Abstract
Background
Animal trypanosomosis caused by Trypanosoma evansi is known as “surra” and is a widespread neglected tropical disease affecting wild and domestic animals mainly in South America, the Middle East, North Africa and Asia. An essential necessity for T. evansi infection control is the availability of reliable and sensitive diagnostic tools. While DNA-based PCR detection techniques meet these criteria, most of them require well-trained and experienced users as well as a laboratory environment allowing correct protocol execution. As an alternative, we developed a recombinase polymerase amplification (RPA) test for Type A T. evansi. The technology uses an isothermal nucleic acid amplification approach that is simple, fast, cost-effective and is suitable for use in minimally equipped laboratories and even field settings.
Methodology/Principle findings
An RPA assay targeting the T. evansi RoTat1.2 VSG gene was designed for the DNA-based detection of T. evansi. Comparing post-amplification visualization by agarose gel electrophoresis and a lateral flow (LF) format reveals that the latter displays a higher sensitivity. The RPA-LF assay is specific for RoTat1.2-expressing strains of T. evansi as it does not detect the genomic DNA of other trypanosomatids. Finally, experimental mouse infection trials demonstrate that the T. evansi specific RPA-LF can be employed as a test-of-cure tool.
Conclusions/Significance
Compared to other DNA-based parasite detection methods (such as PCR and LAMP), the T. evansi RPA-LF (TevRPA-LF) described in this paper is an interesting alternative because of its simple read-out (user-friendly), short execution time (15 minutes), experimental sensitivity of 100 fg purified genomic T. evansi DNA, and ability to be carried out at a moderate, constant temperature (39°C). Therefore, the TevRPA-LF is an interesting tool for the detection of active T. evansi infections.
Author summary
Neglected tropical diseases (NTDs) affecting humans and/or domestic animals severely impair the socio-economic development of endemic areas. One of these diseases, animal trypanosomosis, affects livestock and is caused by the parasites of the Trypanosoma genus. The most widespread causative agent of animal trypanosomosis is T. evansi, which is found in large parts of the world (Africa, Asia, South America, Middle East, and the Mediterranean). Proper control and treatment of the disease requires the availability of reliable and sensitive diagnostic tools. DNA-based detection techniques are powerful and versatile in the sense that they can be tailored to achieve a high specificity and usually allow the reliable detection of low amounts of parasite genetic material. However, many DNA-based methodologies (such as PCR) require trained staff and well-equipped laboratories, which is why the research community has actively investigated in developing amplification strategies that are simple, fast, cost-effective and are suitable for use in minimally equipped laboratories and field settings. In this paper, we describe the development of a diagnostic test under a dipstick format for the specific detection of T. evansi, based on a DNA amplification principle (Recombinase Polymerase Amplification aka RPA) that meets the above-mentioned criteria.
Introduction
Trypanosoma evansi is a haemoflagellate parasite which is closely related to T. brucei, the causative agent of human sleeping sickness and nagana in animals [1]. T. evansi is the causative agent of “surra” or “mal de caderas”, which is the most common and widespread trypanosomal disease of domestic and wild animals and is characterized by high morbidity and mortality. The parasite is mechanically transmitted by biting flies and is found in many regions around the globe [2–6]. Outbreaks of surra have been reported in all types of ungulates (camels, cattle, buffaloes, horses, pigs, and deer) in Africa [7], Asia [8–10], Latin America [11–13] and recently Europe [14–16]. While T. evansi is commonly known as non-infective to humans, human infections were recently reported and confirmed in India and Vietnam, indicating that T. evansi may be emerging as a potential human pathogen [17–20]. Control of T. evansi trypanosomosis is mainly accomplished by drug treatment, but resistance of T. evansi to trypanocidal compounds has been reported in Africa [21, 22] and in the far east of Asia [23].
T. evansi parasites are classified into two groups based on their kDNA minicircle type [24], which are characterised by the presence (Type A) or absence (Type B) of the gene encoding the RoTat1.2 variant surface glycoprotein (VSG) [25, 26]. T. evansi Type B are less commonly found and have only been reported to occur in certain regions in Africa [27–32]. In contrast, T. evansi Type A are widespread. Many diagnostic methods are available to detect T. evansi infections and include parasitological, serological, and molecular assays [33]. While some methods detect both T. evansi Types A and B, others are specific to one of both types. Conventional blood smear examination technique is widely used in the field and detects both T. evansi Type A and B. However, it can only diagnose clinical stages of infection and not latent or chronic infection [34]. In addition, it is time consuming and requires both the presence of microscopy equipment and specifically trained personnel at the screening site. To overcome these shortcomings, the T. evansi card agglutination test (CATT/T. evansi) was developed. It is a standard test for epidemiological field studies of T. evansi Type A since it is based on the use of the T. evansi RoTat 1.2 VSG antigen as an agglutination agent for host antibodies [35]. The advantage of this technique is that it is fast, easy to execute and suitable for field diagnosis. The main disadvantage of the technique is the lack of discrimination between previous exposure and current infections. Indeed, the host antibodies that drive the reaction can be a result of an active infection, a past infection, repeated exposure without necessarily initiation of successful infection, or even polyclonal B cell activation by other infectious agents such as helminths [36].
The diagnosis of trypanosomosis has been improved by the development and application of DNA-based techniques such as PCR, which is a very sensitive and effective method for the detection of chronic infections or prepatent period of disease [37, 38]. The DNA of killed trypanosomes does not remain in the blood for more than 24 to 48 hours, thus PCR-based assays are highly suitable for the detection of active infections [39]. Several genes have been investigated as targets for the PCR-based diagnosis of T. evansi; these include the RoTat1.2 VSG gene (Type A specific) [40–42], ribosomal DNA [43], a region from r-RNA internal transcribed spacer 1 (ITS-1) [44], the gene encoding the invariant surface glycoprotein ISG-75 [45], and the VSG JN 2118Hu gene (Type B specific) [26, 28, 46, 47]. The drawback of PCR-based methods is that they require well-trained and experienced personnel and a laboratory environment suitable for correct protocol execution. Hence, they are difficult to deploy and maintain under most field conditions. An interesting alternative to PCR is the so-called Recombinase Polymerase Amplification (RPA) [48]. The reaction mechanism of RPA has been reviewed elsewhere [49, 50] and is summarized in Fig 1 (the figure legend contains a detailed explanation of the RPA reaction). This isothermal nucleic acid amplification technology is simple, fast, cost-effective and is suitable for minimally equipped laboratories as well as for use in the field [51]. Hence, RPA is especially useful in infectious disease diagnostics and epidemiological studies [52–55]. The RPA reaction can be completed in 10 to 20 minutes at temperatures between 24°C to 45°C [56]. The amplification product can be visualized by gel electrophoresis or in real-time by the inclusion of a nucleic acid dye. The specificity and sensitivity of RPA are typically enhanced by probe-based methods, which (depending on the type of probe) allow amplicon detection based on fluorescence or a lateral flow (LF) assay [48]. To date, RPA has been successfully applied for the detection of bacteria [57, 58], foodborne pathogens [59, 60], parasites [61, 62], and viruses [63, 64].
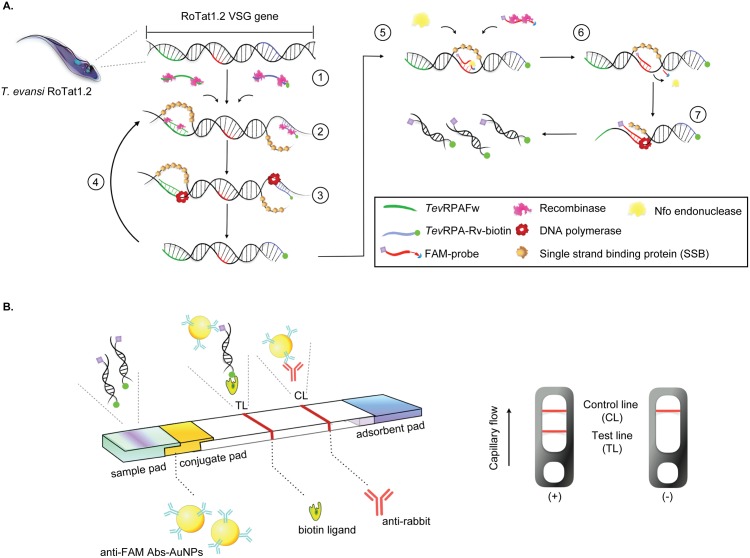
Fig 1. Schematic representation of the TevRPA-LF.
A: RPA-based generation of a T. evansi specific RoTat1.2 VSG amplicon for detection by a lateral flow (LF) assay. Step 1: two oligonucleotide primers (TevRPA-Fw and TevRPA-Rv-biotin) form a complex with the recombinase. Step 2: the primer-recombinase complexes invade the homologous sequences on the target DNA. Step 3: A DNA polymerase with a strand displacement activity performs amplification of the target sequence under isothermal conditions, resulting in the generation of a biotinylated amplicon. Step 4: the generated amplicons are again invaded by primer-recombinase complexes in a self-perpetuating cycle fueled in ATP by creatine kinase. Step 5: an oligonucleotide (FAM-probe) carrying a 5’ FAM tag, a spacer sequence and a 3’ blocking group forms a complex with the recombinase and invades the biotinylated amplicon generated in the previous steps. Step 6: only when the FAM-probe has successfully invaded the biotinylated amplicon and bound its complementary sequence, can the Nfo endonuclease bind and cleave the spacer region and 3’ blocking group. Step 7: after removal of the 3’ region of the FAM probe, the Nfo endonuclease dissociates. This allows the DNA polymerase to employ the cleaved FAM-probe as a forward primer. Together with the biotinylated reverse primer (TevRPA-Rv-biotin) this leads to the formation of an amplicon bearing both the FAM and biotin tags. B: Read-out of the RPA via LF. The FAM- and biotin-tagged RPA product is mixed with the LF buffer, loaded onto the sample pad and is transported to the adsorbent pad through capillary flow. The RPA product is first bound by gold-labeled rabbit anti-FAM antibodies and later captured by a streptavidin-coated test line (TL). The control line (CL) is coated with anti-rabbit antibodies. While a valid negative test only contains a reddish band at the CL, a valid positive test will display bands at both the TL and CL.
In this present study, we describe the development of the first recombinase polymerase amplification lateral flow assay for the detection of active Type A T. evansi infections (TevRPA-LF). The T. evansi RoTat1.2 VSG gene was chosen as the target for the TevRPA-LF for the following reasons: i) to ensure high specificity of the TevRPA-LF for T. evansi as this parasite is closely related to T. brucei, ii) T. evansi Type A are most commonly encountered and widespread, and iii) to allow comparison with the previously described PCR targeting the T. evansi RoTat1.2 VSG gene [33]. We demonstrate that the TevRPA-LF assay is highly specific for T. evansi since no cross-reactions with the closely related parasite T. brucei could be observed. In addition, we have tested the TevRPA-LF in an experimental mouse model and demonstrate that it can be used as a test-of-cure tool. The TevRPA-LF described here has a processing time of 15 minutes and can be performed at a constant temperature of 39°C. Combined with the simplicity, robustness and reliability of the RPA-FL principle, the findings presented in this paper show that the TevRPA-LF can be a promising tool for the detection of active T. evansi infections.
Materials and methods
Ethics statement
All experiments, maintenance and care of the mice complied with the European Convention for the Protection of Vertebrate Animals (ECPVA) used for Experimental and Other Scientific Purposes guidelines (CETS n° 123) and were approved by the Ethical Committee for Animal Experiments (ECAE) at the Vrije Universiteit Brussel (Permit Number: 14-220-31).
Preparation of purified genomic DNA
Total genomic DNA of the different parasites used in this study (Table 1) was extracted and purified from infected mouse whole blood using a DNeasy Blood & Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions. The DNA was eluted in 50 μl nuclease-free water and stored at -20°C until further use. The concentration and quality of the purified DNA were determined by gel electrophoresis (1% agarose gel run in TBE buffer at 110 V for 30 min) and spectrophotometric analysis (measurement of the absorbance at 260 nm, A260; examination of the ratio of the absorbances at 260 nm and 280 nm, A260/A280; performed on a NanoDrop-2000/2000c).
Table 1. Characteristics of trypanosomatid parasites used in this study.
| Strain | Host | Country |
|---|---|---|
| T. evansi RoTat1.2 | Water buffalo | Indonesia |
| T. evansi STIB816 | Camel | China |
| T. evansi ITMAS180697 | Water buffalo | Vietnam |
| T. evansi 020499B | Horse | Columbia |
| T. evansi CAN86K | Dog | Brazil |
| T. evansi ITMAS060297 | Camel | Kazakhstan |
| T. evansi ITMAS050399C | Camel | Morocco |
| T. congolense Tc13 | Cow | Kenya |
| T. vivax TV700 | Cattle | Nigeria |
| T. brucei AnTat1.1 | Bushbuck | Uganda |
| L. donovani Ldl82 | Human | Ethiopia |
Preparation of crude genomic DNA
Genomic DNA was robustly extracted by boiling. Briefly, 50 μl of blood was mixed with 10 μl nuclease-free water (Thermofisher). The sample was heated at 100°C for 5 minutes followed by centrifugation at 20000 g for 5 minutes, and the supernatant was applied as a crude DNA template. The DNA template was kept at -20°C until use.
RPA primers and probes design
The primers and probes were manually designed based on the gene sequence of the Rode Trypanozoon antigenic type 1.2 VSG (RoTat 1.2 VSG) of T. evansi (GenBank accession code: AF317914.1). The NCBI’s nucleotide BLAST tools combined with Primer 5 were used to search for primers specific to T. evansi without significant overlap with other genomes. The TwistAmp LF Probe oligonucleotide backbone includes a 5’-antigenic label FAM group, an internal abasic nucleotide analogue ‘dSpacer’ and a 3’-polymerase extension blocking group C3-spacer. The details of the primers and probes used are given in Table 2.
Table 2. Primers and probes employed in this study.
| Assay type | Primer name | Oligonucleotide (5’-3’) | Reference |
|---|---|---|---|
| TevRPA |
TevRPA-Fw TevRPA-Rv |
CACCGAAGCAAGCGCAGCAAGAGGGTTAGCA GTAGCTGTCTCCTGGGGCCGAGGTGTCATAG |
This study |
| TevRPA-LF |
TevRPA-Rv-biotin FAM-Probe 1 FAM-Probe 2 |
[Biotin]GTAGCTGTCTCCTGGGGCCGAGGTGTCATAG [6F]TCTGCCCGCAGTTGCCTATGGCGGCGAAGT[dS]GCAGGGGCGATTTCAT[C3] [6F]CTAAAATTTCTAAAGCACGCGGTTGGCAACA[dS]CAAGTTTGTGTGGGC[C3] |
This study |
| PCR | RoTat1.2 Fw RoTat1.2 Rv |
GCGGGGTGTTTAAAGCAATA ATTAGTGCTGCGTGTGTTCG |
[40] |
6F stands for 6FAM, dS for dSpacer, and C3 for C3-spacer.
Development and optimization of the TevRPA assay
The RPA reactions were conducted with the TwistAmp Basic kit (TwistDx, Cambridge, UK). A 47.5 μl reaction mixture containing the following components was prepared in a 1.5 ml tube: 2.4 μl of both forward and reverse primers (final concentration: 480 nM), 29.5 μl rehydration buffer supplied by the TwistAmp Basic kit, 12.2 μl nuclease-free water and 1 μl T. evansi purified genomic DNA (concentration of 120 ng μl−1). The reaction mixture was then transferred to the kit’s reaction tubes containing lyophilized enzyme pellet. Next, 2.5 μl magnesium acetate (MgAc; final concentration of 14 nM) was carefully pipetted onto the reaction tube lids. This was followed by a brief vortex and spin to mix MgAc with the RPA reaction mixture. The tubes were incubated in a thermocycler. To pinpoint the most optimal conditions for the TevRPA, the samples were incubated at different reaction temperatures (25°C, 30°C, 35°C, 37°C, 39°C, 41°C, 43°C, 45°C, and 50°C) and for different durations (5 minutes, 10 minutes, 15 minutes, 20 minutes, 25 minutes, 30 minutes, 35 minutes and 40 minutes). Reactions were halted by placing the tubes on ice. The amplified products were first purified using the GenElute PCR Clean-Up kit (Sigma-Aldrich) and visualized on a 2% agarose gel.
Development and optimization of the TevRPA-LF
LF-RPA assays were performed following the indications provided in the TwistAmp nfo kit (TwistDx, Cambridge, UK). Briefly, the RPA reaction was assembled as described above (Materials and Methods subsection ‘Development and optimization of the TevRPA assay’) with the exception of the addition of 2.1 μl of both forward and reverse primers (final concentration: 420 nM) and 0.6 μl probe (final concentration: 120 nM) to the reaction mixture. The amplified DNA was detected using LF strips (Milenia Hybridtech 1, TwistDx, Cambridge, UK) following the instructions indicated in the kit. Briefly, 1 μl of the amplified product was diluted with 99 μl LF buffer. Ten μl of this diluted sample was then loaded on the sample application area according to the manufacturer’s instructions. The final result was visually read out after incubation for 2 minutes at room temperature. A testing sample was considered positive when both the detection line (biotin-ligand line) and the control line (anti-rabbit antibody line) were visible. A testing was considered negative when only the control line was visible (Fig 1). The amplicons could be analyzed on a 2% agarose gel after purification with the GenElute PCR Clean-Up kit (Sigma-Aldrich) to further confirm the testing result.
Evaluation of sensitivity and specificity of the TevRPA-LF
The specificity of the TevRPA-LF was assessed by employing 20 ng of purified genomic DNA isolated from various parasites (Table 1). Samples containing only nuclease-free water were used as negative controls.
The sensitivity of the TevRPA-LF was tested by employing the following concentrations of T. evansi purified genomic DNA as templates for the RPA reaction: 10 ng μl−1, 1 ng μl−1, 100 pg μl−1, 10 pg μl−1, 1 pg μl−1, 100 fg μl−1, 10 fg μl−1 and 1 fg μl−1. The results were analyzed by lateral flow and agarose gel electrophoresis.
Comparison between TevPCR and TevRPA-LF in an experimental mouse infection model
C57BL6/C mice (bred in-house, 8 weeks old) were divided in two groups of six individuals. In each group, five mice were inoculated intraperitoneally with 2000 T. evansi (Rotat 1.2 strain) parasites in 200 μl of PSG buffer (36.4 mM NaCl, 3.12 mM NaH2PO4, 47.5 mM Na2HPO4 and 85.2 mM glucose, pH 8). The remaining mouse in each group was used as a negative control and was not infected. The mice were bled at different times post-infection. The mice in Group 1 were bled at days 1, 3, 5 and 6 post-infection. The animals in Group 2 were bled at days 0, 2, 4, 6, 8, 10 and 12 post-infection. All individuals from Group 2 were treated with Berenil (40 mg per kg), administered intraperitoneally at day 5 post-infection. For both groups, at each time point, 102.5 μl of whole blood was collected from the tail of each individual using nuclease-free tubes with 30 ml heparinized saline (10 units/ml; Sigma-Aldrich) to prevent coagulation. 2.5 μl of the collected blood was used to follow-up mice parasitemia by diluting the sample 200-fold (during high parasitemia periods) and 100-fold (during low parasitemia periods) in PSG buffer and counting the parasites under the light microscope. The rest of the collected blood (100 μl) was split into two parts to evaluate the samples using the TevPCR and TevRPA-LF. Fifty μl of collected blood was employed to prepare purified genomic DNA for the TevPCR, whereas the remaining 50 μl of collected blood was used to obtain crude genomic DNA for the TevRPA-LF. The TevPCR was performed as described in [40] with the following modifications: the amount of purified genomic DNA as starting material (250 ng vs. 3000 ng) and the addition of 10% DMSO to the reaction mixture.
Results and discussion
Development and optimization of the TevRPA
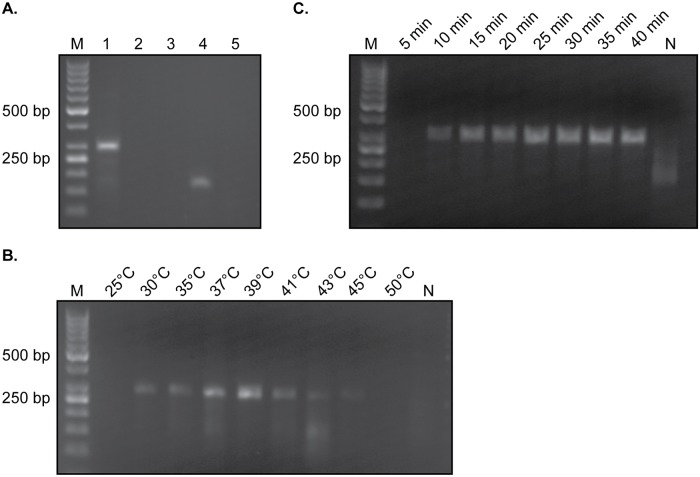
The first requirement of the TevRPA-LF is a high specificity for the detection of T. evansi. This parasite is closely related to T. brucei and thus the selection of an appropriate nucleotide sequence that is unique to T. evansi is crucial. This is the case for a specific region (bp 1 to bp 1300) of the T. evansi RoTat1.2 VSG gene [40–42], which forms the target of the TevRPA-LF for T. evansi detection (Fig 1). This limits the use of the TevRPA-LF described here to the detection of Type A T. evansi, and not Type B. Based on this particular region, a primer pair was designed for the TevRPA such that the resulting amplicon does not exceed 500 bp (as suggested by the RPA manufacturer instructions). As can be seen from Fig 2A, an RPA with this primer pair (initially incubated at 37°C for 30 minutes) on T. evansi purified genomic DNA extracted from infected mice blood yields an amplicon of around 289 bp. The reaction was also performed on genomic DNA purified from a naive mouse to exclude the possible lack of specificity due to cross-reactivity. No amplification could be observed in this negative control sample (Fig 2A).
Fig 2. Optimization of the TevRPA.
A: Initial RPA incubated at 37°C for 30 minutes on various samples. Lane 1, T. evansi purified genomic DNA; Lane 2, naïve mouse purified genomic DNA; Lane 3, sample without any template; Lane 4, RPA kit positive control; Lane 5, RPA kit negative control. B: RPA reaction on T. evansi purified genomic DNA incubated at different temperatures for a constant time of 30 minutes. C: RPA reaction on T. evansi purified genomic DNA incubated at a constant temperature of 39°C for various times. In all panels Lane M indicates the molecular mass marker, whereas Lane N in panels B and C represents a negative control sample (no template DNA).
Next, the assay conditions were optimized by allowing the RPA reaction to proceed at various incubation temperatures and amplification times. First, a range of incubation temperatures between 25°C and 50°C were tested at a constant amplification time of 30 minutes. As can be seen from Fig 2B, 39°C represents the most optimal incubation temperature as it produces the highest amount of amplicon. In a second phase, the RPA was performed at a constant incubation temperature of 39°C while varying the amplification times from 5 to 40 minutes in 5 minute increments (Fig 2C). Although the TevRPA can be performed within 10 minutes, longer incubation times clearly yield a higher signal. The amplification time of 15 minutes was selected in an effort to maintain a balance between providing maximum sensitivity and obtaining a minimal reaction time. In conclusion, these experiments demonstrate that the TevRPA may be reliably performed with an amplification time of 15 minutes and an incubation temperature of 39°C. These conditions were maintained for all subsequent experiments.
The TevRPA can be translated into a specific and sensitive TevRPA-LF
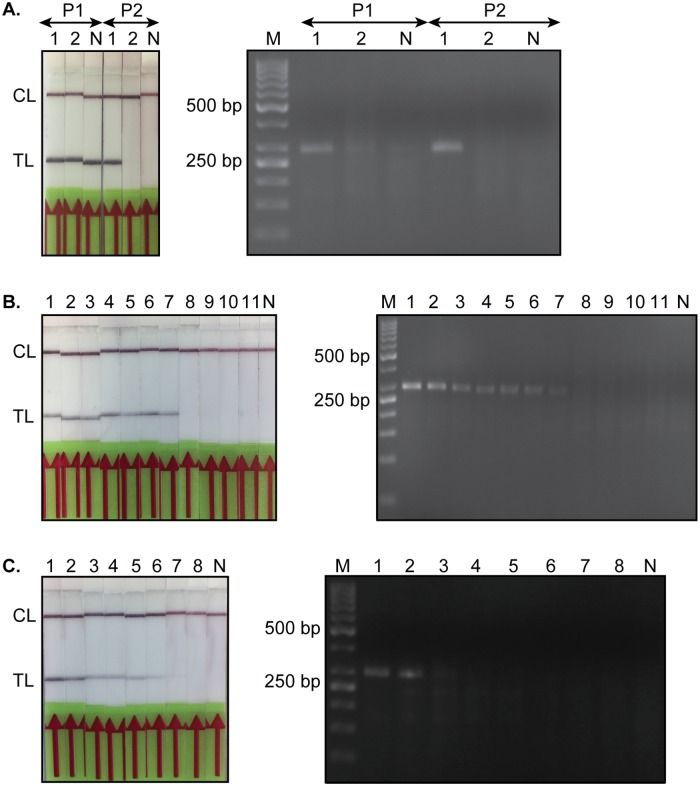
The visualization of the RPA amplicon via agarose gel electrophoresis requires an additional purification step to avoid smeared bands on the gel due to the presence of enzymes and crowding agents [50]. This additional handling step is not necessary if the assay’s read-out is performed via a lateral flow (LF) device [48, 49]. However, the translation of an RPA to an RPA-LF necessitates the addition of a labeled probe to the RPA reaction mixture and the biotinylation of the RPA reverse primer (Fig 1). Two candidate probes were screened for their potential to generate an RPA-LF for T. evansi detection (from here on referred to as TevRPA-LF). Although both probes gave rise to positive signals when tested on T. evansi purified genomic DNA in both agarose gel electrophoresis and lateral flow detection formats, probe 1 clearly generates false positives while probe 2 does not (Fig 3A, right and left panels, respectively). Therefore, probe 2 was selected to be incorporated in the RPA assay to allow post-amplification detection of the amplicon via the TevRPA-LF.
Fig 3. Read-out of the TevRPA via a lateral flow assay (TevRPA-LF) and agarose gel electrophoresis.
A: Selection of a suitable probe for the development of the TevRPA-LF. P1 and P2 refer to FAM probes 1 and 2, respectively. Lane 1, T. evansi purified genomic DNA; Lane 2, naïve mouse purified genomic DNA. B: Assessment of the specificity of the TevRPA-LF. Lanes 1-7, various T. evansi strains as listed in Table 1; Lane 8, T. congolense; Lane 9, T. vivax; Lane 10, T. brucei; Lane 11, L. donovani. C: Comparison of the sensitivities of the TevRPA by a lateral flow assay and agarose gel electrophoresis. Lanes 1-8, 10-fold dilution series of T. evansi purified genomic DNA starting at 10 ng μl−1 (1 μl was loaded onto the gel). Lane 1, 10 ng; Lane 2, 1 ng; Lane 3, 100 pg; Lane 4, 10 pg; Lane 5, 1 pg; Lane 6, 100 fg; Lane 7, 10 fg; Lane 8,1 fg. All panels display the read-out of the TevRPA by a lateral flow assay (left) and agarose gel electrophoresis (right). In all panels Lane M indicates the molecular mass marker, whereas Lane N represents a negative control sample (no template DNA). CL and TL refer to the control and test lines, respectively.
Next, the specificity of the TevRPA-LF was evaluated by employing purified genomic DNA of various Trypanosoma and one Leishmania species as starting material for the amplification reaction. Only T. evansi genomic DNA resulted in visible bands at the test line, while the genomic material of other trypanosomatids did not result in any detection (Fig 3B).
Finally, the detection limit of the TevRPA-LF was compared to the sensitivity of amplicon visualization via agarose gel electrophoresis by performing the TevRPA on a 10-fold dilution series ranging from 10 ng to 1 fg T. evansi purified genomic DNA per reaction (Fig 3C). When visualized using agarose gel electrophoresis, the lowest amount of genomic DNA that produces an amplicon that can be detected is 100 pg. In contrast, the TevRPA-LF allows amplicon detection at an amount of 100 fg genomic DNA, which is 1000-fold more sensitive compared to agarose gel electrophoresis. The loss of sensitivity during post-amplification visualization via agarose gel electrophoresis is most probably related to the additional required purification step [65]. Hence, for the TevRPA, the extra purification step comes at the cost of sensitivity, which advocates the use of the TevRPA-LF over the TevRPA followed by agarose gel electrophoresis.
The TevRPA-LF can detect active T. evansi infections in an experimental mouse model
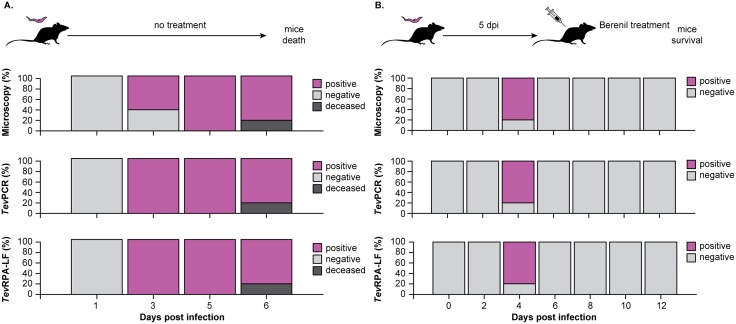
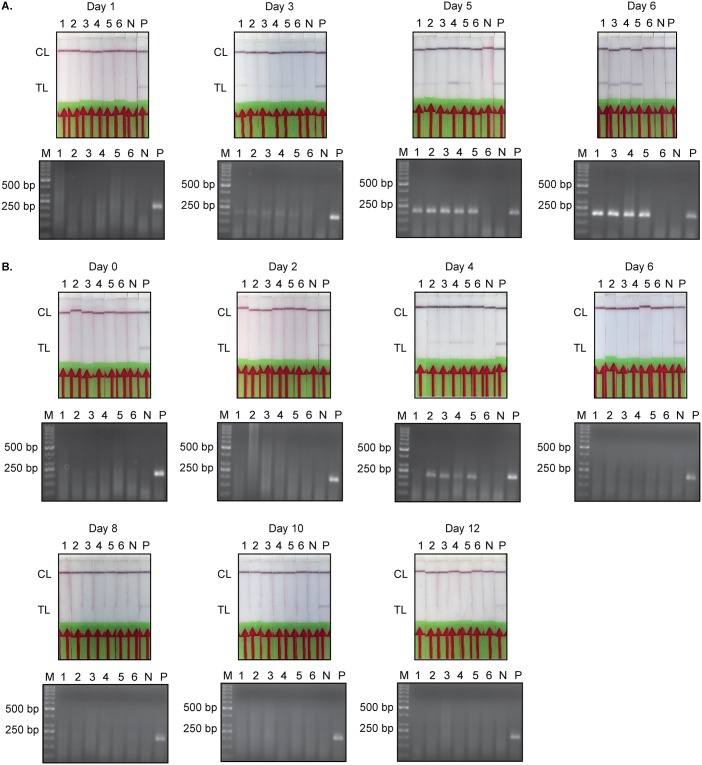
Next, the TevRPA-LF was evaluated for its potential to differentiate between ongoing and past infections in an experimental mouse model. In this experiment, C57BL/6 mice infected with T. evansi RoTat1.2 were divided into two groups and the presence of parasites was analyzed by microscopy, the previously described TevPCR [40] and the TevRPA-LF at various time points. Group 1 was left untreated, while Group 2 was treated with Berenil at 5 days post-infection.
As shown in Figs 4 and 5, all three techniques yielded identical results for most of the collected samples. A discrepancy between the detection methods was only observed at 3 days post-infection in Group 1; while parasites could only be detected in 3 out of 5 mice by microscopy, all samples were found to be positive when tested by the TevPCR and TevRPA-LF (Figs 4A and 5A). It is noteworthy to mention that in Group 1 only 4 samples from infected mice were available for testing at day 6 post-infection due to the premature death of one mouse. As expected, all infected mice in Group 1 succumbed to the infection at 7 days post-infection. In contrast, the mice in Group 2 survived day 7 post-infection indicating successful parasite clearance after Berenil treatment at day 5 post-infection. One mouse in Group 2 did not display any signs of infection (4 days post-infection) and was scored as negative by all three methods. Importantly, no amplicons could be detected post-treatment by either the previously validated TevPCR [40–42] or the TevRPA-LF described in this work (Figs 4B and 5B). This demonstrates that the TevRPA-LF is a suitable ‘test-of-cure’ assay. While both the TevPCR and TevRPA-LF display identical positive and negative score rates under these experimental conditions, the advantage of the TevRPA-LF is that it is effective when performed with crude genomic DNA, whereas execution of the TevPCR requires additional purification of the isolated genomic DNA.
Fig 4. Evaluation of the TevRPA-LF as a test-of-cure tool in T. evansi infections in mice.
A: C57BL/6 mice were infected with T. evansi RoTat1.2 (n = 5) and the presence of parasites was monitored over the course of the infection by microscopy (top panel), the TevPCR (middle panel, performed on parasite genomic DNA purified from the collected blood samples), and TevRPA-LF (bottom panel, executed on crude parasite genomic DNA extracted from the collected blood). The results are displayed as the percentages of mice that scored positive or negative as determined by the above-mentioned techniques. B: C57BL/6 mice infected with T. evansi RoTat1.2 (n = 5) were treated with Berenil at 5 days post-infection. The presence of parasites was followed by microscopy, the TevPCR and the TevRPA-LF throughout the experiment. The panels and color codes are the same as for panel A. The TevPCR and TevRPA-LF read-outs are shown in Fig 5.
Fig 5. TevPCR and TevRPA-LF read-outs.
The TevPCR (bottom panels) and TevRPA-LF (upper panels) read-outs displayed in Fig 4. A: TevPCR and TevRPA-LF results for the mouse infection trial of Group 1 mice (corresponds to the data set shown in Fig 4A). B: TevPCR and TevRPA-LF results for the mouse infection trial of Group 2 mice (corresponds to the data set shown in Fig 4B). In all panels Lane M indicates the molecular mass marker, Lanes 1-6 indicate the individual mice (mouse 6 was used as a negative control within each data set and was not infected), Lane N is a negative control sample (no template DNA) and Lane P is the positive control (T. evansi purified genomic DNA). CL and TL refer to the control and test lines, respectively.
Conclusion
T. evansi is the one of the most widespread causative agents of animal trypanosomosis in the world [6]. An essential part of parasite control is the availability of reliable, quick, and user-friendly diagnostic methods. In this paper, we have described the development of a TevRPA-LF, a test that specifically detects active Type A T. evansi infections by amplifying a region in the T. evansi RoTat1.2 VSG gene. While the T. evansi RoTat1.2 VSG is also targeted by the T. evansi CATT [35] and TevPCR [40–42] at the protein and DNA levels, respectively, the TevRPA-LF presents some interesting advantages: i) compared to antibody-based tests (RoTat 1.2 CATT, Surra Sero K-Set, and T. evansi trypanolysis) the TevRPA-LF can be employed to detect active parasitaemia and also serves as a test-of-cure tool since it is not hampered by the presence of infection-induced antibodies that could be the result of past infections or repeated parasite exposure without active infection and ii) the TevRPA-LF combines the RPA format with a dipstick read-out, which outperforms a regular PCR in terms of user-friendliness and field applicability. While it can be argued that LAMP [66] offers the same advantage, the proposed LF format offers an advantage in terms of user friendliness as it visually resembles an antibody-test format that is already in place, while offering the advantage of detecting active infections. Based on the above-mentioned findings, the newly developed TevRPA-LF presented in this paper provides a proof-of-concept with the potential of becoming a valid alternative for currently used screening tools. Its further development will require an additional evaluation of its performance in both experimental and clinical animal infection models.
Acknowledgments
The authors wish to thank Prof. dr. Guy Caljon (LMPH, University of Antwerp) for providing samples of L. donovani genomic DNA.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by a grant of the China Scholarship Council (CSC), a research grant of the University of Antwerp (DOCPRO1, FFB190197), a research grant of the Foundation for Scientific Research / Fonds voor Wetenschappelijk Onderzoek – Vlaanderen (G013518N) and a UGent BOF startkrediet (01N01518). This work was performed in frame of an Interuniversity Attraction Pole Program (PAI-IAP N. P7/41) and was supported by the Strategic Research Program (SRP3, VUB). BS was supported by the Strategic Research Program (SRP3 and SRP47, VUB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukes J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc Natl Acad Sci U S A. 2008;105(6):1999–2004. 10.1073/pnas.0711799105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monzón CM, Mancebo OA, Roux JP. Comparison between six parasitological methods for diagnosis of Trypanosoma evansi in the subtropical area of Argentina. Vet Parasitol. 1990;36(1-2):141–6. 10.1016/0304-4017(90)90102-h [DOI] [PubMed] [Google Scholar]
- 3. Veer V, Parashar B, SJCs P. Tabanid and muscoid haematophagous flies, vectors of trypanosomiasis or surra disease in wild animals and livestock in Nandankanan Biological Park, Bhubaneswar (Orissa, India). Current Science. 2002;82(5):500–503. [Google Scholar]
- 4. Truc P, Büscher P, Cuny G, Gonzatti MI, Jannin J, Joshi P, et al. Atypical human infections by animal trypanosomes. PLoS Negl Trop Dis. 2013;7(9):e2256 10.1371/journal.pntd.0002256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yadav SC, Kumar R, Kumar S, Tatu U, Singh RK, Gupta AK. Identification and characterization of cysteine proteinases of Trypanosoma evansi. Parasitol Res. 2011;109(3):559–65. 10.1007/s00436-011-2284-9 [DOI] [PubMed] [Google Scholar]
- 6. Desquesnes M, Holzmuller P, Lai DH, Dargantes A, Lun ZR, Jittaplapong S. Trypanosoma evansi and surra: a review and perspectives on origin, history, distribution, taxonomy, morphology, hosts, and pathogenic effects. Biomed Res Int. 2013;2013:194176 10.1155/2013/194176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diall O, Bocoum Z, Diarra B, Sanogo Y, Coulibaly Z, Waïgalo Y. Epidemiology of trypanosomiasis caused by T. evansi in camels in mali: results of parasitological and clinical survey. Rev Elev Med Vet Pays Trop. 1993;46(3):455–61. [PubMed] [Google Scholar]
- 8. Hoare C. The trypanosomes of mammals A zoological monograph. Blackwell Scientific Publications; 1972. [Google Scholar]
- 9. Nurulaini R, Jamnah O, Adnan M, Zaini CM, Khadijah S, Rafiah A, et al. Mortality of domesticated java deer attributed to Surra. Trop Biomed. 2007;24(2):67–70. [PubMed] [Google Scholar]
- 10. Adrian MS, Sani RA, Hassan L, Wong MT. Outbreaks of trypanosomiasis and the seroprevalence of T. evansi in a deer breeding centre in Perak, Malaysia. Trop Anim Health Prod. 2010;42(2):145–50. 10.1007/s11250-009-9406-8 [DOI] [PubMed] [Google Scholar]
- 11. Silva RA, Arosemena NA, Herrera HM, Sahib CA, Ferreira MS. Outbreak of trypanosomosis due to Trypanosoma evansi in horses of Pantanal Mato-grossense, Brazil. Vet Parasitol. 1995;60(1-2):167–71. 10.1016/0304-4017(94)00757-4 [DOI] [PubMed] [Google Scholar]
- 12. Gutierrez C, Corbera JA, Juste MC, Doreste F, Morales I. An outbreak of abortions and high neonatal mortality associated with Trypanosoma evansi infection in dromedary camels in the Canary Islands. Vet Parasitol. 2005;130(1-2):163–8. 10.1016/j.vetpar.2005.02.009 [DOI] [PubMed] [Google Scholar]
- 13. Desquesnes M. Livestock Trypanosomoses and their Vectors in Latin America OIE (World organisation for animal health); 2004. [Google Scholar]
- 14. Garcia H, Garcia ME, Perez H, Mendoza-Leon A. The detection and PCR-based characterization of the parasites causing trypanosomiasis in water-buffalo herds in Venezuela. Ann Trop Med Parasitol. 2005;99(4):359–70. 10.1179/136485905X36271 [DOI] [PubMed] [Google Scholar]
- 15. Desquesnes M, Bossard G, Patrel D, Herder S, Patout O, Lepetitcolin E, et al. First outbreak of Trypanosoma evansi in camels in metropolitan France. Vet Rec. 2008;162(23):750–2. 10.1136/vr.162.23.750 [DOI] [PubMed] [Google Scholar]
- 16. Tamarit A, Gutierrez C, Arroyo R, Jimenez V, Zagalá G, Bosch I, et al. Trypanosoma evansi infection in mainland Spain. Vet Parasitol. 2010;167(1):74–6. 10.1016/j.vetpar.2009.09.050 [DOI] [PubMed] [Google Scholar]
- 17. World Health Organization: A new form of human trypanosomiasis in India Description of the first human case in the world caused by Trypanosoma evansi. vol. 80; 2005. [PubMed] [Google Scholar]
- 18. Truc P, Gibson W, Herder S. Genetic characterization of Trypanosoma evansi isolated from a patient in India. Infect Genet Evol. 2007;7(2):305–7. 10.1016/j.meegid.2006.07.004 [DOI] [PubMed] [Google Scholar]
- 19. Joshi PP, Shegokar VR, Powar RM, Herder S, Katti R, Salkar HR, et al. Human trypanosomiasis caused by Trypanosoma evansi in India: the first case report. Am J Trop Med Hyg. 2005;73(3):491–5. 10.4269/ajtmh.2005.73.491 [DOI] [PubMed] [Google Scholar]
- 20. Van Vinh Chau N, Buu Chau L, Desquesnes M, Herder S, Phu Huong Lan N, Campbell JI, et al. A Clinical and Epidemiological Investigation of the First Reported Human Infection With the Zoonotic Parasite Trypanosoma evansi in Southeast Asia. Clin Infect Dis. 2016;62(8):1002–1008. 10.1093/cid/ciw052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boid R, Jones TW, Payne RC. Malic enzyme type VII isoenzyme as an indicator of suramin resistance in Trypanosoma evansi. Exp Parasitol. 1989;69(4):317–23. 10.1016/0014-4894(89)90080-5 [DOI] [PubMed] [Google Scholar]
- 22. El Rayah IE, Kaminsky R, Schmid C, El Malik KH. Drug resistance in Sudanese Trypanosoma evansi. Vet Parasitol. 1999;80(4):281–7. 10.1016/s0304-4017(98)00221-0 [DOI] [PubMed] [Google Scholar]
- 23. Zhou J, Shen J, Liao D, Zhou Y, Lin J. Resistance to drug by different isolates Trypanosoma evansi in China. Acta Trop. 2004;90(3):271–5. 10.1016/j.actatropica.2004.02.002 [DOI] [PubMed] [Google Scholar]
- 24. Masiga DK, Gibson WC. Specific probes for Trypanosoma (Trypanozoon) evansi based on kinetoplast DNA minicircles. Mol Biochem Parasitol. 1990;40(2):279–83. 10.1016/0166-6851(90)90049-r [DOI] [PubMed] [Google Scholar]
- 25. Ngaira JM, Njagi ENM, Ngeranwa JJN, Olembo NK. PCR amplification of RoTat 1.2 VSG gene in Trypanosoma evansi isolates in Kenya. Vet Parasitol. 2004;120(1-2):23–33. 10.1016/j.vetpar.2003.12.007 [DOI] [PubMed] [Google Scholar]
- 26. Birhanu H, Gebrehiwot T, Goddeeris BM, Büscher P, Van Reet N. New Trypanosoma evansi Type B Isolates from Ethiopian Dromedary Camels. PLoS Negl Trop Dis. 2016;10(4):e0004556 10.1371/journal.pntd.0004556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Borst P, Fase-Fowler F, Gibson WC. Kinetoplast DNA of Trypanosoma evansi. Mol Biochem Parasitol. 1987;23(1):31–8. 10.1016/0166-6851(87)90184-8 [DOI] [PubMed] [Google Scholar]
- 28. Ngaira JM, Olembo NK, Njagi ENM, Ngeranwa JJN. The detection of non-RoTat 1.2 Trypanosoma evansi. Exp Parasitol. 2005;110(1):30–8. 10.1016/j.exppara.2005.01.001 [DOI] [PubMed] [Google Scholar]
- 29. Birhanu H, Fikru R, Said M, Kidane W, Gebrehiwot T, Hagos A, et al. Epidemiology of Trypanosoma evansi and Trypanosoma vivax in domestic animals from selected districts of Tigray and Afar regions, Northern Ethiopia. Parasit Vectors. 2015;8:212 10.1186/s13071-015-0818-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ashenafi H, Yilkal K, Esayass T, Alemu T, Gari FR, Feseha G, et al. Parasitological and serological survey on trypanosomis (surra) in camels in dry and wet areas of Bale Zone, Oromyia Region, Ethiopia. Revue de médecine vétérinaire. 2009;160(12):569–573. [Google Scholar]
- 31. Salim B, Bakheit MA, Kamau J, Nakamura I, Sugimoto C. Molecular epidemiology of camel trypanosomiasis based on ITS1 rDNA and RoTat 1.2 VSG gene in the Sudan. Parasit Vectors. 2011;4:31 10.1186/1756-3305-4-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boid R. Isoenzyme characterisation of 15 stocks of Trypanosoma evansi isolated from camels in the Sudan. Trop Med Parasitol. 1988;39(1):45–50. [PubMed] [Google Scholar]
- 33. Tehseen S, Jahan N, Qamar MF, Desquesnes M, Shahzad MI, Deborggraeve S, et al. Parasitological, serological and molecular survey of Trypanosoma evansi infection in dromedary camels from Cholistan Desert, Pakistan. Parasit Vectors. 2015;8:415 10.1186/s13071-015-1002-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terkawi MA, Thekisoe OMM, Katsande C, Latif AA, Mans BJ, Matthee O, et al. Serological survey of Babesia bovis and Babesia bigemina in cattle in South Africa. Vet Parasitol. 2011;182(2-4):337–42. 10.1016/j.vetpar.2011.05.047 [DOI] [PubMed] [Google Scholar]
- 35. Bajyana Songa E, Hamers R. A card agglutination test (CATT) for veterinary use based on an early VAT RoTat 1/2 of Trypanosoma evansi. Ann Soc Belg Med Trop. 1988;68(3):233–40. [PubMed] [Google Scholar]
- 36. Harris N, Gause WC. To B or not to B: B cells and the Th2-type immune response to helminths. Trends Immunol. 2011;32(2):80–8. 10.1016/j.it.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dávila AMR, Herrera HM, Schlebinger T, Souza SS, Traub-Cseko YM. Using PCR for unraveling the cryptic epizootiology of livestock trypanosomosis in the Pantanal, Brazil. Vet Parasitol. 2003;117(1-2):1–13. 10.1016/j.vetpar.2003.08.002 [DOI] [PubMed] [Google Scholar]
- 38. Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22(7):909–18. 10.1016/0020-7519(92)90047-o [DOI] [PubMed] [Google Scholar]
- 39.Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. vol. 1, 2 and 3. 8th ed. Office International des Epizooties (World Organization for Animal Health; ISBN 978-92-95108-18-9); 2018.
- 40. Claes F, Radwanska M, Urakawa T, Majiwa PA, Goddeeris B, Büscher P. Variable Surface Glycoprotein RoTat 1.2 PCR as a specific diagnostic tool for the detection of Trypanosoma evansi infections. Kinetoplastid Biol Dis. 2004;3(1):3 10.1186/1475-9292-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verloo D, Magnus E, Büscher P. General expression of RoTat 1.2 variable antigen type in Trypanosoma evansi isolates from different origin. Vet Parasitol. 2001;97(3):183–9. 10.1016/s0304-4017(01)00412-5 [DOI] [PubMed] [Google Scholar]
- 42. Urakawa T, Verloo D, Moens L, Büscher P, Majiwa PA. Trypanosoma evansi: cloning and expression in Spodoptera fugiperda insect cells of the diagnostic antigen RoTat1.2. Exp Parasitol. 2001;99(4):181–9. 10.1006/expr.2001.4670 [DOI] [PubMed] [Google Scholar]
- 43. Ijaz MK, Nur-E-Kamal MS, Mohamed AI, Dar FK. Comparative studies on the sensitivity of polymerase chain reaction and microscopic examination for the detection of Trypanosoma evansi in experimentally infected mice. Comp Immunol Microbiol Infect Dis. 1998;21(3):215–23. 10.1016/s0147-9571(98)00002-2 [DOI] [PubMed] [Google Scholar]
- 44. Taylor TK, Boyle DB, Bingham J. Development of a TaqMan PCR assay for the detection of Trypanosoma evansi, the agent of surra. Vet Parasitol. 2008;153(3-4):255–64. 10.1016/j.vetpar.2008.01.045 [DOI] [PubMed] [Google Scholar]
- 45. Rudramurthy GR, Sengupta PP, Balamurugan V, Prabhudas K, Rahman H. PCR based diagnosis of trypanosomiasis exploring invariant surface glycoprotein (ISG) 75 gene. Vet Parasitol. 2013;193(1-3):47–58. 10.1016/j.vetpar.2012.11.045 [DOI] [PubMed] [Google Scholar]
- 46. Njiru ZK, Constantine CC, Masiga DK, Reid SA, Thompson RCA, Gibson WC. Characterization of Trypanosoma evansi type B. Infect Genet Evol. 2006;6(4):292–300. 10.1016/j.meegid.2005.08.002 [DOI] [PubMed] [Google Scholar]
- 47. Njiru ZK, Ouma JO, Enyaru JC, Dargantes AP. Loop-mediated Isothermal Amplification (LAMP) test for detection of Trypanosoma evansi strain B. Exp Parasitol. 2010;125(3):196–201. 10.1016/j.exppara.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 48. Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204 10.1371/journal.pbio.0040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase Polymerase Amplification for Diagnostic Applications. Clin Chem. 2016;62(7):947–58. 10.1373/clinchem.2015.245829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: basics, applications and recent advances. Trends in Analytical Chemistry. 2018;98:19–35. 10.1016/j.trac.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J, Liu L, Wang J, Sun X, Yuan W. Recombinase Polymerase Amplification Assay-A Simple, Fast and Cost-Effective Alternative to Real Time PCR for Specific Detection of Feline Herpesvirus-1. PLoS One. 2017;12(1):e0166903 10.1371/journal.pone.0166903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tu PA, Shiu JS, Lee SH, Pang VF, Wang DC, Wang PH. Development of a recombinase polymerase amplification lateral flow dipstick (RPA-LFD) for the field diagnosis of caprine arthritis-encephalitis virus (CAEV) infection. J Virol Methods. 2017;243:98–104. 10.1016/j.jviromet.2017.01.023 [DOI] [PubMed] [Google Scholar]
- 53. Abd El Wahed A, Patel P, Faye O, Thaloengsok S, Heidenreich D, Matangkasombut P, et al. Recombinase Polymerase Amplification Assay for Rapid Diagnostics of Dengue Infection. PLoS One. 2015;10(6):e0129682 10.1371/journal.pone.0129682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Paz HD, Brotons P, Muñoz-Almagro C. Molecular isothermal techniques for combating infectious diseases: towards low-cost point-of-care diagnostics. Expert Rev Mol Diagn. 2014;14(7):827–43. 10.1586/14737159.2014.940319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. James A, Macdonald J. Recombinase polymerase amplification: Emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev Mol Diagn. 2015;15(11):1475–89. 10.1586/14737159.2015.1090877 [DOI] [PubMed] [Google Scholar]
- 56. Castellanos-Gonzalez A, White AC Jr, Melby P, Travi B. Molecular diagnosis of protozoan parasites by Recombinase Polymerase Amplification. Acta Trop. 2018;182:4–11. 10.1016/j.actatropica.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. del Río JS, Yehia Adly N, Acero-Sánchez JL, Henry OYF, O’Sullivan CK. Electrochemical detection of Francisella tularensis genomic DNA using solid-phase recombinase polymerase amplification. Biosens Bioelectron. 2014;54:674–8. 10.1016/j.bios.2013.11.035 [DOI] [PubMed] [Google Scholar]
- 58. Ahmed A, van der Linden H, Hartskeerl RA. Development of a recombinase polymerase amplification assay for the detection of pathogenic Leptospira. Int J Environ Res Public Health. 2014;11(5):4953–64. 10.3390/ijerph110504953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Murinda SE, Ibekwe AM, Zulkaffly S, Cruz A, Park S, Razak N, et al. Real-time isothermal detection of Shiga toxin-producing Escherichia coli using recombinase polymerase amplification. Foodborne Pathog Dis. 2014;11(7):529–36. 10.1089/fpd.2013.1663 [DOI] [PubMed] [Google Scholar]
- 60. Liu HB, Zang YX, Du XJ, Li P, Wang S. Development of an isothermal amplification-based assay for the rapid visual detection of Salmonella bacteria. J Dairy Sci. 2017;100(9):7016–7025. 10.3168/jds.2017-12566 [DOI] [PubMed] [Google Scholar]
- 61. Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Multiplexed Recombinase Polymerase Amplification Assay To Detect Intestinal Protozoa. Anal Chem. 2016;88(3):1610–6. 10.1021/acs.analchem.5b03267 [DOI] [PubMed] [Google Scholar]
- 62. Castellanos-Gonzalez A, Saldarriaga OA, Tartaglino L, Gacek R, Temple E, Sparks H, et al. A Novel Molecular Test to Diagnose Canine Visceral Leishmaniasis at the Point of Care. Am J Trop Med Hyg. 2015;93(5):970–5. 10.4269/ajtmh.15-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang Y, Qin X, Wang G, Zhang Y, Shang Y, Zhang Z. Development of a fluorescent probe-based recombinase polymerase amplification assay for rapid detection of Orf virus. Virol J. 2015;12:206 10.1186/s12985-015-0440-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Faye O, Faye O, Soropogui B, Patel P, El Wahed AA, Loucoubar C, et al. Development and deployment of a rapid recombinase polymerase amplification Ebola virus detection assay in Guinea in 2015. Euro Surveill. 2015;20(44). 10.2807/1560-7917.ES.2015.20.44.30053 [DOI] [PubMed] [Google Scholar]
- 65. Jaroenram W, Owens L. Recombinase polymerase amplification combined with a lateral flow dipstick for discriminating between infectious Penaeus stylirostris densovirus and virus-related sequences in shrimp genome. J Virol Methods. 2014;208:144–51. 10.1016/j.jviromet.2014.08.006 [DOI] [PubMed] [Google Scholar]
- 66. Tong Q, Chen R, Kong Q, Goossens J, Radwanska M, Lou D, et al. DNA detection of Trypanosoma evansi: Diagnostic validity of a new assay based on loop-mediated isothermal amplification (LAMP). Vet Parasitol. 2018;250:1–6. 10.1016/j.vetpar.2017.12.006 [DOI] [PubMed] [Google Scholar]