Abstract
Avian adenoviruses (AdVs) are a very diverse group of pathogens causing diseases in poultry and wild birds. Wild birds, endangered by habitat loss and habitat fragmentation in the tropical forests, are recognised to play a role in the transmission of various AdVs. In this study, two novel, hitherto unknown AdVs were described from faecal samples of smooth-billed ani and tropical screech owl. The former was classified into genus Aviadenovirus, the latter into genus Atadenovirus, and both viruses most probably represent new AdV species as well. These results show that there is very limited information about the biodiversity of AdVs in tropical wild birds, though viruses might have a major effect on the population of their hosts or endanger even domesticated animals. Surveys like this provide new insights into the diversity, evolution, host variety, and distribution of avian AdVs.
Introduction
Urbanisation, land conversion and road-building pose a risk to wildlife by habitat fragmentation and also by exposing the animals to the risk of roadkill [1]. Habitat loss and fragmentation modify the landscape and have consequences for biodiversity conservation [2,3]. The highway ES-060 in Brazil crosses three important environmental reserves: the Jacarenema Ecological Reserve (307 ha), the Setiba Environmental Protection Area (12,960 ha) and the Paulo César Vinha State Park (1,500 ha). The vegetation of these reserves is mainly composed of restinga, a distinct type of tropical moist broadleaf forest, regarded as a key area for biodiversity [4]. The rural expansion is intensive adjacent to the highway.
Among the negative anthropic actions, the establishment of new roads is among the most impacting changes of environment [5]. Beyond the risk of roadkill, roads inhibit the movement of many species and act as barriers (total or partial), isolating populations [2,3], with few species immune to this threat [6]. The likelihood of roadkill depends on the animal movement patterns and the landscape characteristics [7,8]. Animal movement patterns are the result of behavioural trade-offs, and are influenced by the individual’s internal state and also by the environment [9]. When roads or railroads cross movement routes, this creates areas of greater risk for the wildlife, for human safety and also for domestic animals [7].
Adenoviruses (AdVs) are DNA viruses with an icosahedral capsid and a double-stranded, linear genome. The presence of AdVs is described in many species of vertebrate animals, including mammals, birds, reptiles, amphibians, and fish [10–14]. It is known that birds are common hosts for AdVs, a fact that has been mirrored by the large number of avian AdVs [15–25]. The host bird species often live in crowded flocks, and migrate large distances by flying, and this predisposes to a rapid viral dissemination in the environment.
The International Committee on Taxonomy of Viruses recognises five genera belonging to the family Adenoviridae. Birds can be infected by highly divergent AdVs classified into three different genera: genus Aviadenovirus, Siadenovirus and Atadenovirus [11]. The pathogenicity of these viruses is not always clear, they can cause latent infections but diseases as well, depending on the virulence of the strain and also on cospeciation time [26]. The deeper evolutionary history of the family Adenoviridae still needs to be resolved, and the discovery of new AdVs in new hosts provides more accurate phylogenetic trees and better understanding of the co-evolution and host switches of these viruses.
Wild birds play an important role in the transmission and as reservoirs of various AdVs [27–35]. Habitat loss and fragmentation cause novel interactions between pathogens, hosts and the environment. This creates new routes for disease transmission, which results in the possible dispersion and adaptation of pathogens to new hosts. To address this risk, we investigated the occurrence of AdVs in tropical screech owls (Megascops choliba), guira cuckoos (Guira guira) and smooth-billed anis (Crotophaga ani) found dead along the highway ES-060.
Materials and methods
1. Study area
The ES-060 highway crosses the municipalities of Vitória, Vila Velha and Guarapari. The northern end of the highway is located at the coordinates 20°18'48.50'' S– 40°17'32.48'' W, and the southern at the coordinates 21°18'5.44'' S– 41°0'7.77" W.
2. Origin of samples
The highway is monitored every 90 minutes for accidents or roadkill occurrence. The animals found alive or dead are collected, recorded and transferred for veterinary treatment or disposal. Faecal samples were collected from 19 dead birds in 2017: five tropical screech owls, four guira cuckoos and ten smooth-billed anis. Samples were collected during necropsies directly from the rectal ampulla of the birds.
For faecal suspension, approximately 200 mg of faecal sample was diluted to a concentration of 20% in Tris-calcium buffer (Tris 0.01 M, CaCl2 1.5 mM, pH 7.2), homogenised and centrifuged at 2000 g for 10 min. Nucleic acid was extracted by the method of Boom et al. [36], using guanidine thiocyanate and silica particles (product numbers: 50983 and 107536, respectively, both from Merck, Darmstadt, Germany).
3. PCR and sequencing
Extracted DNA samples were screened for the presence of AdVs using a pan-adenovirus PCR, targeting the gene of the viral DNA polymerase and detecting all known AdVs [10,37]. The 321-bp-long PCR products were purified using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel; Düren, Germany), and sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific; Waltham, Massachusetts, United States of America) on both strands according to the manufacturers’ protocol.
4. Phylogenetic analysis
The acquired sequences were assembled and translated to amino acid sequences using Geneious 9.1.8. For phylogenetic tree inference, the multiple alignment was conducted using MAFFT [38], the length of the multiple alignment was 90 amino acids. The evolutionary model selection and the phylogenetic calculation were performed using RAxML 8.2.10 [39], the best-scoring model was LG with empirical base frequencies. The robustness of the tree was determined with a non-parametric bootstrap calculation using 1,000 repeats. The phylogenetic tree was visualised using MEGA 7 [40], the tree was rooted on the midpoint, and bootstrap values were given as percentages if they reached 75%. The obtained sequences were compared to entries of the NCBI Protein database using BlastX [41] on 20/09/2019.
Results and discussion
PCR products were gained from three smooth-billed ani samples and one tropical screech owl sample. The GenBank accession numbers for the sequences are MN540447 and MN540448.
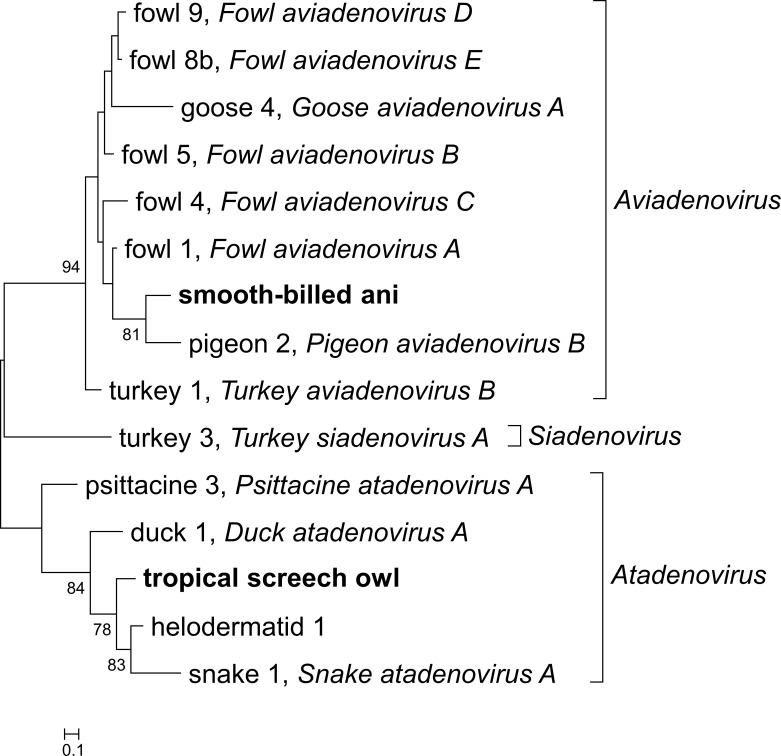
The result of the phylogenetic analysis is displayed in Fig 1. The three smooth-billed ani AdV strains shared identical nucleic acid sequences on the conserved stretch of the DNA polymerase, the virus strain clustered into genus Aviadenovirus. The BlastX hit with the highest pairwise amino acid sequence identity was pigeon AdV-2 with 75.6% (E value: 4.22 x 10−43, query coverage: 100%). The screech owl AdV clustered into genus Atadenovirus, and shared 80.0% sequence identity with the helodermatid AdV-1 (syn. lizard AdV-1 or gila monster AdV; E value: 2.31 x 10−47, query coverage: 100%). This level of sequence divergence suggests that these virus strains represent novel AdV species, as the primary species demarcation criterion for AdVs is 10–15% sequence divergence on the DNA-dependent DNA polymerase amino acid sequence [11]. However, further description of the virus is required to support this hypothesis, primarily, more genomic information—including the complete sequence of the DNA polymerase—would be essential.
Fig 1. Phylogenetic analysis of the smooth-billed ani and the tropical screech owl adenoviruses.
The tree is based on derived amino acid sequence of partial DNA polymerase gene sequences. Adenovirus strains are represented using the host name and the serotype ordinal number, and viral species names are also applied if available. The tree was rooted on the midpoint. Accession numbers: duck 1: NP_044702, fowl 1: AAC54904, fowl 4: AEK64762, fowl 5: YP_007985646, fowl 8b: ANJ02558, fowl 9: derived from AC_000013, goose 4: YP_006383556, helodermatid 1: AAS89696, pigeon 2: APO40944, psittacine 3: YP_009112716, smooth-billed ani: QHA24662, snake 1: YP_001552247, tropical screech owl: QHA24661, turkey 1: YP_003933581, turkey 3: NP_047384.
As identical virus sequences were detected in the three smooth-billed anis, the identified AdV is most probably infectious for this bird species, and it is not a foodborne contaminant. A long-term co-evolution is hypothesised between this aviadenovirus and its host as aviadenoviruses infect a wide range of bird species [12,27,31–34,42]. Though infectious, it may not be evidently pathogenic, as co-evolving viruses are often non-pathogenic or facultatively pathogenic to their host species [26,43–46]. No pathological findings were observed apart from trauma due to vehicular damage neither in these anis nor in the screech owl.
The tropical screech owl AdV was detected in one bird only, and clustered into genus Atadenovirus. Atadenoviruses are thought to have coevolved with squamate reptiles [47,48], but several bird species are also infected by atadenoviruses [17,23,49]. As this owl species is a bird of prey, it is equally possible that this virus had already adapted to this host and replicated in this bird, or that this virus is a foodborne contaminant. Tropical screech owls are omnivorous and known to prey on small reptiles, rodents, and amphibians [50], so the detected atadenovirus might originate from a prey reptile. If the latter will be supported by future results, the designation tropical screech owl associated AdV is recommended for the strain.
The tropical screech owl (family Strigidae) and the smooth-billed ani (family Cuculidae) are species widely distributed in Brazil but also endemic from Costa Rica to Paraguay and northern Argentina [51,52]. The two species of birds act directly on the population dynamics of the prey populations and are able to contribute to maintaining the diversity of these communities [53,54] and to produce secondary effects in plant communities [55].
These samples show us that there is very limited information about the biodiversity of AdVs in tropical wild birds, though viruses might have a major effect on the population of their hosts or endanger even domesticated animals. Several studies have already been published about poultry AdVs [56,57], but further investigations are required to shed light on the diversity and pathogenicity of AdVs in tropical host animals.
Rural expansion causes irreversible damages to wildlife, all 19 birds investigated were found dead because of road traffic accidents. As human activity intensifies, an increasing number of pet animals and livestock are raised in the close proximity of previously unharmed natural habitats. This enhances the risk of viral spread and host changes, but this phenomena was not observed in this investigation: the detected virus strains are not of domestic animal origin, nor have been found in domesticated or pet animals yet.
Due to the ability of avian AdVs—mainly aviadenoviruses—to cause subclinical infections, the diversity of AdVs in birds is probably much more extensive than thought before [17]. The identification of two novel AdVs in specimens of two Brazilian tropical bird species suggests that numerous further unknown AdVs are circulating in other tropical bird species. Surveys like this provide new insights into the pathogenicity, diversity, evolution, host variety, and distribution of avian AdVs; thus, similar studies should be conducted in different geographical locations too.
Acknowledgments
The authors would like to thank the concessionaire of the Rodovia do Sol System and the Sinhá Laurinha Society.
Data Availability
Sequences were submitted to the NCBI GenBank under accession numbers MN540447 and MN540448.
Funding Statement
The research of APJO is supported by the Espírito Santo Research and Innovation Support Foundation (438/2016, fapes.es.gov.br) and University of Vila Velha (201672442, www.uvv.br). GLK is supported by the OMA Foundation (101öu6, omaa.hu), and he is also the recipient of the János Bolyai Research Scholarship of the Hungarian Academy of Sciences (https://mta.hu/english). The research of MZV and BH is supported by the National Research, Development and Innovation Office (NN128309, nkfih.gov.hu/english-nkfih).
References
- 1.Rosa AO, Mauhs J. Atropelamento de animais silvestres na rodovia RS-040 [Wildlife roadkill on the RS-040 highway]. Cad Pesqui Série Biol. 2004;16: 35–42. [Google Scholar]
- 2.Forman RTT, Alexander LE. Roads and their major ecological effects. Annu Rev Ecol Syst. 1998;29: 207–231. 10.1146/annurev.ecolsys.29.1.207 [DOI] [Google Scholar]
- 3.Richard T, Forman T, Deblinger RD. The ecological road-effect zone of a Massachusetts (U.S.A.) suburban highway. Conserv Biol. 2000;14: 36–46. 10.1046/j.1523-1739.2000.99088.x [DOI] [Google Scholar]
- 4.iema.es.gov.br. Parque Estadual Paulo Cesar Vinha [Internet]. 2019 [cited 26 Sep 2019]. Available: https://iema.es.gov.br/PEPCV
- 5.Pinheiro BF, Turci LCB. Vertebrados atropelados na estrada da Variante (BR-307), Cruzeiro do Sul, Acre, Brasil [Vertebrates roadkill on the Variante road (BR-307), Cruzeiro do Sul, Acre, Brazil]. Nat online. 2013;11: 68–78. [Google Scholar]
- 6.Trombulak SC, Frissell CA. Review of ecological effects of roads on terrestrial and aquatic communities. Conserv Biol. 2000;14: 18–30. 10.1046/j.1523-1739.2000.99084.x [DOI] [Google Scholar]
- 7.Gunson KE, Mountrakis G, Quackenbush LJ. Spatial wildlife-vehicle collision models: a review of current work and its application to transportation mitigation projects. J Environ Manage. 2011;92: 1074–82. 10.1016/j.jenvman.2010.11.027 [DOI] [PubMed] [Google Scholar]
- 8.Lewis JS, Rachlow JL, Horne JS, Garton EO, Wakkinen WL, Hayden J, et al. Identifying habitat characteristics to predict highway crossing areas for black bears within a human-modified landscape. Landsc Urban Plan. 2011;101: 99–107. 10.1016/j.landurbplan.2011.01.008 [DOI] [Google Scholar]
- 9.Nathan R, Getz WM, Revilla E, Holyoak M, Kadmon R, Saltz D, et al. A movement ecology paradigm for unifying organismal movement research. Proceedings of the National Academy of Sciences of the United States of America. 2008. pp. 19052–19059. 10.1073/pnas.0800375105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellehan JFX, Johnson AJ, Harrach B, Benkő M, Pessier AP, Johnson CM, et al. Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. J Virol. 2004;78: 13366–13369. 10.1128/JVI.78.23.13366-13369.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrach B, Benkő M, Both GW, Brown M, Davison AJ, Echavarría M, et al. Family Adenoviridae In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus Taxonomy: IXth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2011. pp. 125–141. [Google Scholar]
- 12.Harrach B, Kaján GL. Aviadenovirus In: Tidona CA, Darai G, editors. The Springer Index of Viruses. 2nd ed New York: Springer-Verlag; 2011. pp. 13–28. [Google Scholar]
- 13.Doszpoly A, Harrach B, LaPatra S, Benkő M. Unconventional gene arrangement and content revealed by full genome analysis of the white sturgeon adenovirus, the single member of the genus Ichtadenovirus. Infect Genet Evol. 2019;75: 103976 10.1016/j.meegid.2019.103976 [DOI] [PubMed] [Google Scholar]
- 14.Harrach B, Tarján ZL, Benkő M. Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett. 2019;n/a. 10.1002/1873-3468.13687 [DOI] [PubMed] [Google Scholar]
- 15.Katoh H, Ohya K, Kubo M, Murata K, Yanai T, Fukushi H. A novel budgerigar-adenovirus belonging to group II avian adenovirus of Siadenovirus. Virus Res. 2009;144: 294–297. 10.1016/j.virusres.2009.04.012 [DOI] [PubMed] [Google Scholar]
- 16.Wellehan JFJ, Greenacre CB, Fleming GJ, Stetter MD, Childress AL, Terrell SP. Siadenovirus infection in two psittacine bird species. Avian Pathol. 2009;38: 413–417. 915082004 [pii] 10.1080/03079450903183660 [DOI] [PubMed] [Google Scholar]
- 17.Phalen DN, Agius J, Vaz FF, Eden J-S, Setyo LC, Donahoe S. A survey of a mixed species aviary provides new insights into the pathogenicity, diversity, evolution, host range, and distribution of psittacine and passerine adenoviruses. Avian Pathol. 2019;48: 437–443. 10.1080/03079457.2019.1617835 [DOI] [PubMed] [Google Scholar]
- 18.Kovács ER, Benkő M. Complete sequence of raptor adenovirus 1 confirms the characteristic genome organization of siadenoviruses. Infect Genet Evol. 2011;11: 1058–1065. S1567-1348(11)00102-X [pii] 10.1016/j.meegid.2011.03.021 [DOI] [PubMed] [Google Scholar]
- 19.Ballmann MZ, Vidovszky MZ. Tág gazdaspektrumú psittacine adenovírus (PsAdV-2) kimutatása különböző papagájfajok hazai egyedeiben. [Detection of broad-host-range psittacine adenovirus (PsAdV-2) in representatives of different parrot species]. Magy Állatorvosok Lapja. 2013;135: 78–84. [Google Scholar]
- 20.Bodewes R, van de Bildt MW, Schapendonk CM, van Leeuwen M, van Boheemen S, de Jong AA, et al. Identification and characterization of a novel adenovirus in the cloacal bursa of gulls. Virology. 2013;440: 84–88. 10.1016/j.virol.2013.02.011 [DOI] [PubMed] [Google Scholar]
- 21.Joseph HM, Ballmann MZ, Garner MM, Hanley CS, Berlinski R, Erdélyi K, et al. A novel siadenovirus detected in the kidneys and liver of Gouldian finches (Erythura gouldiae). Vet Microbiol. 2014;172: 35–43. 10.1016/j.vetmic.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 22.Marek A, Kaján GL, Kosiol C, Harrach B, Schlötterer C, Hess M. Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology. 2014;462–463: 107–114. 10.1016/j.virol.2014.08.031 [DOI] [PubMed] [Google Scholar]
- 23.To KKW, Tse H, Chan W-M, Choi GKY, Zhang AJX, Sridhar S, et al. A novel psittacine adenovirus identified during an outbreak of avian chlamydiosis and human psittacosis: zoonosis associated with virus-bacterium coinfection in birds. Vinetz JM, editor. PLoS Negl Trop Dis. 2014;8: e3318 10.1371/journal.pntd.0003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ballmann MZ, Harrach B. Detection and partial genetic characterisation of novel avi- and siadenoviruses in racing and fancy pigeons (Columba livia domestica). Acta Vet Hung. 2016;64: 514–528. 10.1556/004.2016.047 [DOI] [PubMed] [Google Scholar]
- 25.Cassmann E, Zaffarano B, Chen Q, Li G, Haynes J. Novel siadenovirus infection in a cockatiel with chronic liver disease. Virus Res. 2019;263: 164–168. 10.1016/j.virusres.2019.01.018 [DOI] [PubMed] [Google Scholar]
- 26.Kaján GL, Doszpoly A, Tarján ZL, Vidovszky M, Papp T. Virus–host coevolution with a focus on animal and human DNA viruses. J Mol Evol. 2020;88: 41–56. 10.1007/s00239-019-09913-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schrenzel M, Oaks JL, Rotstein D, Maalouf G, Snook E, Sandfort C, et al. Characterization of a new species of adenovirus in falcons. J Clin Microbiol. 2005;43: 3402–3413. 43/7/3402 [pii] 10.1128/JCM.43.7.3402-3413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovács ER, Jánoska M, Dán Á, Harrach B, Benkő M. Recognition and partial genome characterization by non-specific DNA amplification and PCR of a new siadenovirus species in a sample originating from Parus major, a great tit. J Virol Methods. 2010;163: 262–268. S0166-0934(09)00446-7 [pii] 10.1016/j.jviromet.2009.10.007 [DOI] [PubMed] [Google Scholar]
- 29.Park YM, Kim JH, Gu SH, Lee SY, Lee MG, Kang YK, et al. Full genome analysis of a novel adenovirus from the South Polar skua (Catharacta maccormicki) in Antarctica. Virology. 2012;422: 144–150. 10.1016/j.virol.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SY, Kim JH, Park YM, Shin OS, Kim H, Choi HG, et al. A novel adenovirus in Chinstrap penguins (Pygoscelis antarctica) in Antarctica. Viruses. 2014;6: 2052–2061. 10.3390/v6052052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das S, Fearnside K, Sarker S, Forwood JK, Raidal SR. A novel pathogenic aviadenovirus from red-bellied parrots (Poicephalus rufiventris) unveils deep recombination events among avian host lineages. Virology. 2017;502: 188–197. 10.1016/j.virol.2016.12.031 [DOI] [PubMed] [Google Scholar]
- 32.Teske L, Rubbenstroth D, Meixner M, Liere K, Bartels H, Rautenschlein S. Identification of a novel aviadenovirus, designated pigeon adenovirus 2 in domestic pigeons (Columba livia). Virus Res. 2017;227: 15–22. 10.1016/j.virusres.2016.09.024 [DOI] [PubMed] [Google Scholar]
- 33.Milani A, Zamperin G, Fusaro A, Salviato A, Bano L, Zandonà L, et al. Complete genome sequence of psittacine adenovirus 1, identified from Poicephalus senegalus in Italy. Thrash JC, editor. Microbiol Resour Announc. 2018;7: e01037–18. 10.1128/MRA.01037-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vibin J, Chamings A, Collier F, Klaassen M, Nelson TM, Alexandersen S. Metagenomics detection and characterisation of viruses in faecal samples from Australian wild birds. Sci Rep. 2018;8: 8686 10.1038/s41598-018-26851-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang N, McLelland J, McLelland DJ, Clarke J, Woolford L, Eden P, et al. Psittacid Adenovirus-2 infection in the critically endangered orange-bellied parrot (Neophema chrysogastor): A key threatening process or an example of a host-adapted virus? PLoS One. 2019;14: e0208674 10.1371/journal.pone.0208674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boom R, Sol CJA, Salimans MMM, Jansen CL, Wertheim-van Dillen PM., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaján GL. Poultry adenoviruses In: Liu D, editor. Molecular detection of animal viral pathogens. Boca Raton: CRC Press; 2016. pp. 735–746. [Google Scholar]
- 38.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marek A, Kaján GL, Kosiol C, Benkő M, Schachner A, Hess M. Genetic diversity of species Fowl aviadenovirus D and Fowl aviadenovirus E. J Gen Virol. 2016;97: 2323–2332. 10.1099/jgv.0.000519 [DOI] [PubMed] [Google Scholar]
- 43.Kaján GL, Stefancsik R, Ursu K, Palya V, Benkő M. The first complete genome sequence of a non-chicken aviadenovirus, proposed to be turkey adenovirus 1. Virus Res. 2010;153: 226–233. 10.1016/j.virusres.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 44.Kaján GL, Davison AJ, Palya V, Harrach B, Benkő M. Genome sequence of a waterfowl aviadenovirus, goose adenovirus 4. J Gen Virol. 2012;93: 2457–2465. 10.1099/vir.0.042028-0 [DOI] [PubMed] [Google Scholar]
- 45.Benkő M. Adenoviruses: Pathogenesis In: Caplan M Mitchell R BRML, editor. Reference Module in Biomedical Sciences. Amsterdam: Elsevier; 2014. [Google Scholar]
- 46.Kaján GL, Affranio I, Tóthné Bistyák A, Kecskeméti S, Benkő M. An emerging new fowl adenovirus genotype. Heliyon. 2019;5: e01732 10.1016/j.heliyon.2019.e01732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrach B. Reptile adenoviruses in cattle? Acta Vet Hung. 2000;48: 485–490. 10.1556/004.48.2000.4.11 [DOI] [PubMed] [Google Scholar]
- 48.Papp T, Fledelius B, Schmidt V, Kaján GL, Marschang RE. PCR-sequence characterization of new adenoviruses found in reptiles and the first successful isolation of a lizard adenovirus. Vet Microbiol. 2009;134 10.1016/j.vetmic.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 49.Hess M, Blöcker H, Brandt P. The complete nucleotide sequence of the egg drop syndrome virus: an intermediate between mastadenoviruses and aviadenoviruses. Virology. 1997;238: 145–156. 10.1006/viro.1997.8815 [DOI] [PubMed] [Google Scholar]
- 50.Motta-Junior JC, de Arruda Bueno A. Trophic ecology of the burrowing owl in Southeast Brazil. In: Chancellor RD, Meyburg B-U, editors. Raptors Worldwide. Budapest: World Working Group on Birds of Prey and Owls and MME/BirdLife Hungary; 2004. pp. 763–775. [Google Scholar]
- 51.Sick H. Famílias e espécies: Ordem Cuculiformes. Ornitologia Brasileira. Rio de Janeiro: Nova Fronteira; 1997. pp. 383–392. [Google Scholar]
- 52.Marks JS, Cannings RJ, Mikkola H. Family Strigidae (Typical Owls) In: Hoyo J del, Elliott A, Sargatal J, Christie DA, editors. Handbook of the Birds of the World–Volume 5: Barn-owls to Hummingbirds. Barcelona: Lynx Edicions; 1999. pp. 56–151. [Google Scholar]
- 53.Valkama J, Korpimäki E, Arroyo B, Beja P, Bretagnolle V, Bro E, et al. Birds of prey as limiting factors of gamebird populations in Europe: A review. Biol Rev Camb Philos Soc. 2005;80: 171–203. 10.1017/s146479310400658x [DOI] [PubMed] [Google Scholar]
- 54.Norrdahl K, Korpimäki E. Effects of predator removal on vertebrate prey populations: birds of prey and small mammals. Oecologia. 1995;103: 241–248. 10.1007/BF00329086 [DOI] [PubMed] [Google Scholar]
- 55.Estes JA, Terborgh J, Brashares JS, Power ME, Berger J, Bond WJ, et al. Trophic downgrading of planet earth. Science. 2011;333: 301–306. 10.1126/science.1205106 [DOI] [PubMed] [Google Scholar]
- 56.Zadravec M, Slavec B, Krapez U, Kaján GL, Racnik J, Juntes P, et al. Inclusion body hepatitis associated with fowl adenovirus type 8b in broiler flock in Slovenia—a case report. Slov Vet Res. 2011;48: 107–113. [Google Scholar]
- 57.Schachner A, Matos M, Grafl B, Hess M. Fowl adenovirus-induced diseases and strategies for their control—a review on the current global situation. Avian Pathol. 2018;47: 111–126. 10.1080/03079457.2017.1385724 [DOI] [PubMed] [Google Scholar]