Abstract
The constantly evolving nature of genomics provides new challenges for students in Public Health as they try to understand how genomic information relates to health and disease. As Public Health curricula attempt to keep pace with the most recent advances in genomics, students should gain experience with analyzing genomic data and applying genomic tools to the study of health-related issues. To advance undergraduate and graduate student education and provide a more comprehensive view of genomics, we developed an educational project including both pedagogic and research components to characterize skin microbial communities (microbiomes) using targeted amplicon sequencing of their genomes (metataxonomy). All students completed the lab procedures, analyzed 16S rRNA genomic data (formative assessments), and wrote a five-page scientific report summarizing and discussing their results (summative assessment). Student grades for the summative assessment ranged from 31.5 to 40 (out of 40) points. They also successfully completed two practicums (problem sets) focused on microbiome sequence data and responded to 12 minute-papers related to genomic topics covered in class. In all these exercises the 2019 students outperformed 2018 students, who did not participate in this educational lab project. By fulfilling all the requirements of this project-based learning experience, students better understood the complexity of genomics and acquired a valuable set of marketable experience and skills in molecular technologies, bioinformatics and statistics (quantitative skills). Additionally, students were able to generate new valuable microbial 16S rRNA genomic data and test hypotheses about the composition and diversity of the microbes living on our skin (microbiota).
INTRODUCTION
Background
Genomics is an interdisciplinary field of science involving the sequencing and analysis of full or partial genomes through the use of high-throughput DNA sequencing technologies and bioinformatics. Graduate students from George Washington University (GWU) enrolled in the Public Health Genomics (PHG) course learn about the structure, function, evolution, mapping, and editing of microbial genomes. They then use computational tools to search, retrieve, and analyze available genomic data (e.g., DNA amplicons and genomes) from public databases (e.g., National Center for Biotechnology and Information). Finally, through practicums (problem sets and case exercises), students apply those concepts and computational skills to the investigation of human health questions involving microbial diversity, pathogen identification, host-microbe interactions, and community disease outbreaks.
Students in PHG are taught theoretical concepts (e.g., central dogma of molecular biology; cell structure and genome regulation) and shown past and modern genomic technologies and some of their concomitant software to analyze genomic data. However, the complexity and constantly changing nature of genomics makes it challenging to deliver those concepts and master those skills effectively. Additionally, students have trouble visualizing and comprehending the different phases of a genomic experiment; they often fail to see the interconnections in the research pipeline going from bench to a public health application and intervention. Consequently, they end up with a disjointed view of genomics and its interactions with other disciplines (e.g., epidemiology, forensics, or medicine).
To alleviate this problem and give students a comprehensive view of genomics, as well as to improve their learning experiences and skills, in 2019 we initiated a new educational approach that brings students to the lab to complement their classroom learning with a direct engagement in applied genomics research through Project-Based Learning (PBL). Students carried out several molecular procedures in the lab (e.g., sample collection, DNA extraction, and PCR amplification) and bioinformatic analyses of genomic data. They also wrote up their final results in a summative scientific report. We motivated PHG students by direct engagement in the research enterprise through a driving question centering on what we have termed the “grandma hypothesis.” Many of us have loving grandmothers who, with the best of intentions, explain the finer art of personal hygiene. Caring grandmothers often propose to wash specifically between the toes, behind the ears, and in the belly button (navel), as target areas that are often neglected by young children. Thus, we proposed to our PHG class to test the grandma hypothesis, namely, that microbiome composition is different in the “grandma hotspots” of behind the ears, between the toes, and in the belly button, compared with other areas of the body that receive more regular washing attention.
Here we report the results of our pilot educational activity carried out at GWU, including formative and summative assessments throughout the PBL approach (Fig. 1). Our study provides data on the positive impact of PBL on learning outcomes in higher education.
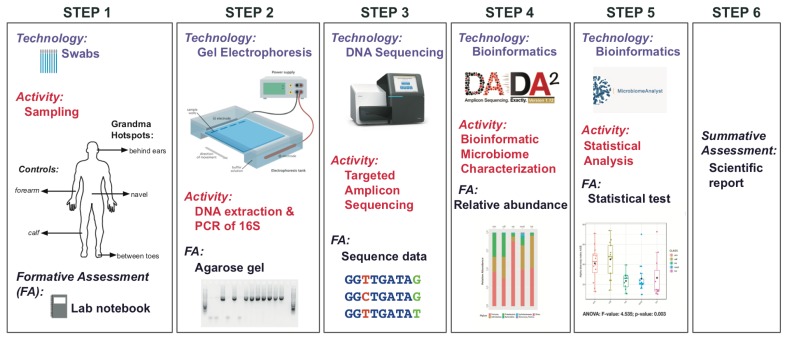
FIGURE 1.
Different sections of our educational project including technologies, activities, and formative assessments.
Intended audience
This curriculum activity is designed for students interested in developing foundational knowledge, skills, and lab experience in the generation, analysis, and applications of genomic data. Students apply and use molecular biology, next-generation sequencing technology, bioinformatics, and statistics. The activity is intended for upper division undergraduate students and/or beginning graduate students pursuing degrees in Public Health, Microbiology, Biology, Bioinformatics, or Biotechnology (or related areas). This lab activity is currently offered as part of a three-credit-hour genomics course to GWU graduate students in the MS Public Health Microbiology and Emerging Infectious Disease program at the Milken Institute School of Public Health. We are currently developing plans to implement the lab activity as part of an undergraduate genomics course offering that will serve students in Biology, Neuroscience, Bioinformatics, Public Health, and Biomedical Engineering and serve as a core course in our minor in Bioinformatics at GWU. This course can be adjusted to small or large student cohorts (e.g., 100 students), since lab protocols and high-throughput amplicon sequencing runs can accommodate multiplexing of hundreds of samples.
Learning time
The grandma hypothesis educational project was conducted in fifteen two-hour laboratory sessions taught once a week (Appendix 1) for a total of 30 hours of hands-on lab experience (including both wet lab and computational activities). The PBL activity is embedded within two three-credit-hour courses also taught weekly at GWU: Introduction to Genomics for undergraduate students and Public Health Genomics for graduate students. Both courses include one-hour lectures on diverse genomics topics covering omic technologies (e.g., genome sequencing, RNA-Seq, etc.) and core genomic concepts (e.g., the Human Genome Project, microbiome, transcriptome, etc.). Students also give class presentations, attend computer demonstrations, and complete problem sets covering diverse genomics topics. Assuming all prerequisite student knowledge is met and students complete the other requirements of the course, this curriculum and PBL activity could be completed in one semester.
Prerequisite student knowledge
Students are required to complete an introductory first-year biology course covering basic principles of molecular biology, evolution, microbiology, and biostatistics. In combination with this activity and as part of the curriculum materials, students review principles of genomics, previous genome projects, molecular techniques, analytical methods, genomic applications, and computational tools for genome research. All applied laboratory methods and bioinformatic and statistical tools are not assumed knowledge and are covered during the project.
Learning objectives
In this PBL experience, students characterize their own skin microbial communities through targeted amplicon sequencing (i.e., metataxonomics). By the end of this activity students are able to:
Acquire and apply hands-on research experience with lab methods of DNA extraction, PCR, and electrophoresis
Work independently to choose and apply appropriate bioinformatic and statistical tools to analyze and integrate microbiome amplicon sequence data and clinical information
Communicate experimental results using text and data visualization formats and compare them with existing literature in a peer-review scientific manner
PROCEDURE
Below we describe the different sections and methodologies of our educational project (Fig. 1). We detail the genomics concepts being implemented in the PBL approach, the scaffolding provided for student learning, and the formative and summative assessments for each section.
Materials
Sampling
We followed the same research procedures used in the Human Microbiome Project (https://hmpdacc.org), summarized in Figure 1 and described in detail in Appendix 2. Specimens were self-collected by the students at the beginning of the experiment using aseptic technique. Each student swabbed five different external skin areas of his/her body corresponding to regions less frequently washed (according to grandma)—or grandma hotspots (behind the ears, between the toes, and on the navel)—and regions more frequently washed—or controls (forearms and calves). All students confirmed that hotspots were less frequently washed than control areas. The null hypothesis is that microbiotas on grandma hotspots will not show different composition and structure from those on control areas. All samples were stored at −80°C until used for DNA extraction.
Molecular methods
Total DNA was extracted using the ZymoBIOMICS DNA MINIPREP KIT and protocol (https://files.zymoresearch.com/pdf/d4300t_d4300_d4304_zymobiomics_dna_miniprep_kit_1-3-0.pdf) from Zymo Research. DNA extractions were then prepared for amplification and sequencing using the Schloss’ MiSeq_WetLab_SOP protocol (https://github.com/SchlossLab/MiSeq_WetLab_SOP) in Kozich et al. (1). We targeted the V4 region (~250 bp) of the 16S rRNA gene, a region commonly used for bacterial characterization of human microbiotas during health and disease (2–4). Students performed PCR amplifications as indicated in Appendix 3. PCR products for each of the five body sites were randomly selected and checked via gel electrophoresis (Appendix 4). Genomic libraries were prepared and sequenced by the GWU Genomics Core (http://www.gwgenomics.org) on a single run of the Illumina MiSeq sequencing platform. Students first received a lecture on these topics, then watched the following instructive videos on next-generation sequencing on YouTube (https://www.youtube.com/watch?v=_yC0Bzw3WbQ&t=71s, https://www.youtube.com/watch?v=fCd6B5HRaZ8, and https://www.youtube.com/watch?v=t0akxx8Dwsk), and finally attended a demonstration by our technician. These educational activities provide scaffolding to enhance student learning outcomes.
Statistical analysis
MiSeq FASTQ sequence files were processed and reads clustered into Amplicon Sequence Variants (ASVs) using the dada2 pipeline (5) as explained in this tutorial https://benjjneb.github.io/dada2/tutorial.html. An ASV table is a higher-resolution analogue of the traditional Operational Taxonomic Units table, which records the number of times each exact ASV was observed in each sample. ASV and taxonomy tables and student metadata were then loaded into MicrobiomeAnalyst (6) for further microbiome analyses. MicrobiomeAnalyst is an open access web-based tool (https://www.microbiomeanalyst.ca/faces/home.xhtml) for comprehensive statistical, visual, and meta-analysis of microbiome data. The MicrobiomeAnalyst website also includes several detailed tutorials and datasets for learning microbiome analyses.
Student instructions
Students characterized the diversity of the bacterial communities living on their skin by extracting, sequencing, and analyzing their microbial DNA (Fig. 1). First they self-collected their own samples, following instructions in Appendix 2, and processed them in the lab, following instructions in Appendices 3 and 4. Each student individually completed most of the core lab procedures of the project (DNA extraction, 16S rRNA PCR amplification, and gel electrophoresis) and witnessed others (library preparation and high-throughput sequencing) with assistance from the instructor and the GWU Genomics Core. Once the lab procedures were completed and the 16S rRNA sequences generated, students analyzed the new data on their own laptops using dada2 and MicrobiomeAnalyst. All the computational tools and statistical methods used in this PBL exercise were also explained and demonstrated by the instructor in class. Students also used these tools to analyze datasets available online from the website developers and those provided by the instructors in the two problem sets (Appendix 5). Students were encouraged to compare and discuss their individual results with their peers (group work). Finally, students presented their results in the format of an individual scientific report as described in Appendix 6 (summative assessment).
Faculty instructions
Instructors first discussed the conceptual underpinnings of each procedure, demonstrated them if needed according to the instructions above, and supervised students performing the same task. Instructors must have a basic understanding of the R language to run the dada2 pipeline and have expertise in microbiome analyses to demonstrate MicrobiomeAnalyst. In their MicrobiomeAnalyst demonstrations, instructors need to cover data normalization and visual exploration, community profiling, clustering and correlation, and univariate analyses. All these topics are explained in detail in the MicrobiomeAnalyst tutorials. Instructors collected student information (metadata) and de-identified student names.
Suggestions for determining student learning
An essential component of authentic research is the communication of scientific findings in a format consistent with professional scientific standards. Accordingly, the main assessment task for the grandma hypothesis microbiome project revolved around an individual peer-reviewed report following the structural conventions of a scientific publication. Therefore, once students completed their metataxonomic analyses, they presented their results in the form of an individual scientific report according to the directions given by the instructor (Appendix 6). This provided familiarity and practice with scientific writing and independent search of the literature. A similar assessment approach has been used in previous microbiome research experiences similar to ours (7). The requested report was five pages long: the first two pages included the title, background (including aims and hypothesis), methods, results, discussion, and conclusions; the next two pages included tables and figures, and the last page was for references. The integration of this assessment task with the learning activities in this project directly aligns with the three learning objectives above.
The marking rubric for the scientific report (see Appendix 5) spanned the following criteria: title originality, knowledge of background, effective introduction of the project aims and hypotheses, clear presentation of the results, explanation, interpretation, and critical evaluation of trends according to own and previous results, text and graphical summary of the results in Tables and Figures, and the importance of selected references. Students turned in a first draft of the scientific report by Session 14 (Appendix 1) to gain 10 points and the final peer-reviewed version within two weeks of completing Session 15 to gain another 30 points (total credit = 40 points).
Additionally, students also completed two problem sets of ten questions each using the same bioinformatic and statistical tools applied in this educational experience but using data examples available at the MicrobiomeAnalyst website. These same problem sets were given to graduate students enrolled in the 2018 Public Health Genomics course. The 2018 PHG students, however, did not have the opportunity to carry out the project-based learning experience described here.
After each class session, students were asked to turn in “minute-papers,” where they could list concepts or topics that they found difficult to understand that day (formative assessment and student reflection on learning). A total of 12 minute-papers per student were turned in throughout the semester.
Sample data
Examples of student results are presented in Figures 2 and 3 and Appendix 4. Figure plots summarize alpha- and beta-diversity results of the skin microbiomes across the entire student cohort. Students were expected to interpret gel electrophoresis results (Appendix 4) to determine the quality of DNA extractions and outcome of the PCR on skin samples. They also estimated relative proportions of microbes across samples (see microbial profiles below).
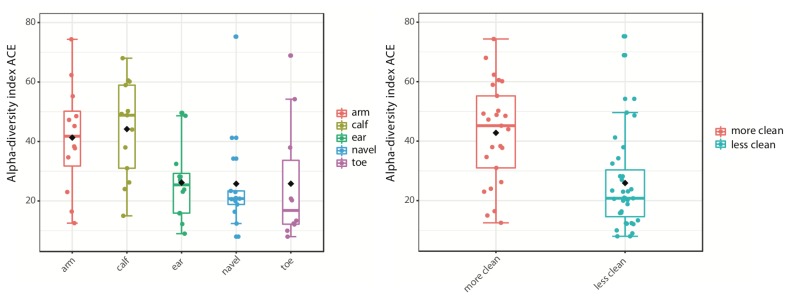
FIGURE 2.
Abundance-based coverage estimator of alpha-diversity across skin locations alone and combined. Less clean: behind the ears, between the toes, and in the navel; more clean: forearms and calves.
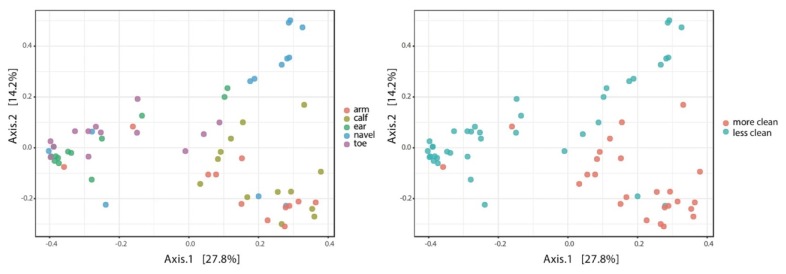
FIGURE 3.
PCoA analysis of Bray-Curtis distances across skin locations alone and combined. Less clean: behind the ears, between the toes, and on the navel; more clean: forearms and calves.
Safety issues
This curriculum experience can be run safely in any BSL2 lab designed to conduct routine molecular techniques. Institutions lacking in-house sequencing capacity could ship the DNA extractions or library products to other academic institutions or biotech companies. All laboratory procedures and practices outlined here adhere to the ASM Guidelines for Biosafety in Teaching Laboratories. Students are required to maintain aseptic technique and wear lab coats, goggles, and gloves during experimentations. Students are also instructed on how to handle human swabs (see Appendix 2), gel electrophoresis safety (e.g., handling of hazardous chemicals), and the operation of electrophoresis equipment.
DISCUSSION
In this new pedagogic and research experience, students had the opportunity to characterize their own skin microbial communities through targeted amplicon sequencing (metataxonomics) approaches (8, 9). This high-throughput sequencing approach has greatly advanced the field of microbial ecology in the past decade (10). We thought this type of experiential learning (11) would give students a more comprehensive view of the field of genomics through a PBL approach (12).
Field testing
We have developed an educational opportunity for graduate and undergraduate students to learn and directly participate in applied authentic genomic research. As we described before, this PBL opportunity was offered for the first time in 2019 to graduate students enrolled in the three-credit-hour Public Health Genomics (PHG) course. A similar two-credit-hour course was offered in 2018, but it did not include the lab component.
Written consent (via e-mail) was obtained from all participants using the GWU IRB-approved informed consent documents (IRB# 180703). This educational project complies with all relevant federal guidelines and institutional policies. All samples and resulting data were de-identified.
In this PBL activity, students tested the following overarching hypothesis (i.e., the grandma hypothesis): skin sites we wash less frequently (behind the ears, between the toes, and in the navel) have different microbial composition and structure than those we wash more often (forearms and calves). A similar pedagogic experience was suggested by Gibbens et al. (13) as a possible modification to their Biology Course to study microbes from environmental samples using metagenomics.
To test the grandma hypothesis, ten students and three extra volunteers successfully self-swabbed their skin, extracted total DNA and performed a PCR amplification of the bacterial 16S rRNA V4 gene region (250 bp). The GWU Genomics Core then sequenced all of the 65 samples plus two negative controls (no DNA) and one positive control (mock community) in their Illumina MiSeq platform. One sample gave low yields (431 sequence reads) compared with the other samples (> 1,500 reads) and was discarded in further analyses. The sequence data generated from this project has been deposited in GenBank under SRA submission PRJNA553551.
Evidence of student learning
All PHG-2019 students completed the lab procedures, analyzed the metataxonomic data, and wrote a five-page scientific report summarizing and discussing their results (summative assessment). They also completed two problem sets using the MicrobiomeAnalyst platform, the same bioinformatic tool used in the report (formative assessments), and filled out 12 minute-papers (see above) related to genomic topics covered in class.
Student performance in the individual scientific report is the primary indicator of learning gains resulting from the grandma hypothesis microbiome project. The scientific report accounted for 40 points, and student grades ranked from 31.5 to 40 (mean = 34.1). No student failed to complete the report. All students correctly analyzed the genomic data using new bioinformatic and statistical tools, created data tables and data figures, addressed the basic research questions, and presented their results in the form of a succinct and intelligible written report. Although all the students have to address the same research questions using the same genomic and clinical data and bioinformatic tools, they had the freedom to choose among the plethora of statistical tests available in the MicrobiomeAnalyst platform.
Unanimously, student reports showed that sites cleaned more frequently (calves and forearms) have significantly (ANOVA F-value > 3.42; p < 0.014) higher alpha-diversity (intra-sample) than sites we clean less frequently (behind the ears, between the toes, and in the navel)—see Figure 2. Based on this and similar analyses, students concluded that there is significant variation in both richness and evenness among the sampled skin microbiomes. Student analyses also showed that sites we clean more frequently differ significantly (PERMANOVA F-value > 6.017; p < 0.001) in beta-diversity (inter-sample) from those we potentially clean less frequently (see Fig. 3). Finally, students also found significant differences in the proportions of specific taxa (i.e., biomarkers) between more and less frequently washed regions. For example, two phyla (Proteobacteria and Firmicutes) and ten genera (Escherichia/Shigella, Lactobacillus, Bacillus, Streptococcus, Micrococcus, Pseudomonas, Lawsonella, Acinetobacter, Enhydrobacter, and Staphylococcus) varied significantly (Log LDA score > 1; p < 0.05) in their relative mean proportions between groups. These results are highly interesting and seem to suggest that cleaning behavior may have an impact on the diversity of skin microbiotas. The grandma hotspots (behind the ears, between the toes, and in the navel) seem to host distinct microbial communities compared with the control areas (forearms and calves). Future research will confirm whether our washing habits (as grandma suggested) and/or other factors (e.g., skin exposure) are driving the diversity of our skin microbiota.
PHG-2019 graduate students also completed two problem sets related to microbiome research using genomic data and bioinformatic tools. These two problem sets were also given to students in 2018 (PHG-2018), who did not have the chance to participate in the grandma hypothesis microbiome project. A comparison of their grades (Table 1) shows that PHG-2019 students outperformed PHG-2018 students, as indicated by their significantly higher mean grades in both problem sets. Grade means for PS4 and PS5 increased by a factor of 1.12 and 1.14 in 2019, respectively.
TABLE 1.
Statistical comparison of grades from students who participated in the grandma hypothesis microbiome project (2019) and students who did not (2018).
| Student* | PHG2018 | PHG2019 | PHG2018 | PHG2019 |
|---|---|---|---|---|
| PS4 | PS4 | PS5 | PS5 | |
| Mean | 7.64 | 8.55 | 7.59 | 8.65 |
| Median | 7.5 | 8.75 | 7.5 | 8.75 |
| SD | 0.92 | 1.01 | 1.04 | 0.92 |
| MW-test | 82 (p=0.050) | 85 (p=0.036) | ||
| T-test | 2.16 (p=0.040) | 2.40 (p=0.027) |
The grades are for two problem sets given in the PHG course and focused on the analysis of metataxonomic data using bioinformatic tools.
PHG = Public Health Genomics; PS = problem set; MW = Mann-Whitney U test; SD = standard deviation.
Additionally, we also noticed that PHG-2019 students showed greater confidence in interpreting and presenting genomic results in class and a greater appreciation for research after completing their lab experience, despite having very limited a priori knowledge of the topics covered in this course. This was also confirmed by personal written feedback (i.e., 12 minute-papers) from the students. When 2018 and 2019 PHG students were asked at the end of each class, “What was the most difficult or unclear topic for you during class?,” a total of 82 (62.1%) out of 132 responses (11 students x 12 minute-papers) in 2018 included a genomic topic that was not clear, while only 35 (29.2%) out of 120 responses (10 students x 12 minute papers) in 2019 included a genomic topic. This seems to indicate that our project-based learning activity encouraged active participation in the learning process and helped students better understand key concepts in genomics. We think this is likely the result of a greater engagement of students in the lab, due to the excitement and relevance of studying their own skin microbial communities, which may carry over and keep students motivated to study and perform well throughout the course. Similar results have been seen in other inquiry-based and research-based laboratory projects in the fields of chemistry, biochemistry, and microbiology (7, 14–16). These results were also supported by school post-course anonymous evaluation surveys, in which PHG-2019 students gave more positive responses about the course than PHG-2018 students. For example, in response to the question about “How much learned?,” PHG-2018 student responses averaged a 4.1 score on a scale of 1 to 5, while PHG-2019 student responses averaged a 4.8 score.
In conclusion, the above results combined suggest that after participating in this authentic genomics research experience and successfully completing all the learning objectives, students better understood the complexity of genomics and the multiple steps of a genomic project going from bench to public health applications. They also indicate that the students performed significantly better on formative assessments, had less confusion about core topics, and had an overall better learning experience through the incorporation of project-based learning. Additionally, we also feel that by completing this pedagogic exercise, students acquired a valuable set of marketable tools and skills in genomics, bioinformatics, and statistics (quantitative skills). They also addressed interesting questions in microbiome research (inquiry-based learning) and generated valuable genomic data and new insights about the diversity of the microbial communities living on our skin.
Possible modifications
We recommend including field professionals in the audience during student presentations of individual short scientific reports, which would further strengthen the project.
Our targeted 16S rRNA amplicon approach can be applied to other areas of human skin or even to environmental samples. Additionally, extensive de-identified raw data to run the activity in silico is accessible on the HMP (https://hmpdacc.org) and NCBI-SRA (https://www.ncbi.nlm.nih.gov/sra) websites. Amplicon sequencing is limited in that it only surveys known bacterial taxa and is subject to the vagaries of PCR. Alternative more powerful methods such as shotgun metagenomics can survey the entire microbiome, including viruses, fungi, and bacteria, simultaneously. Fungal components might be particularly important in locations such as between the toes and yeast components behind the ears.
SUPPLEMENTAL MATERIALS
ACKNOWLEDGMENTS
We dedicate this work to our grandmothers, who inspire and educate us on personal hygiene and a wide variety of other topics. We thank Hayley DeHart and Bryan Nguyen for their valuable help with this project. We also thank Peter LaPuma for his mentorship and Manya Magnus for her help with the IRB application. We thank the thoughtful and supportive reviewers and editor for helping improve our manuscript. This project was partially supported by a Fellowship Award from the GWU Milken Institute School of Public Health’s Academy of Master Teachers and grants from the Milken Institute School of Public Health Pilot Fund Program, the Margaret Q. Landenberger Research Foundation, Award Number UL1TR001876 from the NIH National Center for Advancing Translational Sciences, and the Fundação para a Ciência e a Tecnologia (T495756868-00032862). The authors declare that there are no conflicts of interest.
Footnotes
Supplemental materials available at http://asmscience.org/jmbe
REFERENCES
- 1.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhai W, Huang Y, Zhang X, Fei W, Chang Y, Cheng S, Zhou Y, Gao J, Tang X, Zhang X, Yang S. Profile of the skin microbiota in a healthy Chinese population. J Dermatol. 2018;45:1289–1300. doi: 10.1111/1346-8138.14594. [DOI] [PubMed] [Google Scholar]
- 3.Ross AA, Muller KM, Weese JS, Neufeld JD. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc Natl Acad Sci U S A. 2018;115:E5786–E5795. doi: 10.1073/pnas.1801302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardiner M, Vicaretti M, Sparks J, Bansal S, Bush S, Liu M, Darling A, Harry E, Burke CM. A longitudinal study of the diabetic skin and wound microbiome. Peer J. 2017;5:e3543. doi: 10.7717/peerj.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JTH, Daly JN, Willner DL, Patil J, Hall RA, Schembri MA, Tyson GW, Hugenholtz P. Do you kiss your mother with that mouth? An authentic large-scale undergraduate research experience in mapping the human oral microbiome. J Microbiol Biol Educ. 2015;16:50–60. doi: 10.1128/jmbe.v16i1.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilton SK, Castro-Nallar E, Perez-Losada M, Toma I, McCaffrey TA, Hoffman EP, Siegel MO, Simon GL, Johnson WE, Crandall KA. Metataxonomic and metagenomic approaches vs. culture-based techniques for clinical pathology. Front Microbiol. 2016;7:484. doi: 10.3389/fmicb.2016.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boers SA, van der Reijden WA, Jansen R. High-throughput multilocus sequence typing: bringing molecular typing to the next level. PLOS One. 2012;7:e39630. doi: 10.1371/journal.pone.0039630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolb AY, Kolb DA. Learning styles and learning spaces: enhancing experiential learning in higher education. Acad Manage Learn Educ. 2005;4:193–212. doi: 10.5465/amle.2005.17268566. [DOI] [Google Scholar]
- 12.Bell S. Project-based learning for the 21st century: skills for the future. Clearing House. 2010;83:39–43. doi: 10.1080/00098650903505415. [DOI] [Google Scholar]
- 13.Gibbens BB, Scott CL, Hoff CD, Schottel JL. Exploring metagenomics in the laboratory of an introductory biology course. J Microbiol Biol Educ. 2015;16:34–40. doi: 10.1128/jmbe.v16i1.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanauer DI, Jacobs-Sera D, Pedulla ML, Cresawn SG, Hendrix RW, Hatfull GF. Inquiry learning. Teaching scientific inquiry. Science. 2006;314:1880–1881. doi: 10.1126/science.1136796. [DOI] [PubMed] [Google Scholar]
- 15.Weaver GC, Russell CB, Wink DJ. Inquiry-based and research-based laboratory pedagogies in undergraduate science. Nat Chem Biol. 2008;4:577–580. doi: 10.1038/nchembio1008-577. [DOI] [PubMed] [Google Scholar]
- 16.Wang JTH, Schembri MA, Ramakrishna M, Sagulenko E, Fuerst JA. Immersing undergraduate students in the research experience. Biochem Mol Biol Educ. 2012;40:37–45. doi: 10.1002/bmb.20572. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.