (a known target of HuR) (28) in HuR RIP were also examined and served as a positive control, while Gapdh mRNA, found in all samples as a low-level contaminating transcript (not a target of HuR), served to monitor the evenness of sample input, as reported previously (21).
FIG 4.
HuR regulates ATG16L1 translation by directly interacting with the 3′-UTR of Atg16l1 mRNA. (A) Association of endogenous HuR with endogenous Atg16l1 mRNA in IECs as examined by RIP using either anti-HuR antibody (Ab) or control IgG followed by Q-PCR analysis. (a) Levels of the Atg16l1 and Atg5 mRNAs in HuR IP; (b) levels of total input mRNAs. Values are the means ± SEM (n = 3). *, P < 0.05 compared with IgG IP. (B) HuR immunoblots using the pulldown materials for biotinylated transcripts of the Atg16l1 5′-UTR, CR, and 3′-UTR. Left, schematic representation of various fractions of biotinylated Atg16l1 transcripts used in this study. Right, cytoplasmic lysates were incubated with 6 μg of biotinylated Atg16l1 5′-UTR, CR, or 3′-UTR for 30 min at 25°C, and the resulting RNP complexes were pulled down by streptavidin-coated beads. The presence of HuR and other RBPs (CUGBP1 and AUF1) in the pulldown material was assayed by Western blotting. GAPDH in the pulldown material was also detected and served as a negative control. (C) Left, schematic of plasmids of different chimeric firefly Luc-Atg16l1 reporters. Right, levels of luciferase (Luc) reporter activities as measured by analysis of the Luc-Atg16l1 5′-UTR (Luc-5′UTR), CR (Luc-CR), and 3′-UTR (Luc-3′UTR) after HuR silencing. *, P < 0.05 compared with C-siRNA.
To determine if HuR directly interacts with the 5′-UTR, CR, or 3′-UTR of Atg16l1 mRNA, HuR/Atg16l1 mRNA complexes were further tested by using biotinylated transcripts spanning different regions of Atg16l1 mRNA (Fig. 4B, schematic). As shown, HuR bound only to the Atg16l1 3′-UTR and not to the 5′-UTR or CR. Neither CUGBP1 nor AUF1 interacted with Atg16l1 mRNA. To investigate the functional consequence of HuR interactions with Atg16l1 mRNA in IECs, we used a firefly luciferase (FL) reporter gene construct containing the Atg16l1 5′-UTR, CR, or 3′-UTR and negative-control vector pGL3-Luc (Fig. 4C, schematic). To distinguish translational output from changes in mRNA turnover, luciferase activities were normalized to luciferase mRNA levels to calculate the translational efficiency (the “translation index”). HuR silencing for 48 h decreased luciferase reporter activity when cells were transfected with the Luc-3′-UTR (containing the full-length Atg16l1 3′-UTR) but not with the Luc-5′-UTR or Luc-CR. These results indicate that HuR enhances ATG16L1 translation by directly interacting with Atg16l1 mRNA via its 3′-UTR rather than its 5′-UTR or CR.
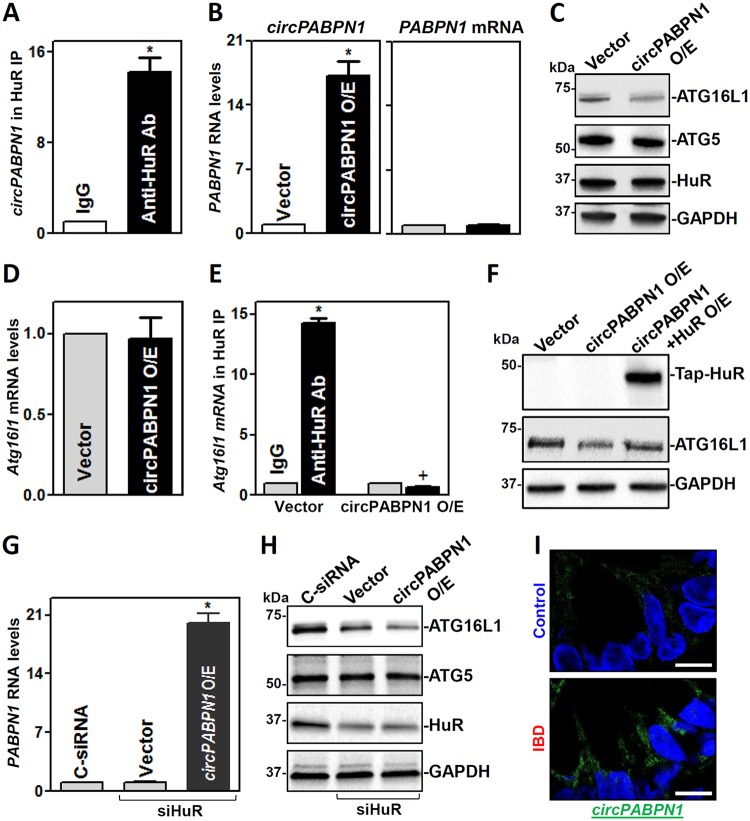
circPABPN1 suppresses HuR binding to Atg16l1 mRNA and lowers ATG16L1 levels.
circPABPN1 is a prominent HuR target circRNA, and elevation of circPABPN1 levels prevented HuR binding to its target transcript PABPN1 mRNA in human cervical carcinoma HeLa cells (37). To test the possibility that sequestration of HuR by circPABPN1 regulates ATG16L1 expression levels, we first examined the association of HuR with circPABPN1 in IECs. Consistent with the findings in HeLa cells, RIP analysis verified that HuR associated with circPABPN1 in Caco-2 cells (Fig. 5A). Second, we determined whether increasing the levels of circPABPN1 inhibited the expression of ATG16L1. Transient transfection with a vector to express circPABPN1 (pCircPABPN1) dramatically increased the levels of cellular circPABPN1 (Fig. 5B, left), but it did not alter the levels of PABPN1 mRNA (Fig. 5B, right). Increasing the levels of circPABPN1 by transfection with pCircPABPN1 specifically inhibited the expression of ATG16L1 without affecting the expression levels of ATG5 or HuR (Fig. 5C); moreover, ectopically expressed circPABPN1 failed to alter total Atg16l1 mRNA levels (Fig. 5D). Third, we examined the role of the circPABPN1-HuR complex in the regulation of ATG16L1 expression. As shown in Fig. 5E, the ectopic rise in circPABPN1 levels abolished the binding of HuR to Atg16l1 mRNA. There were no differences in the levels of Atg16l1 mRNA in pulldown materials between anti-HuR antibody and IgG when cells were transfected with pCircPABPN1.
FIG 5.
circPABPN1 inhibits ATG16L1 expression by preventing HuR association with Atg16l1 mRNA. (A) Association of endogenous HuR with endogenous circPABPN1 in IECs as examined by RIP assays using anti-HuR antibody (Ab). Values are the means ± SEM (n = 3). *, P < 0.05 compared with IgG IP. (B) Levels of circPABPN1 (left) and PABPN1 mRNA (right) at 48 h after transfection with the circPABPN1 expression vector (pCircPABPN1). *, P < 0.05 compared with control vector. (C) Immunoblots of ATGs and HuR in cells treated as described for panel B. (D) Levels of Atg16l1 mRNA in cells treated as described for panel B. (E) Effect of increasing circPABPN1 levels on HuR association with Atg16l1 mRNA. Cells were transfected with pCircPABPN1 or control vector, and the levels of Atg16l1 mRNA in the material pulled down by anti-HuR Ab were examined 48 h thereafter. *, P < 0.05 compared with IgG; +, P < 0.05 compared with anti-HuR Ab in cells transfected with vector. (F) Immunoblots of TAP-HuR and ATG16L1 in cells transfected with pCircPABPN1 alone or cotransfected with pCircPABPN1 and HuR expression vectors; Western blot analysis was carried out 48 h after transfection. (G) Levels of circPABPN1 in cells transfected with siHuR alone or cotransfected with siHuR and pCircPABPN1. *, P < 0.05 compared with vector (n = 3). (H) Immunoblots of ATGs and HuR in cells treated as described for panel G. Three separate experiments were performed and showed similar results. (I) In situ hybridization of circPABPN1 with fluorescent LNA-RNA detection probe in the human intestinal mucosa. Green, circPABPN1; blue, nuclei stained by DAPI. Scale bars, 25 μm. Experiments were repeated in colonic tissue samples obtained from 3 patients with ulcerative colitis or controls and showed similar results.
In accordance with these effects, ectopically expressed HuR partially rescued ATG16L1 expression in cells overexpressing circPABPN1 (Fig. 5F), whereas HuR silencing and circPABPN1 overexpression synergistically inhibited ATG16L1 expression (Fig. 5G and H). The levels of ATG16L1 in cells cotransfected with pCircPABPN1 and siHuR were lower than those observed in cells transfected with siHuR alone. Interestingly, human mucosal tissues obtained from patients with UC exhibited a significant increase in the levels of circPABPN1 compared with those observed in controls (Fig. 5I), as measured by RNA-fluorescent in situ hybridization (FISH) analysis (3). The increased levels of mucosal circPABPN1 in UC patients were associated with a decrease in ATG16L1, as shown in Fig. 2B. Taken together, these findings indicate that HuR and circPABPN1 regulate ATG16L1 expression antagonistically and that increasing circPABPN1 levels reduces ATG16L1 abundance predominantly by suppressing HuR binding to Atg16l1 mRNA.
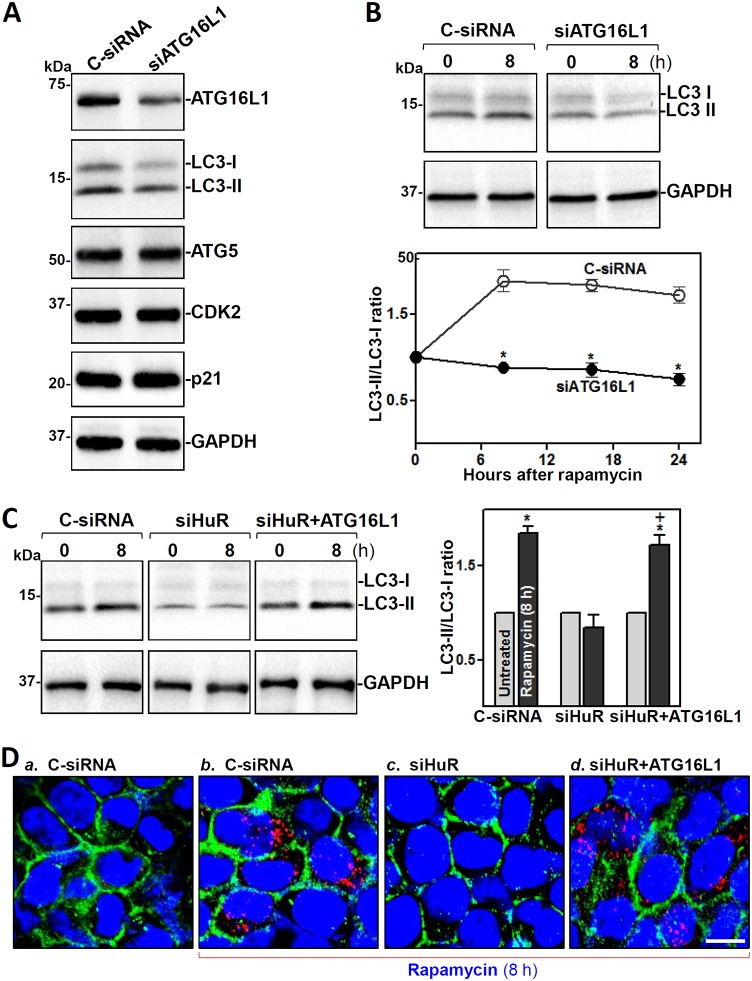
HuR-regulated ATG16L1 plays an essential role in autophagy activation in cultured cells.
Analysis of the role of HuR-mediated ATG16L1 production in the defense of the intestinal epithelium revealed that inhibition of ATG16L1 by HuR silencing prevented the autophagy activation induced by the pharmacologic inducer rapamycin in cultured IECs. Decreasing ATG16L1 levels by transfection with a specific siRNA targeting ATG16L1 (siATG16L1) lowered the basal levels of LC3-I and LC3-II, but it did not alter the levels of ATG5 or proliferation-associated proteins such as CDK2 and p21 (Fig. 6A). ATG16L1 silencing also inhibited autophagy activation when cells were exposed to rapamycin (Fig. 6B). As shown, treatment with rapamycin activated autophagy in control cells, as indicated by increased levels of LC3-II, but this activation was prevented in ATG16L1-silenced cells. As expected, HuR silencing by transfection with siHuR also decreased basal levels of LC3-I and LC3-II and abolished rapamycin-induced autophagy activation (Fig. 6C, left). Importantly, ectopically expressed ATG16L1 in HuR-silenced cells restored the activation of autophagy in response to rapamycin (Fig. 6C, right). The levels of rapamycin-induced LC3-II in HuR-silenced cells transfected with ATG16L1 expression vector increased significantly compared with those observed in cells transfected with siHuR alone. Immunohistochemical staining assays further revealed that the cytoplasmic fluorescence intensity of LC3-II was relatively low before rapamycin treatment in control cells but increased remarkably when cells were exposed to rapamycin (Fig. 6D). In HuR-silenced cells, there were no detectable LC3-II immunoreactive signals regardless of rapamycin treatment. However, when HuR-silenced cells were transfected with ATG16L1 expression vector, the rapamycin-induced LC3-II signals returned almost to the levels of control cells. On the other hand, neither ATG16L1 overexpression nor treatment with rapamycin affected cell viability, as measured by trypan blue staining (data not shown). These data indicate that the reduction in ATG16L1 expression following HuR inhibition disrupts autophagy, thus compromising the intestinal epithelial defense.
FIG 6.
ATG16L1 silencing prevents autophagy activation in vitro. (A) Immunoblots of ATGs and proliferation-related proteins CDK2 and p21 in IECs transfected with siATG16L1 or C-siRNA. Western blot analysis was carrierd out 48 h after the transfection. (B) Effect of ATG16L1 silencing on autophagy activation induced by rapamycin. Forty-eight hours after transfection with siATG16L1 or C-siRNA, cells were exposed to rapamycin (50 ng/ml). Whole-cell lysates were harvested at different times after exposure to rapamycin. Top, representative LC3 immunoblots 8 h after adding rapamycin; bottom, LC3 activity as quantified by examining the ratio of LC3-II and LC3-I. Values are the means ± SEM (n = 3). *, P < 0.05 compared with C-siRNA. (C) Changes in rapamycin-induced LC3 activity in cells transfected with siHuR alone or cotransfected with siHuR and ATG16L1 expression vector. *, P < 0.05 compared with untreated groups; +, P < 0.05 compared with siHuR-treated cells exposed to rapamycin for 8 h. (D) Immunohistochemical staining of LC3-II in cells treated as described for panel C. Red, LC3-II; green, E-cadherin; blue, DAPI. Scale bars, 25 μm. Three experiments were performed and showed similar results.
DISCUSSION
Autophagy enhances host defense of the intestinal epithelium against invading bacteria and other pathogens (5), and the defective autophagy occurs commonly in patients with IBD and other mucosal inflammation-associated diseases (14, 16). However, the exact mechanism that controls autophagy activation at the posttranscriptional level is poorly understood. Using mice bearing an intestinal epithelium-specific deletion of HuR, we found strong genetic evidence supporting the view that HuR regulates autophagy at least in part by controlling ATG16L1 translation. Targeted deletion of HuR in mice specifically decreased the levels of ATG16L1 protein in the intestinal mucosa without affecting tissue mRNA abundance. HuR bound directly to the Atg16l1 mRNA and promoted its translation. In contrast, circPABPN1 inhibited ATG16L1 expression by repressing the association of HuR with Atg16l1 mRNA. These findings advance our understanding of the biological function of HuR in the intestinal epithelium and highlight a key role for HuR loss and ATG16L1 inhibition in defective autophagy in pathologies. Because the levels of both HuR and ATG16L1 decrease in human intestinal mucosae from patients with IBD, our results provide a strong rationale for considering therapeutic strategies directed at the HuR/ATG16L1 pathway to augment the intestinal mucosal defense in clinical settings.
The results reported here indicate that targeted deletion of HuR in mice decreased the expression levels of ATG16L1 in the intestinal epithelium, which is consistent with the findings observed in cultured HK-2 and HCC cells in vitro (30, 31). On the other hand, HuR knockout in mice failed to alter ATG5 levels in the intestinal mucosa, although HuR silencing inhibits ATG5 expression in cultured HCC cells (31). This difference in the levels of ATG5 expression between IE-HuR−/− mice and HuR-silenced HCC cells is not surprising, because observations from mouse gene deletion studies, in some circumstances, are distinct from those in cultured cells (38–40). In support of our present findings, several studies using mice with conditional tissue-specific HuR deletion have demonstrated that HuR is essential for maintaining homeostasis in the intestinal epithelium. Mice lacking intestinal expression of HuR displayed reduced IEC proliferation (15, 19, 27), increased sensitivity of the mucosa to injury and inflammation (18), and delayed repair after acute injury (29). We have recently reported that HuR regulates Paneth cell function in the intestinal epithelium, whereas targeted deletion of HuR in IECs decreases the numbers of Paneth cells (lysozyme-positive cells) and inhibits Paneth cell function, primarily by disrupting subcellular Toll-like receptor 2 (TLR2) localization via posttranscriptional suppression of chaperone protein CNPY3 levels (15). In addition, HuR is required for the nuclear mobilization of the GTP-binding protein RAC1 in the intestinal epithelium, as the levels of cytoplasmic RAC1 in the small intestinal mucosa increase markedly in IE-HuR−/− mice (29).
The results represented here also show that HuR enhances ATG16 translation by directly interacting with the Atg16l1 3′-UTR rather than its 5′-UTR or CR. Through the use of various partial biotinylated Atg16l1 mRNA transcripts, we found that HuR did not associate with the 5′-UTR or CR of the Atg16l1 mRNA, but it interacted with the Atg16l1 3′-UTR, which contains predicted HuR-binding motifs. The association of HuR with the Atg16l1 3′-UTR mediates HuR actions, because the repression of the reporter activity by HuR silencing occurred only when cells were transfected with the Atg16l1-3′-UTR luciferase reporter constructs and not with the Atg16l1-5′-UTR or Atg16l1-CR reporter constructs. These observations are consistent with our previous results (41, 42) and work from other groups (43, 44) showing that HuR commonly interacts with many of its target mRNAs via their 3′-UTRs, in turn affecting their stability and/or translation. In fewer reported instances, HuR binds target mRNAs in the CR and regulates their expression. In this regard, HuR increases the stability and translation of Cnpy3 mRNA by interacting with the Cnpy3 CR (15), and it stabilizes Xiap mRNA via association with both the CR and 3′-UTR of Xiap mRNA (41). Because neither HuR silencing in cultured cells nor HuR gene deletion in vivo changed total Atg16l1 mRNA levels, HuR/Atg16l1 mRNA association only regulates ATG16L1 translation and has no effect on Atg16l1 mRNA stability.
In this study, we also showed that circPABPN1 inhibits HuR binding to Atg16l1 mRNA and thus represses ATG16 L1 translation. Ectopically overexpressed circPABPN1 did not alter the levels of HuR protein or Atg16l1 mRNA in IECs, but it decreased cellular ATG16L1 abundance by preventing HuR association with Atg16l1 mRNA. Because there are no predicted sites of complementarity between circPABPN1 and the Atg16l1 3′-UTR, we did not examine the association of circPABPN1 with the Atg16l1 mRNA in this study. circRNAs are a vast and diverse class of endogenous RNAs that are often expressed in a tissue- and developmental stage-specific manner (45). Unlike linear RNAs, circRNAs are covalently closed loop structures without 5′ or 3′ ends. circRNAs can function as decoys or sponges that reduce the number of freely available miRNAs (46, 47) and interact with RBPs to jointly regulate gene expression (48, 49). HuR was found to interact extensively with many circRNAs, particularly showing the strongest binding to circPABPN1. Consistent with our findings observed in IECs, high levels of circPABPN1 also inhibited HuR binding to PABPN1 mRNA and lowered PABPN1 production in HeLa cells (37). Several studies have shown that HuR binding to target mRNAs is modulated by numerous factors at multiple levels, including HuR phosphorylation by different kinases (such as Chk2, protein kinase C [PKC], AMP-activated protein kinase [AMPK], and JAK3) (22, 50), methylation by CARM1 (51), and interaction with ncRNAs and other RBPs (52, 53). Since these posttranslational modifications affect the subcellular localization of HuR and its binding affinity for target RNAs, it will be interesting to further investigate if they may also modulate the levels and function of HuR/circPABPN1 complexes in the intestinal epithelium.
Finally, our results reported here also provide translational evidence and point to a crucial role of disruption of the HuR/ATG16L1 pathway in intestinal mucosal pathologies. We establish for the first time a cause-effect relationship between HuR and posttranscriptional control of ATG16L1 expression in in vivo and in vitro models and show that human intestinal mucosae with injury/erosions and inflammation from patients with IBD exhibit decreased levels of both HuR and ATG16L1. Autophagy is believed to have a generally favorable impact on cell, tissue, and organ homeostasis and participates in the intestinal mucosal defense and barrier function (5, 13). Moreover, Paneth cells secrete lysozyme through secretory autophagy during bacterial infection of the intestine (54), and autophagy is also involved in the control of goblet cell function (55). Deficiency of autophagy proteins and defects in autophagy activation reduce the numbers of Paneth and goblet cells and render them dysfunctional (9, 15, 54). Defective autophagy is commonly observed in mice with destructive mucosal inflammatory erosions (56), and disrupted expression of autophagy genes such as Atg16l1, Atg5, and Atg7 also frequently occurs in patients with IBD, including UC and Crohn’s disease (16). Taken together, the findings of our current study strongly support the notion that HuR regulates autophagy at least in part by modulating ATG16L1 translation through interaction with circPABPN1. These findings suggest that decreased levels of ATG16L1 and subsequent autophagy defects in the HuR-deficient intestinal epithelium are implicated in the pathogenesis of human IBD and other mucosal disorders.
MATERIALS AND METHODS
Animal studies.
All animal experiments were performed in accordance with NIH guidelines and were approved by the Institutional Animal Care and Use Committee of the University of Maryland School of Medicine and Baltimore VA hospital. The HuRfl/fl and villin-Cre mice on the C57BL/6 background were purchased from the Jackson Laboratory, and intestinal epithelial tissue-specific HuR deletion (IE-HuR−/−) mice were generated by crossing the HuRfl/fl mice with villin-Cre mice, as described in our previous studies (27, 28). HuRfl/fl-Cre− mice developed and served as control littermates. Both IE-HuR−/− mice and control littermates were housed and handled in a specific-pathogen-free breeding barrier and cared for by trained technicians and veterinarians. Animals were deprived of food but allowed free access to tap water for 24 h before experiments. Two portions of the small intestine taken 0.5 cm distal to the ligament of Trietz or the middle colon were removed, one for histological examination and the other for extraction of protein and RNA. The mucosa was scraped with a glass slide for various measurements as described previously (23, 34).
Histology and immunohistochemistry.
Human tissue samples were obtained from surplus discarded tissue from the Department of Surgery, University of Maryland Health Science Center, and commercial tissue banks. The study was approved by the University of Maryland Institutional Review Board. Dissected and opened intestines were mounted onto a solid surface and fixed in formalin and paraffin. Sections 5 μm thick were stained with hematoxylin and eosin (H&E) for general histology. The immunofluorescence staining procedure was carried out according to the method described in our previous publications (40, 57). Briefly, the slides were fixed in 3.7% formaldehyde in phosphate-buffered saline, rehydrated, and then incubated with the primary antibody against ATGs, HuR, or E-cadherin in blocking buffer overnight. After incubation with secondary antibody conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, OR), the slides were washed, mounted, and viewed through a Zeiss confocal microscope (model LSM700). Images were processed using Photoshop software (Adobe, San Jose, CA). Slides were examined in a blinded fashion by coding them, and only after examination was complete were they decoded.
Plasmid construction.
The vectors expressing wild-type (WT) HuR-tandem affinity purification (HuR-Tap) fusion proteins were generated as described previously (32). Cloning for circPABPN1 overexpression (pCircPABPN1) was performed as described previously (37). An expression vector containing WT full-length Atg16l1 cDNA under the control of the pCMV promoter was purchased from Origene (Rockville, MD) and used to increase cellular ATG16L1 as described previously (11, 33). The chimeric firefly luciferase reporter construct containing the Atg16l1 mRNA was constructed as described previously (21, 58). The full-length Atg16l1 5′-UTR, CR, and 3′-UTR fragments were subcloned into the pmirGLO dual-luciferase miRNA target expression vector (Promega, Madison, WI) to generate the pmirGLO-Luc-ATG16L1-5′-UTR, pmirGLO-Luc-ATG16L1-CR, and pmirGLO-ATG16L1-3′-UTR reporter constructs. This vector is based on the Promega Dual-Luciferase technology, with luc2 used as the primary reporter to monitor mRNA regulation and Renilla luciferase (hRluc-neo) acting as a control reporter for normalization. Transient transfections were performed using the Lipofectamine reagent following the manufacturer’s recommendations (Invitrogen). Cells were harvested for analysis 48 h after transfection, and the levels of firefly luciferase activity were measured using the Dual-Luciferase assay system. To measure translational changes (translation index), the firefly-to-Renilla luciferase ratio was further normalized with RNA levels. All of the primer sequences for generating these constructs are provided in Table S2 in the supplemental material.
Assays of newly translated protein and polysome analysis.
New synthesis of nascent ATG16L1 protein was detected with the Click-iT protein analysis detection kit (Life Technologies, Grand Island, NY) and conducted following the company’s instructions (57). Briefly, cells were incubated in methionine-free medium and then exposed to l-azidohomoalanine (AHA). After mixing cell lysates with the reaction buffer for 20 min, the biotin-alkyne/azide-modified protein complex was pulled down using paramagnetic streptavidin-conjugated Dynabeads. The pulldown material was resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western immunoblotting using antibodies against ATG16L1 or GAPDH. Polysome analysis was carried out as described previously (32). Briefly, cells at ∼70% confluence were incubated in 0.1 mg/ml cycloheximide and then lifted by scraping in polysome extraction lysis buffer. Nuclei were pelleted, and the resulting supernatant was centrifuged through a 15 to 60% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weight. The eluted fractions were prepared with a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm using a UV-6 detector (ISCO, Louisville, KY). After RNA in each fraction was extracted, the levels of each individual mRNA were quantified by reverse transcription (RT) followed by quantitative real-time PCR (Q-PCR) analysis in each of the fractions.
Biotin pulldown assays and RIP analysis.
The synthesis of biotinylated transcripts and measurement of HuR bound to biotinylated RNA were performed as previously described (21, 32). cDNA from Caco-2 cells was used as a template for PCR amplification of 5′-UTR, CR, and 3′-UTR segments of Atg16l1 mRNA. The 5′ primers contained the T7 RNA polymerase promoter sequence (T7; CCAAGCTTCTAATACGAC-TCACTATAGGGAGA). All sequences of oligonucleotides for synthesizing the full-length Atg16l1 5′-UTR, CR, or 3′-UTR are described in Table S1 in the supplemental material. PCR-amplified products were used as templates to transcribe biotinylated RNAs by using T7 RNA polymerase in the presence of biotin-cytidine 5′-triphosphate as described previously (33). Biotinylated transcripts were incubated with cytoplasmic lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) and analyzed by Western blotting using anti-HuR antibody. To assess the association of endogenous HuR with endogenous Atg16l1 mRNA, immunoprecipitation of RNP complexes (RIP) was performed as described previously (21). Twenty million cells were collected per sample, and lysates were used for RIP for 4 h at room temperature in the presence of excess (30 μg) IP antibody (IgG, anti-HuR). RNA extracted from RIP samples was used in RT reactions, followed by PCR and Q-PCR analysis to detect the levels of Atg16l1 and Gapdh mRNAs.
Q-PCR and immunoblotting analyses.
Total RNA was isolated by using the RNeasy minikit (Qiagen, Valencia, CA) and used in reverse transcription (RT) and PCR amplification reactions as described previously (23). Q-PCR analysis was performed using Step-One-Plus systems with specific primers, probes, and software (Applied Biosystems, Foster City, CA). To measure circRNA levels, total RNA was digested with RNase R to remove all linear RNAs, and the primer pairs that spanned the circularization junction (Table S1) were employed in Q-PCR analysis. To examine protein levels, whole-cell lysates were prepared using 2% sodium dodecyl sulfate, sonicated, and centrifuged. The supernatants were boiled and size fractionated by sodium SDS-PAGE. After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibody, washed, and incubated with secondary antibody before enhanced chemiluminescence (ECL) detection.
Chemicals and cell culture.
Culture medium and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA), and biochemicals were from Sigma (St. Louis, MO). Antibodies recognizing HuR, ATG16L1, ATG5, ATG7, and LC3 were purchased from Thermo Fisher Scientific (Waltham, MA) and Santa Cruz Biotechnology (Santa Cruz, CA). The secondary antibody conjugated to horseradish peroxidase was obtained from Sigma. The siRNAs targeting ATG16L1 and HuR were made by Santa Cruz Biotechnology. Caco-2 and IEC-6 cells were purchased from the American Type Culture Collection (Manassas, VA) and were maintained under standard culture conditions (24, 52).
Statistical analysis.
All values were expressed as the means ± standard errors of the means (SEM) from five animals or three separate experiments. The unpaired, two-tailed Student t test was used when indicated, with a P value of <0.05 considered statistically significant. When assessing multiple groups, one-way analysis of variance (ANOVA) was utilized with Tukey’s post hoc test (59). The statistical software used was Instat Prism 5 (GraphPad, San Diego, CA).
Supplementary Material
ACKNOWLEDGMENTS
Xiao-Xue Li and Lan Xiao performed most experiments and summarized data. Hee Kyoung Chung, Xiang-Xue Ma, Xiangzheng Liu, Jia-Le Song, and Cindy Z. Jin participated in in vivo experiments and RNA-binding assays. Jaladanki N. Rao and Myriam Gorospe participated in experiments using human tissues and data analysis. Jian-Ying Wang designed experiments, analyzed data, prepared figures, and drafted the manuscript.
This work was supported by Merit Review Awards (to J.-Y.W. and J.N.R.) from the U.S. Department of Veterans Affairs, by grants from National Institutes of Health (NIH) (DK57819, DK61972, and DK68491 to J.-Y.W.), and by funding from the National Institute on Aging Intramural Research Program, NIH (to M.G.).
Jian-Ying Wang is a senior research career scientist at the Biomedical Laboratory Research and Development Service, U.S. Department of Veterans Affairs. The remaining authors disclose no conflicts.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Bankaitis ED, Ha A, Kuo CJ, Magness ST. 2018. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology 155:1348–1361. doi: 10.1053/j.gastro.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krndija D, El Marjou F, Guirao B, Richon S, Leroy O, Bellaiche Y, Hannezo E, Matic Vignjevic D. 2019. Active cell migration is critical for steady-state epithelial turnover in the gut. Science 365:705–710. doi: 10.1126/science.aau3429. [DOI] [PubMed] [Google Scholar]
- 3.Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, Gorospe M, Wang JY. 2018. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology 154:599–611. doi: 10.1053/j.gastro.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin JL, Sumpter R Jr, Levine B, Hooper LV. 2013. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe 13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iida T, Yokoyama Y, Wagatsuma K, Hirayama D, Nakase H, Iida T, Yokoyama Y, Wagatsuma K, Hirayama D, Nakase H. 2018. Impact of autophagy of innate immune cells on inflammatory bowel disease. Cells 8:7. doi: 10.3390/cells8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boya P, Reggiori F, Codogno P. 2013. Emerging regulation and functions of autophagy. Nat Cell Biol 15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. 2016. Mammalian autophagy: how does it work? Annu Rev Biochem 85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 9.Gukovskaya AS, Gukovsky I, Algul H, Habtezion A. 2017. Autophagy, inflammation, and immune dysfunction in the pathogenesis of pancreatitis. Gastroenterology 153:1212–1226. doi: 10.1053/j.gastro.2017.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slowicka K, Serramito-Gómez I, Boada-Romero E, Martens A, Sze M, Petta I, Vikkula HK, De Rycke R, Parthoens E, Lippens S, Savvides SN, Wullaert A, Vereecke L, Pimentel-Muiños FX, van Loo G. 2019. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat Commun 10:1834. doi: 10.1038/s41467-019-09667-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aden K, Tran F, Ito G, Sheibani-Tezerji R, Lipinski S, Kuiper JW, Tschurtschenthaler M, Saveljeva S, Bhattacharyya J, Hasler R, Bartsch K, Luzius A, Jentzsch M, Falk-Paulsen M, Stengel ST, Welz L, Schwarzer R, Rabe B, Barchet W, Krautwald S, Hartmann G, Pasparakis M, Blumberg RS, Schreiber S, Kaser A, Rosenstiel P. 2018. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med 215:2868–2886. doi: 10.1084/jem.20171029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, Beal A, Harris PA, Bertin J, Liu C, Ding Y, van den Brink MRM, Cadwell K. 2017. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 214:3687–3705. doi: 10.1084/jem.20170558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pott J, Kabat AM, Maloy KJ. 2018. Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe 23:191–202. doi: 10.1016/j.chom.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 14.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW IV. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao L, Li XX, Chung HK, Kalakonda S, Cai JZ, Cao S, Chen N, Liu Y, Rao JN, Wang HY, Gorospe M, Wang JY. 2019. RNA-binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3. Gastroenterology 157:731–743. doi: 10.1053/j.gastro.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khor B, Gardet A, Xavier RJ. 2011. Genetics and pathogenesis of inflammatory bowel disease. Nature 474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao L, Wang JY. 2014. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol 19:46–53. doi: 10.1016/j.coph.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giammanco A, Blanc V, Montenegro G, Klos C, Xie Y, Kennedy S, Luo J, Chang SH, Hla T, Nalbantoglu I, Dharmarajan S, Davidson NO. 2014. Intestinal epithelial HuR modulates distinct pathways of proliferation and apoptosis and attenuates small intestinal and colonic tumor development. Cancer Res 74:5322–5335. doi: 10.1158/0008-5472.CAN-14-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garneau NL, Wilusz J, Wilusz CJ. 2007. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 20.Yu TX, Wang PY, Rao JN, Zou T, Liu L, Xiao L, Gorospe M, Wang JY. 2011. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res 39:8472–8487. doi: 10.1093/nar/gkr567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdelmohsen K, Pullmann R Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. 2007. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell 25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L, Rao JN, Cao S, Liu L, Chung HK, Zhang Y, Zhang J, Liu Y, Gorospe M, Wang JY. 2016. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell 27:617–626. doi: 10.1091/mbc.E15-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, Xu Y, Gorospe M, Wang JY. 2016. H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol 36:1332–1341. doi: 10.1128/MCB.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. 2013. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res 41:7905–7919. doi: 10.1093/nar/gkt565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mubaid S, Ma JF, Omer A, Ashour K, Lian XJ, Sanchez BJ, Robinson S, Cammas A, Dormoy-Raclet V, Di Marco S, Chittur SV, Tenenbaum SA, Gallouzi IE. 2019. HuR counteracts miR-330 to promote STAT3 translation during inflammation-induced muscle wasting. Proc Natl Acad Sci U S A 116:17261–17270. doi: 10.1073/pnas.1905172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Zhuang R, Xiao L, Chung HK, Luo J, Turner DJ, Rao JN, Gorospe M, Wang JY. 2017. HuR enhances early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol 37:e00574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Christodoulou-Vafeiadou E, Rao JN, Zou T, Xiao L, Chung HK, Yang H, Gorospe M, Kontoyiannis D, Wang JY. 2014. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell 25:3308–3318. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Xiao L, Chung HK, Kwon MS, Li XX, Wu N, Rao JN, Wang JY. 2019. RNA-binding protein HuR regulates Rac1 nucleocytoplasmic shuttling through nucleophosmin in the intestinal epithelium. Cell Mol Gastroenterol Hepatol 8:475–486. doi: 10.1016/j.jcmgh.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palanisamy K, Tsai TH, Yu TM, Sun KT, Yu SH, Lin FY, Wang IK, Li CY. 2019. RNA-binding protein, human antigen R regulates hypoxia-induced autophagy by targeting ATG7/ATG16L1 expressions and autophagosome formation. J Cell Physiol 234:7448–7458. doi: 10.1002/jcp.27502. [DOI] [PubMed] [Google Scholar]
- 30.Ji E, Kim C, Kang H, Ahn S, Jung M, Hong Y, Tak H, Lee S, Kim W, Lee EK. 2019. RNA binding protein HuR promotes autophagosome formation by regulating expression of autophagy-related proteins 5, 12, and 16 in human hepatocellular carcinoma cells. Mol Cell Biol 39:e00508-18. doi: 10.1128/MCB.00508-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T. 2009. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest 119:3530–3543. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. 2009. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell 20:4885–4898. doi: 10.1091/mbc.e09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zou T, Rao JN, Liu L, Xiao L, Yu TX, Jiang P, Gorospe M, Wang JY. 2010. Polyamines regulate the stability of JunD mRNA by modulating the competitive binding of its 3′ untranslated region to HuR and AUF1. Mol Cell Biol 30:5021–5032. doi: 10.1128/MCB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JY, Johnson LR. 1990. Luminal polyamines stimulate repair of gastric mucosal stress ulcers. Am J Physiol 259:G584–G592. doi: 10.1152/ajpgi.1990.259.4.G584. [DOI] [PubMed] [Google Scholar]
- 35.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. 2006. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem 281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang Y, Xiao L, Yu TX, Li JZ, Rao JN, Turner DJ, Gorospe M, Wang JY. 2017. Cooperative repression of insulin-like growth factor type 2 receptor translation by microRNA 195 and RNA-binding protein CUGBP1. Mol Cell Biol 37:e00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M. 2017. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL. 2009. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol 182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 39.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. 2009. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung HK, Wang SR, Xiao L, Rathor N, Turner DJ, Yang P, Gorospe M, Rao JN, Wang J-Y, Chung HK, Wang SR, Xiao L, Rathor N, Turner DJ, Yang P, Gorospe M, Rao JN, Wang J-Y. 2018. α4 coordinates small intestinal epithelium homeostasis by regulating stability of HuR. Mol Cell Biol 38:e0631-17. doi: 10.1128/MCB.00631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Zou T, Rao JN, Liu L, Xiao L, Wang PY, Cui YH, Gorospe M, Wang JY. 2009. Stabilization of XIAP mRNA through the RNA binding protein HuR regulated by cellular polyamines. Nucleic Acids Res 37:7623–7637. doi: 10.1093/nar/gkp755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. 2007. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol Biol Cell 18:4579–4590. doi: 10.1091/mbc.e07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Wang H, Cheng X, Fan X, Yang F, Zhang M, Chen Y, Tian Y, Liu C, Shao D, Jiang B, Dou Y, Cong Y, Xing J, Zhang X, Yi X, Songyang Z, Ma W, Zhao Y, Wang X, Ma J, Gorospe M, Ju Z, Wang W. 2018. HuR regulates telomerase activity through TERC methylation. Nat Commun 9:2213. doi: 10.1038/s41467-018-04617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Yang L, Chen LL. 2018. The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 46.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 47.Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N, Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kühn R, Rosenmund C, Birchmeier C, Rajewsky N. 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M, Wang X, Huang K, Zheng L, Tong Q. 2019. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ 26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, Liu H, Yang F. 2019. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 45:155–167. doi: 10.1016/j.ebiom.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. 2008. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem J 409:389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grammatikakis I, Abdelmohsen K, Gorospe M, Grammatikakis I, Abdelmohsen K, Gorospe M. 2017. Posttranslational control of HuR function. Wires RNA 8:e1372. doi: 10.1002/wrna.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, Wu J, Donahue JM, Gorospe M, Wang JY. 2015. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell 26:1797–1810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. 2013. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell 24:85–99. doi: 10.1091/mbc.E12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, Hooper LV. 2017. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science 357:1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, Dinauer MC, Virgin HW, Stappenbeck TS. 2013. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 32:3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, Wang C, Ziel JW, van Rooijen N, Nunez G, Finlay BB, Mysorekar IU, Cadwell K. 2013. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe 14:216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao L, Rao JN, Zou T, Liu L, Cao S, Martindale JL, Su W, Chung HK, Gorospe M, Wang JY. 2013. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell 24:3038–3046. doi: 10.1091/mbc.E13-05-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN, Gorospe M, Wang JY. 2018. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol 38:e00010-18. doi: 10.1128/MCB.00010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harter HL. 1960. Critical values for Duncan’s new multiple range test. Biometrics 16:671–685. doi: 10.2307/2527770. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.