Recent advancements in genetic and proteomic technologies have revealed that more of the genome encodes proteins than originally thought possible. Specifically, some putative long noncoding RNAs (lncRNAs) have been misannotated as noncoding. Numerous lncRNAs have been found to contain short open reading frames (sORFs) which have been overlooked because of their small size. Many of these sORFs encode small proteins or micropeptides with fundamental biological importance. These micropeptides can aid in diverse processes, including cell division, transcription regulation, and cell signaling.
KEYWORDS: lncRNA, mRNA, circRNA, coding potential, micropeptides
ABSTRACT
Recent advancements in genetic and proteomic technologies have revealed that more of the genome encodes proteins than originally thought possible. Specifically, some putative long noncoding RNAs (lncRNAs) have been misannotated as noncoding. Numerous lncRNAs have been found to contain short open reading frames (sORFs) which have been overlooked because of their small size. Many of these sORFs encode small proteins or micropeptides with fundamental biological importance. These micropeptides can aid in diverse processes, including cell division, transcription regulation, and cell signaling. Here we discuss strategies for establishing the coding potential of putative lncRNAs and describe various functions of known micropeptides.
INTRODUCTION
The human genome harbors protein-coding and noncoding regions, with less than 2% annotated as protein coding (1). New advancements in genetic and proteomic technologies have allowed for the genome, its transcripts, and corollary proteins to be studied more extensively. Recent findings now suggest that the division between coding and noncoding transcripts may not be so clearly defined (2–4). Typically, translation of a protein begins with the start codon within an mRNA’s open reading frame (ORF). Traditionally, an ORF contains codons for at least 100 amino acids in eukaryotes or 50 amino acids in bacteria (5) and ends with a stop codon that causes the ribosome to disassemble and terminate protein synthesis (6). However, these arbitrary criteria of what makes a transcript protein coding have led to the misannotation of many putative noncoding RNAs (ncRNAs) that contain ORFs smaller than the traditional cutoff; there is growing evidence that some putative ncRNAs encode small proteins, or micropeptides (7–29).
One subclass of putative ncRNAs with an increasing number of transcripts misannotated as noncoding are long noncoding RNAs (lncRNAs). lncRNAs are longer than ∼200 nucleotides (nt) (30), and like mRNAs, they are transcribed by RNA polymerase II (RNAPII) and often undergo 5′ capping, polyadenylation, and splicing (31, 32). The subcellular localization of an lncRNA determines its function. Generally, lncRNAs are localized in the nucleus and/or cytoplasm (33). lncRNAs retained in the nucleus can, directly or indirectly, control transcription (e.g., chromatin remodeling or functioning as enhancer RNA), regulate pre-mRNA splicing, and act as scaffolds for the formation of protein complexes and subnuclear domains (34). Some lncRNA transcripts which localize to the cytoplasm can be potentially translated into micropeptides (35). Many of these lncRNA-encoded micropeptides have been shown to perform vital biological functions within organisms ranging from bacteria to flies to humans. In this review, we describe the various techniques and strategies used to study the coding potential of lncRNAs, the challenges to these approaches, and examples of putative lncRNAs that code for endogenous micropeptides.
TECHNIQUES FOR STUDYING TRANSLATION OF PUTATIVE lncRNAs
In this section, we discuss techniques that are used to determine if a putative lncRNA can be translated. It should be noted that evidence supporting translation of a putative lncRNA does not necessarily mean that the expressed micropeptide has a function.
Ribosome profiling.
Recent experimental approaches have been designed to identify small ORFs (sORFs) that have the potential to be translated. One such approach is ribosome profiling, a technique that relies on the fundamental principle that actively translating ribosomes can protect ∼30-nt-long segments of RNA from nuclease digestion (4). These ribosome-protected fragments, or “ribosome footprints,” can be used to study translational activity as well as changes in translation in response to environmental stress (4). Using this technology, many putative lncRNAs have been shown to be potentially translated (36). Additionally, this technique has shown that translation, especially in upstream ORFs, can be initiated at alternative initiation start sites (36). Lee et al. improved the ribosome profiling technique to distinguish between ribosome initiation and elongation by using global translation initiation sequencing (GTI-Seq) and also found alternative translation initiation start sites (2). However, even though an lncRNA may associate with ribosomes, it does not necessarily mean that the transcript is being actively translated into a protein (6). For example, the H19 mouse transcript is associated with polysomes, yet it is a bona fide ncRNA that regulates insulin-like growth factor 2 mRNAs (36, 37). Therefore, ribosome occupancy cannot be the only tool used to determine if an lncRNA is protein coding.
To combat this, more recent tools like RibORF, a support vector machine classifier, have been trained to distinguish in-frame ORFs from overlapping ORFs as well as RNA that is not associated with ribosomes (35). Additional metrics, like the ribosome release score (RRS), have also been designed to differentiate between coding and noncoding transcripts. This metric is based on the fundamental principle that protein-coding transcripts will be released from the ribosome when a stop codon is reached. This process should not occur for noncoding transcripts, so they are not detected by this metric (6). Using this technique, Guttman et al. argued that large lncRNAs are not protein coding (6). However, more recent methods that use the entire 3′ untranslated region (UTR) for both coding and noncoding transcripts are more accurate in determining the RRS for coding transcripts (6, 38). Using the new technique, Popa et al. found that over one-third of lncRNAs in murine embryonic stem (ES) cells could be translated (38).
To further improve upon prior methods of sORF detection, Aspden et al. developed Poly-Ribo-Seq (39). This technique is based on the ability of multiple ribosomes to bind to the same RNA transcript during translation and form polysomes. Poly-Ribo-Seq enriches for small polysomes, which are more likely to form during sORF translation than the translation of longer, canonical mRNAs (39). Using Poly-Ribo-Seq, Aspden et al. identified two classes of sORFs (39). The first category includes sORFs which code for functional micropeptides at least 80 amino acids in length, are translated as frequently as transcripts from larger ORFs, and are well conserved between species. The second group encompasses sORFs which are much smaller and code for micropeptides around 20 amino acids long. This group neither is translated into micropeptides as frequently as larger sORFs nor is well conserved between species. Consequently, studying the translational abilities of lncRNAs can be challenging; however, proteomics-based technology has helped overcome some of the obstacles in predicting if an lncRNA encodes a micropeptide.
Mass spectrometry, proteomics, and proteogenomics.
Mass spectrometry (MS)-based proteomics is the gold standard for protein detection. This technique measures the mass-to-charge ratio of ionized peptides or proteins in a gaseous state, thus allowing for the study of protein expression and interactions (40). More recently, MS has been used to validate the presence of micropeptides encoded by putative lncRNAs, thus providing strong evidence as to whether a sORF codes for a micropeptide.
To provide a more robust approach to the study of micropeptides, MS proteomics is frequently used in tandem with genomic analysis such as transcriptome sequencing (RNA-Seq). Proteogenomic approaches help identify uncharacterized novel micropeptides. Bánfai et al. used tandem mass spectrometry (MS/MS) and RNA-Seq to determine which lncRNAs in ENCODE are translated into micropeptides (41). They compared MS/MS data with poly(A)+ and poly(A)− RNA-Seq data from ENCODE for the human cell lines K562 and GM12878 to measure transcript abundance for the genes in GENCODE v7 (41). A random forest model, RuleFit3, was used to compare RNA expression with translated peptides to predict translation (41). This machine-learning technique rarely predicted an lncRNA as protein coding (41). However, the smallest ORF in their data set corresponded to 23 amino acids, and this minimum length may have influenced their results (41). Ji et al. showed that only ∼40% of lncRNA-encoded micropeptides are longer than 10 amino acids (35). Consequently, many micropeptides were likely overlooked by Bánfai et al. (35, 41). In a proteogenomics study, Slavoff et al. were able to discover previously unidentified micropeptides encoded by sORFs (24). They created a custom database that included all potential polypeptides greater than 8 amino acids from the human genome (RefSeq), the Sequest database (which is a database of peptides with MS/MS spectra), and liquid chromatography and tandem mass spectroscopy (LC-MS/MS) (24). Using this approach, they discovered 86 previously uncharacterized micropeptides in K562 cells (24). Overall, this technique has been effective at discovering novel micropeptides through the combination of proteomics and genomics.
Typical MS analysis is performed using a reference database of peptide sequences. This approach limits the ability of the technique to predict novel peptides. To combat this, Karunratanakul et al. developed SMSNet, a de novo peptide sequencing method that utilizes deep learning algorithms to predict a peptide sequence from an MS spectrum (42). Using this SMSNet framework, they were able to identify over 10,000 uncharacterized human leukocyte antigens and 4,000 novel phosphopeptides (42). This de novo approach has the potential to be used in the discovery of novel micropeptides that are overlooked by reference databases.
Despite these advancements, there are some weaknesses in MS-based proteomics. For example, extraordinarily small micropeptides are nearly impossible to detect by MS (43), likely because small peptides can be lost in the sample preparation process. Additionally, the digestion protease used during sample preparation largely determines how a micropeptide will be fragmented (44). If the fragments after digestion of the micropeptide are too small, they may not produce a large enough signal (44), making it difficult to distinguish noise from small peptides (43). Conversely, if the fragments after digestion of the micropeptide are larger than a few kilodaltons, then they likely cannot be analyzed. Additionally, when micropeptide concentrations are low, competition between other peptides can make it impossible for MS spectra to be produced for some small peptides (43).
Overall, MS is an extremely powerful tool that allows for the discovery and verification of an endogenously expressed micropeptide. The evidence of a micropeptide on the spectra strongly supports the presence of the micropeptide. However, if a micropeptide does not appear in the MS spectra, it is not definitive that the micropeptide is not present in the cell. Further analysis that combines proteogenomics and in vitro assay techniques is required to analyze the presence of the micropeptide.
Validation of sORF translation.
A common way to determine if a sORF is translated into a micropeptide is by in vitro translation. Using this technique, the double-stranded cDNA encoding the micropeptide is inserted into a vector which includes a phage polymerase promoter (44). The construct is then expressed in cell extracts with the [35S]methionine radioisotope, which allows for the peptide to be visualized via gel electrophoresis and autoradiography (44). Although this technique provides evidence that a sORF can be translated into a micropeptide in vitro, additional experiments are required to establish the expression of the micropeptide in a given cell.
To better understand if an endogenous micropeptide is expressed in a given cell, an antibody against the peptide of interest can be generated. The antibody can be a very effective tool for identifying the presence of a micropeptide because it allows for the study of its natural contexts. Once the antibody is generated, it is important to verify that it is specific to the desired micropeptide. To do this, the gene encoding the micropeptide can be silenced using small interfering RNAs (siRNAs), and Western blotting can be performed to make sure that the antibody is specific to the micropeptide. Overexpression of the micropeptide in cells using an expression vector can be used as a positive control in these experiments. However, in some cases, this technique can be difficult for micropeptides because epitope design may be challenging for short peptides. As many micropeptides are localized to membranes, this further restricts potential epitope sites (45). Because some micropeptides are produced at low levels, it can be difficult for antibodies to interact with enough micropeptides to allow for detection (45). Therefore, the inability to detect the endogenous micropeptide using an antibody does not necessarily mean that the micropeptide is not expressed.
In the case that generating a specific antibody against the micropeptide proves difficult, another way to detect micropeptide levels within cells is epitope tagging. An epitope tag can be added to the micropeptide directly by inserting the gene of interest into a tagged expression vector. The tagged gene can then be expressed in a stable cell line via a lentiviral expression system. Usually, the tag is added to either the C or N terminus of the micropeptide of interest. A more effective way to determine if a micropeptide is translated in vivo is to use the CRISPR/Cas9 technology. This gene-editing approach allows an epitope tag to be inserted into the locus of the micropeptide via homology-directed repair. Although the efficiency of CRISPR/Cas9 is highly dependent on the cell line used, this strategy can be an effective way to determine the localization and endogenous expression of a micropeptide within a cell (61).
These techniques should be performed with caution because the addition of a tag to the N terminus could disrupt a localization signal. However, the approach is sometimes beneficial because it can increase protein solubility and proper folding (44, 47). Both constructs should be tested to determine if the tag disrupts the localization and function of the micropeptide. Because micropeptides are small and many have transmembrane domains, adding an epitope tag of equal or greater size has the potential to disrupt the charge, folding, and protein interactions of the micropeptide (45). Therefore, appropriate controls should be performed, and experimental design constraints should be considered to minimize unwanted effects.
It is important to note that the prediction algorithms for mRNA translation and methods of protein detection provide evidence in support of sORF translation; however, every translational event does not necessarily produce a functional protein. Further experimentation needs to be performed to determine if a micropeptide is functional.
POSTTRANSCRIPTIONAL REGULATION AND FUNCTIONS OF MICROPEPTIDES
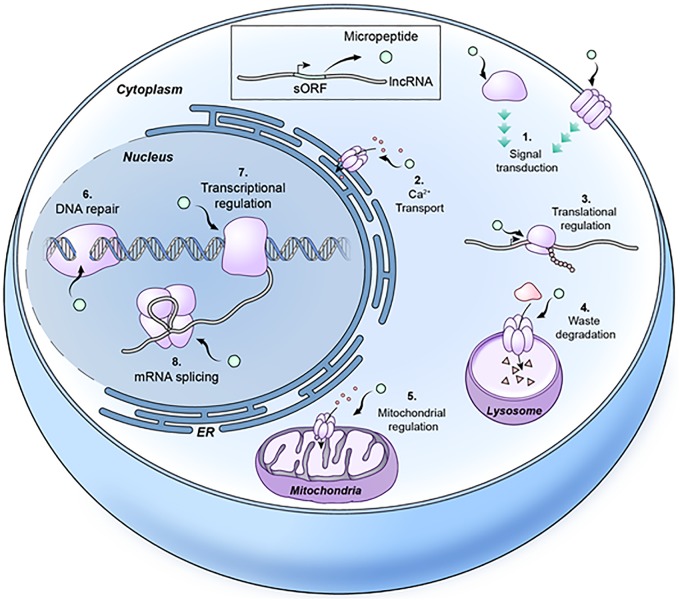
Some micropeptides encoded by putative lncRNAs are conserved between numerous species ranging from prokaryotic bacteria to eukaryotes like Drosophila, mice, and humans. However, micropeptides can be cell type or tissue specific and help cells and tissues perform specialized functions. They can also aid in the regulation of diverse cellular processes, including, but not limited to, waste degradation, transcription, DNA repair, and signaling pathways (Fig. 1).
FIG 1.
Various biological roles of micropeptides. Micropeptides are involved in various cellular processes, like signal transduction (e.g., Toddler), calcium transport (e.g., myoregulin), translational regulation (e.g., STORM), waste degradation (e.g., hemotin), mitochondrial regulation (e.g., Mtln), DNA repair (e.g., MRI-2), transcriptional regulation (e.g., Pgc), and mRNA splicing (HOXB-AS3).
Translational regulation and degradation of putative lncRNAs.
Translation of putative lncRNAs undergoes posttranscriptional regulation. In addition to being spliced and polyadenylated, lncRNA translation is controlled by regulatory proteins. An example of this is seen in the activation of eukaryotic initiation factor 4E (eIF4E) via phosphorylation by mammalian Ste20-like kinase (MST1) (19). Once active, eIF4E binds to the 5′ cap of a subset of mRNAs (eIF2-α, eukaryotic translation elongation factor 2 [eEF2], and CCT2) to inhibit their translation (19). This allows for the translation of the lncRNA linc00689, which codes for the stress- and tumor necrosis factor alpha (TNF-α)-activated ORF micropeptide (STORM), which competes with the SRP19, a ribonucleoprotein, for 7SL RNA, which may prevent proper localization of translation products to the endoplasmic reticulum (19). Thus, the expression of some micropeptides can be induced under specific conditions.
Nonsense-mediated decay (NMD) is another way to perform quality control on mRNA (26). mRNAs with abnormal termination of translation or too-long 3′ UTRs are subject to NMD (48, 49). This process can also occur with coding lncRNAs which are bound to ribosomes. Using ribosome profiling, Wery et al. found that actively translated lncRNA sORFs with long 3′ UTRs were sensitive to NMD (26). Therefore, putative lncRNAs do undergo quality control processes like mRNAs.
Cell division, differentiation, and development.
lncRNA-encoded micropeptides in bacteria have been found to regulate cell division. One such example is MciZ, a 40-amino-acid-long micropeptide (13). During cell division, the cell’s machinery forms a divisome, a structure of 10 core proteins, including the tubulin homolog FtsZ, which anchor to the membrane and facilitates its contraction. To better understand the proteins interacting with FtsZ, Handler et al. used a yeast two-hybrid screen and found that MciZ directly binds to FtsZ in mother cells during sporulation (13). Similarly, lncRNAs have been shown to play an important role in cell movements during tissue morphogenesis in Drosophila. One of the best-studied micropeptide-encoding genes is mille-pattes. This gene was originally identified in Tribolium as coding for four micropeptides involved in segmentation (46). Its Drosophila homolog, polished rice/tarsal-less, was initially reported as an ncRNA (50, 51). It was later reidentified as a micropeptide-encoding gene by Kondo et al. and Galindo et al. concurrently (12, 52). Galindo et al. found that sORFs from a tarsal-less (tal) gene, originally thought to be noncoding, code for small peptides that control gene expression and tarsal development (12). The authors found through rescue and ectopic expression experiments that the 11-amino-acid-long micropeptides are responsible for tal function. Because the sORF is translated, the lncRNA was eventually reclassified as an mRNA (16). Another example of a misannotated lncRNA is pgc (polar granule component). The Drosophila transcript was originally thought to be noncoding, and it was believed that pgc localized to polar granules to support normal germ line development (20). However, genetic analysis showed that this lncRNA contained a sORF which potentially codes for a micropeptide of 71 amino acids (20). More recent studies revealed that the lncRNA does encode the micropeptide Pgc. Hanyu-Nakamura et al. predicted that positive transcription elongation factor b (P-TEFb) was a target of Pgc because Pgc knockout cells were unable to inhibit the phosphorylation of RNAPII (14). Pgc inhibited the transcription of somatic genes in germ line cells by preventing P-TEFb from phosphorylating the carboxy-terminal domain of RNAPII to promote proper germ line development (14). lncRNA-encoded micropeptides have also been shown to regulate the function and growth of muscle cells in mammals, like mice. It has also been shown that the micropeptide minion, encoded by a putative lncRNA, works in tandem with the micropeptide myomixer to form syncytial myotubes and promote normal muscle development (27, 53). These findings suggest that many lncRNAs likely contain undiscovered sORFs which code for functional micropeptides important in regulating cell differentiation and development.
Metabolism.
lncRNA-encoded micropeptides have also been shown to play important roles in calcium and mitochondrial metabolism. With regard to calcium metabolism, Magny et al. showed that sarcolamban (scl) codes for two micropeptides involved in cardiac contraction in Drosophila (17). Knockout experiments that removed this gene and nearby CG13283 and CG13282 genes caused flies to express more cardiac arrhythmias than wild-type flies. Localization experiments showed that the Scl micropeptides localize to dyadic space, which is important in ionic signaling. Therefore, these micropeptides are important for Ca2+ movement in cardiac cells (17). Predicted homologs of Scl are the vertebrate micropeptides phospholamban (PLN) and sarcolipin (SLN) (17).
Anderson et al. found that in human tissues, a muscle-specific lncRNA encodes the micropeptide myoregulin (MLN) (7). Fluorescence microscopy of murine tissues and coimmunoprecipitation experiments confirmed that MLN, along with the PLN and SLN, directly interacts with the membrane pump, SERCA, to inhibit its ability to transport Ca2+ into the sarcoplasmic reticulum (SR) of skeletal muscle cells, an organelle in muscle cells that stores calcium ions (7). Similar micropeptides have also been found in non-muscle tissue cells. The micropeptides endoregulin (ELN) and another-regulin (ALN) have also been shown to inhibit isoforms of SERCA (7). Additionally, a previously unrecognized sORF within a putative, muscle-specific lncRNA was also found to code for a micropeptide that localizes to the SR (21). Using the comparative genomics method, PhyloCSF, a dwarf open reading frame, Dworf, was discovered. Dworf encodes a micropeptide which enhances SERCA activity by controlling the effects of inhibitory peptides (21). These evidences suggest that some putative lncRNAs can encode micropeptides that are important for regulating vital cellular functions such as metabolism.
Micropeptides also regulate mitochondrial metabolism. One example is mitoregulin (Mtln), a 56-amino-acid-long micropeptide encoded by a putative lncRNA predominantly expressed in skeletal and cardiac muscle (54). Mtln localizes to the inner mitochondrial membrane (IMM), and binding assays indicated that the micropeptide binds to cardiolipin, a phospholipid important in the regulation of membrane integrity (54). Knockdown of Mtln in HeLa cells exhibited decreased mitochondrial respiration and increased the generation of reactive oxygen species (54). These findings were confirmed in CRISPR/Cas 9 Mtln knockout mice because fasted mice showed decreased fatty acid oxidation and increased Ca2+ retention (54). These micropeptides highlight the fundamental role of micropeptides in the production of cellular energy and homeostasis.
Waste degradation.
Putative lncRNAs have also been shown to encode various micropeptides that localize to other cytoplasmic organelles, such as the lysosome. In the lysosome, micropeptides are involved in waste and toxin degradation. One such example is the Drosophila micropeptide hemotin (23). The gene hemotin has been found to encode an 88-amino-acid-long micropeptide involved in the regulation of endosomal maturation during phagocytosis (23). Further experimentation shows that Hemotin interacts with 14-3-3ζ proteins to inhibit the function of various phosphatidylinositol enzymes (23). Pueyo et al. used a bioinformatics pipeline and discovered that there is a human homolog to hemotin, stannin, a micropeptide which mediates organometallic toxicity (23). Pueyo et al. argued that these micropeptides could have played a role in the first microphage-like cells, thus suggesting that these sORF-encoded micropeptides may have been conserved over hundreds of millions of years (23). These studies underscore just how biologically important these micropeptides are.
Another example of a putative mammalian lncRNA encoding a lysosomal micropeptide is LINC00961, which codes for SPAR (small regulatory polypeptide of amino acid response), a micropeptide involved in amino acid signaling response (18). SPAR was found to negatively regulate mammalian target of rapamycin complex 1 (mTORC1) activation via association with v-ATPase to prevent muscle regeneration from occurring (18). These findings emphasize how many micropeptides are conserved across species and that their role in biological functions like waste degradation should not be overlooked.
DNA repair and transcriptional regulation by micropeptides.
Putative lncRNAs have also been shown to code for micropeptides that localize within the nucleus. A micropeptide involved in nonhomologous end joining (NHEJ) is the modulator of retrovirus infection homolog 2 (MRI-2), a 69-amino-acid-long micropeptide (55). Using techniques like coimmunoprecipitation and an electrophoretic mobility shift assay, Slavoff et al. determined that MRI-2 directly binds to subunits of Ku (i.e., Ku70 and Ku80), a heterodimeric DNA end-binding protein complex involved in DNA repair via NHEJ (55). A double-stranded DNA ligation assay showed that MRI-2 stimulates double-strand breaks in the DNA via its interaction with Ku heterodimers (55).
lncRNA-encoded micropeptides can also regulate transcription. Cai et al. found that lncRNA-Six1-ORF2 encodes a micropeptide that works in tandem with lncRNA-Six1 to activate the Six homeobox 1 (Six1) gene, a gene important for muscle growth (9). Dual-luciferase reporter assays designed to measure Six1 promoter activity displayed increased luciferase activity when lncRNA-Six1-ORF2 or lncRNA-Six1 was overexpressed (9). When lncRNA-Six1 was knocked down, the luciferase activity decreased (9). This suggested that the micropeptide is most likely necessary for the cis mechanisms of lncRNA-Six1, thus demonstrating how micropeptides perform important roles in DNA repair and gene expression.
Signaling.
In bacteria, micropeptides have been shown to regulate signaling kinases and signal transduction. One example is Sda, a 46-amino-acid-long micropeptide which inhibits the first kinase, KinA, in the histidine kinase signaling pathway for genes involved in sporulation in Bacillus subtilis (8). This pathway is normally activated under times of stress or starvation (8). In vitro assays confirmed that Sda directly binds to and inhibits KinA by inducing a conformational change in KinA’s dimerization/histidine-phosphotransfer (DHp) domain (10). Some putative lncRNAs have also been shown to code for micropeptides which are involved in extracellular signaling. One such micropeptide is Toddler, a secreted motogen in zebrafish (22). Loss-of-function experiments produced zebrafish without functional hearts and no blood circulation, thus exemplifying Toddler’s importance in the embryogenesis of zebrafish (22). To test whether Toddler interacts with the predicted APJ/apelin receptor, Pauli et al. used receptor internalization experiments to confirm that the apelin receptor is internalized and therefore activated when Toddler is bound (22). Activation of the APJ/apelin receptor signaling pathway subsequently promotes gastrulation in zebrafish (22). Thus, even micropeptides can regulate important cellular processes like signaling pathways.
Inflammation.
Putative lncRNAs also play a role in regulating inflammation. van Solingen et al. found that a putative lncRNA, lncVLDLR, encodes a 44-amino-acid-long micropeptide named inflammation-modulating micropeptide (IMP) (25). This putative lncRNA is known to be dysregulated in individuals with type II diabetes and cardiovascular disease (25). Using sequence homology, van Solingen et al. found that IMP exhibited high sequence homology with transcription factors involved in inflammation and immune response, like NF-κB and c-myb (25). THP1 macrophages overexpressing IMP exhibited higher levels of expression for inflammatory genes, like those for cytokines and chemokines, thus suggesting that IMP may interact with transcriptional coactivators to regulate genes involved in an inflammatory response (25). This study reveals how micropeptides can act as targets for therapeutic approaches for inflammatory diseases or even cancer.
Cancer.
Putative lncRNAs have also been shown to play a key role in diseases like cancer. Huang et al. discovered that the putative lncRNA HOXB-AS3 encodes a micropeptide with a length of 53 amino acids (15). The HOXB-AS3 micropeptide was found to suppress colon cancer (CRC) cell line growth by competitively binding to hnRNP A1 (15). This interaction disrupts the ability of hnRNPA1 to mediate pyruvate kinase M (PKM) pre-mRNA splicing, thus decreasing the formation of the isoform pyruvate kinase M2 and suppressing glucose metabolism reprogramming in CRC cells (15). Huang et al. argued that this gives the HOXB-AS3 micropeptide tumor-suppressive properties (15).
Another putative lncRNA involved in tumor suppression is the lncRNA LINC01420 (11). This putative lncRNA was identified by D’Lima et al. as coding for NoBody, a micropeptide composed of 68 amino acids (11). NoBody interacts with proteins involved in mRNA decapping and decay by localizing to mRNA processing bodies (P-bodies) (11). P-bodies are highly enriched in proteins involved in NMD, like EDC4 (11). D’Lima et al. tested for NoBody’s role in mRNA decay and found that expression levels of NoBody were inversely proportional to the number of P-bodies and the steady-state levels of NMD substrates present in Calu-6 cells (11). Therefore, it is likely that NoBody negatively regulates mRNA decay and is inversely related to the expression levels of mutant oncogenes (11). These studies emphasize how micropeptides could provide new opportunities for cancer therapeutic targets.
CIRCULAR RNAs ENCODING MICROPEPTIDES
Alternative splicing can produce a variety of noncanonical processed transcripts. One example is circular RNAs (circRNAs). These transcripts are highly conserved, abundant products of alternative RNA splicing (62). circRNAs are composed of back-spliced exons, meaning that a splice donor and upstream splice acceptor are joined together, thus scrambling the order of exons (56). Because of its circular shape, a circRNA is unable to undergo further processing and lacks ends [it has neither a 5′ cap nor a poly(A) tail]. These transcripts were first discovered in plant viroids (57), and recently, mammalian transcripts have also been discovered (28, 29). Due to their nontraditional shape, circRNAs were not predicted to undergo the classical mechanism of translation; however, these transcripts have the potential to be protein coding. Pamudurti et al. (29) proposed that many circRNAs are translated by membrane-associated ribosomes at internal ribosome entry sites (IRESs). Supporting this, Legnini et al. found that the UTR of circ-ZNF609, a circRNA involved in myoblast proliferation, functioned as an IRES (28). The authors used CRISPR/Cas9 to insert a 3×FLAG tag into the ZNF609 locus and found that when circ-ZNF609 is endogenously overexpressed in murine ES cells, the transcript is translated into small peptides in a cap-independent manner (28). Another example of a translated circRNA is circMbl3 (29). Pamudurti et al. used MS to detect the presence of small endogenous peptides encoded by the circular RNAs from the muscleblind locus of Drosophila (29). Therefore, circRNAs have the potential to be translated in a cap- and splice-independent manner. Because the ends of circRNAs are protected from nuclease digestion, the transcripts exhibit a long half-life, thus allowing for the production of significant amounts of small peptides (58). These alternatively spliced transcripts increase the complexity of protein-coding genes and provide new, potential therapeutic targets.
CONCLUSION
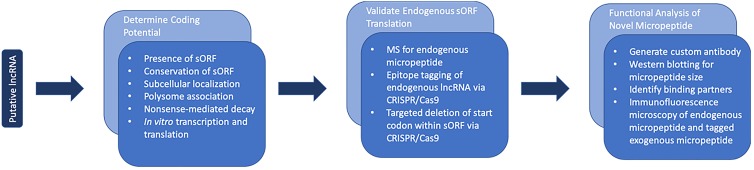
Overall, various tools have been developed to aid in the study of putative lncRNAs that are protein coding. Some lncRNA-encoded micropeptides have been demonstrated to be key regulators of vital cell functions like muscle development, metabolism, and cell signaling in vivo (Table 1). However, identifying and functionally characterizing these micropeptides are challenging. As depicted in Fig. 2, there are many steps involved in the process, and not every experiment is appropriate for every putative lncRNA and its encoded micropeptide. Variations to experimental design should be made appropriately, as a micropeptide may be too small for MS, or the CRISPR/Cas9 system may not work effectively in the desired cell line. Nonetheless, these techniques provide a comprehensive method for the identification of novel micropeptides encoded by putative lncRNAs.
TABLE 1.
Characteristics of various noncoding RNA-encoded micropeptides shown to be endogenously expressed
| Gene | Micropeptide | Putative ncRNA class | Species | Length (aaa ) | Function |
|---|---|---|---|---|---|
| ENSG00000227877 | Myoregulin | lncRNA | Human | 46 | Muscle development |
| ENSG00000175701 | Mtln | lncRNA | Human | 56 | Metabolism |
| ENSMUSG00000103476 | DWORF | lncRNA | Mouse | 34 | Muscle contraction |
| ENSDARG00000094729 | Toddler | lncRNA | Zebrafish | 58 | Embryonic signal |
| BSU23616 | MciZ | lncRNA | B. subtilis | 40 | Cell division |
| BSU25690 | Sda | lncRNA | B. subtilis | 46 | Sporulation |
| ENSG00000180357 | circ-ZNF609 | circRNA | Human | 250 | Myogenesis |
aa, amino acids.
FIG 2.
Representative workflow for identifying a micropeptide encoded by a putative lncRNA.
Now that the study of micropeptides is well established, it is important to delve deeper into the functional analysis of these micropeptides. What would happen if lncRNA transcripts contained mutations? Future directions should investigate the effects of structural changes in micropeptides on the risk and origins of disease. Additional work should also be done to further develop exome sequencing. Currently, exome sequencing has been performed for known protein-coding exons within traditional mRNA transcripts. Now that it is widely accepted that putative lncRNAs can be protein coding, exome sequencing should be updated to include micropeptides. Because the distinction between coding and noncoding can be ambiguous, it is also important to determine if the genes encoding micropeptides are bifunctional (59). The first ncRNA identified as both coding and noncoding was the steroid RNA activator (SRA), a functional ncRNA that also encodes an endogenous protein (60). Like SRA, lncRNAs have also been identified as bifunctional. LncRNA-Six1 is a good example of a bifunctional lncRNA that regulates the Six1 gene in cis and encodes a micropeptide (9). Thus, it is important to confirm that the micropeptides, and not the RNA transcripts, are producing the observed phenotypes. Considering how important these micropeptides are to fundamental cellular processes, future research should focus on studying the coding potential of putative lncRNAs and identifying and classifying lncRNA-encoded micropeptides. Further identification of the biological functions of micropeptides will likely elucidate the key roles of micropeptides in cellular functioning and the pathology of diseases.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program (C.C.R.H. and A.L.) of the National Cancer Institute (NCI), Center for Cancer Research (CCR), NIH.
We thank Emily Dangelmaier from the Lal lab (NCI, NIH) for her comments on the manuscript. We also thank the NIH Medical Arts section for help generating figures. We apologize to those whose work we were unable to cite due to space limitations.
REFERENCES
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See L-H, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR. 2012. Landscape of transcription in human cells. Nature 489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A 109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson DM, Makarewich CA, Anderson KM, Shelton JM, Bezprozvannaya S, Bassel-Duby R, Olson EN. 2016. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci Signal 9:ra119. doi: 10.1126/scisignal.aaj1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkholder WF, Kurtser I, Grossman AD. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 9.Cai B, Li Z, Ma M, Wang Z, Han P, Abdalla BA, Nie Q, Zhang X. 2017. LncRNA-Six1 encodes a micropeptide to activate six1 in cis and is involved in cell proliferation and muscle growth. Front Physiol 8:230. doi: 10.3389/fphys.2017.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham KA, Burkholder WF. 2009. The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to Spo0F. Mol Microbiol 71:659–677. doi: 10.1111/j.1365-2958.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- 11.D’Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke-Andersen J, Saghatelian A, Slavoff SA. 2017. A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 13:174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol 5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handler AA, Lim JE, Losick R. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol 68:588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. 2008. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J-Z, Chen M, Chen D, Gao X-C, Zhu S, Huang H, Hu M, Zhu H, Yan G-R. 2017. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 68:171–184.e176. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Li LJ, Leng RX, Fan YG, Pan HF, Ye DQ. 2017. Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res 361:1–8. doi: 10.1016/j.yexcr.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. 2013. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116–1120. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, Pandolfi PP. 2017. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 19.Min KW, Davila S, Zealy RW, Lloyd LT, Lee IY, Lee R, Roh KH, Jung A, Jemielity J, Choi EJ, Chang JH, Yoon JH. 2017. eIF4E phosphorylation by MST1 reduces translation of a subset of mRNAs, but increases lncRNA translation. Biochim Biophys Acta Gene Regul Mech 1860:761–772. doi: 10.1016/j.bbagrm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. 1996. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science 274:2075–2079. doi: 10.1126/science.274.5295.2075. [DOI] [PubMed] [Google Scholar]
- 21.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN. 2016. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, Ma J, Dubrulle J, Reyon D, Tsai SQ, Joung JK, Saghatelian A, Schier AF. 2014. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343:1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pueyo JI, Magny EG, Sampson CJ, Amin U, Evans IR, Bishop SA, Couso JP. 2016. Hemotin, a regulator of phagocytosis encoded by a small ORF and conserved across metazoans. PLoS Biol 14:e1002395. doi: 10.1371/journal.pbio.1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. 2013. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Solingen C, Sharma M, Bijkerk R, Afonso MS, Koelwyn GJ, Scacalossi KR, Holdt LM, Maegdefessel L, van Zonneveld AJ, Moore K. 2019. A novel micropeptide, IMP, directs inflammation through interaction with transcriptional co-activators. Arterioscler Thromb Vasc Biol 39:A544. doi: 10.1161/atvb.38.suppl_1.027. [DOI] [Google Scholar]
- 26.Wery M, Descrimes M, Vogt N, Dallongeville AS, Gautheret D, Morillon A. 2016. Nonsense-mediated decay restricts lncRNA levels in yeast unless blocked by double-stranded RNA structure. Mol Cell 61:379–392. doi: 10.1016/j.molcel.2015.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Vashisht AA, O’Rourke J, Corbel SY, Moran R, Romero A, Miraglia L, Zhang J, Durrant E, Schmedt C, Sampath SC, Sampath SC. 2017. The microprotein Minion controls cell fusion and muscle formation. Nat Commun 8:15664. doi: 10.1038/ncomms15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. 2017. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22–37.e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. 2017. Translation of circRNAs. Mol Cell 66:9–21.e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 31.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen LL. 2016. Linking long noncoding RNA localization and function. Trends Biochem Sci 41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. 2015. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisler S, Coller J. 2013. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Z, Song R, Regev A, Struhl K. 2015. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li YM, Franklin G, Cui HM, Svensson K, He XB, Adam G, Ohlsson R, Pfeifer S. 1998. The H19 transcript is associated with polysomes and may regulate IGF2 expression in trans. J Biol Chem 273:28247–28252. doi: 10.1074/jbc.273.43.28247. [DOI] [PubMed] [Google Scholar]
- 38.Popa A, Lebrigand K, Barbry P, Waldmann R. 2016. Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells. BMC Genomics 17:52. doi: 10.1186/s12864-016-2384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aspden JL, Eyre-Walker YC, Phillips RJ, Amin U, Mumtaz MA, Brocard M, Couso JP. 2014. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife 3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X, Aslanian A, Yates JR III. 2008. Mass spectrometry for proteomics. Curr Opin Chem Biol 12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karunratanakul K, Tang HY, Speicher DW, Chuangsuwanich E, Sriswasdi S. 2019. Uncovering thousands of new peptides with sequence-mask-search hybrid de novo peptide sequencing framework. Mol Cell Proteomics 18:2478–2491. doi: 10.1074/mcp.TIR119.001656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma B. 2010. Challenges in computational analysis of mass spectrometry data for proteomics. J Comput Sci Technol 25:107–123. doi: 10.1007/s11390-010-9309-1. [DOI] [Google Scholar]
- 44.Makarewich CA, Olson EN. 2017. Mining for micropeptides. Trends Cell Biol 27:685–696. doi: 10.1016/j.tcb.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zordan RE, Beliveau BJ, Trow JA, Craig NL, Cormack BP. 2015. Avoiding the ends: internal epitope tagging of proteins using transposon Tn7. Genetics 200:47–58. doi: 10.1534/genetics.114.169482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savard J, Marques-Souza H, Aranda M, Tautz D. 2006. A segmentation gene in tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell 126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 47.Dyson MR, Shadbolt SP, Vincent KJ, Perera RL, McCafferty J. 2004. Production of soluble mammalian proteins in Escherichia coli: identification of protein features that correlate with successful expression. BMC Biotechnol 4:32. doi: 10.1186/1472-6750-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muhlrad D, Parker R. 1994. Premature translational termination triggers mRNA decapping. Nature 370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 49.Muhlrad D, Parker R. 1999. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. 2005. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci U S A 102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inagaki S, Numata K, Kondo T, Tomita M, Yasuda K, Kanai A, Kageyama Y. 2005. Identification and expression analysis of putative mRNA-like non-coding RNA in Drosophila. Genes Cells 10:1163–1173. doi: 10.1111/j.1365-2443.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 52.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. 2007. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol 9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 53.Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sanchez-Ortiz E, Bassel-Duby R, Olson EN. 2017. Control of muscle formation by the fusogenic micropeptide myomixer. Science 356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, Anderson EJ, Elrod JW, Boudreau RL. 2018. Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep 23:3710–3720.e3718. doi: 10.1016/j.celrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slavoff SA, Heo J, Budnik BA, Hanakahi LA, Saghatelian A. 2014. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J Biol Chem 289:10950–10957. doi: 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lasda E, Parker R. 2014. Circular RNAs: diversity of form and function. RNA 20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Granados-Riveron JT, Aquino-Jarquin G. 2016. The complexity of the translation ability of circRNAs. Biochim Biophys Acta 1859:1245–1251. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 58.Tatomer DC, Wilusz JE. 2017. An unchartered journey for ribosomes: circumnavigating circular RNAs to produce proteins. Mol Cell 66:1–2. doi: 10.1016/j.molcel.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Nam JW, Choi SW, You BH. 2016. Incredible RNA: dual functions of coding and noncoding. Mol Cells 39:367–374. doi: 10.14348/molcells.2016.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E. 2004. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 61.Zhang F, Wen Y, Guo X. 2014. CRISPR/Cas9 for genome editing: progress, implications and challenges. Hum Mol Genet 23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 62.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]