Abstract
BACKGROUND AND PURPOSE:
Our aim was to use 2D convolutional neural networks for automatic segmentation of the spinal cord and traumatic contusion injury from axial T2-weighted MR imaging in a cohort of patients with acute spinal cord injury.
MATERIALS AND METHODS:
Forty-seven patients who underwent 3T MR imaging within 24 hours of spinal cord injury were included. We developed an image-analysis pipeline integrating 2D convolutional neural networks for whole spinal cord and intramedullary spinal cord lesion segmentation. Linear mixed modeling was used to compare test segmentation results between our spinal cord injury convolutional neural network (Brain and Spinal Cord Injury Center segmentation) and current state-of-the-art methods. Volumes of segmented lesions were then used in a linear regression analysis to determine associations with motor scores.
RESULTS:
Compared with manual labeling, the average test set Dice coefficient for the Brain and Spinal Cord Injury Center segmentation model was 0.93 for spinal cord segmentation versus 0.80 for PropSeg and 0.90 for DeepSeg (both components of the Spinal Cord Toolbox). Linear mixed modeling showed a significant difference between Brain and Spinal Cord Injury Center segmentation compared with PropSeg (P < .001) and DeepSeg (P < .05). Brain and Spinal Cord Injury Center segmentation showed significantly better adaptability to damaged areas compared with PropSeg (P < .001) and DeepSeg (P < .02). The contusion injury volumes based on automated segmentation were significantly associated with motor scores at admission (P = .002) and discharge (P = .009).
CONCLUSIONS:
Brain and Spinal Cord Injury Center segmentation of the spinal cord compares favorably with available segmentation tools in a population with acute spinal cord injury. Volumes of injury derived from automated lesion segmentation with Brain and Spinal Cord Injury Center segmentation correlate with measures of motor impairment in the acute phase. Targeted convolutional neural network training in acute spinal cord injury enhances algorithm performance for this patient population and provides clinically relevant metrics of cord injury.
The natural history of recovery after spinal cord injury (SCI) is highly variable and often difficult to predict.1,2 Early objective neurologic assessment of injury severity in these patients is challenging due to several confounding factors, such as traumatic brain injury, severe pain, intubation, spinal shock, and sedative medications, among others.2,3 Early and accurate classification of patients with SCI as close to the time of injury as possible is imperative for the guidance of triage, acute management, prognostication, and for patient selection in the design of clinical trials in which novel therapies are tested.2,3 MR imaging is the criterion standard imaging technique for evaluation of the injured spinal cord (SC) and has been extensively studied for its ability to assess injury severity and predict outcome.2,4–6 However, to date, there are mixed results with respect to the accuracy of MR imaging biomarkers for injury prognostication.4,5,7–9
Recent advances in SC imaging analysis have led to the development of a robust anatomic template and atlas incorporated into an open-source platform referred to as the Spinal Cord Toolbox (SCT; https://sourceforge.net/projects/spinalcordtoolbox/).10,11 Similar tools have greatly advanced brain image analysis in recent years, offering accurate and less biased methods for analyzing quantitative and semiquantitative multiparametric MR imaging data.12,13 Such tools applied to the SC offer great potential for a unique MR imaging biomarker identification.11,14,15 SC segmentation is a first step in atlas-based SC analysis.16 Manual SC segmentation techniques are not conducive to clinical workflow or high-volume batch analysis because they are time-consuming and have interrater variability. Advances in automated SC segmentation algorithms have been made during the past decade, though currently available algorithms have primarily been tested in healthy controls or nontraumatic pathologies.16–18 The latest generation segmentation algorithms in acute SCI are challenged by coexisting spinal pathology such as canal stenosis, SC compression, and intrinsic SC signal abnormalities, leading to gross segmentation errors. A model specifically targeted to deal with the challenges of the acute blunt SCI population is needed for application of advanced MR imaging analysis tools in traumatic SCI.
Leveraging a robust, prospectively maintained clinical and radiologic data base of patients with acute blunt traumatic SCI as part of the ongoing prospective clinical trial entitled Transforming Research and Clinical Knowledge in Spinal Cord Injury (TRACK-SCI), we present 3 models using 2D convolutional neural networks (CNNs) to segment the SC from T2-weighted axial images of patients with acute SCI. These networks, named Brain and Spinal Cord Injury Center segmentation (BASICseg1–3), are fully automated and use axial image augmentation and a network architecture. Furthermore, we present preliminary data using the same network architecture to automatically segment intramedullary contusion in patients with acute SCI on T2WI. To assess the clinical validity of injury volumes segmented by this network, we correlated the contusion volumes segmented by BASICseg with lower extremity motor scores. Targeted 2D convolutional neural network training with a cohort of patients with acute SCI, in whom whole-cord and contusion-injury segmentation is particularly challenging, enhances the performance of automated spinal cord and contusion injury segmentation for this patient population compared with currently available state-of-the-art algorithms and also provides clinically relevant metrics of cord injury.
Materials and Methods
Study Population
The study population included all consenting patients admitted for an acute traumatic SCI at Zuckerberg San Francisco General Hospital between June 2015 and January 2017. This study was in compliance with the Health Insurance Portability and Accountability Act and was approved by the institutional review board for human research. The medical imaging and clinical records included in this study are compiled as part of the TRACK-SCI research trial conducted under the auspices of the Brain and Spinal Injury Center at Zuckerberg San Francisco General Hospital. This prospective study consecutively enrolled all consenting patients with acute traumatic spinal cord injury at Zuckerberg San Francisco General Hospital between June 2015 and January 2017, with the collection of imaging, acute neurologic examinations, intraoperative and intensive care unit monitoring, and long-term neurologic and functional outcome assessment. Inclusion criteria were the following: 1) blunt acute cervical or thoracic SCI, 2) 18 years of age or older, 3) presurgical cervical or thoracic spine MR imaging performed within 24 hours after injury, and 4) documented motor score assessment and American Spinal Injury Association Impairment Scale score obtained during first hospitalization. Exclusion criteria were the following: 1) penetrating SCI, 2) surgical decompression and/or fusion before MR imaging, 3) MR imaging that was too degraded by motion or other artifacts so that images were nondiagnostic, 4) pre-existing surgical hardware, and 5) incomplete clinical data for the outcome (American Spinal Injury Association Impairment Scale score) or relevant confounders. We identified 56 patients, of whom 47 met all inclusion criteria and were enrolled in the study. More detailed patient demographics and injury characteristics are summarized in Table 1. Penetrating injuries were excluded due to their unique clinical, imaging, and pathophysiologic characteristics.
Table 1:
Summary of patient demographics and injury
| Patient Demographics and Injury Characteristics (N = 47) | |
|---|---|
| Age (mean) (SD) (yr) | 55 (19) |
| Sex (M/F) | 32:15 |
| Time from injury to MRI (mean) (SD) (hr) | 5.4 (4.8) |
| Admission ASIA Grade | |
| A | 10 |
| B | 3 |
| C | 5 |
| D | 21 |
| Unknowna | 8 |
| Admission lower extremity motor score (mean) (SD) | 25 (23) |
| Discharge lower extremity motor score (mean) (SD) | 29 (21) |
Note:—ASIA indicates American Spinal Injury Association.
We could not perform full formal ASIA grade assessment at admission.
MR Imaging Acquisition Parameters
All MR imaging studies were performed on a single 3T Magnetom Skyra scanner with software Version E11 (Siemens, Erlangen, Germany). Only the axial and sagittal T2 fast spin-echo sequences from the routine cervical spine trauma protocol were used for image analysis and were performed with the following parameters: axial T2; TR = 3870 ± 400 ms, TE = 96 ± 4 ms, slice thickness = 3 mm, echo-train length = 16, FOV = 240 × 120 mm, nominal in-plane pixel size = 0.47 mm2. Additional sequences performed as part of the routine clinical spine MR imaging protocol were not evaluated for this study.
Image Annotation
Images from patients with SCI were divided into training,19 validation,5 and testing14 groups for analysis. Because our model is a 2D neural network, 1120 axial slices were used in the training set, 200 axial slices were used in the validation set, and 560 axial slices were used in the test set. Two fellowship-trained neuroradiologists (J.F.T. and J.N.) performed image annotation independently and then reached consensus together on manual segmentation in FSL (http://www.fmrib.ox.ac.uk/fsl) for both the whole SC and areas of intramedullary T2 signal abnormality related to the cord contusion referred to as the SC lesion. Sagittal T2WI was used only to cross-validate axial annotations.
Image Preprocessing
We used the Optic algorithm18 from the SCT to first detect the SC centerline and create a 26 × 26 mm square mask around the centerline. No denoising, smoothing, or inhomogeneity or bias correction was applied. All raw images were resampled at 1 × 1 mm2 to be robust to native image resolution. This mask dimension was sufficient to include the entire spinal cord and spinal canal within the FOV for optimal segmentation. The raw images were then cropped using this mask. Images were then resampled to have a pixel size of 0.2 mm2 in the axial plane, with no changes made to slice thickness. Because all images had to be of the same dimensions before entering the network for batch-wise processing, this resampling and mask cropping resulted in an image matrix size of 128 × 128 voxels.
BASICseg-1 Network Architecture
On-line Fig 1 graphically depicts the deep learning architectures for BASICseg-1 and BASICseg-2. These network architectures were used for both whole SC and intramedullary lesion segmentation and are based on the U-net architecture (https://lmb.informatik.uni-freiburg.de/people/ronneber/u-net/).20 The U-net architecture for segmentation consists of contracting and expansive paths. The contracting path follows the standard CNN architecture wherein the image is processed into a series of feature maps. This pathway consists of 4 layers, each involving the following: 1) 3 × 3 zero-padded convolutions followed by a rectified linear unit,21 2) 20% drop-out22 of the previous convolution, 3) 3 × 3 zero-padded convolutions followed by a rectified linear unit, and 4) 2 × 2 max pooling operation (https://www.quora.com/What-is-max-pooling-in-convolutional-neural-networks) with Stride 2 for downsampling. At each iteration of the contracting layer, the number of feature maps was doubled (first layer: 8 features; second layer: 16 features; third layer: 32 features; fourth layer: 64 features). The fifth layer consisted of the same processes without drop-out (aforementioned processes 1 and 3) creating 128 feature channels. The expansive path consists of 4 layers to rebuild a mask image to match the dimensions of the input image. Upsampling and concatenation processes were followed by two 3 × 3 upconvolutions using zero-padding, which halves the number of feature channels each followed by rectified linear unit. A 1 × 1 convolution with sigmoid activation was used in the final layer to map each 8-component feature vector as either SC tissue (value set to 1) or not SC tissue (value set to 0). In total, the network comprises 19 convolutional layers. Stochastic gradient descent was used with a learning rate of 1e-5 with an Adam optimizer.23 All hyperparameters were the same for each of the BASICseg architectures. Programming was performed in Python with the TensorFlow framework (https://www.tensorflow.org/). Training was performed on 2 GeForce-1080ti GPUs (NVIDIA, Santa Clara, California) with a combined random-access memory of 22G.
BASICseg-2 Network Architecture
To complement the first network architecture, we created a second architecture to study the impact of integrating batch normalization24 between every convolution and rectified linear unit (https://www.kaggle.com/dansbecker/rectified-linear-units-relu-in-deep-learning activation) layer as shown in On-line Fig 1. Batch normalization involves subtracting each value of the feature map from the feature map mean and dividing by the feature map SD, or
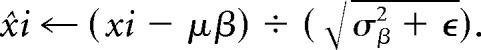 |
This process helps prevent interval covariate shift. With batch normalization, the distribution of each feature map is the same before activation, which leads to better performance and faster convergence.
We removed the drop-out of the feature maps from the contracting path, replaced them with batch normalization layers, and added them in the expanding path. Additionally, we doubled the number of feature maps created at each layer. All other elements of the architecture remained the same. This network was applied to both spine- and lesion-segmentation tasks.
BASICseg-3 Network Architecture and Noise-Adaption Layer
We expected that in areas of SC compression, where there is notable damage and substantially less signal delineation between the SC tissue and adjacent extramedullary tissues, the labeling mask will be imperfect and therefore prone to label noise. To address potentially erroneous labels, we introduced an additional “noise-adaptation layer” into the network between the final feature maps per pixel (in this case, a tensor of 128 × 128 × 16) and the 1 × 1 convolution-to-sigmoid activation layer (On-line Fig 2).25 Thus, each pixel feature map (1 × 16) is augmented by a 16 × 16 weight matrix. This weight matrix can be thought of as an additional Softmax layer (https://stats.stackexchange.com/questions/79454/softmax-layer-in-a-neural-network), which modifies the feature vectors for each pixel before compression to 128 × 128 × 1 and calculation of the loss. The weights of this noise layer are estimated as part of the training process to reduce the overall loss of the system. This process is illustrated in On-line Fig 2.
Data Augmentation
To avoid overfitting, we implemented data augmentation of the images using the Keras framework (https://keras.io/). We also augmented T2WI and the corresponding masks pair-wise in batches of 32. Additionally, data are augmented by the drop-out after each convolution in the contracting path by randomly removing feature channels during training.
Model Performance and Statistical Analysis
Spine-segmentation and lesion-segmentation performances were evaluated using the Dice coefficient.26,27 The Dice coefficient is a measure of overlapping defined as
Where T is the criterion standard segmentation performed by a fellowship-trained neuroradiologist and P is the predicted segmentation of the SC. The proposed segmentation method (BASICseg) was compared with 2 state-of-the-art methods: Propseg (https://sourceforge.net/p/spinalcordtoolbox/wiki/sct_propseg/)16 and Deepseg,28 both included in the SCT.11 Deepseg (sct_deepseg_sc module in SCT; https://sourceforge.net/p/spinalcordtoolbox/discussion/help/thread/c9972de78a/) is a recently described deep-learning-based spinal cord segmentation using a succession of 2 CNNs, the first one to detect the SC and the second one to perform the segmentation.28
Dice coefficient results for the 3 evaluated methods (BASICseg, Propseg, and Deepseg) were compared using linear mixed modeling. This initial analysis included all axial images from the test set (including both injured and normal-appearing SC on the T2WI). We restricted the analysis to include injured axial slices only and used the same linear mixed modeling to evaluate differences in segmentation quality based on the Dice coefficient among the evaluated segmentation methods. To determine whether there were relative differences in the models for damaged slice-versus-nondamaged slice segmentation, we used a linear mixed model with an interaction term for model by lesion.
Simple linear regressions were conducted to determine the association between volumes of lesions segmented by the BASICseg and with lower extremity motor scores at day 0 (initial injury) and at a subacute period at the time of patient discharge from the hospital. Data that did not show linearity were appropriately transformed, and R2 was compared with linear regression; if there was no improvement, a Spearman correlation was used. All statistical analyses were performed in R statistical and computing software (http://www.r-project.org/)19; the significance threshold was α < .05. Because our cohort of patients had injuries at a variety of cervical and upper thoracic levels (Table 1), which confounds evaluation of upper extremity motor scores, outcome correlations with injury volume were focused on lower extremity motor scores, which are less likely to be confounded by the level of injury for cervical and upper thoracic spinal cord injuries.
Model Comparison
The 3 models (BASICseg-1 using drop-out, BASICseg-2 using batch normalization, BASICseg-3 using batch normalization and a noise-adaptation layer) were compared for their segmentation performance in the test set data. Models were selected on the basis of the highest validation accuracy, not total completion of epochs; these models were applied to the test sets. For SC segmentation, the model with the highest Dice coefficient was used in statistical analysis to compare with Propseg16 and Deepseg.28 Propseg is based on iterative propagation of deformable models for SC segmentation, while Deepseg uses a machine learning–based automatic SC segmentation approach with convolutional neural networks. For lesion segmentation, the results for the model with the best performance were modeled with lower extremity motor scores.
Results
Comparison of BASICseg with Existing Models for Traumatic SC Segmentation
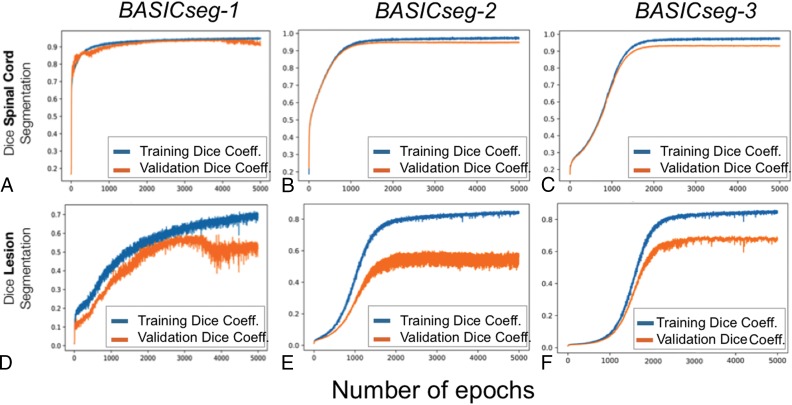
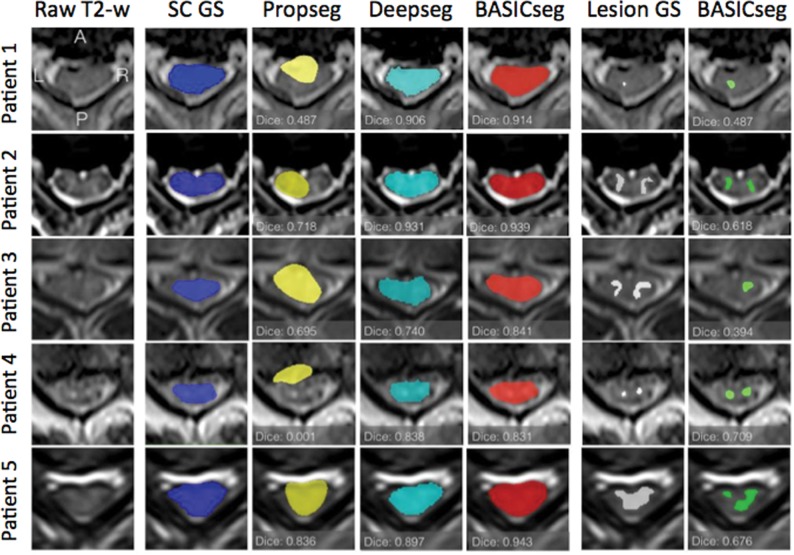
The average SC segmentation time for each test patient was 5 seconds. The whole SC Dice coefficient by epoch for each model is plotted and shown in Fig 1A–C. For SC segmentation, BASICseg-1 performed best with a Dice coefficient of 0.93 in the test set versus 0.91 for BASICseg-2 and 0.90 for BASICseg-3. The SC segmentation output at a representative injury level is shown in Fig 2 for 1) the criterion standard manual segmentation, 2) the BASICseg-1 model, 3) Propseg segmentation, and 4) Deepseg segmentation in 5 different subjects from the test set cohort with respective Dice coefficients for each slice. Furthermore, the mean dice coefficients across patients for each segmentation model with corresponding 95% confidence intervals overall and stratified by injury subgroup are presented in On-line Table 1. Linear mixed-effects modeling of Dice coefficients in the test set for segmentation across all axial slices (both normal-appearing and lesion levels on axial T2WI) showed a significantly higher Dice coefficient for BASICseg-1 compared with Propseg (estimate difference = 0.12, P < .001) and borderline significance compared with Deepseg (estimate difference = 0.03, P < .056).
Fig 1.
Whole spinal cord and lesion segmentation performance by epoch for BASICseg algorithms for the whole spinal cord (A–C) and the lesion (D–F).
Fig 2.
Sample segmentation outputs for the criterion standard manual segmentation and each model for whole spinal cord (columns 2–5) and lesions (columns 6 and 7) in 5 sample patients. The first column contains the sample axial T2WI on which segmentation was performed.
When analysis of the SC segmentation was restricted to slices of SC lesions on axial T2WI, the BASICseg-1 had significantly higher Dice coefficients compared with Propseg (difference = 0.220, P < .001) and Deepseg (difference = 0.102, P = .020; On-line Fig 3). In undamaged regions of the cord, no statistically significant difference in segmentation performance was found between BASICseg-1 and Deepseg (difference = 0.013, P = .43), but there was a significant difference compared with Propseg (difference = 0.0953, P < .001; On-line Fig 3). The relative variance in segmentation performance in areas of damage versus no damage among the models showed a significant difference (Deepseg: estimate = 0.087, P < .026; Propseg: estimate = 0.122, P < .001). Violin plots illustrate the Dice distribution by model and by lesion in On-line Fig 3. Table 2 shows the results from linear mixed modeling.
Table 2:
Results from linear mixed modeling
| Model | Estimate | SE | df | T Value | P Value |
|---|---|---|---|---|---|
| Overall spine segmentation | |||||
| BASICseg-1 vs Propseg | −0.12166 | 0.01765 | 484 | −6.893 | <.001 |
| BASICseg-1 vs Deepseg | −0.03382 | 0.01765 | 484 | −1.916 | .056 |
| Spine segmentation at levels of lesion | |||||
| BASICseg-1 vs Propseg | −0.22028 | 0.04410 | 102.62 | −4.995 | <.001 |
| BASICseg-1 vs Deepseg | −0.10254 | 0.04347 | 102.62 | −2.359 | .020 |
| Spine segmentation at normal-appearing level | |||||
| BASICseg-1 vs Propseg | −0.09525 | 0.01687 | 367.5 | −5.645 | <.001 |
| BASICseg-1 vs Deepseg | −0.01338 | 0.01694 | 367.5 | −0.790 | .43 |
| Relative difference in model segmentation between lesion and normal-appearing levels | |||||
| Lesion category difference in Propseg compared with BASICseg-1 | −0.12249 | 0.03928 | 484.4 | −3.119 | <.001 |
| Lesion category difference in Deepseg compared with BASICseg-1 | −0.08689 | 0.03889 | 484.4 | −2.234 | .026 |
Note:—SE indicates standard error.
Intramedullary Lesion Segmentation and Correlation with Motor Impairment
BASICseg-3 demonstrated the best lesion segmentation performance as shown in Fig 1D–F. All models showed overfitting of the training set. Univariate analysis (On-line Table 2) of volumes of intramedullary injury segmented by BASICseg-3 showed significant association with day 0 lower extremity motor scores (estimate = −4.583, P = .002), indicating that for each increase in lower extremity motor score, the volume of damage decreases by approximately 6 mm3. Similarly, BASICseg-3 segmented volumes showed a significant association with lower extremity motor scores at the time of patient discharge: estimate = −4.030, P = .009. Additionally, application of BASICseg-3 to segment lesions from the test set images took approximately 3 seconds per patient.
Discussion
In the present study, we show results for automated whole-SC and traumatic-SC lesion segmentation from axial T2WI performed acutely after blunt SCI. Specifically, we applied a customized image-analysis and processing pipeline integrating 3 different novel 2D-CNN architectures for both whole-SC and traumatic lesion segmentation. Segmentation results from these CNNs were compared with each other and with the criterion standard manual segmentation as well as with current state-of-the-art SC segmentation algorithms.
The BASICseg-1 model, which uses drop-out, showed slightly better performance than models using batch normalization (BASICseg-2) and batch normalization with a noise-adaptation layer (BASICseg-3) for automated segmentation of the SC in this cohort. Because drop-out randomly sets a certain proportion of network weights to zero during training and scales the weight activations by proportion during testing, it may be that better testing segmentation occurs with drop-out because of more distributed learning of the features that map to SC tissue. This result is surprising because many deep learning architectures for image classification have adopted batch normalization; however, our results may be due to the network architecture being comparatively shallow. Additionally, the performance differences between BASICseg-1 and BASICseg-2 and -3 are not large.
BASICseg-1 also demonstrates significantly better performance for overall SC segmentation compared with Propseg and borderline significance compared with Deepseg, the 2 state-of-the-art methods for SC segmentation presently included in the SCT. Additionally, our model shows significantly better SC segmentation in areas of injury and better overall adaptivity as measured by an interaction term using linear mixed-effects modeling. Deepseg, another CNN algorithm developed for automated SC segmentation, was trained primarily with a combination of healthy control subjects in addition to a more heterogeneous cohort of patients with diverse spinal pathologies, primarily including patients with multiple sclerosis, but also those with neuromyelitis optica, amyotrophic lateral sclerosis, degenerative cervical myelopathy, syringomyelia, and 4 patients with traumatic spinal cord injury.28 Our cohort, enhanced with patients with acute traumatic SCIs, is more specifically focused on image analysis in this patient population, and segmentation performance for other SC disease was not tested. SC segmentation for acute SCI is particularly challenging given the high frequency of SC distortion related to compression and associated geometric distortion as well as heterogeneous intramedullary signal abnormality.2,29,30 Our targeted, disease-specific approach to network training likely, in part, explains performance differences between BASICseg and Deepseg algorithms for our SCI cohort. All CNN-based algorithms (BASICseg1–3 and Deepseg) outperformed Propseg; this difference highlights the value of CNN applications for SC segmentation.
Current standard of care for MR imaging evaluation of traumatic SCI largely relies on subjective and qualitative descriptions of MR imaging findings such as the presence or absence of SC edema and hemorrhage.2,4,31 Thus, few validated MR imaging biomarkers for SC injury stratification and prognosis have been described, despite 4 decades of MR imaging clinical application for SCI.5,6 In addition to development of improved quantitative imaging sequences, advanced, nonbiased, and automated image-analysis techniques may prove useful to accelerate robust MR imaging biomarker identification.32 As an important step toward achieving this goal, current data show that CNNs can segment the SC in a population with acute SCI with an accuracy close to that of criterion standard manual segmentation by neuroradiologists.
For traumatic SC lesion segmentation, the BASICseg-3 algorithm performed best. The difference in performance for the network using a noise-adaptation layer with batch normalization and the other models may be due to the particularly noisy pixel label data for lesions. During training, a high volume of erroneous label data can impede adequate learning. The noise-adaptation layer can absorb this noise and allows the network to correctly map network features to labeled areas. We show that volumetric measures of SC injury derived from the BASICseg-3 correlate with acute and subacute lower extremity motor scores, validating this approach for injury-severity stratification. Ongoing studies with long-term neurologic assessment and functional outcome measures are currently underway to determine the potential prognostic value of CNN-derived biomarkers for acute SCI.
Similar to SC segmentation, automated injury segmentation based on T2WI is an important advance toward enhanced MR imaging biomarker identification for acute traumatic SCI. Manual traumatic lesion segmentation is a time-consuming process requiring an experienced technician and dedicated software packages. In this study, we have shown that volumetric measures of lesions derived from the BASICseg CNN correlate with acute and subacute lower extremity motor scores. To our knowledge, this is the first demonstration of clinically relevant automated MR imaging biomarker extraction in the context of SCI. Most important, accurate and automated SC and traumatic lesion segmentation from T2WI enables rapid image processing for registration with existing anatomic atlases so that whole-cord and subdomain-specific ROIs (for example, specific white matter tracts or gray matter subregions) can be analyzed. We have recently demonstrated the added value of atlas-based volumetric analysis of traumatic SCI lesions using manual injury-segmentation methods.33 With rapid and automated SC and injury segmentation using conventional T2WI, atlas-based analysis tools could be feasibly integrated into the radiologist's workflow without the need for time-consuming image postprocessing. Furthermore, large multi-institutional studies incorporating high-volume MR imaging data would potentially benefit from our proposed image-processing pipeline, which is conducive to batch processing with few errors in automated segmentation. Application of radiomic and texture feature-analysis techniques may also potentially benefit from the proposed image-segmentation techniques.34
Limitations of this study primarily relate to the relatively small sample size of patients with SCI, which both lowers the power of statistical tests using biomarkers and precludes the use of 3D convolutional neural networks. Despite application of data-augmentation techniques to ameliorate feature learning particular to the training set, differences in Dice values between training and validation datasets also suggest some degree of data overfitting for lesion segmentation. 3D convolutional networks may improve both lesion and SC segmentation by using features in the z-direction. Despite these limitations, the excellent Dice coefficient of the BASICseg algorithms for SC and traumatic injury segmentation and the correlation of CNN-based lesion volumes with motor scores are reassuring for the application of these tools in the SCI population. As an additional limitation, the current study only evaluates the use of CNNs to segment tissues from T2WI in the axial plane, whereas automated segmentation of the SC and lesions from multiplanar, multiparametric data will allow more robust analysis.28 In addition, our study cohort is derived from a single institution with all imaging performed on a single MR imaging scanner using similar parameters, thus potentially biasing our results. Future multi-institutional studies will be needed with more diverse datasets to validate the current findings. Furthermore, similar to the Softmax layer to correct for possible bias in the labels, clinical data can also be integrated into the final layer infrastructure to modulate segmentation on the basis of clinical variables. These limitations will be addressed in our future work.
Conclusions
This study demonstrates state-of-the-art performance for SC segmentation after traumatic injury using CNNs. Our model performs favorably in our cohort of patients with acute traumatic SCI compared with currently available algorithms for SC segmentation in areas of damaged cord and shows better overall adaptability with its ability to segment both damaged and undamaged areas. Additionally, we show that training a similar network architecture with the addition of a noise-adaptation layer can successfully segment areas of traumatic SC lesion identified on T2WI. Volumes extracted from lesion segmentation were significantly associated with patient motor scores. Ultimately, the application of these tools will potentially help to advance modernized SC MR image analysis for both research and clinical application. Integration with currently available SC atlases and associated tools as part of the SCT will potentially enhance MR imaging biomarker identification for predictive modeling.
ABBREVIATIONS:
- BASICseg
Brain and Spinal Cord Injury Center segmentation
- CNN
convolutional neural network
- SC
spinal cord
- SCI
spinal cord injury
- SCT
Spinal Cord Toolbox
Footnotes
Disclosures: Charley Gros—RELATED: Grant: IVADO Labs.* Adam Ferguson—RELATED: Consulting Fee or Honorarium: University of Texas Medical Branch; Burke Neurological Institute, Weill Cornell School of Medicine, Comments: I have received honoraria for talks at medical schools; Support for Travel to Meetings for the Study or Other Purposes: University of Texas Medical Branch; Burke Neurological Institute, Weill Cornell School of Medicine; European Neurotrauma Summer School; International Symposium on Neural Regeneration; National Neurotrauma Symposium, Comments: I travel to give talks; Fees for Participation in Review Activities such As Data Monitoring Boards, Statistical Analysis, Endpoint Committees, and the Like: National Institutes of Health study section; Swiss National Science Foundation; Wings for Life Foundation; UNRELATED: Grants/Grants Pending: National Institutes of Health; Veterans Administration; Department of Energy; Wings for Life Foundation; Craig H. Neilsen Foundation, Comments: I serve as Principal Investigator or Co-Investigator on grant applications to federal agencies related to this work.* Vineeta Singh—RELATED: Grant: Craig H. Neilsen Foundation grant.* Lisa Pascual—RELATED: Grant: Department of Defense, Comments: “Early Critical Care Decisions and Outcomes after Spinal Cord Injury.”* Michael S. Beattie—RELATED: Grant: Department of Defense Congressionally Directed Medical Research Programs Spinal Cord Injury Research Program, Comments: Department of Defense award to support the TRACK-SCI study. I am Principal Investigator.* Jacqueline C. Bresnahan—RELATED: Grant: Department of Defense.* William Whetstone—RELATED: Grant: Department of Defense.* Jason F. Talbott—UNRELATED: Expert Testimony: Tindall Bennett & Shoup; RELATED: Grant: Department of Defense Grant SC120259. Russel J. Huie—RELATED: Grant: Department of Defense Grant SC120259.* Leigh Thomas—RELATED: Grant: Department of Defense; Craig H. Neilsen Foundation*; UNRELATED: Employment: University of California, San Francisco. Xuan Duong Fernandez—RELATED: Grant: Department of Defense, Craig H. Neilsen Foundation*; UNRELATED: Employment: University of California, San Francisco. *Money paid to the institution.
This work was supported by the Craig H. Neilsen Foundation and the Department of Defense grant SC120259.
REFERENCES
- 1. Ahuja CS, Wilson JR, Nori S, et al. . Traumatic spinal cord injury. Nat Rev Dis Primers 2017;3:17018 10.1038/nrdp.2017.18 [DOI] [PubMed] [Google Scholar]
- 2. Talekar K, Poplawski M, Hegde R, et al. . Imaging of spinal cord injury: acute cervical spinal cord injury, cervical spondylotic myelopathy, and cord herniation. Semin Ultrasound CT MR 2016;37:431–47 10.1053/j.sult.2016.05.007 [DOI] [PubMed] [Google Scholar]
- 3. Burns AS, Marino RJ, Flanders AE, et al. . Clinical diagnosis and prognosis following spinal cord injury. Handb Clin Neurol 2012;109:47–62 10.1016/B978-0-444-52137-8.00003-6 [DOI] [PubMed] [Google Scholar]
- 4. Bozzo A, Marcoux J, Radhakrishna M, et al. . The role of magnetic resonance imaging in the management of acute spinal cord injury. J Neurotrauma 2011;28:1401–11 10.1089/neu.2009.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kurpad S, Martin AR, Tetreault LA, et al. . Impact of baseline magnetic resonance imaging on neurologic, functional, and safety outcomes in patients with acute traumatic spinal cord injury. Global Spine J 2017;7:151S–74S 10.1177/2192568217703666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fehlings MG, Martin AR, Tetreault LA, et al. . A clinical practice guideline for the management of patients with acute spinal cord injury: recommendations on the role of baseline magnetic resonance imaging in clinical decision making and outcome prediction. Global Spine J 2017;7:221S–30S 10.1177/2192568217703089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fehlings MG, Tetreault LA, Wilson JR, et al. . A clinical practice guideline for the management of patients with acute spinal cord injury and central cord syndrome: recommendations on the timing (≤24 hours versus>24 hours) of decompressive surgery. Global Spine J 2017;7:195S–202S 10.1177/2192568217706367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elizei SS, Kwon BK. The translational importance of establishing biomarkers of human spinal cord injury. Neural Regen Res 2017;12:385–38 10.4103/1673-5374.202933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farhadi HF, Kukreja S, Minnema A, et al. . Impact of admission imaging findings on neurological outcomes in acute cervical traumatic spinal cord injury. J Neurotrauma 2018;35:1398–406 10.1089/neu.2017.5510 [DOI] [PubMed] [Google Scholar]
- 10. Fonov VS, Le Troter A, Taso M, et al. . Framework for integrated MRI average of the spinal cord white and gray matter: the MNI-Poly-AMU template. Neuroimage 2014;102(Pt 2):817–27 10.1016/j.neuroimage.2014.08.057 [DOI] [PubMed] [Google Scholar]
- 11. De Leener B, Lévy S, Dupont SM, et al. . SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage 2017;145:24–43 10.1016/j.neuroimage.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 12. Jenkinson M, Beckmann CF, Behrens TE, et al. . FSL. Neuroimage 2012;62:782–90 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 13. Fischl B. FreeSurfer. Neuroimage 2012;62:774–81 10.1016/j.neuroimage.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin AR, De Leener B, Cohen-Adad J, et al. . Clinically feasible microstructural MRI to quantify cervical spinal cord tissue injury using DTI, MT, and T2*-weighted imaging: assessment of normative data and reliability. AJNR Am J Neuroradiol 2017;38:1257–65 10.3174/ajnr.A5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCoy DB, Talbott JF, Wilson M, et al. . MRI atlas-based measurement of spinal cord injury predicts outcome in acute flaccid myelitis. AJNR Am J Neuroradiol 2017;38:410–17 10.3174/ajnr.A5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Leener B, Kadoury S, Cohen-Adad J. Robust, accurate and fast automatic segmentation of the spinal cord. Neuroimage 2014;98:528–36 10.1016/j.neuroimage.2014.04.051 [DOI] [PubMed] [Google Scholar]
- 17. De Leener B, Cohen-Adad J, Kadoury S. Automatic segmentation of the spinal cord and spinal canal coupled with vertebral labeling. IEEE Trans Med Imaging 2015;34:1705–18 10.1109/TMI.2015.2437192 [DOI] [PubMed] [Google Scholar]
- 18. Gros C, De Leener B, Dupont SM, et al. . Automatic spinal cord localization, robust to MRI contrasts using global curve optimization. Med Image Anal 2018;44:215–27 10.1016/j.media.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 19. The R Core Team. R: a language and environment for statistical computing. 2013. https://www.researchgate.net/publication/221943918_R_A_Language_And_Environment_for_Statistical_Computing_Reference_Index_2152. Accessed on January 11, 2018.
- 20. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, et al., eds. Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany October 5–9, 2015;234–41 [Google Scholar]
- 21. Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: Proceedings of the 27th International Conference on Machine Learning, Haifa, Israel June 21–24, 2010; 807–14 [Google Scholar]
- 22. Srivastava N, Hinton G, Krizhevsky A, et al. . Dropout: a simple way to prevent neural networks from overfitting. Journal of Machine Learning Research 2014;15:1929–58 [Google Scholar]
- 23. Kinga D, Adam JB. A method for stochastic optimization. In: Proceedings of the International Conference on Learning Representations, San Diego, California May 7–9, 2015 [Google Scholar]
- 24. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. 2015. https://arxiv.org/abs/1502.03167. Accessed on January 11, 2018.
- 25. Sukhbaatar S, Bruna J, Paluri M, et al. . Training Convolutional Networks with Noisy Labels. 2014. https://arxiv.org/abs/1406.2080. Accessed on January 11, 2018.
- 26. Dice R. Measures of the amount of ecologic association between species. Ecology 1945;26:297–302 10.2307/1932409 [DOI] [Google Scholar]
- 27. Zou KH1, Warfield SK, Bharatha A, et al. . Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 2004;11:178–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gros C, De Leener B, Badji A, et al. . Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. Neuroimage 2019;184:901–15 10.1016/j.neuroimage.2018.09.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haefeli J, Mabray MC, Whetstone WD, et al. . Multivariate analysis of MRI biomarkers for predicting neurologic impairment in cervical spinal cord injury. AJNR Am J Neuroradiol 2017;38:648–55 10.3174/ajnr.A5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pandit P, Talbott JF, Pedoia V, et al. . T1ρ and T2 -based characterization of regional variations in intervertebral discs to detect early degenerative changes. J Orthop Res 2016;34:1373–81 10.1002/jor.23311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shah LM, Ross JS. Imaging of spine trauma. Neurosurgery 2016;79:626–42 10.1227/NEU.0000000000001336 [DOI] [PubMed] [Google Scholar]
- 32. Martin AR, Aleksanderek I, Cohen-Adad J, et al. . Translating state-of-the-art spinal cord MRI techniques to clinical use: a systematic review of clinical studies utilizing DTI, MT, MWF, MRS, and fMRI. Neuroimage Clin 2016;10:192–238 10.1016/j.nicl.2015.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCoy D, Huie R, Dupont S, et al. . Atlas-based volumetric assessment of T2 abnormality in acute spinal cord injury predicts motor outcomes: a transforming research and clinical knowledge in SCI (TRACK-SCI) pilot study. In: Proceedings of the International Society of Magnetic Resonance in Medicine, Paris, France June 16–21, 2018 [Google Scholar]
- 34. Gillies RJ, Kinahan, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]