Abstract
Most bacteria accomplish cell division with the help of a dynamic protein complex called the divisome, which spans the cell envelope in the plane of division. Assembly and activation of this machinery is coordinated by the tubulin-related GTPase FtsZ, which was found to form treadmilling filaments on supported bilayers in vitro1 and in live cells where they circle around the cell division site2,3. Treadmilling of FtsZ is thought to actively move proteins around the cell thereby distributing peptidoglycan synthesis and coordinating the inward growth of the septum to form the new poles of the daughter cells4. However, the molecular mechanisms underlying this function are largely unknown. Here, to study how FtsZ polymerization dynamics are coupled to downstream proteins, we reconstituted part of the bacterial cell division machinery using its purified components FtsZ, FtsA and truncated transmembrane proteins essential for cell division. We found that the membrane-bound cytosolic peptides of FtsN and FtsQ co-migrated with treadmilling FtsZ-FtsA filaments, but despite their directed collective behavior, individual peptides showed random motion and transient confinement. Our work suggests that divisome proteins follow treadmilling FtsZ filaments by a diffusion-and-capture mechanism, which can give rise to a moving zone of signaling activity at the division site.
In a treadmilling filament, monomers are added to one end of the filament and detach from the opposite end, resulting in the apparent forward movement of the polymer while the individual monomers remain static. While FtsZ filament treadmilling was found to power the directed movement of transmembrane cell division proteins such as the PG synthases PBP3 (FtsI) in E. coli3 and PBP2b in B. subtilis2, these studies could not provide a mechanistic model for how stationary subunits within the treadmilling FtsZ protofilament can give rise to the directed motion of PG synthases5. Accordingly, the role of treadmilling for the spatiotemporal organization of cell division proteins as well as the underlying molecular processes are still unknown.
FtsZ filaments are attached to the inner leaflet of the cytoplasmic membrane via the membrane anchors ZipA and FtsA. Of those two proteins, the latter appears to be most important as the requirement for ZipA can be bypassed6,7, while FtsA is essential in E. coli. This indicates that FtsZ-FtsA co-filaments trigger later steps in cytokinesis by recruiting about 20 additional proteins including several peptidoglycan (PG) synthases and hydrolases8,9. In fact, FtsA was previously suggested to interact with late cell division proteins including FtsL, FtsN, FtsQ and PBP3 in the Z-ring (Fig. 1a, left side)7,8,10–12. PBP3 is an essential cell division specific peptidoglycan synthase, FtsQ and FtsL form a complex with FtsB in the inner membrane which, like FtsN, is thought to regulate the PG synthase activity of PBP313,14. These proteins all contain a short N-terminal cytoplasmic tail, a single transmembrane-helix and a larger periplasmic domain. As FtsA binds peripherally to the cytoplasmic membrane, it is ideally oriented to interact with the cytoplasmic peptides of these proteins thereby linking cell wall synthesis to Z-ring dynamics15. In fact, recent evidence indicated that in some mutants the interaction between FtsA and FtsN becomes essential for the initiation of septal PG synthesis and cell constriction16,17. However, only the interaction between FtsN and FtsA in solution was so far confirmed in vitro18, while most other data were obtained in vivo and hence they could not reveal whether or not direct protein-protein interactions were involved in the recruitment of these proteins to the site of division. Accordingly, despite the information obtained from in vivo studies, important questions about the mechanism of cell division remain unanswered: first, which of these cytoplasmic tails interact with FtsA and second, does FtsZ treadmilling control the spatiotemporal dynamics of FtsA-binding proteins and if yes, what is the mechanism of this coupling?
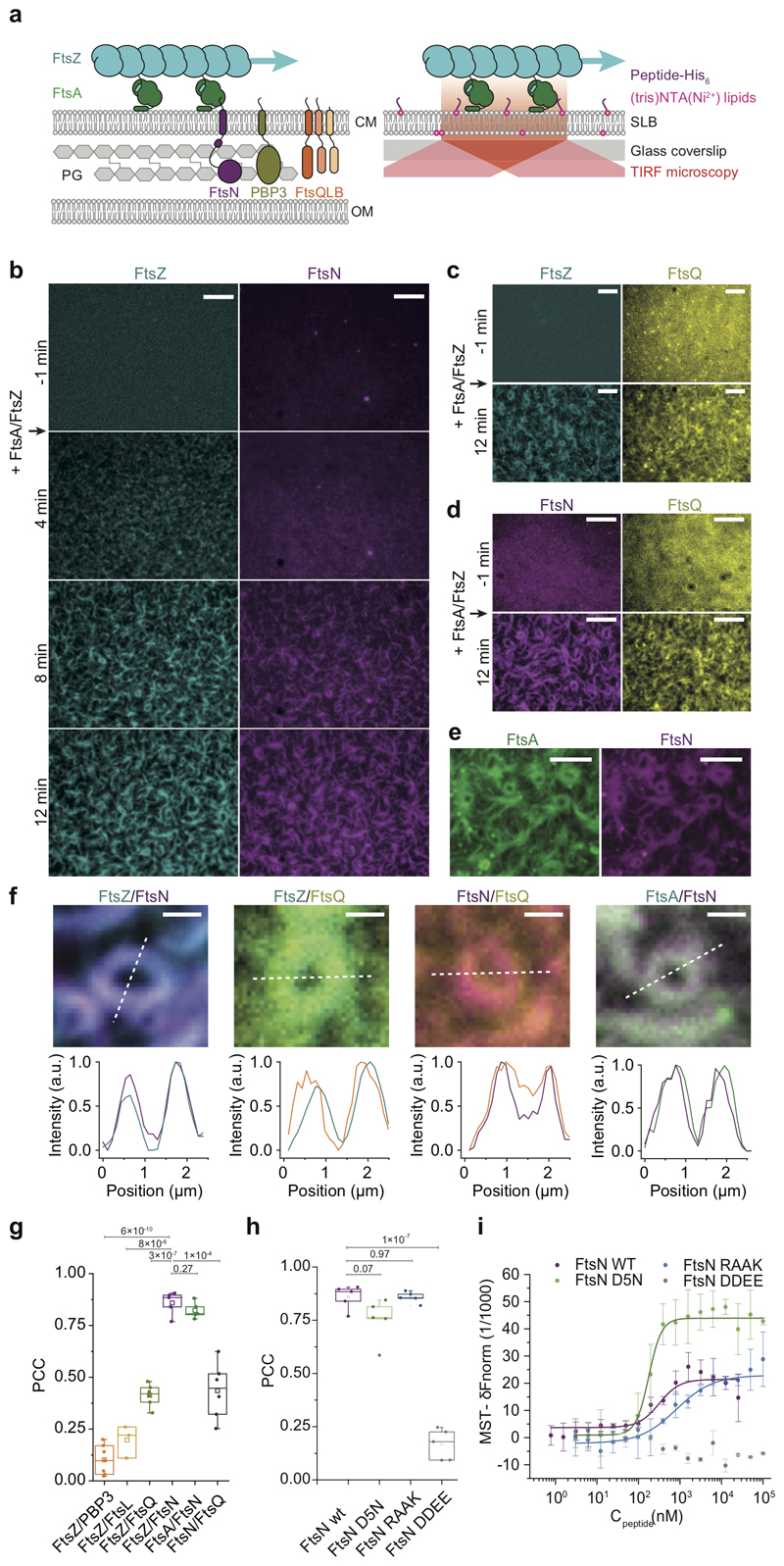
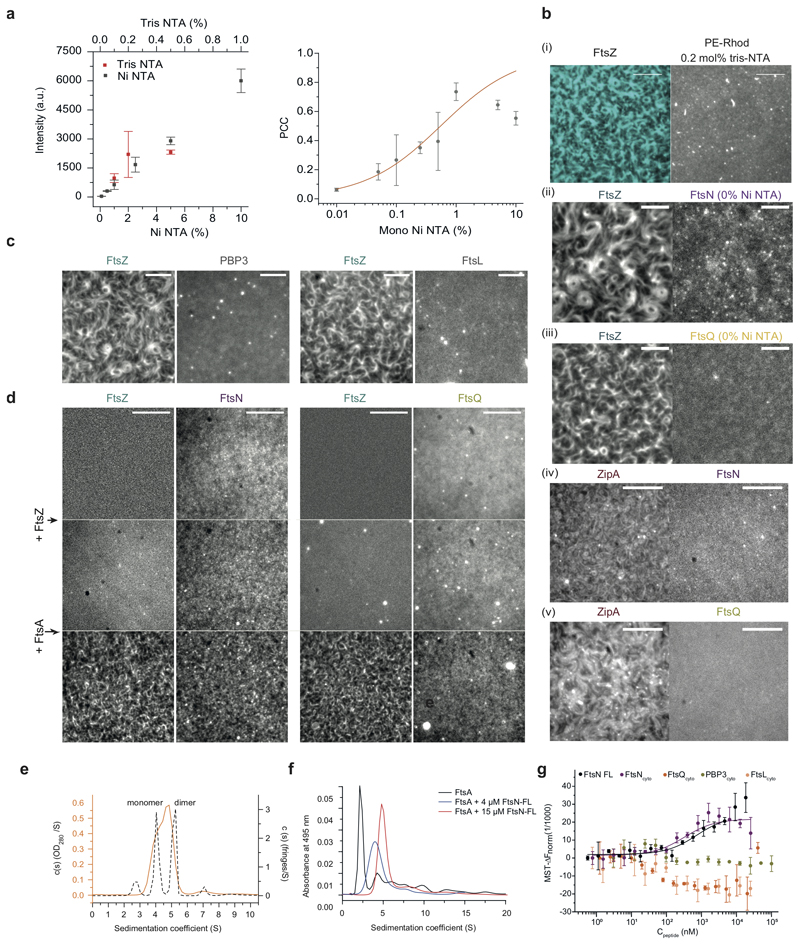
Figure 1. FtsZ-FtsA filaments co-localize with cytoplasmic tails of FtsN and FtsQ.
a. Left: Selected proteins of the E. coli cell division machinery. FtsZ-FtsA co-filaments on the cytoplasmic membrane (CM) interact with the N-terminal cytoplasmic tails of FtsN, PBP3 and FtsQLB. These proteins act on the peptidoglycan (PG) layer in the periplasm between CM and outer membrane (OM). Right: In vitro reconstitution approach. N-terminal peptides of candidate proteins were attached to the surface of glass-supported lipid bilayers (SLBs). Arrows indicate treadmilling direction of FtsZ filaments.
b. From a homogeneous distribution, CF488-FtsNcytoHis (magenta) colocalizes with FtsZ filaments on the membrane after addition of Cy5-FtsZ (cyan) and FtsA at t = 0 min. Representative micrograph from n=5 independent experiments. Scale bars are 5 μm. Supplementary Video 1.
c. Representative micrograph of Alexa488-FtsZ (cyan) and Cy5-FtsQcytoHis (yellow) with unlabeled FtsA from n=5 independent experiments. Scale bars are 5 μm. Supplementary Video 2.
d. Representative micrograph of Alexa488-FtsNcytoHis (magenta) and Cy5-FtsQcytoHis (yellow) with unlabeled FtsA and FtsZ (12 min) from n=6 independent experiments. Scale bars are 5 μm. Supplementary Video 3.
e. Representative micrograph of Cy5-FtsNcytoHis (magenta) on TMR-FtsA (green) from n=4 independent experiments Scale bars are 5 μm. Supplementary Video 4.
f. Top: Ring-like structures formed by Cy5-FtsZ/CF488-FtsNcytoHis (n=5), Alexa 488-FtsZ/Cy5-FtsQcytoHis (n=5), Alexa488-FtsNcytoHis /Cy5- FtsQcytoHis (n=6) and TMR-FtsA/Cy5- FtsNcytoHis (n=4). Bottom: Corresponding intensity profiles along dashed lines. Scale bars are 1 μm.
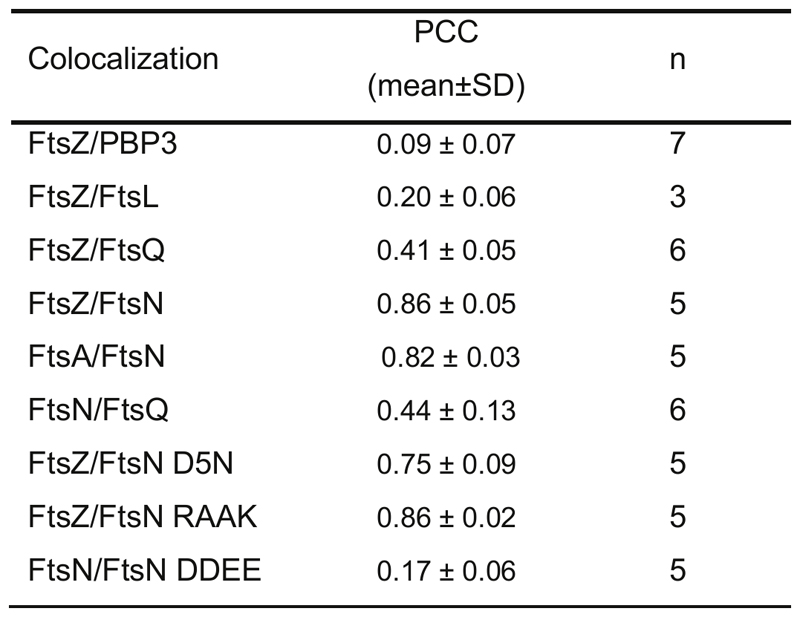
g Colocalization of membrane-bound peptides quantified by the Pearson Correlation coefficient (PCC, mean±SD and n are specified in ED Fig.9). Each dot represents an independent experiment.
h. Colocalization efficiency of different versions of FtsN with FtsZ. Each dot represents an independent experiment.
i. FtsN peptides bind to FtsA with different affinities as measured by Microscale Thermophoresis. wild-type FtsN peptide. FtsNcyto-DDEEHis shows no interaction. Solid lines indicate fit of Hill equation. Error bars represent standard deviation between n=3 independent experiments.
To address these questions, we developed an in vitro assay that recapitulates early stages of cytokinesis dynamics in Escherichia coli. In this assay, His-tagged, fluorescently labeled cytoplasmic peptides of our candidate divisome proteins FtsL, FtsN, FtsQ or PBP3 were attached to supported lipid bilayers containing defined concentrations of Ni2+-chelating lipids (Fig. 1a, right, ED Fig. 1a). In the absence of other proteins, the membrane-bound peptides were homogeneously distributed and diffused on the bilayer. This situation is similar as in the cell where cell division proteins are distributed homogeneously in the absence of a Z-ring19. When we allowed FtsZ and FtsA to polymerize on these membranes, the fluorescent signal for FtsNcytoHis and FtsQcytoHis increasingly overlapped with the cytoskeletal structures formed by FtsA and FtsZ, even when both peptides were simultaneously present (Fig. 1b-d, Supplementary Videos 1, 2 and 3). In all cases, the overlap for FtsQcytoHis was lower than for FtsNcytoHis, as quantified by the Pearson Correlation Coefficient (PCC) (Fig. 1g, Extended Data 9). In contrast, FtsLcytoHis and PBP3cytoHis did not colocalize with FtsZ-FtsA cofilaments and continued to exhibit a homogeneous membrane-distribution (ED Fig. 1c).
To rule out any direct binding of FtsNcytoHis and FtsQcytoHis to FtsZ, we confirmed that the membrane-bound peptides did not recruit any FtsZ filaments in the absence of FtsA or when FtsA was replaced by ZipA (ED Fig. 1b, d). Furthermore, even with FtsA, colocalization of FtsZ with FtsNcytoHis or FtsQcytoHis was nearly abolished when these peptides were not attached to the membrane due to the absence of Ni2+-chelating lipids (ED Fig. 1b).
Interestingly, the monomeric peptide FtsNcytoHis, strongly colocalized with FtsA on the membrane (Fig. 1e, Supplementary Video 4), while previous observations suggested that only a dimerized form of FtsNcyto interacted with FtsA in solution18. To test this apparent discrepancy, we studied the interactions between FtsA and full-length FtsN or the cytoplasmic peptides of divisome proteins by analytical ultracentrifugation and microscale thermophoresis (MST). Full-length FtsN formed oligomers in solution (ED Fig. 1e) and interacted with FtsA with a similar affinity as FtsNcytoHis (ED Fig. 1 f,g, Extended Data 10). In contrast, FtsQcytoHis, FtsLcytoHis and PBP3cytoHis showed no interaction with FtsA in solution (ED Fig. 1g).
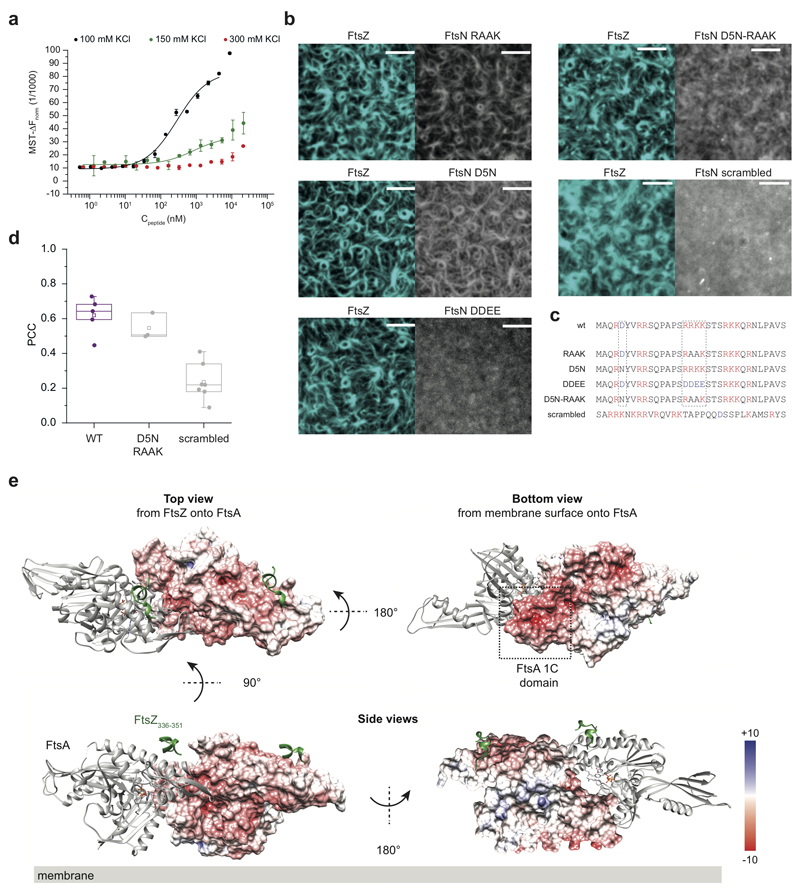
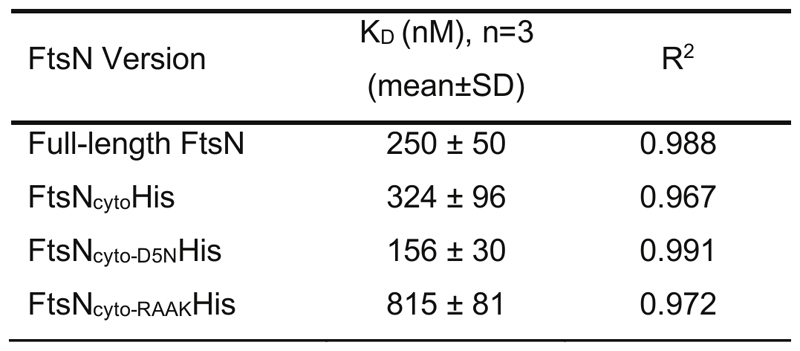
We observed that the binding of full-length FtsN to FtsA was impaired at high salt concentrations suggesting an ionic interaction (ED Fig. 2a). To test the ionic nature of the FtsN-FtsA interaction in more detail, we studied previously reported mutants of FtsN in which the conserved positively charged residues at positions 16-19 (RRKK) were either replaced by negatively charged (DDEE) or hydrophobic (RAAK) residues16,18. We also produced a version of FtsN in which the conserved negatively charged aspartate at position 5 was replaced by asparagine (D5N)7. Both motifs are highly conserved and were suggested to be critical for the interaction of FtsN with FtsA. However, only FtsN D5N was found to produce a significant phenotype in cells where the FtsN-FtsA interaction becomes essential20, while FtsN RAAK had no effect. FtsN DDEE was not tested under the same conditions, most likely due to its instability when expressed in vivo7,20. Surprisingly, in our assay FtsNcyto-RAAKHis and FtsNcyto-D5NHis showed similar colocalization with FtsZ compared to the wildtype peptide (Fig. 1h) and both peptide versions bound to FtsA in solution (Fig. 1i). In contrast, the version of FtsN with inversed charge (FtsNcyto-DDEEHis) showed much weaker overlap with FtsZ on the membrane and no interaction with FtsA in solution (Fig. 1h,i, ED Fig. 2b, and Extended Data 10). Next, we created peptide variant, FtsNcytoD5N-RAAKHis, where both conserved motifs of FtsN were altered. This version also colocalized with FtsZ filaments on the membrane (ED Fig. 2d). Therefore, as only a charge inversion at positions 16-19 (RRKK to DDEE) inhibited binding, the interaction is likely dominated by electrostatic interactions involving the RRKK motif. Indeed, when we analyzed the surface charge of FtsA, we found that its 1C domain contains a highly negatively charged patch facing towards the membrane (ED Fig. 2e). To further test for the specificity of the interaction between FtsA and FtsN, we produced a version of its cytoplasmic peptide, where the RRKK and D5 motifs are lost while the overall charge of the peptide is conserved (ED Fig. 2c). Interestingly, the signal for this mutant did not overlap with FtsZ filaments indicating that there is a certain degree of specificity in the interaction between FtsNcyto and FtsA despite its electrostatic nature and that both conserved motifs have to be at a specific distance from the membrane to efficiently interact with FtsA.
In sum, we confirmed a direct interaction between FtsA and the cytoplasmic tail of FtsN and provided evidence that FtsA directly binds to FtsQ. As a consequence, both membrane-bound peptides colocalize with FtsZ filaments on the membrane. In contrast, the D5 residue in FtsNcyto was not essential for the interaction with FtsA. Therefore, the D5N mutation must disrupt another function of FtsN giving rise to the observed phenotype in vivo.
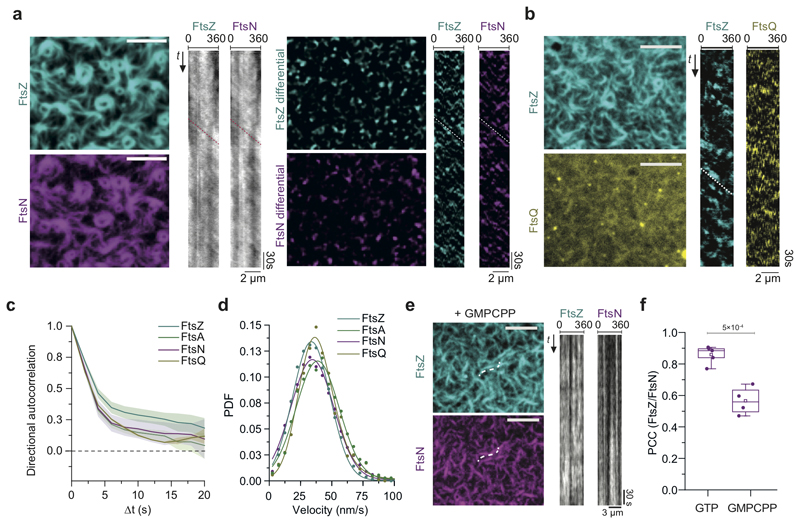
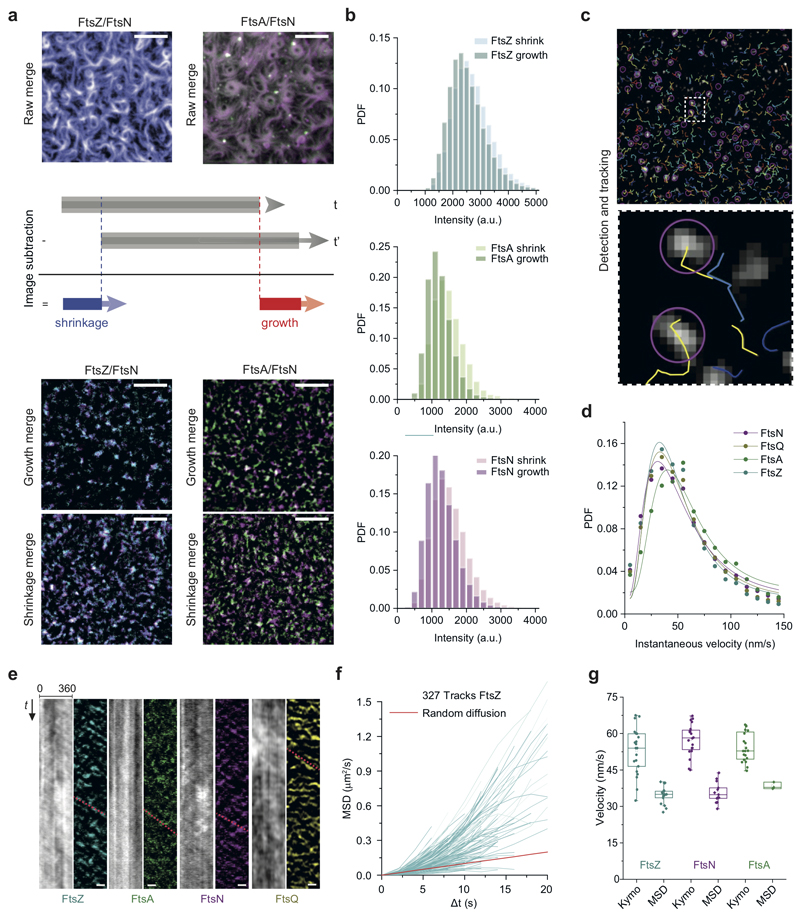
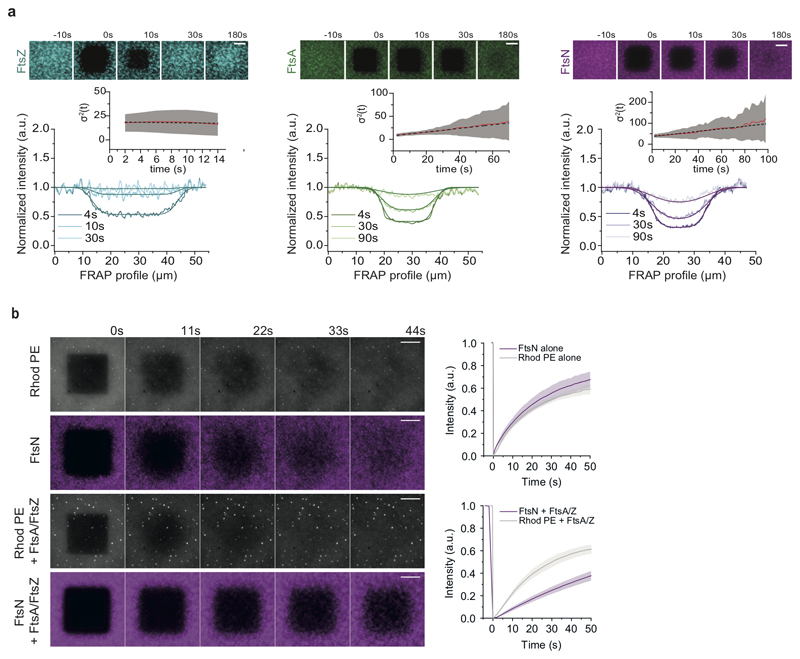
Next, we studied the dynamic coupling between FtsZ treadmilling and FtsNcytoHis and FtsQcytoHis. By acquiring dual-color time-lapse movies we simultaneously imaged the dynamics of FtsZ and membrane-bound cytoplasmic peptides. Both peptides were not only recruited to FtsZ filaments, but closely followed their treadmilling dynamics (Fig. 2a-b). To better visualize protein dynamics, we constructed differential time-lapse movies, which show the intensity difference between two frames separated by a constant time delay (Fig. 2a, ED Fig. 3a). This processing step removed the non-moving background and produced fluorescent spots that represent the growth or shrinkage of an FtsZ filament bundle and the collective motion of associated peptides (Supplementary Video 5). We found that spots corresponding to FtsZ filament growth and shrinkage had similar intensity distributions, consistent with a constant steady-state length of filaments during treadmilling (ED Fig. 3b). Interestingly, spots corresponding to FtsA and FtsNcytoHis also showed this property, demonstrating that these proteins closely mirror the behavior of FtsZ and that the amount of the protein associated with FtsZ filaments is constant. The tight dynamic coupling between these proteins was also evident from kymographs along the circumference of rotating rings (Fig. 2a,b and ED Fig. 3a).
Figure 2. Cytoplasmic tails of FtsN and FtsQ co-migrate directionally with treadmilling FtsZ-FtsA cofilaments.
a. FtsNcytoHis follows treadmilling of FtsZ filaments. Left: Micrographs of A488-FtsZ (cyan) and Cy5-FtsNcytoHis (magenta) and representative kymographs taken along the circumference of a rotating FtsZ ring. Right: Micrographs showing differential images from the corresponding micrographs on the left and representative differential kymographs of treadmilling dynamics. Scale bars are 5 μm, n=20
b. FtsQcytoHis follows treadmilling of FtsZ filaments. Left: Micrographs of Alexa488-FtsZ (cyan) and Cy5-FtsQcytoHis (yellow) and representative differential kymographs taken along the circumference of a rotating FtsZ ring. Right: Scale bars are 5 μm, n= 20.
c. On the ensemble level all proteins move directionally, as revealed by directional autocorrelation analysis. Positive values confirm directional motion for FtsZ (cyan), FtsA (green), FtsNcytoHis (magenta) and FtsQcytoHis (dark yellow). Line represents the mean and error bars represent the standard deviation from n(FtsN) = 13, n(FtsA) = 3; n(FtsQ) = 3, n(FtsZ) = 16 independent experiments.
d. Velocity histograms and Gaussian distribution fit (solid line) did not reveal any significant difference in the velocity FtsZ (v = 33.7 ± 15 nm/s; n = 16; cyan), FtsA (v = 38.9 ± 17.2 nm/s; n = 3; green), FtsQ (v = 37.4 ± 13.7 nm/s; n = 3; dark yellow and FtsNcytoHis (v = 34.5 ± 17.1 nm/s; n = 13, magenta). The velocity values represent mean and SD from the corresponding Gaussian fits (R2 = 0.96-0.99). PDF stands for Probability density function.
e. FtsZ treadmilling and FtsNcytoHis co-migration is GTP-dependent while colocalization is not. TIRF micrographs are revealing colocalized Cy5-FtsZ and CF488-FtsN patterns with GMPCPP (left) and static kymographs (right). Scale bars are 5 μm, n=4. Supplementary Video 6.
f. Treadmilling increases the FtsZ/FtsNcytoHis colocalization. In the presence of GMPCPP the colocalization efficiency is 1.5-fold lower comparing to the presence of GTP (PCC = 0.57 ± 0.09, mean±SD; grey box, n = 4, p-value = 4.9×10−4).
Our differential movies allowed us to use particle-tracking methods to quantitatively analyze the migration trajectories of fluorescent spots in an automated, non-biased manner (ED Fig. 3c, Supplementary Table 2). We first performed a directional autocorrelation analysis of treadmilling trajectories for FtsZ, FtsA, FtsNcytoHis and FtsQcytoHis, which provided information about the local directional persistence of treadmilling. The correlations for all four proteins were always positive and followed a similar decay, even for longer time delays, confirming their directional motion (Fig. 2c). Next, we computed the mean-squared displacement (MSD) for every trajectory and estimated the corresponding velocity by fitting a quadratic equation (ED Fig. 3f). We then fitted a Gaussian function to the distribution of velocities to obtain the mean velocity for FtsZ, FtsA, FtsN and FtsQ, comparable within the standard deviation (Fig. 2d). From this analysis and the instantaneous velocity distribution (ED Fig. 3g), we concluded that there is a tight coupling between FtsZ treadmilling and proteins recruited to the filaments such that on the ensemble level all four proteins moved at the same velocity.
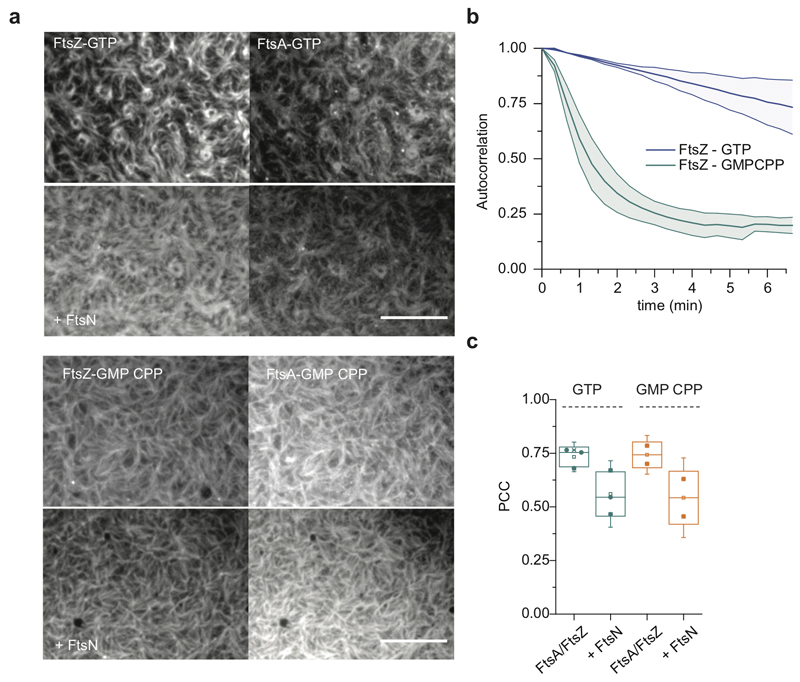
Importantly, any directed collective motion of membrane-bound peptides was abolished when we replaced GTP by the slowly-hydrolyzable analog GMPCPP, which did not allow for FtsZ treadmilling (Fig. 2e, Supplementary Video 6). Interestingly, the colocalization of FtsZ with FtsNcytoHis, but not with FtsA, was slightly but significantly decreased with GMPCPP compared to GTP (Fig. 2f, ED Fig. 4a-c), suggesting that FtsZ filament dynamics can influence the recruitment of FtsN to FtsA.
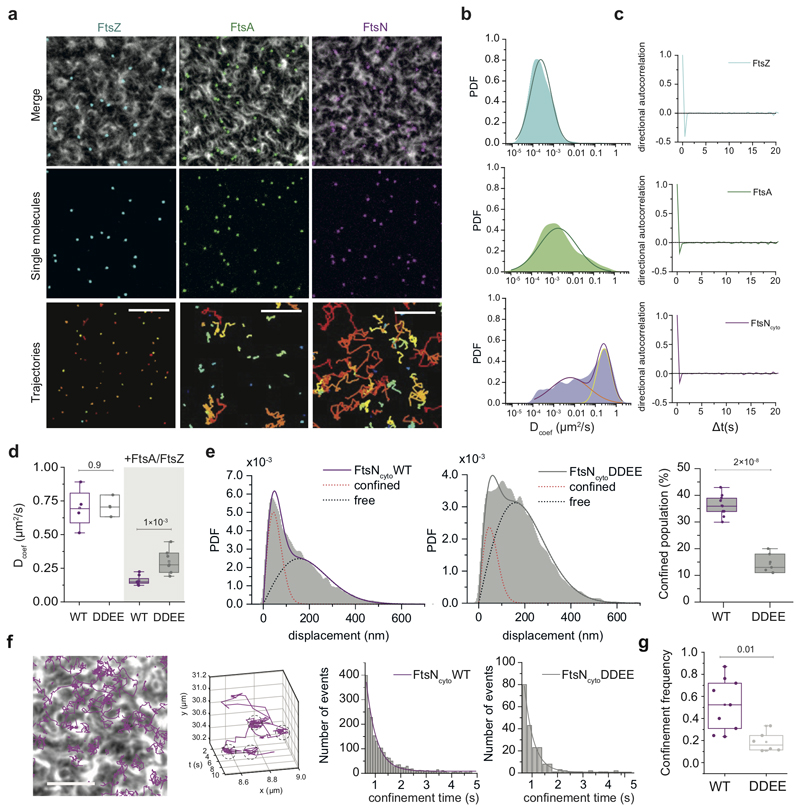
Next, we focused on the characterization FtsNcytoHis behavior as its colocalization with FtsA was stronger than that of FtsQcytoHis. To better understand the tight dynamic coupling between FtsZ treadmilling and FtsN and FtsA we performed single-molecule experiments. By reducing the fraction of fluorescently labeled proteins, we were able to monitor and track the dynamics of individual proteins on the membrane (Fig. 3a, Supplementary Video 8). While FtsZ molecules were stationary as expected for treadmilling filaments, FtsA molecules showed slow diffusion with a broad distribution of mobilities. The behavior of FtsNcytoHis peptides showed a pronounced mobile fraction next to a broad distribution of immobile particles (Fig. 3b). We confirmed this behavior on the ensemble level in fluorescence recovery after photobleaching (FRAP) experiments (ED Fig. 5a,b, Supplementary Video 7). Importantly, we found no evidence for directed motion for any of the three proteins. Consistently, and in contrast to what we found for the ensemble level, the directional autocorrelation for individual proteins dropped to zero, indicating random, uncorrelated behavior (Fig. 3c).
Figure 3. FtsNcytoHis follows treadmilling FtsZ-FtsA cofilaments via a diffusion-and-capture mechanism.
a. Single-molecule imaging reveals different protein behavior. Top rows: Small amounts of Cy5-labelled proteins were added to follow individual FtsZ (cyan, n=5), FtsA (green, n=8) and FtsNcytoHis (magenta, n=6)) molecules in the presence of an Alexa488-FtsZ pattern. Scale bars are 5 μm. Images were subjected to background subtraction and Gaussian smoothing. Bottom row: Corresponding trajectories showing different diffusive behavior of FtsZ, FtsA, and FtsNcytoHis. Scale bars are 2 μm. Supplementary Video 8.
b. Histograms of diffusion coefficients (Dcoef) of single trajectories for FtsZ, FtsA and FtsNcytoHis. A single Gaussian (cyan) can be fitted to the probability density distribution for FtsZ with a mean diffusion constant below 1x10-4 μm2/s, which corresponds to immobile molecules. FtsA has an average Dcoef of 0.0016 ± 0.0003 μm2/s (n = 3, mean±SD) approximated with a single Gaussian (green). Two populations can be observed for FtsNcytoHis: mobile (yellow line) and immobile particles (red line) (n = 4, mean±SD), n represents independent experiments.
c. Directional autocorrelations of single-molecule trajectories of FtsZ, FtsA and FtsNcytoHis (averaged between independent experiments, n = 3).
d. Diffusion coefficients are comparable between FtsNcytoHis (n=2, N=4) and FtsNcyto-DDEEHis (n=2, N=5) without FtsA and FtsZ proteins present (white boxes). FtsA/FtsZ filaments on the membrane slow down diffusion of both peptides (grey boxes), but to a different extent: diffusion of FtsNcytoHis is 4.4-fold slower (n=3 experiments, N=8 movies), while diffusion of FtsNcyto-DDEEHis (n=3, N=7) is slowed down only 2.4-fold. The p-value was calculated using a two-tailed Student’s t-test. Boxplots encompasses the 25-75th percentile, the whiskers indicate outliers, the midline indicates the median and squares the mean.
e. Step-size distribution of mobile fraction for FtsNcytoHis (left) and FtsNcyto-DDEEHis (middle). The weighted contribution of the confined population is 2-fold higher for FtsNcytoHis in comparison to FtsNcyto-DDEEHis (right). Each dot represents the weighted contribution of the confined population for all detected trajectories in a randomly selected region of interest (55x55 μm), n=3. Boxplots encompasses the 25-75th percentile, the whiskers indicate outliers, the midline indicates the median and squares the mean.
f. From left to right: An overlay of the FtsZ pattern and single particle trajectories of FtsNcytoHis, scale bar 5 μm, n=3. Example for an individual trajectory of FtsNcytoHis. Histograms of confinement times as determined from cluster analysis and monoexponential fits (combined data from 3 independent experiments, total 4702 trajectories), FtsNcytoHis (0.41±0.02s, Nclusters=1827, mean±SD, R2= 0.985) and FtsNcyto-DDEEHis (0.36±0.02s, Nclusters=283, mean±SD, R2= 0.987).
g. The number of identified confinement events divided by the number of trajectories corresponds to the frequency of confinement, which was about 3 times higher for FtsNcytoHis than for FtsNcyto-DDEEHis, n=3. The p-value was calculated using a two-tailed Student’s t-test. Boxplots encompasses the 25-75th percentile, the whiskers indicate outliers, the midline indicates the median and squares the mean.
How can FtsNcytoHis co-migrate with treadmilling FtsZ-FtsA cofilaments if single molecules do not show directed motion? To answer this question, we further analyzed the behavior of single molecules of FtsNcytoHis and compared it to FtsNcyto-DDEEHis. As this version of the peptide showed only weak overlap with FtsZ-FtsA cofilaments and weak binding to free FtsA (Fig.1h and ED Fig. 2b, 5a), we expected a clear difference in their behavior at a single molecule level. For our analysis, we first excluded particles that were immobile during the time of observation as they most likely arise from non-specific attachments. First, we quantified the mean diffusion constants of FtsNcytoHis and FtsNcyto-DDEEHis with and without FtsZ filaments on the membrane. We found that the wildtype peptide diffused significantly slower than FtsNcyto-DDEEHis, but only in the presence of FtsZ and FtsA (Fig. 3d). Next, we calculated displacement lengths of mobile molecules, which showed two sub-populations of step sizes in their histograms (Fig. 3e), indicating that the effective diffusion constant is the result of a mix of different behaviors. Interestingly, the weight of the population showing short displacements was about 3-fold higher for FtsNcytoHis than for FtsNcyto-DDEEHis. Together, these observations suggest transient capturing of wildtype FtsN by FtsZ-FtsA co-filaments.
Indeed, a detailed inspection revealed that most of the FtsNcytoHis trajectories included short periods of confined motion (Fig. 3f, Supplementary Video 9-10). Using computer-aided spatial cluster identification, we found an average duration of these periods of 0.41 ± 0.02 s for FtsNcytoHis (n = 3 experiments with 4702 trajectories) and a slightly lower value for the DDEE peptide (0.36 ± 0.02 s, n = 3 experiments with 1763 trajectories). However, the frequency of confinement events was 3-fold higher for wildtype FtsNcytoHis than for the DDEE version (Fig. 3g) consistent with the difference in step size distribution and the decreased colocalization of this version with FtsZ-FtsA co-filaments on the membrane (Fig. 1h, ED Fig. 2b).
Our approach cannot precisely determine interaction kinetics due to photobleaching and a possible contribution of molecular crowding to peptide confinement. Nevertheless, the analysis reveals transient confinement of FtsNcytoHis at FtsZ-FtsA co-filament bundles for durations about 10-fold shorter than the residence time of FtsZ monomers (Fig. 4d)1,21, and that wildtype FtsNcytoHis was trapped more often than FtsNcyto-DDEEHis. Thus, our single-molecule analysis, in combination with our MST (Fig. 1i) and FRAP (ED Fig. 5a) experiments revealed that FtsNcytoHis is transiently interacting with the treadmilling scaffold provided by the FtsZ-FtsA co-filament on the membrane. Importantly, the observed migration of proteins corresponds to an emergent property of this self-organized system: while on the ensemble level all proteins move directionally (Fig. 2c), the individual molecules show uncorrelated diffusive or stationary behavior (Fig. 3c).
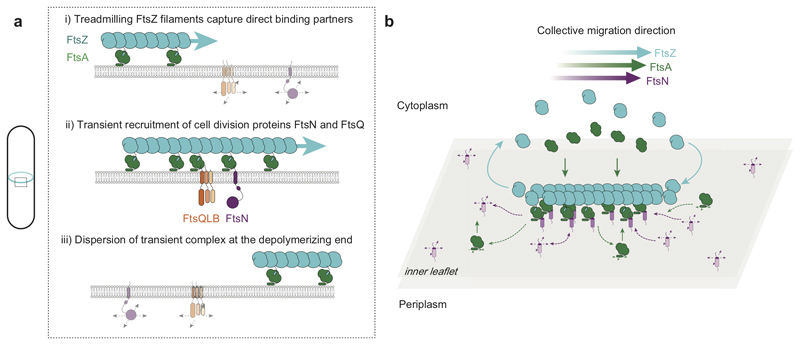
Figure 4. Schematic illustration of coupling between treadmilling FtsZ filaments and cell division proteins.
a. At early stages of cell division, treadmilling FtsZ filaments in the Z-ring act as a dynamic scaffold to recruit transmembrane proteins freely diffusing in the membrane. Both, FtsN and FtsQ directly bind to the FtsZ-FtsA co-filament to form a transient, stationary complex, which can then recruit other division proteins to the division site via interactions in the membrane or periplasm. Due to the local increase in concentration, weak interactions between proteins can be sufficient to initiate assembly of the division machinery. b. Illustration of proposed mechanism of co-migration. Freely diffusing FtsN in the membrane binds to the treadmilling FtsZ-FtsA filament on the membrane surface. While proteins reversibly bind along the whole length of the filaments, there is a net accumulation of proteins at the growing end. In contrast, proteins rapidly disperse at the depolymerizing end. This dynamic coupling to the treadmilling filaments permits a collective co-migration on the ensemble level while individual proteins show uncorrelated behavior.
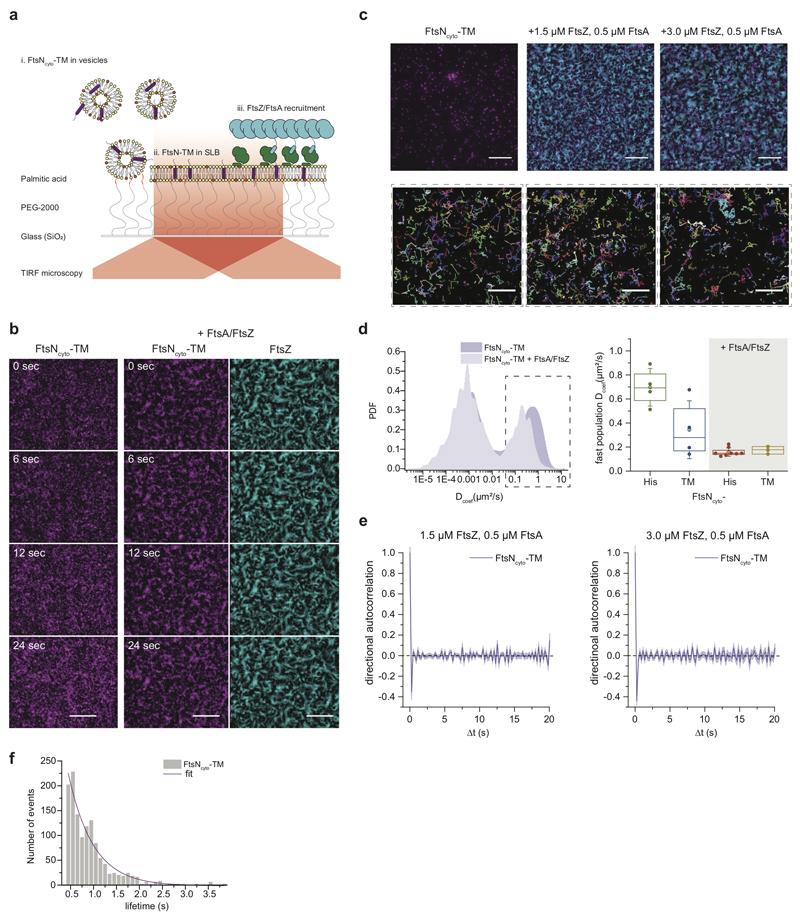
So far, we showed that the behavior of FtsNcytoHis is governed by rapid diffusion and transient confinement to FtsZ-FtsA co-filaments. However, the presence of the transmembrane domain in the full-length protein might slow down diffusion and as a consequence, inhibit co-migration with treadmilling FtsZ-FtsA co-filaments. To test the influence of lateral mobility on the behavior observed in vitro, we purified the cytoplasmic peptide of FtsN together with its transmembrane helix (FtsNcyto-TM) and reconstituted this protein into polymer-supported lipid membranes, where a PEG-layer provides a spacer between the cover slip and membrane (ED Fig. 6a)22. Indeed, using single molecule tracking, we found that the diffusion constant of FtsNcyto-TM was about two-fold lower than that of FtsNcytoHis (ED Fig. 6d), but FtsNcyto-TM still co-localized with treadmilling FtsZ filaments, albeit less efficiently than FtsNcytoHis. This difference is likely because it is not possible to fully control the orientation and density of transmembrane proteins in a polymer-supported lipid membrane22. Next, we analyzed the behavior of single FtsNcytoTM molecules in the presence of FtsA and FtsZ. Again, we found no evidence of directed motion (ED Fig. 6e, Supplementary Video 11), instead FtsNcyto-TM showed transient capturing like the lipid-attached FtsNcyto-His peptide as well as a similar confinement time (ED Fig. 6f).
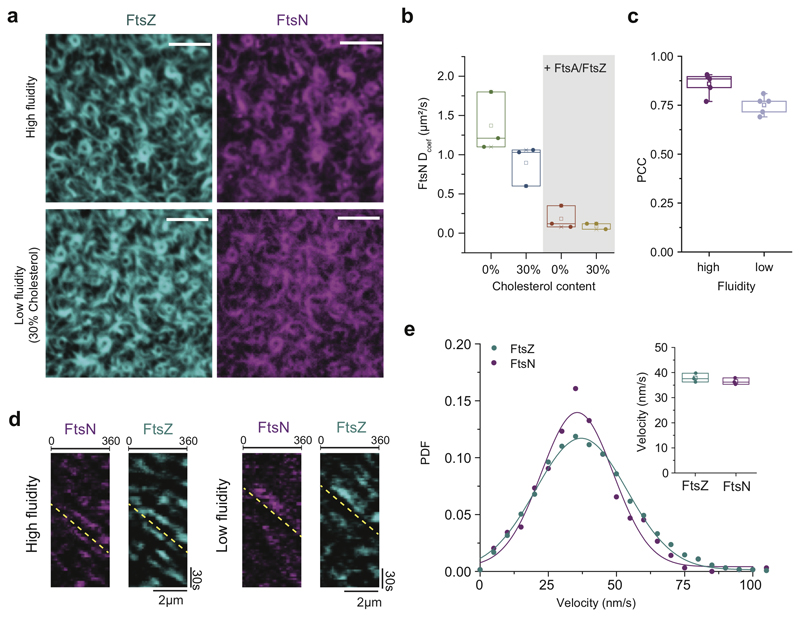
Due to the low density of FtsNcyto-TM in polymer-supported bilayers, an analysis of the dynamic ensemble behavior was problematic. To address how a decreased lateral diffusion affects co-migration of FtsN with FtsZ-FtsA filaments, we prepared a bilayer containing 30% cholesterol in addition to Ni2+-lipids to create low-fluidity membranes with a high density of correctly oriented peptides (ED Fig. 7). Diffusion of FtsNcytoHis on these membranes was about 1.5-fold decreased (ED Fig. 7b), but neither its colocalization efficiency nor co-migration with FtsZ were affected compared to high-fluidity membranes (ED Fig. 7c-e). These experiments demonstrate that FtsZ treadmilling is slow enough to be robustly followed by transmembrane proteins randomly diffusing on the membrane.
Our findings corroborate the proposed signaling role of the Z-ring, according to which FtsZ filaments act as dynamic binding platform that locally increases the density of downstream interaction partners23 to trigger a downstream response. First, we identified the molecular link between FtsA and the cytoplasmic peptides of the transmembrane cell division proteins FtsN and FtsQ using purified components. We thereby provide evidence for the direct interaction between the cytoskeleton located in the cytoplasm and transmembrane proteins mainly acting on the periplasmic side of the cell membrane. The fact that no direct interaction could be detected between FtsA and PBP3cytoHis suggests that the directed motion of individual PBP3 molecules observed in vivo3 originates from a more complex mechanism, presumably involving additional interactions with other cell division proteins such as FtsW or FtsN, or the cell wall24,25. Next, we found that FtsZ treadmilling gives rise to a collective directional migration of FtsQ and FtsN. Using single molecule imaging, we determined that this behavior originates from weak, transient interactions with the treadmilling FtsZ-FtsA scaffold. While proteins rapidly localize to the growing end of filaments, they rapidly disperse at the depolymerizing end of the treadmilling polymer. This mechanism establishes a dynamic accumulation of peptides that closely follows the translational motion of the membrane-bound filament and therefore establishes a moving zone of signaling activity (Fig. 4). Compared to static interactions, a moving, dynamic assembly via transient interactions allows to quickly respond to repositioning and constriction of the Z-ring without the need to disassemble stable complexes. This mechanism would also avoid local clustering of stable complexes at the septum, and thereby facilitating homogeneous synthesis of the new cell poles. Furthermore, we found that ionic interactions play an important role for the binding of FtsN to FtsA. As electrostatic interactions between highly charged peptides and protein filaments were also found to be important for the hetero-polymerization of ESCRT-III proteins26, this suggests that ionic interactions with weak residue-to-residue specificity might be a common mechanism for dynamic protein assemblies on a membrane surface.
Finally, although FtsN has long been thought to be the last protein to appear at the division site27, it is now evident that it is recruited by two consecutive, independent interactions8,28. At an early stage of divisome assembly, which we have reconstituted in this work, FtsN can be localized to midcell via the interaction of its cytoplasmic tail with FtsA in the Z-ring7,8,16,18 where it, together with ZipA, interacts with PG synthases, PBP1A and PBP1B, to initiate a pre-septal phase of PG synthesis17. After the onset of septation, FtsN can bind via its periplasmic SPOR domain to PG processed by division specific amidases, which leads to further accumulation of FtsN at the division site29. At the same time, FtsN interacts with the PG synthases PBP1B and PBP3 in the periplasm, and stimulates PG synthesis activity of PBP1B in vitro25,30. This situation would be reminiscent of the situation in S. aureus, where the divisome is first assembled in an FtsZ treadmilling-dependent step, before PG synthesis provides the contractile force for complete septum formation31.
The FtsZ-FtsA-FtsN/Q system is an example for a reaction-diffusion process in which the behavior of individual components is different to their emergent ensemble properties. It will be interesting to see if co-migration of proteins with dynamic filaments by transient binding is a general mechanism to organize and transmit spatiotemporal information in the living cell.
Methods
Reagents
All reagents, chemical, peptides and software used are listed in Supplementary Table 1.
Protein biochemistry
Purification and fluorescence labelling of FtsZ
The gene coding for FtsZ (UP P0A9A6) was cloned into a modified pML45 vector containing an N terminal His6-SUMO fusion protein plus seven residues (AEGCGEL) for maleimide-coupling of thiol-reactive dyes and further fluorescence detection (hereafter pML45–GCG-FtsZ vector)1. FtsZ was expressed in Escherichia coli C41 (DE3) cells, which were grown at 37°C in Terrific Broth (TB) medium supplemented with 100μg/ml ampicillin. Cultures were induced at an OD600 of 0.8 with 1 mM isopropyl-β-thiogalactopyranoside (IPTG) and incubated for 5 h at 37°C.
Purification of FtsZ was performed as follows. After centrifugation, the pellet was resuspended in buffer A (50 mM Tris-HCl pH 8.0, 500 mM KCl, 2 mM β-mercaptoethanol, 10% glycerol) plus 20 mM Imidazole and supplemented with EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics). The cell pellet was frozen and stored at −80°C until further processing. Cells were lysed using a cell disrupter (Constant Systems Cell TS 1.1) at a pressure of 1.36 kbar. The resulting lysate was incubated with 2.5 mM MgCl2 and 1 mg/ml DNase for 15 min, and the cell debris was removed by centrifugation at 60,000 × g for 30 min at 4°C. The clarified lysate was incubated with nickel-nitrilotriacetic (Ni-NTA) resin (HisPurTN Ni-NTA resin, ThermoFisher Scientific) for 1h at 4 °C. The resin was washed with buffer A containing 10 mM imidazole, followed by buffer A with 20 mM imidazole. The fusion protein was eluted with buffer A containing 250 mM imidazole. Subsequently, peak FtsZ fractions were dialyzed overnight at 4°C against buffer B (50 mM Tris-HCl pH 8.0, 300 mM KCl, 10% glycerol) in the presence of His6-tagged SUMO protease (Ulp1) at a protease to sample molar ratio of 1:100. This cleavage left the peptide AEGCGEL at the N terminus of the protein. The digested sample was passed several times through Ni-NTA resin (HisPurTN Ni-NTA resin, ThermoFisher Scientific) previously equilibrated with buffer B to remove His6-containing molecules. The flow-through was collected, and active to polymerize fraction of FtsZ was enriched by CaCl2-induced polymerization. To this end, the sample buffer was exchanged to buffer C (50 mM PIPES pH 6.7, 10 mM MgCl2) using a PD10 desalting column and the sample was warmed to room temperature. After addition of 10 mM CaCl2 and 5 mM GTP the sample was incubated for 20 min at room temperature. To pellet polymeric FtsZ the mixture was centrifuged at 15,000 × g for 2 min, a gel-like pellet was collected and resuspended in buffer D (50 mM Tris-HCl pH 7.4, 50 mM KCl, 1 mM EDTA, 10% glycerol).
In all cases, unless otherwise stated, protein identity and purity were assessed by 12.5% Glycine-SDS-PAGE stained with Coomassie blue, proteins were concentrated by ultrafiltration with Vivaspin filter devices of 5-KDa cut-off (Sartorius) and protein concentration was determined by Bradford assay.
For specific labelling of FtsZ, thiol-reactive dyes Alexa Fluor488 C5 Maleimide (ThermoFisher Scientific) or Sulfo-cyanine 5-maleimide (Lumiprobe) were dissolved in DMSO according to the manufacturer’s instructions. FtsZ was incubated first with 100 times molar excess of TCEP for 20 min at room temperature, and then with 10 times molar excess of the thiol-reactive dye Alexa Fluor488 C5 Maleimide or Sulfo-cyanine 5-maleimide (diluted in DMSO). Immediately after, the reaction was extensively dialyzed against buffer D overnight at 4°C and loaded on a PD10 desalting column to remove CaCl2, GTP and free dye. Peak fractions were collected and after determining the protein concentration, FtsZ was aliquoted, flash frozen in liquid nitrogen and stored at -80°C.
Purification and fluorescence labelling of FtsA
The gene coding for FtsA (UP P0ABH0) was cloned into a modified pTB146 vector as described elsewhere2. The resulting vector, pML60, contains an N terminal His6-SUMO fusion protein plus a pentaglycine tag for fluorescent labelling. FtsA was expressed in E. coli C41 (DE3) cells, which were grown at 37 °C in 2xYT medium supplemented with 100μg/ml ampicillin. Cultures were induced at an OD600 of 0.6 with 1mM IPTG and incubated overnight at 18 °C.
Cells were harvested by centrifugation (5,000 × g, 30 min at 4 °C), frozen in liquid nitrogen and stored at -80 °C until further use. The pellet was resuspended in buffer A (50 mM Tris-HCl pH 8.0, 500 mM KCl, 10 mM MgCl2) supplemented with 1 mg/ml lysozyme, EDTA-free protease inhibitor cocktail tablets, 1mg/ml DNase I, 0.5 mM DTT and 0.5 mM ADP (DTT and ADP were freshly added). Cells were lysed using a cell disrupter at a pressure of 1.36 kbar. Cell debris was removed by two successive centrifugation steps at 24,000 × g for 1 h and 35,000 × g for 30 min at 4°C. The clarified lysate was incubated with nickel-(tris-carboxymethyl ethylene diamine) resin (Protino Ni-TED, Macherey Nagel) for 1h at 4°C. The resin was washed with buffer A and buffer A supplemented with 5 mM imidazole. The fusion protein was eluted using buffer A with 250 mM imidazole. Immediately after the affinity purification buffer A was exchanged to buffer B (50 mM HEPES-KOH pH 7.5, 500 mM KCl, 10 mM MgCl2, 0.5 mM ADP, 20% glycerol) using a PD10 column, and the protein was stored at 4°C overnight. The sample was incubated at 30°C for 1.5 h in the presence of Ulp1 at a protease:sample molar ratio of 1:100. The cleaved sample was passed several times through Protino Ni-TED resin, previously equilibrated with buffer B to remove His6-containing molecules and FtsA was collected in the flow-through.
For TIRF microscopy and single molecule tracking, FtsA was labeled with fluorescent dyes using sortagging32 where a pentaglycine at the N-terminus of FtsA was conjugated to the TMR-dye labelled peptide by Sortase 7M from S. aureus (Addgene plasmid number #51141). The reaction was performed by mixing 0.5 -1 mM LPETG/A-containing probe (from 5 mM stock in DMSO) with 10 μM target protein and 10 μM Sortase 7M during overnight dialysis into buffer B at 4°C. The protein was further purified by size-exclusion chromatography on a HiLoad Superdex 200 10/300, previously equilibrated with buffer B (supplemented with 10% of glycerol) during which free labeled peptide and His6-sortase were removed. The final protein corresponding to labelled monomeric FtsA was collected.
For MST and analytical ultracentrifugation experiments, FtsA was purified as described before1, but with 10 mM CHAPS added during tag-cleavage. The detergent was later removed using a Hi-Trap Desalting column, before storing the protein in storage buffer (50 mM HEPES-KOH pH 7.5, 500 mM KCl, 10mM MgCl2, 1mM ADP, 10% glycerol). The protein was labelled with amine-reactive Alexa647-NHS ester or Alexa488-NHS ester following the instructions by the manufacturer. Briefly, FtsA was equilibrated in storage buffer and then mixed with the probe in DMSO at a 1:1 protein:probe molar ratio for 30 min on ice with stirring. Unbound probe was removed by desalting into the same buffer using a 5 ml HiTrap desalting column. FtsA concentration was quantified by Bradford Assay and dye concentrations were determined by absorbance measurements at 690 nm and 490 nm for AlexaFluor647 and AlexaFluor488, respectively.
Purification of full length FtsN
Full-length FtsN (UP P29131) modified with a C-terminal oligohistidine tag was expressed in E. coli BL21(DE3) pFE4233 and purified as described previously30 with minor modifications. Briefly, overproduction was induced by the addition of 1 mM IPTG and cells were incubated for 2 h at 37°C. Cells were harvested by centrifugation (7000×g, 15 min, 4°C), and then resuspended in buffer A (25 mM sodium phosphate pH 6.0, 1 M NaCl) in the presence of protease inhibitor cocktail (Sigma, USA) and 100 μM phenylmethylsulfonylfluoride (PMSF). After disruption by sonication (Branson Digital, USA), the sample was centrifuged (130 000×g, 1 h, 4°C) and the resulting membrane pellet was resuspended in buffer B (25 mM sodium phosphate pH 6.0, 1 M NaCl, 40 mM imidazole, 1% dodecyl maltoside (DDM)) and incubated overnight at 4°C. After centrifugation (130 000×g, 1 h, 4°C), the supernatant was applied to a 5 ml HisTrap HP column using an FPLC system. The column was washed with four column volumes of buffer C (25 mM sodium phosphate pH 6.0, 1 M NaCl, 40 mM imidazole, 0.05% DDM). Bound protein was eluted step-wise with buffer D (25 mM sodium phosphate pH 6.0, 1 M NaCl, 400 mM imidazole, 0.05% DDM). Full length FtsN (His6) was dialyzed into buffer E (25 mM sodium phosphate pH 6.0, 500 mM NaCl, 0.05% DDM, 10% glycerol) and stored in aliquots at −80°C.
Purification and labelling of transmembrane FtsNcyto (FtsNcyto-TM)
A truncation of full length FtsN (FtsN1-54) was cloned into vector pTB146. The resulting vector (pML170_CysFtsN-TMD) contains an N-terminal His6-SUMO tag followed by Gly-Cys-Gly-Gly used for labelling. FtsNcyto-TM construct was expressed in E. coli BL21 cells, which were grown in TB medium supplemented with 100 μg/ml ampicillin at 37°C; and induced at OD of 0.8, followed by 4h of expression at 37°C. Cells were harvested at 10000×g for 15 min at 4°C. The pellet was collected and resuspended in a buffer A (25 mM Tris, 1 M NaCl, pH 6.0) supplemented with 1 mg/ml lysozyme, EDTA-free protease inhibitor cocktail tablets, 1mg/ml DNase I, 1 mM DTT. Cells were lysed using a cell disrupter at a pressure of 1.36 kbar. The lysate was centrifuged at 130000×g for 1h, at 4°C; afterwards resuspended in a buffer B (25 mM Tris, pH 6.0, 1 M NaCl, 5 mM imidazole, 1% Tx100, 1mM DTT). To extract the transmembrane protein the suspension was incubated in buffer B overnight at 4°C upon gentle mixing, followed by centrifugation at 130000×g for 1h at 4°C. The obtained supernatant was incubated with a Protino resin for 1h at 4°C. The resin was washed with buffer C (25 mM Tris, 1 M NaCl, 0.25% Tx100, pH 6.0) and buffer C supplemented with 5 mM imidazole. The fusion protein was eluted with 250 mM imidazole buffer C. For labelling, the eluted protein was incubated with a thiol-reactive Sulfo-Cy5 dye at 4°C overnight, and simultaneously dialysed into imidazole-free buffer C. Afterwards, a second affinity purification was performed to remove an excess of free dye. To cleave off the SUMO-tag, a 10x excess of Ulp1 was added to the labelled FtsN-TM and incubated at 4°C overnight during dialysis into a storage buffer (25 mM Tris/HCl, 500 mM NaCl, 0.25% Triton X-100,10% glycerol, pH 6.0). The cleaved tag and Ulp-1 (both His6-tagged) were removed by reverse affinity purification using Thermo Ni-NTA resin. The concentration of the purified FtsN-TM was measured using BCA assay against the BSA standard.
Peptide reconstitution and labelling
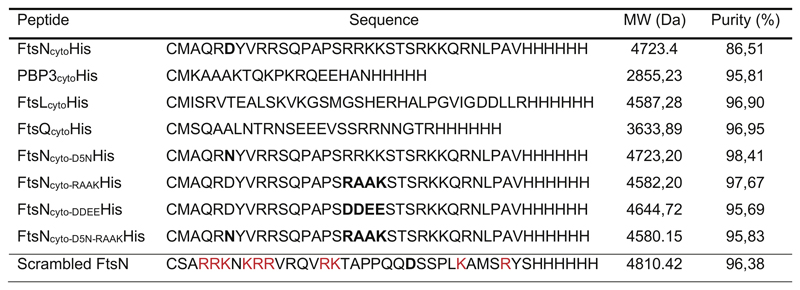
The cytoplasmic peptides of FtsN, PBP3, FtsL and FtsQ as well as the FtsN mutants D5N, DDEE and RAAK, containing a 6-His tag and an N-terminal cysteine residue were purchased from Biomatik (Extended Data 8).
For labelling the lyophilized peptides were dissolved in buffer A (50 mM HEPES-KOH pH 7.4, 150 mM KCl, 10% glycerol) to 10 mg/ml. Prior to the labelling step, a 20x molar excess of TCEP was added and incubated for 20 min at ambient temperature. Afterwards a 10 times molar excess of thiol-reactive maleimide dyes was added, and the sample was incubated either overnight at 4°C or for 4 h at room temperature. The free dye was removed by affinity purification using Ni-NTA resin (HisPurTN Ni-NTA resin, ThermoFisher Scientific). The beads were washed extensively with 100 column volumes of buffer A, 100 column volumes of buffer A containing 10 mM Imidazole and eluted with 850 μL of buffer A containing 250 mM Imidazole. The peptides were dialyzed into storage buffer B (20 mM HEPES-KOH pH 6.0, 150 mM KCl, 10% glycerol). The concentration was determined using the Pierce BCA Protein assay kit. Samples were aliquoted and stored at -80 °C, and used within 4 weeks.
Microscale thermophoresis
For MST experiments, 25 nM FtsA labelled with AlexaFluor647 was titrated with increasing concentrations of unlabeled full-length FtsN, PBP3cytoHis, FtsQcytoHis, FtsLcytoHis or FtsNcytoHis. Titrations with full-length FtsN were equilibrated in 10 mM HEPES-KOH pH 7.4, 2 mM MgCl2, 0.2 mM TCEP, 0.19% DDM and either 100 or 300 mM KCl. Titrations with the cytoplasmic tail peptides were equilibrated in 50 mM Tris-HCl pH 7.4, 150 mM KCl, 5 mM MgCl2 and 0.2 mM TCEP. After 10 min incubation at room temperature, samples were loaded in premium coated capillary tubes (NanoTemper, Germany) and measured in a Monolith NT.115 (NanoTemper, Germany) equipped with a blue and a red filter set. Data were acquired using 60% MST and 50% LED settings. Red fluorescence was measured for 3 s before applying a thermal gradient for 20 s. Binding curves were obtained by plotting the normalized change in fluorescence intensity after the 20 s gradient against the concentration of titrated protein or peptide. Binding curves were then fitted using a Hill Equation.
Analytical ultracentrifugation
Sedimentation velocity (SV) experiments were performed to study the oligomerization of full length FtsN in the presence of detergent as well as its association with FtsA. For the latter, FtsA labelled with Alexa488 was used in order to track changes in FtsA sedimentation upon addition of FtsN. Samples were thoroughly dialyzed in the indicated buffers before the SV experiments. FtsN was loaded at 9-20 μM and equilibrated in buffer A (10 mM HEPES-KOH pH 7.4, 2 mM MgCl2, 0.2 mM TCEP, 0.19% DDM and 100, 200 or 300 mM KCl). For interaction assays samples were equilibrated in the buffer A with 100 mM KCl. An FtsA-Alexa488 was loaded at 4 μM with full length FtsN present either at 1:1 or 1:4 FtsA:FtsN molar ratios. Experiments were carried out in a XL-I analytical ultracentrifuge (Beckman-Coulter Inc., USA) equipped with UV-visible and interference detection systems. Sedimentation profiles were registered by Raleigh interference and by absorbance at 280 nm in experiments with FtsN only or at 495 nm when FtsN was mixed with FtsA-Alexa488. Sedimentation coefficient distributions were calculated by least-squares boundary modeling of sedimentation velocity data using the c(s) method as implemented in SEDFIT 34. Absorbance and Raleigh interference measurements were combined to study FtsN solubilized in DDM and to quantify the amount of bound DDM per molecule of FtsN 35
Surface preparation
Glass coverslips were cleaned in piranha solution (30% H2O2 mixed with concentrated H2SO4 at a 1:3 ratio) for 30 min, and extensively washed and sonicated for 10 min in double distilled H2O. Cleaned coverslips were stored for no longer than one week under water. Before each experiment, glass coverslips were dried with compressed air and cleaned in air plasma for 10 min. In the experiments with the transmembrane FtsN, glass coverslips were additionally modified with 2000kDa-PEG-palmitic acid film36. The reaction chamber was prepared by attaching a 0.5 ml Eppendorf tube, with its conical end removed, on a coverslip using ultraviolet glue (Norland optical adhesive 63), and exposing the chambers to UV light for 5 min.
Preparation of small unilamellar vesicles (SUVs) and proteoliposomes
For initial experiments a mixture of DOPC, DOPG, DOGS-Ni-NTA at a ratio of 66:33:1 mol%37 was used. To improve peptide attachment to lipid membrane commercial mono-Ni-NTA lipid was replaced with 0.2 mol% of DODA-tris-Ni-NTA, which results in the same peptide density on the membrane surface and comparable results. To decrease membrane fluidity a standard mixture of DOPC/DOPG/DODA-tris-Ni-NTA was supplemented with cholesterol at final ratio of 36:34:0.2:30 mol%. For the experiments with the transmembrane peptide a mixture of DOPC/DOPG or an extract of E. coli polar lipids were used.
To prepare SUVs lipids were mixed in a glass vial at the desired ratio, afterwards blown-dried with filtered N2 to form a thin homogeneous film. To remove residual chloroform the vials were kept under vacuum for 2-3 h. The lipid film was rehydrated in a swelling buffer (50 mM Tris-HCl pH 7.4, 300 mM KCl) for 30 min at room temperature to a total lipid concentration of 5 mM.
The mixture was vortexed rigorously and the resulting dispersion of multilamellar vesicles was repeatedly freeze-thawed (8-10 times) in liquid N2. To obtain small unilamellar vesicles, the vesicle dispersion was tip-sonicated for 25 min on ice. The vesicles were centrifuged for 5 min at 10 000 × g and the supernatant stored at -20 °C and used within two weeks. In the experiments with cholesterol the mixture was kept at RT during sonication.
In the experiments with transmembrane proteins, SUVs (DOPC/DOPG mixture or an extract from E. coli polar lipids) were prepared in a swelling buffer supplemented with 1.12% Triton-x100. To reconstitute FtsNcyto-TM the transmembrane protein was first incorporated into a mixture of SUVs and detergent. To this end, FtsNcyto-TM (in 0.25% Triton-x100 buffer) was mixed with SUVs/Triton-x100 in a ratio 500 phospholipids per protein. After 10 min of incubation at RT, Triton-x-100 was removed by addition of 2-times excess of beta-cyclodextrin and subsequent incubation for 5 min at RT. Prepared proteoliposomes were diluted 10-times in the reaction buffer directly in the microscopy chambers.
Formation of supported lipid bilayers (SLBs) and reconstitution of transmembrane protein
Supported lipid membranes were formed by diluting the SUV dispersion in the reaction buffer (50 mM Tris-HCl pH 7.4, 150 mM KCl, 5 mM MgCl2) to a lipid concentration of 0.5 mM. Formation of SLBs on the glass surface was induced by addition of 4 mM CaCl2. For the low fluidity experiments with cholesterol SLBs were formed at 37°C, but cooled down to RT during microscopy. After 30 min of incubation at room temperature, the surface was washed 10 times with an excess of reaction buffer to remove non-fused vesicles. The supported lipid membranes were used shortly after preparation.
Membranes with the FtsNcyto-TM were prepared by incubation of fresh proteoliposomes for 30 min at 37°C on coverslips modified with the PEG-palmitic acid films. To rupture surface-bound vesicles 10% of 8 kDa PEG solution was added, for 15 min at 37°C followed by extensive washing.
TIRF microscopy
All experiments were performed using a total internal reflection fluorescence (TIRF) microscope, iMIC (Till Photonics, Germany), equipped with a 100x Olympus TIRF NA 1.49 DIC objective. The fluorophores were excited using laser lines at 488, 561 and 640 nm. The emitted fluorescence from the sample was filtered using an Andromeda quad-band bandpass filter (FF01-446-523-600-677). For dual-color experiments, an Andor TuCam beam splitter, equipped with a spectral long pass of 580 and 640nm and band pass filter of 525/50, 560/25, 710/80 (Semrock) was used. Time series were recorded using iXon Ultra 897 EMCCD Andor Cameras (X-8499 & X-8533) operating at 5 Hz frequency for standard acquisition and at 10 Hz for single molecule tracking.
Peptide-filament interaction experiments
For the self-organization of FtsA and FtsZ into cytoskeletal networks of treadmilling filaments on supported lipid membranes, we used FtsA (0.45-0.6 μM) and FtsZ (1.5-1.8 μM) in 100 μl of reaction buffer (50 mM Tris-HCl pH 7.4, 150 mM KCl, 5 mM MgCl2) in the presence of 4 mM ATP and 4mM GTP. The dynamic protein pattern was monitored by time-lapse TIRF microscopy for about 30 min, at 2 frames/s and 50 ms exposure time. The system reached steady state dynamics (defined by a constant intensity of membrane-bound proteins) within 10 min after the initiation of the reaction. To prevent photobleaching during acquisition, 30 mM D-glucose, 0.050 mg/ml Glucose Oxidase, 0.016 mg/ml Catalase, 1-10 mM DTT and 1 mM Trolox were added to the reaction buffer.
For colocalization experiments 1 μM of fluorescently labeled His-tagged peptides were added to a pattern of FtsZ-FtsA proteins undergoing steady-state reorganization on either 1 mol% Ni-NTA or 0.2 mol% tris-Ni-NTA SLBs, which gave comparable results. After addition of peptides, the time-lapse movies were recorded for another 20 min to estimate efficiency of colocalization during reorganization of the pattern. In these experiments FtsZ and His6-tagged peptides (FtsNcytoHis, FtsQcytoHis, FtsLcytoHis, PBP3cytoHis, FtsNcyto-D5NHis, FtsNcyto-RAAKHis and FtsNcyto-DDEEHis) were labeled with Cy5 or Alexa488/CF488 dyes. To study colocalization of FtsNcytoHis peptide with FtsA, a protein pattern was formed on 1 mol% Ni-NTA SLBs by mixing unlabeled FtsZ with TMR-FtsA (about 30% of labelled protein) in the presence of 1 μM of Cy5 labelled FtsNcytoHis. Efficient colocalization of peptides with FtsZ or FtsA was only observed on membranes containing Ni2+-chelating lipids.
To assess if formation of FtsZ-FtsA patterns on SLBs can recruit homogeneously distributed peptides attached to the membrane, Cy5-FtsNcytoHis (or CF488-) and Cy5-FtsQcytoHis were first added to SLBs in the absence of FtsA and FtsZ. We confirmed homogeneous distribution peptides in time-lapse movies, before nucleotides, unlabeled FtsA/labelled FtsZ were added to the reaction. For simultaneous sorting of two peptides FtsNcytoHis and FtsQcytoHis were labeled with Alexa488 and Cy5 dyes respectively, mixed in a molar ratio 1:3 and attached to 0.5 mol% tris-Ni-NTA SLBs. The homogeneously distributed signal for both peptides was confirmed before adding unlabeled FtsA/ FtsZ proteins and nucleotides.
To study the interaction of single Cy5-labeled FtsNcytoHis peptides with membrane-bound filaments we used membranes containing 0.2 % tris-Ni-NTA lipids to ensure a stable peptide immobilization.
To test the influence of treadmilling on the colocalization efficiency between FtsZ and FtsNcytoHis GTP was replaced with 1 mM GMPCPP. Movies of FtsZ-GMPCPP pattern were acquired at a slower frame rate of 5 s/frame to prevent excessive photobleaching.
Image processing and analysis
For image processing, image stacks were imported using FIJI software38. For data analysis raw, unprocessed time-lapse movies were used. For visualization purposes and to remove noise, all micrographs shown in the manuscript were processed with the “Walking average” plugin of ImageJ averaging the signal of 4 consecutive frames.
Colocalization analysis
For colocalization analysis, we first normalized the intensity of the time-lapse movie to correct for photobleaching and enhanced the contrast of both channels for visualization purposes. Next, movies were aligned with the “Linear Stack Alignment with SIFT” plugin prior to analysis to eliminate XY drift. To minimize the effect on non-specifically bound aggregates, regions of interest (20 x 20 μm) without protein aggregates were chosen for the colocalization analysis. The alignment of the two channels was adjusted with the 3TP align plugin (J. Anthony Parker, Beth Israel Deaconess Medical Center Boston). Finally we used the ImageJ plugin “Image CorrelationJ 1o” to quantify colocalization between the proteins and candidate peptides39. This plugin measures the crosscorrelation coefficient (Pearson’s correlation coefficient, PCC) values between -1 and 1. In our experiments, we reached maximal PCC values of ~0.9, which indicates a high degree of colocalization, while the minimum value varied was 0.05, due to some residual signal of non-colocalized peptides diffusing on the membrane.
Treadmilling analysis
To quantify treadmilling dynamics, we developed an automated image analysis protocol based on ImageJ and custom Python scripts.
For this method, we first applied a low pass Gaussian filter in space (0.5 pixels) and time (1.5 frames) to all frames of a time-lapse movie and subtracted consecutive frames from one another using ImageJ ImageCalculator command. This creates a new image moving fluorescent spots, corresponding to growth or shrinkage of filaments, which can be identified and tracked using ImageJ’s TrackMate40 toolbox. We used the LoG (Laplacian Gaussian) detector with an estimated diameter of 0.8 μm and discarded spots with a signal-to-noise ratio lower than 1 and with a track displacement distance smaller than 0.3 μm. To build the final trajectories we used the “Simple LAP tracker” with a “Max Linking Distance” of 0.3 μm, a “Maximal gap-closing distance” of 0 μm and “Max frame Gap” of 0 frames, and we only considered trajectories longer than 6 sec for analysis. To obtain velocity and directionality of the moving fluorescent spots, we further analyzed trajectories using a custom python script. We computed the mean-square displacement (MSD) for each individually trajectory and estimated their velocity by fitting a quadratic equation to 50% of the total track duration. A Gaussian function was fitted to the distribution of velocities to extract the mean velocity at each condition. In addition, we calculated the directional autocorrelation as a function of an increasing time delay (Δt).
To measure the intensity of growing and shrinking spots for FtsZ, FtsA and FtsNcytoHis, no pre-processing was performed, as normalizing the intensity would lead to a loss of information. The intensity values for each spot produced during growing and shrinking polymerization dynamics were extracted from the corresponding differential movie using TrackMate.
For the co-migration analysis of growing and shrinking spots (Extended Fig. 3c) dual-color movies with an acquisition rate of 1 frame/s. The alignment of the two channels was adjusted with the 3TP align plugin using raw images. Both channels were then subjected to the image subtraction and the PCC was measured as described above.
FRAP (Fluorescence recovery after photobleaching)
For FRAP experiments we allowed proteins to self-organize on supported lipid membranes containing either 0.2 mol-% of tris-Ni-NTA or 1 mol-% of mono-Ni-NTA. For experiments on FtsNcytoHis, the peptide was added to steady state FtsZ patterns, as described above, using membranes with tris-Ni-NTA lipids. FRAP experiments were performed with a frame rate of 2s/frame, and bleached area of about 15 μm x 15 μm. To obtain the half-time of fluorescence recovery, we fitted a mono-exponential function to the intensity trace using a Python-based Jython macro (Image Processing School Pilsen 2009).
To discriminate between fluorescence recovery via protein exchange or lateral diffusion a rectangular area was bleached, and fitted a 1D diffusion equation to the corresponding intensity profile41,42: where erf denotes the Gaussian error function, and The initial height of the well was given by the intensity profile at time zero after bleaching, normalized by the background signal. In case of FtsZ, only the first 10 s of the recovery movie were fitted, while for FtsA and FtsN first 100 s were taken into consideration. The mode of recovery is revealed by a linear function σ2=4Dt, where an increase in the slope σ2 in time allows to determine the lateral diffusion coefficient. The comparison of the recovery half-time at the edge vs center revealed much faster recovery at edge, confirming that our immobilization strategy allows us to probe only lateral diffusion. Accordingly, σ2 increased with time consistent with recovery solely due lateral diffusion.
Single molecule diffusion and confinement time analysis
Single-molecule localization was done in SLIMfast software using a localization module described elsewhere43. In this software, the tracking was performed in 2 phases: the first phase linking nearest-neighbors, while in the next phase fragments were linked into final trajectories. A Kalman filter was used to estimate the search radius within the user-defined parameters. The search radius was adapted to local density for frame-to-frame linking and final track assembly. To quantify the diffusion coefficient an MSD was calculated for each individual trajectory. Only trajectories longer than 10 frames were considered. The probability density distribution of diffusion coefficients calculated for FtsZ and FtsA was fitted with a Gaussian distribution. In case of FtsN a double-Gaussian fit was applied. To a steady-state pattern of FtsZ-FtsA, unlabeled FtsNcytoHis was added from bulk solution at a concentration of 1 μM supplemented with Cy5-labelled peptides FtsNcytoHis or FtsNcyto-DDEEHis at 10-20 pM. Images were acquired every 32 or 51 ms, with exposure times of 30 and 50 ms respectively. The trajectory set produced by SLIMfast revealed heterogeneous behavior of the FtsNcytoHis peptide within a single trajectory, where fast motions were alternating with the confinement to FtsZ-FtsA filaments. Such capturing behavior was shorter for FtsNcyto-DDEEHis. The distribution of diffusion coefficients was analysed using a two-component Gaussian function to estimate weighted contribution for each population. The slow population of FtsNcytoHis had more confined trajectories in comparison to FtsNcyto-DDEEHis. To discard contribution of non-specifically bound molecules, only trajectories of the fast population were considered for further analysis. The displacement distribution of the fast population for FtsNcytoHis and FtsNcyto-DDEEHis peptides was fitted with two-component Rayleigh distribution and the weighted contribution of short and long displacements was estimated for each peptide (Fig. 3e).
The confined regions within each trajectory were identified by spatial clustering, based on DBSCAN algorithm44 implemented into SLIMfast software45. To this end a spatial cluster with a search area of 200 nm, minimal time window of 10 frames and initial window of 100 frames (at 51ms/frame acquisition rate) was built. Afterwards, a single exponential including an y-offset was fitted to the lifetime distribution of the identified clusters to quantify an apparent confinement time.
Surface charge representation of FtsA
The surface charge of FtsA was obtained using a model generated by I-TASSER46 from the sequence of E. coli FtsA (UniProt P0ABH0). This model was than superimposed with the structure of a FtsA dimer in complex with the C-terminal peptide of FtsZ47 (PDB: 4A2A) using UCSF Chimera (www.cgl.ucsf.edu/chimera/).
Statistics and reproducibility
Statistical details of experiments are reported in the figure legends. For all box plots throughout this work, boxes indicate the 25–75th percentiles, whiskers the outlier, the midline indicates the median and square indicates the mean. P values are reported in figures and were calculated using a two-tailed Student’s t-test for parametric distributions.
Sample size is reported in the figure legends with n corresponding to independent experiments, and N replicates within the same experiment, in a different region of interest. No statistical test was used to determine sample size. The biological replicate (n) is defined as the number of independent experiments where a new protein pattern was assembled. Independent experiments in some cases were performed on the same coverslip, which can fit up to 6 reaction chambers. Unless differently stated in the figure legends, the graphs show mean ± sd, and the error bars are calculated and shown based on the number of independent experiments, as indicated. The distribution assumed to be normal for all biological replicates.
Extended Data
Extended Data Fig. 1. Colocalization of FtsNcytoHis and FtsQcytoHis with FtsZ filaments depends on their interaction with FtsA.
a. Left: The surface density of FtsNcytoHis scales linearly with density of mono-Ni-NTA (black dots) and tris-NTA-NTA lipids (red dots, mean±SD, n=3 or higher). Right: Colocalization efficiency of FtsNcytoHis to FtsA-FtsZ filaments at increasing concentrations of mono-Ni-NTA (mean±SD, n=3 or higher).
b.(i) The FtsZ pattern is not affected at 0.2 mol% tris-Ni-NTA (or mono-Ni-NTA at 1 mol%), n=5. Scale bars are 5 μm. (ii) FtsNcytoHis (0.5 μM) in solution does not affect the FtsZ-FtsA cofilament pattern and shows only weak binding to FtsA/FtsZ filaments, n=5. Scale bars are 10 μm. (iii) FtsQcytoHis shows no colocalization to FtsZ filaments (cyan) on membranes without Ni-NTA. Scale bars are 10 μm, n=3. (iv-v) FtsNcytoHis and FtsQcytoHis do not colocalize with FtsZ filaments when FtsA is replaced by ZipA, n=3. Scale bars are 5 μm.
c. PBP3cytoHis and FtsLcytoHis attached to 0.2 mol% tris-Ni-NTA lipid membrane remained homogeneously distributed in the presence of FtsA-FtsZ filaments. Representative micrographs from n=7 (PBP3) and n=3 (FtsL) independent experiments. Scale bars are 5 μm.
d. FtsNcytoHis does not interact with FtsZ. After addition of 0.5 μM FtsA in the same experiment FtsNcytoHis and FtsZ are organized into cytoskeleton patterns. A similar experiment was performed with FtsQcytoHis, n=3. Scale bars are 5 μm.
e. Sedimentation velocity (SV) characterization of protein-detergent complexes formed by the solubilization of 16 μM full-length FtsN in 3.72 mM DDM. Sedimentation velocity c(s) distributions obtained from the analysis of the absorbance (orange line) and Raleigh interference (grey dashed line) signals of FtsN complexes using the program SEDFIT. Main peaks at 4.1 ± 0.1 and 5.2 ± 0.1 S detected by Raleigh interference (short dash line, n=3) are compatible with FtsN monomers and dimers.
f. SV analysis of AlexaFluor488FtsA in the presence of full-length FtsN at a 1:1 FtsA:FtsN molar ratio (blue) and a ~1:4 FtsA:FtsN molar ratio (red), or in the absence of FtsN (black). The main peak of FtsA at ~2 S shifts to a higher s-value upon addition of FtsN, indicating the formation of higher molecular weight species, n=2.
g. Binding curves obtained by MST of Alexa647-FtsA titrated with corresponding peptides. Binding was observed only for FtsNcytoHis and FtsN-FL, n=3. See Extended Data 10 for fitting results.
Extended Data Fig. 2. Binding between FtsN and FtsA is dominated by electrostatic interactions.
a. Binding curves obtained from the MST thermograms of AlexaFluor647 FtsA titrated with full length FtsN at 100 (black symbols), 150 (green symbols) or 300 mM KCl (red symbols). The binding curves at the lower ionic strengths are biphasic with a first transition from 100 to 1000 nM FtsN and a second transition that could not be saturated. The interaction was significantly weaker in the presence of 300 mM KCl (red symbols). The Hill equation, with n fixed to 1 was used to calculate the midpoint of the first transition for the curves at 100 mM (black line) or 150 mM KCl (green line), resulting in apparent KD values of 250 ± 51 nM at 100 mM KCl or 734 ± 23 nM at 150 mM KCl, n=3.
b. The mutant peptides D5N (n=5), RAAK (n=5), D5N-RAAK (n=3) efficiently colocalized with the FtsZ filaments similar to the wildtype peptide, while FtsNcytoDDEEHis (n=5) mutant and the scrambled FtsN peptide (n=6) show only weak colocalization Scale bars are 5 μm.
c. Sequences of tested peptides. Dashed boxes correspond to the conserved D5 and the RRKK motifs, which are lost in the scrambled peptide.
d. The colocalization efficiency of the scrambled peptide (n = 7, 0.24 ± 0.1, mean±SD) was significantly decreased in comparison to FtsNcytoHis (n = 5, 0.62 ± 0.1) and FtsNcytoD5N-RAAKHis (n = 3, 0.55 ± 0.06) (p-value = 1.15*10-4 and 1.19*10-3 respectively). Each dot corresponds to an independent experiment acquired within the same batch of proteins. The boxes indicate the 25–75th percentiles, whiskers the outliers, the midline indicates the median and square indicates the mean. P-values were calculated using a two-tailed Student’s t-test for parametric distributions.
d. The colocalization efficiency of the scrambled peptide (n = 7, 0.24 ± 0.1, mean±SD) was significantly decreased in comparison to FtsNcytoHis (n = 5, 0.62 ± 0.1) and FtsNcytoD5N-RAAKHis (n = 3, 0.55 ± 0.06) (p-value = 1.15*10-4 and 1.19*10-3 respectively). Each dot corresponds to an independent experiment acquired within the same batch of proteins. The boxes indicate the 25–75th percentiles, whiskers the outliers, the midline indicates the median and square indicates the mean. P-values were calculated using a two-tailed Student’s t-test for parametric distributions.
e. Structure of the FtsA dimer in complex with the C-terminal peptide of FtsZ46. The surface charge of one FtsA monomer is shown. The 1C domain (dashed square) contains a large negatively charged patch, which is oriented towards the membrane surface, where it can interact directly with positively charged FtsN peptide.
Extended Data Fig. 3. Differential imaging of protein patterns.
a. Top: Representative pre-processed micrographs showing the pattern of FtsZ and FtsNcytoHis as well as FtsA and FtsNcytoHis. Middle: Illustration of image subtraction process to obtain a differential image. Bottom: Fluorescent spots corresponding to growing and shrinking ends of FtsZ filament bundles and associated proteins. Micrographs show representative results from n=16 independent experiments with FtsZ/FtsN and n=3 for FtsA/FtsN. Scale bars are 5 μm.
b. Fluorescence intensity histograms for spots of FtsZ, FtsA and FtsNcytoHis from one representative experiment. The histograms for growth and shrinkage showed similar distributions and intensities, suggesting symmetric behavior with a small shift towards brighter values for the shrinking ends.
c. Differential image of fluorescent spots overlaid with result from detection (purple circles) and tracking (coloured lines). Represent image from more more than 20 independent experiments.
d. Instantaneous velocities of fluorescent spots (growth) of FtsNcytoHis and FtsQcytoHis, FtsZ and FtsA and lognormal fits. FtsZ (v = 42.6 ± 32.1 nm/s, mean±SD; n = 16; cyan), FtsA (v = 50.1 ± 33.5 nm/s; n = 3; green, p-value = 0.10), FtsQcytoHis (v = 46.8 ± 37.1 nm/s; n = 3; dark yellow, p-value = 0.21) and FtsNcytoHis (v = 44.3 ± 38.2 nm/s; n = 13, magenta, p-value = 0.94). P-values were calculated using a two-tailed Student’s t-test for parametric distributions.
e. Kymographs obtained from raw and differential movies of FtsZ, FtsA, FtsNcytoHis and FtsQcytoHis, illustrating that image subtraction simplifies detection of the dynamic behavior, representative kymographs from n=20 rings.
f. Mean-squared displacement calculated for individual trajectories. Positive slope corresponds to directed motion, red straight line illustrates diffusive behavior.
g. Comparison between different methods for treadmilling speed quantification: directly from kymographs (Kymo, n = 20 from 3 (FtsA) and 5 (FtsN and FtsZ) independent experiments) and from automated treadmilling analysis via differential imaging (MSD, n(FtsZ) = 16 independent experiments with 30398 tracks total, n(FtsN) = 13 independent experiments with 20809 tracks total, n(FtsA) = 3 independent experiments with 3964 tracks total). The boxes indicate the 25–75th percentiles, whiskers the outliers, the midline indicates the median and square indicates the mean.
Extended Data Fig. 4. GMP CPP slows down filament reorganization dynamics, but does not affect colocalization between FtsZ and FtsA.
a. Filament patterns formed by FtsZ and FtsA in the presence of either GTP (top, n=3) or GMP CPP (bottom, n=2) before and after FtsNcyto-His addition. Scale bars are 10μm.
b. Reorganization dynamics of the FtsZ filament pattern is slowed down in the presence of GMPCPP compared to pattern formed with GTP. Graph shows temporal autocorrelation of fluorescence time lapse movies, fast decay indicates rapid reorganization. n=3 (GTP), n=2 (GMPCPP). Line represents the mean, error bars the standard deviation.
c. PCC quantification reveals comparable colocalization between FtsZ and FtsA in the presence of different nucleotides. The presence of FtsN decreases PCC value, however this decrease is nucleotide-independent (n=3 (GTP), n=2 (GMPCPP). The boxes indicate the 25–75th percentiles, whiskers the outliers, the midline indicates the median and square indicates the mean.
Extended Data Fig. 5. FRAP analysis.
a. Micrographs of FRAP experiments (top) and corresponding intensity profiles across photobleached regions of Cy5-FtsZ (left, cyan, n=5), TMR-FtsA (middle, green, n=3) and CF488-FtsNcytoHis (right, magenta, n=6) at different time points. The first micrograph shows the area before bleaching, micrographs at 0s correspond to the pattern at the first frame after bleaching. Scale bars are 10 μm. Fitting a Gaussian error function to the profiles to obtain σ2(t) points reveals changes in the profile shape over time (inset plots). For FtsZ, σ2 remains constant, showing that fast recovery of the photobleached region is dominated by a homogenous exchange of FtsZ monomers instead of lateral diffusion. For FtsA and FtsNcytoHis, σ2 increases with time, consistent with a strong contribution of lateral diffusion for recovery. Shaded areas represent the standard deviation. This can also be seen by the delayed recovery of fluorescence in the center of the bleached area compared to its edges. See Supplementary Video 7.
b. Recovery of FtsNcytoHis and rhodamine-DOPE are comparable, within the error bar, in the absence of FtsA and FtsZ. The recovery of FtsNcytoHis (magenta) within the rectangular profile is slowed down significantly after FtsA and FtsZ have been added (FtsN alone: Ƭ0.5 = 14.9 ± 2.6 s, mean±SD, FtsN + FtsA/Z:Ƭ0.5 = 75.4 ± 25.6 s (n = 4)), whereas rhodamine recovery (grey) decreased slightly, presumably due to crowding effects by membrane-attached FtsA/FtsZ co-filaments. (rhodamine-PE: Ƭ0.5 = 12.8 ± 3.4 s, rhodamine-PE + FtsA/Z: Ƭ0.5 = 17.7 ± 0.2 s (n = 3)). Scale bars are 10 μm. Line represents the mean, error bars the standard deviation.
Extended Data Fig. 6. Single-molecule analysis of FtsNcyto-TM in supported bilayers supports a diffusion-and-capture mechanism.
a. Schematic illustration of the reconstitution approach: i. FtsNcyto-TM was incorporated into SUVs by detergent extraction ii. Proteoliposomes were adsorbed on a surface modified with a dense film of PEG2000-palmitic acid and vesicle rupturing and formation of polymer-SLB was induced by addition of PEG 8 kDa. iii. FtsZ and FtsA are then allowed to self-organize on the membrane surface.
b. Micrographs showing colocalization of FtsNcyto-TM-Cy5 with AlexaFluro488-FtsZ. An FtsNcyto-TM pattern similar to FtsZ was observed after addition of FtsZ and FtsA confirming recruitment of the transmembrane peptide to FtsZ-FtsA filaments, n=2. To enhance visualization of the filament bundles, images were processed by subtracting an averaged background over a circular area of 20 px, scale bars are 10 μm.
c. Micrographs and detected trajectories of single FtsNcyto-TM peptides alone, in the presence of 1.5 μM FtsZ/ 0.5 μM FtsA (n=2) and with of 3 μM FtsZ/ 0.5 μM FtsA (n=1). Scale bars are 10 μm for micrographs and 5 μm for corresponding trajectory maps.
d. Left: Quantification of the diffusion coefficients of single molecules reveals presence of two populations for FtsNcyto-TM (left n=2, N=2), indicative for non-specifically stuck particles. Addition of FtsA and FtsZ slows down mobility of fast diffusing population (left). Right: Box plot of obtained diffusion constants. Although FtsNcyto-TM (n=2, N=2) diffuses about 2-fold slower than FtsNcytoHis (n=2, N=4), their diffusivities were comparable after addition of FtsZ and FtsA (left). The diffusion coefficients are 0.70 ±.0.14 for FtsNcyto-His and 0.34 ± 0.21 for FtsN-TM without FtsA/Z; 0.15 ±.0.04 for FtsNcyto-His and 0.21 ± 0.07 for FtsN-TM + in the presence of FtsA/Z; mean ± SD. The boxes indicate the 25–75th percentiles, whiskers the outliers, the midline indicates the median and square indicates the mean.
e. Directional autocorrelation analysis confirms random non-correlated behavior of single FtsNcyto-TM molecules at 1.5 μM (left) and 3.0 μM (right) FtsZ, from all trajectories in one representative experiment. Line represents mean, error bars the standard deviation.
f. Confinement time analysis of FtsNcyto-TM (0.66 ± 0.1s, mean±SD, n=1, N=3) shows similar confinement time as FtsNcyto-His.
Extended Data Fig. 7. Decrease in membrane fluidity does not affect colocalization and comigration with treadmilling FtsZ filaments.
a. The FtsZ filament pattern and colocalization with FtsNcytoHis is not affected on low fluidity membranes (supplemented with 30% cholesterol), n=3, Scale bars are 5 μm.
b. Addition of cholesterol to a standard lipid composition slows down diffusion of FtsNcytoHis, n=3 (bulk diffusion constant obtained from FRAP profile analysis) Dcoef FtsNcyto-His0%Chol = 1.37 ± 0.31 Dcoef FtsNcyto-His30%Chol = 0.90 ± 0.21. After addition of FtsA and FtsZ, FtsNcytoHis shows the same diffusion constant on high and low fluidity membranes as diffusion is limited due to interaction with FtsZ/FtsA cofilaments. Dcoef FtsNcyto-His0%Chol = 0.18 ± 0.12 Dcoef FtsNcyto-His30%Chol = 0.10 ± 0.03; mean ± SD.
c Colocalization analysis reveals that a decrease in membrane fluidity does not affect colocalization efficiency between FtsNcytoHis and FtsZ, n=5.
d. Differential kymographs, n=3 and e. treadmilling analysis (n=3) of FtsZ and FtsN co-migration on low-fluidity membranes with 30% cholesterol. Velocity histograms and fit to a Gaussian distribution (solid line) did not reveal any significant difference in the velocities of FtsZ (v = 35.6 ± 13.2 nm/s; n = 3; cyan) and FtsN (v = 37.1 ± 16.8 nm/s; n = 3; magenta) on low fluidity membranes, p-value = 0.33. The velocity values represent mean and SD from the corresponding Gaussian fits (R2 = 0.97-0.99). The boxes indicate the 25–75th percentiles, whiskers the outliers, the midline indicates the median and square indicates the mean. P-value was calculated using a two-tailed Student’s t-test for parametric distributions.
Extended Data Fig. 8. Peptide sequences.
Sequences of peptides used in this study
Extended Data Fig. 9. Summary of PCC for different peptides (correspond to Fig1g-h).
Extended Data Fig. 10. Summary of the binding constants for different FtsN constructs to FtsA.
Supplementary Material
FtsNcytoHis is homogeneously distributed on a membrane when no other proteins are present (0-30s, magenta, right). After addition of FtsA and FtsZ to the reaction chamber (noisy signal in both channels), FtsZ filaments assemble on the membrane (cyan, left). Within 5 minutes after the addition, FtsNcytoHis is sorted into a pattern resembling the FtsZ filaments. The experiment was performed on 1 mol% mono-Ni-NTA membranes with 1.5 μM Cy5-FtsZ, 0.5 μM FtsA and 1 μM CF488-FtsNcytoHis. The movie was acquired at 2 s/frame and corresponds to the micrographs in Fig. 1b, n=5.
Top: Single molecule dynamics of Cy5-FtsZ, Cy5-FtsA and Cy5-FtsNcytoHis.
Bottom: Single molecule dynamics merged with the corresponding Alexa488-FtsZ pattern.
Single molecules of FtsZ display no diffusive behavior at all, monomers within the filaments are constantly exchanged by binding/unbinding, as expected for treadmilling filaments.
Single molecules of FtsA display a mixed behavior of lateral diffusion and binding/unbinding. Single molecules of FtsNcytoHis move by fast lateral diffusion on the plane of the membrane, showing transient confinement to FtsZ-FtsA co-filaments.
Each experiment was performed on 0.2 mol% Tris-Ni-NTA membranes with 1.5 μM Alexa488-FtsZ, 0.5 μM FtsA and 1 μM FtsNcytoHis present. For visualization of the single molecules, small amounts of Cy5-labeled FtsZ, FtsA or FtsNcytoHis was added (100 pM, 75 pM and 75 pM respectively). The movies were recorded at 0.228 s/frame and correspond to Fig. 3a.
Left. Diffusion of FtsNcyto-TM in a polymer-supported lipid membrane. FtsNcyto-TM shows diffusion and capture behavior at normal protein concentrations (middle, FtsA 0.5 μM, FtsZ 1.5 μM) and at an increased FtsZ concentration of 3 μM. Under no conditions we found directional motion of FtsNcyto-TM.
Representative movie of single molecules of Cy5-FtsNcytoHis in the presence of FtsA-FtsZ filaments acquired at 228 ms. Signle particle FtsN undergoes rapid random motion with the transient confinement to FtsZ filaments, corresponds to Fig. 3f
Representative trajectories showing the differences in the diffusive motion of FtsNcyto-DDEEHis (gray track, left) and FtsNcytoHis (magenta, right). FtsNcyto-DDEEHis covers a large area and does not remain at isolated spots. FtsNcytoHis shows similar diffusive motion in the beginning until it is captured by FtsZ-FtsA co-filaments where it shows confined behavior. The red links represent gaps in trajectory linking.
CF488-FtsNcytoHis shows stronger colocalization to FtsZ in the presence of GTP (top row) than in the presence of GMPCPP (bottom row). Top: CF488-FtsNcytoHis (magenta, right) was added to the reaction chamber with FtsA and FtsZ co-filaments at steady state (cyan, left). Bottom: CF488-FtsNcytoHis (magenta, right) was added to a reaction chamber containing 0.5 μM FtsA and 1.5 μM Cy5-FtsZ (cyan, left). In both experiments, 0.5 μM FtsA, 1.5 μM Cy5-FtsZ and 1 μM CF488-FtsNcytoHis, as well as 4 mM GTP and 2 mM GMPCPP, respectively were used. For GTP experiments, the acquisition rate was kept at 2s/frame, while the rate was decreased in the presence of GMPCPP to 5 s/frame to limit photobleaching. The raw files were processed to show every 5th (GTP) or every 2nd (GMPCPP) frame, resulting in a 10 s/frame movie. The movies correspond to Fig. 2c.
FtsNcytoHis and FtsQcytoHis are simultaneously bound and homogeneously distributed on the membrane before addition of FtsA-FtsZ (0-1min, cyan and yellow respectively). After the addition of FtsA and FtsZ, FtsNcytoHis and to FtsQcytoHis peptides co-localize in a filament-like patterns.
The experiment was performed on 0.5 mol% Tris-Ni-NTA membranes with 0.15 μM Alexa488-FtsNcytoHis, 0.75 μM Cy5-FtsQcytoHis, 0.5 μM FtsA and 1.5 μM FtsZ. The movie was acquired at 2 s/frame and corresponds to Fig. 1d.
FtsNcytoHis shows similar colocalization to FtsZ (left) and FtsA (right).
Top: CF488-FtsNcytoHis (magenta, middle) was added to FtsA/Cy5-FtsZ co-filaments at the steady state (cyan, left). A merge of both channels shows colocalization between CF488-FtsNcytoHis and treadmilling filaments of FtsZ (bottom left).
Bottom: CF488-FtsNcytoHis (magenta, - top right) was added to FtsZ-TMR-FtsA co-filaments at the steady state (green, top left). A merge of both channels shows colocalization between CF488-FtsNcytoHis and co-treadmilling TMR-FtsA (bottom right). The experiments were acquired at 2 s/frame and correspond to data in Fig. 1e-g.
Top left: overlay of FtsZ and FtsNcytoHis pattern, bottom left: overlay of the differential image of FtsZ and FtsNcytoHis. The differential movie shows close coupling between FtsNcytoHis and the directional motion of treadmilling FtsZ filaments. Right: overlay of FtsNcytoHis (cyan) and FtsZ (magenta), moving spots correspond to growing filament bundles move along the filaments. The experiment was performed on a 1 mol% mono-Ni-NTA membrane with 0.5 μM FtsA, 1.5 μM Cy5-FtsZ and 1 μM CF488-FtsNcytoHis and acquired at 2 s/frame. The differential movie was created as described in the methods section. The movie corresponds to Extended Data Fig. 3a.
FtsZ, FtsA and FtsNcytoHis dynamics arise from different processes, as shown by FRAP experiments on supported lipid bilayers. The difference in the recovery times of the patterns is evident, with FtsZ showing the fastest recovery, mainly by exchange with FtsZ in the buffer and no influence of diffusion on the membrane, while fluorescence recovery of FtsA and FtsNcytoHis is slower and dominated by lateral diffusion.
Each experiment was performed in the presence of 1.5 μM FtsZ, 0.5 μM FtsA and 1 μM FtsNcytoHis labelled with Cy5, TMR and CF488CF, respectively. All movies were acquired at 2 s/frame and correspond to Extended Data Fig. 5b.
FtsQcytoHis is homogeneously distributed on a membrane prior to addition of FtsA and FtsZ (0-1min, yellow, middle). After addition of FtsA and FtsZ, FtsZ filaments assemble on the membrane (cyan, left). Within several minutes, membrane-bound FtsQcytoHis is organized into a pattern resembling FtsZ filaments.
The experiment was performed on 0.2 mol% Tris-Ni-NTA membranes with 1.5 μM Alexa488-FtsZ, 0.5 μM FtsA and 1 μM Cy5-FtsQcytoHis. Acquisition rate was 2s/frame. The movie corresponds to the micrographs in Fig. 1c.
Acknowledgements
We acknowledge members of the Loose lab at IST Austria for helpful discussions, in particular Paulo Caldas for help with the treadmilling analysis. Mercedes Jimenez, Ana Raso and Noelia Ropero for providing AlexaFluor488- and AlexaFluor647-labelled FtsA for MST and AUC experiments. We want to thank Changjiang You for providing DODA-tris-NTA phospholipids as well as Jacob Piehler and Christian Richter (Department of Biology, University of Osnabruck, Germany) for the SLIMfast single molecule tracking software and help with the confinement analysis. We thank Jeff Errington and Heath Murray (both Newcastle University, UK) for critical reading of the manuscript and Jan Brugués (MPI-CBG and MPI-PKS, Dresden, Germany) for help with MATLAB programming and reading the manuscript. This work was supported by the European Research Council through grant ERC-2015-StG-679239 to ML, HFSP LT 000824/2016-L4 and EMBO ALTF 1163-2015 to NB, a grant from the Ministry of Economy and Competitiveness of the Spanish Government BFU2016-75471-C2-1-P to CA and GR, and a Wellcome Trust Senior Investigator award (101824/Z/13/Z) and a grant from BBSRC (BB/R017409/1) to WV.
Footnotes
Reporting Summary
Further information on research design is available from the authors upon request.
Code availability
The Fiji macro used for differential image processing and Matlab codes for treadmilling velocity analysis, life-time analysis, FRAP profile analysis are available upon request from the corresponding author. Code source files for our treadmilling analysis are available online: https://github.com/paulocaldas/Treadmilling-Speed-Analysis
Data availability
Source data for all figures have been provided in a data source tables and raw data for the supplementary video are available on the Figshare at https://doi.org/10.6084/m9.figshare.10282046.v1. All other data supporting the findings of this study are available from the corresponding author upon request.
Author contributions
N.B. and M.L. conceptualized experiments, N.B and P.R performed and analysed most of biochemical experiments, P.R. and M.L. performed treadmilling analysis, N.B. performed single molecule analysis, M.L.P, N.B., P.R., and V.H.-R: purified proteins, M.L. performed structural modelling, V.H.-R. performed and analyzed MST experiments. C.A. performed and analyzed SV and AUC experiments. N.B., P.R., and M.L. wrote the original draft. N.B., P.R., V.H.-R., W.V., M.L.P. and M.L. reviewed and edited the manuscript. N.B., G.R., W.V. and M.L. supervised the study. Funding was acquired by W.V. and M.L.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Loose M, Mitchison TJ. The bacterial cell division proteins ftsA and ftsZ self-organize into dynamic cytoskeletal patterns. Nature Cell Biology. 2014;16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisson-Filho AW, et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 2017;355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017;355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haeusser DP, Margolin W. Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol. 2016;14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoenemann KM, Margolin W. Bacterial Division: FtsZ Treadmills to Build a Beautiful Wall. Curr Biol. 2017;27:R301–R303. doi: 10.1016/j.cub.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 6.Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichoff S, Du S, Lutkenhaus J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol Microbiol. 2015;95:971–987. doi: 10.1111/mmi.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichoff S, Du S, Lutkenhaus J. Disruption of divisome assembly rescued by FtsN–FtsA interaction in Escherichia coli. Proc Natl Acad Sci. 2018;115:E6855–E6862. doi: 10.1073/pnas.1806450115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang MJ, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbin BD, Geissler B, Sadasivam M, Margolin W. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Persons L, Lee L, de Boer PAJ. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol. 2015;95:945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan AJF, Vollmer W. The stoichiometric divisome: a hypothesis. Front Microbiol. 2015;6:455. doi: 10.3389/fmicb.2015.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boes A, Olatunji S, Breukink E, Terrak M. Regulation of the peptidoglycan polymerase activity of PBP1b by antagonist actions of the core divisome proteins FtsBLQ and FtsN. MBio. 2019 doi: 10.1128/mBio.01912-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang M-J, Bernhardt TG. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol. 2015;95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busiek KK, Margolin W. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol Microbiol. 2014;92:1212–1226. doi: 10.1111/mmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pazos M, et al. Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07559-2. 5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busiek KK, Eraso JM, Wang Y, Margolin W. The Early Divisome Protein FtsA Interacts Directly through Its 1c Subdomain with the Cytoplasmic Domain of the Late Divisome Protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Ploeg R, et al. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol Microbiol. 2013;87:1074–1087. doi: 10.1111/mmi.12150. [DOI] [PubMed] [Google Scholar]
- 20.Pichoff S, Du S, Lutkenhaus J. Disruption of divisome assembly rescued by FtsN–FtsA interaction in Escherichia coli. Proc Natl Acad Sci. 2018 doi: 10.1073/pnas.1806450115. 201806450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baranova N, Loose M. Methods in Cell Biology. Vol. 137. Elsevier Ltd; 2017. Single-molecule measurements to study polymerization dynamics of FtsZ-FtsA copolymers. [DOI] [PubMed] [Google Scholar]
- 22.Roder F, et al. Reconstitution of membrane proteins into polymer-supported membranes for probing diffusion and interactions by single molecule techniques. Anal Chem. 2011;83:6792–6799. doi: 10.1021/ac201294v. [DOI] [PubMed] [Google Scholar]
- 23.Coltharp C, Xiao J. Beyond force generation: Why is a dynamic ring of FtsZ polymers essential for bacterial cytokinesis? BioEssays. 2017;39:1–11. doi: 10.1002/bies.201600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wissel MC, Weiss DS. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J Bacteriol. 2004;186:490–502. doi: 10.1128/JB.186.2.490-502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller P, et al. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- 26.Banjade S, Tang S, Shah YH, Emr SD. Electrostatic lateral interactions drive ESCRT-III heteropolymer assembly. Elife. 2019;8:e46207. doi: 10.7554/eLife.46207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 28.Du S, Lutkenhaus J. Assembly and activation of the Escherichia coli divisome. Mol Microbiol. 2017;105:177–187. doi: 10.1111/mmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerding MA, et al. Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egan AJF, Biboy J, Van’t Veer I, Breukink E, Vollmer W. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0031. 20150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monteiro JM, et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature. 2018;554:528–532. doi: 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theile CS, et al. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8:1800–7. doi: 10.1038/nprot.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J-C, van den Ent F, Neuhaus D, Brevier J, Löwe J. Solution structure and domain architecture of the divisome protein FtsN. Mol Microbiol. 2004;52:651–660. doi: 10.1111/j.1365-2958.2004.03991.x. [DOI] [PubMed] [Google Scholar]
- 34.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvay AG, Santamaria M, Le Maire M, Ebel C. Analytical ultracentrifugation sedimentation velocity for the characterization of detergent-solubilized membrane proteins Ca++-ATPase and ExbB. J Biol Phys. 2007;33:399–419. doi: 10.1007/s10867-008-9058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piehler J, Brecht A, Valiokas R, Liedberg B, Gauglitz G. A high-density poly(ethylene glycol) polymer brush for immobilization on glass-type surfaces. Biosens Bioelectron. 2000;15:473–481. doi: 10.1016/s0956-5663(00)00104-4. [DOI] [PubMed] [Google Scholar]
- 37.Vecchiarelli AG, Li M, Mizuuchi M, Mizuuchi K. Differential affinities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol Microbiol. 2014 doi: 10.1111/mmi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinga Quantification of paper mass distributions within local picking areas. Nord Pulp Pap Res J. 2007;22:441–446. [Google Scholar]
- 40.Tinevez JY, et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–92. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 42.Goehring NW, Chowdhury D, Hyman AA, Grill SW. FRAP analysis of membrane-associated proteins: Lateral diffusion and membrane-cytoplasmic exchange. Biophys J. 2010;99:2443–2452. doi: 10.1016/j.bpj.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sergé A, Bertaux N, Rigneault H, Marguet D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods. 2008;5:687–694. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 44.Sander J, Ester M, Kriegel HP, Xu X. Density-based clustering in spatial databases: The algorithm GDBSCAN and its applications. Data Min Knowl Discov. 1998;2:169. [Google Scholar]
- 45.Roder F, Wilmes S, Richter CP, Piehler J. Rapid transfer of transmembrane proteins for single molecule dimerization assays in polymer-supported membranes. ACS Chem Biol. 2014;9:2479–2484. doi: 10.1021/cb5005806. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Zhang Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012 doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FtsNcytoHis is homogeneously distributed on a membrane when no other proteins are present (0-30s, magenta, right). After addition of FtsA and FtsZ to the reaction chamber (noisy signal in both channels), FtsZ filaments assemble on the membrane (cyan, left). Within 5 minutes after the addition, FtsNcytoHis is sorted into a pattern resembling the FtsZ filaments. The experiment was performed on 1 mol% mono-Ni-NTA membranes with 1.5 μM Cy5-FtsZ, 0.5 μM FtsA and 1 μM CF488-FtsNcytoHis. The movie was acquired at 2 s/frame and corresponds to the micrographs in Fig. 1b, n=5.
Top: Single molecule dynamics of Cy5-FtsZ, Cy5-FtsA and Cy5-FtsNcytoHis.
Bottom: Single molecule dynamics merged with the corresponding Alexa488-FtsZ pattern.
Single molecules of FtsZ display no diffusive behavior at all, monomers within the filaments are constantly exchanged by binding/unbinding, as expected for treadmilling filaments.
Single molecules of FtsA display a mixed behavior of lateral diffusion and binding/unbinding. Single molecules of FtsNcytoHis move by fast lateral diffusion on the plane of the membrane, showing transient confinement to FtsZ-FtsA co-filaments.
Each experiment was performed on 0.2 mol% Tris-Ni-NTA membranes with 1.5 μM Alexa488-FtsZ, 0.5 μM FtsA and 1 μM FtsNcytoHis present. For visualization of the single molecules, small amounts of Cy5-labeled FtsZ, FtsA or FtsNcytoHis was added (100 pM, 75 pM and 75 pM respectively). The movies were recorded at 0.228 s/frame and correspond to Fig. 3a.
Left. Diffusion of FtsNcyto-TM in a polymer-supported lipid membrane. FtsNcyto-TM shows diffusion and capture behavior at normal protein concentrations (middle, FtsA 0.5 μM, FtsZ 1.5 μM) and at an increased FtsZ concentration of 3 μM. Under no conditions we found directional motion of FtsNcyto-TM.
Representative movie of single molecules of Cy5-FtsNcytoHis in the presence of FtsA-FtsZ filaments acquired at 228 ms. Signle particle FtsN undergoes rapid random motion with the transient confinement to FtsZ filaments, corresponds to Fig. 3f
Representative trajectories showing the differences in the diffusive motion of FtsNcyto-DDEEHis (gray track, left) and FtsNcytoHis (magenta, right). FtsNcyto-DDEEHis covers a large area and does not remain at isolated spots. FtsNcytoHis shows similar diffusive motion in the beginning until it is captured by FtsZ-FtsA co-filaments where it shows confined behavior. The red links represent gaps in trajectory linking.
CF488-FtsNcytoHis shows stronger colocalization to FtsZ in the presence of GTP (top row) than in the presence of GMPCPP (bottom row). Top: CF488-FtsNcytoHis (magenta, right) was added to the reaction chamber with FtsA and FtsZ co-filaments at steady state (cyan, left). Bottom: CF488-FtsNcytoHis (magenta, right) was added to a reaction chamber containing 0.5 μM FtsA and 1.5 μM Cy5-FtsZ (cyan, left). In both experiments, 0.5 μM FtsA, 1.5 μM Cy5-FtsZ and 1 μM CF488-FtsNcytoHis, as well as 4 mM GTP and 2 mM GMPCPP, respectively were used. For GTP experiments, the acquisition rate was kept at 2s/frame, while the rate was decreased in the presence of GMPCPP to 5 s/frame to limit photobleaching. The raw files were processed to show every 5th (GTP) or every 2nd (GMPCPP) frame, resulting in a 10 s/frame movie. The movies correspond to Fig. 2c.
FtsNcytoHis and FtsQcytoHis are simultaneously bound and homogeneously distributed on the membrane before addition of FtsA-FtsZ (0-1min, cyan and yellow respectively). After the addition of FtsA and FtsZ, FtsNcytoHis and to FtsQcytoHis peptides co-localize in a filament-like patterns.
The experiment was performed on 0.5 mol% Tris-Ni-NTA membranes with 0.15 μM Alexa488-FtsNcytoHis, 0.75 μM Cy5-FtsQcytoHis, 0.5 μM FtsA and 1.5 μM FtsZ. The movie was acquired at 2 s/frame and corresponds to Fig. 1d.
FtsNcytoHis shows similar colocalization to FtsZ (left) and FtsA (right).
Top: CF488-FtsNcytoHis (magenta, middle) was added to FtsA/Cy5-FtsZ co-filaments at the steady state (cyan, left). A merge of both channels shows colocalization between CF488-FtsNcytoHis and treadmilling filaments of FtsZ (bottom left).
Bottom: CF488-FtsNcytoHis (magenta, - top right) was added to FtsZ-TMR-FtsA co-filaments at the steady state (green, top left). A merge of both channels shows colocalization between CF488-FtsNcytoHis and co-treadmilling TMR-FtsA (bottom right). The experiments were acquired at 2 s/frame and correspond to data in Fig. 1e-g.
Top left: overlay of FtsZ and FtsNcytoHis pattern, bottom left: overlay of the differential image of FtsZ and FtsNcytoHis. The differential movie shows close coupling between FtsNcytoHis and the directional motion of treadmilling FtsZ filaments. Right: overlay of FtsNcytoHis (cyan) and FtsZ (magenta), moving spots correspond to growing filament bundles move along the filaments. The experiment was performed on a 1 mol% mono-Ni-NTA membrane with 0.5 μM FtsA, 1.5 μM Cy5-FtsZ and 1 μM CF488-FtsNcytoHis and acquired at 2 s/frame. The differential movie was created as described in the methods section. The movie corresponds to Extended Data Fig. 3a.
FtsZ, FtsA and FtsNcytoHis dynamics arise from different processes, as shown by FRAP experiments on supported lipid bilayers. The difference in the recovery times of the patterns is evident, with FtsZ showing the fastest recovery, mainly by exchange with FtsZ in the buffer and no influence of diffusion on the membrane, while fluorescence recovery of FtsA and FtsNcytoHis is slower and dominated by lateral diffusion.
Each experiment was performed in the presence of 1.5 μM FtsZ, 0.5 μM FtsA and 1 μM FtsNcytoHis labelled with Cy5, TMR and CF488CF, respectively. All movies were acquired at 2 s/frame and correspond to Extended Data Fig. 5b.
FtsQcytoHis is homogeneously distributed on a membrane prior to addition of FtsA and FtsZ (0-1min, yellow, middle). After addition of FtsA and FtsZ, FtsZ filaments assemble on the membrane (cyan, left). Within several minutes, membrane-bound FtsQcytoHis is organized into a pattern resembling FtsZ filaments.
The experiment was performed on 0.2 mol% Tris-Ni-NTA membranes with 1.5 μM Alexa488-FtsZ, 0.5 μM FtsA and 1 μM Cy5-FtsQcytoHis. Acquisition rate was 2s/frame. The movie corresponds to the micrographs in Fig. 1c.