Abstract
Background:
Cystic fibrosis (CF) remains without a definitive cure. Novel therapeutics targeting the causative defect in the cystic fibrosis transmembrane conductance regulator (CFTR) gene are in clinical use. Lumacaftor/ivacaftor is a CFTR modulator approved for patients homozygous for the CFTR variant p.Phe508del, but there are wide variations in treatment responses preventing prediction of patient responses. We aimed to determine changes in gene expression related to treatment initiation and response.
Methods:
Whole-blood transcriptomics was performed using RNA-Seq in 20 patients with CF pre- and 6 months post-lumacaftor/ivacaftor (drug) initiation and 20 non-CF healthy controls. Correlation of gene expression with clinical variables was performed by stratification via clinical responses.
Results:
We identified 491 genes that were differentially expressed in CF patients (pre-drug) compared with non-CF controls and 36 genes when comparing pre-drug to post-drug profiles. Both pre- and post-drug CF profiles were associated with marked overexpression of inflammation-related genes and apoptosis genes, and significant under-expression of T cell and NK cell-related genes compared to non-CF. CF patients post-drug demonstrated normalized protein synthesis expression, and decreased expression of cell-death genes compared to pre-drug profiles, irrespective of clinical response. However, CF clinical responders demonstrated changes in eIF2 signaling, oxidative phosphorylation, IL-17 signaling, and mitochondrial function compared to non-responders. Top overexpressed genes (MMP9 and SOCS3) that decreased post-drug were validated by qRT-PCR. Functional assays demonstrated that CF monocytes normalized calcium (increases MMP9 expression) concentrations post-drug.
Conclusions:
Transcriptomics revealed differentially regulated pathways in CF patients at baseline compared to non-CF, and in clinical responders to lumacaftor/ivacaftor.
Keywords: RNA-Seq, CFTR, transcriptomics
1. Introduction
Cystic fibrosis (CF) is an inherited disorder of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. CFTR is widely expressed in epithelial-lined organs and immune cells, and variants in CFTR result in multi-system disease, particularly impacting the respiratory and gastrointestinal tracts [1]. Care for patients with CF has been revolutionized by the development of medications that enhance CFTR function, termed CFTR modulators.[2-7] Unfortunately, clinical responses to CFTR modulators vary by genotype, and can be difficult to predict even within genotypes. Variations in clinical responses are especially difficult to predict for the most common CF variant, p.Phe508del (F508del) [8-10]. Further, long-term biochemical changes elicited by CFTR modulators are relatively unknown. The problems described can limit use of CFTR modulators, both from the physician and patient perspective. These are important considerations as many young children with CF are now eligible to initiate CFTR modulators due to expanded age indications.
To increase our understanding of the impact of CFTR modulators, we recently completed a comprehensive metabolomics analysis in sera from patients with CF and homozygous for F508del initiating treatment with the CFTR modulator lumacaftor/ivacaftor [11]. Selected lipid and amino acid metabolic pathways were significantly altered by lumacaftor/ivacaftor in clinical responders. Concurrent with that analysis, in this study we performed RNA-Seq on separate whole-blood CF patient samples pre- and post-drug initiation and included a cohort of healthy non-CF controls that were used as a reference. We aimed to determine the transcriptional pathways expressed in relation to drug initiation and clinical responses. We hypothesized that differential gene expression would occur in patients with CF post-lumacaftor/ivacaftor treatment, which would reveal novel gene targets corresponding to therapeutic CFTR modulation and clinical responses.
2. Methods
2.1. Subjects
Patients with a confirmed CF diagnosis and homozygous for the F508del variant were recruited from the outpatient CF clinic prior to initiation of lumacaftor/ivacaftor. The diagnosis of CF was defined as 2 disease-causing variants or a sweat chloride test ≥ 60 mmol/L. Human subject recruitment was approved by the Institutional Review Board of Nationwide Children’s Hospital (IRB15-00611). Informed consent and/or assent was obtained for all participants. A parent or guardian of child participants provided informed consent on their behalf. Healthy, age and gender-matched controls were also recruited.
2.2. Clinical measures/samples
Patient clinical characteristics and demographics were recorded and stored in a secured electronic REDCap database. A full description of the cohort methods was previously published [11]. Whole blood was obtained from CF patients at the pre-lumacaftor/ivacaftor initiation visit as well as six-month follow-up visit, and from healthy non-CF patients during research visits. At collection, one ml of whole blood was placed in a RNA stabilization tube and immediately frozen at −20°C for later extraction. CF clinical responses were defined by changes in body mass index (BMI) (≥ 0.3 increase) and forced expiratory volume in one second (FEV1) (≥ 3% increase) via stratification into response and no-response groups. Response changes were based on prior clinical trial results [12].
2.3. RNA-Seq library preparation and sequencing
Whole blood RNA quality was assessed using the Agilent 2100 Bioanalyzer and RNA Nano Chip kit (Agilent Technologies, CA) to ensure that the RNA Integrity Number (RIN) was ≥ 7, and that there were no RIN value outliers. RNA-seq libraries were then generated using TruSeq Stranded Total RNA with Ribo-Zero Globin Complete kit (Illumina, CA). Briefly, ribosomal RNA (rRNA) was removed from 1000 ng of total RNA with biotinylated, target-specific oligos combined with Ribo-Zero rRNA removal beads from the Human/Mouse/Rat Globin kit. This kit depletes samples of cytoplasmic and mitochondrial rRNA and globin mRNA in a single step. Following rRNA removal, mRNA was fragmented using divalent cations under elevated temperature and converted into ds cDNA. Dual same indexed P5 and P7 adapters were ligated to ends of cDNA (Integrated DNA Technologies, Inc., Iowa). Following purification using the AMPure XP System (Beckman Coulter), the adaptor-ligated cDNA was amplified by limit-cycle PCR. Quality of libraries were determined via Agilent 4200 Tapestation using a High Sensitivity D1000 ScreenTape Assay kit, and quantified by KAPA qPCR (KAPA BioSystems). Approximately, 60-80 million paired-end 150 bp reads were generated per sample using Illumina HiSeq4000 platform. Raw data was converted to FASTQ using v2.19.0.316 of Illumina’s bcl2fastq application. Sequencing adapters matching at least 6 bases were then removed from the reads, as well as low-quality bases (< 10) using v1.10 of cutadapt (https://cutadapt.readthedocs.io/en/stable/index.html). An alignment report was also generated, using custom scripts, and manually reviewed to ensure that at least ~ 80% of reads aligned to the expected reference and that at least ~ 50% of the reads aligned to features annotated as protein coding.
2.4. RNA-Seq data analysis
Each sample was aligned to the GRCh38.p9 assembly of the Homo Sapiens reference from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.35/) using version 2.5.2b of the RNA-Seq aligner STAR (http://bioinformatics.oxfordjournals.org/content/early/2012/10/25/bioinformatics.bts635). Features were identified from the general feature format (GFF) file that came with the assembly from NCBI. Feature coverage counts were calculated using HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html). Differentially expressed features were calculated using DESeq2 (http://genomebiology.com/2014/15/12/550). Genes were removed from comparisons if they were not expressed above a background threshold (0.5 reads per million) for the majority of samples within each group. Principal Components Analysis (PCA) was performed with standard methodology.
Functional gene analyses were performed using modular repertoires and Ingenuity Pathway Analysis software (IPA) as described [13-16]. Modular analysis is a data driven approach based on clusters of coordinately expressed genes (modules) that share the same biological function. The data is deposited in the NCBI Gene Expression Omnibus (GEO accession number: GSE124548).
2.5. Gene validation by qRT-PCR
Two of the top overexpressed genes (MMP-9, SOCS3) were selected for quantitative real-time PCR (qRT-PCR) validation as well as one of the top upstream regulator genes (IL-1β). qRT-PCR was then performed using pre-designed TaqMan primer/probe combinations for MMP9 (Thermo Fisher, Hs00957562_m1), SOCS3 (Thermo Fisher, Hs02330328_s1) and IL-1β (Thermo Fisher, Hs01555410_m1). Whole blood RNA was extracted using the preserved RNA purification kit (Norgen Biotek, 43400) in accordance with the manufacturer’s instructions. The first strand of cDNA was synthesized from 1 μg total RNA using a high capacity cDNA reverse transcription kit (Thermo Fisher, 4368814) with 10X RT buffer, 25X dNTP mix (100 mM), 10X RT random primers, MultiScribe reverse transcriptase and RNase Inhibitor (Thermo Fisher, N8080119). Quantitative PCR (qPCR) mixes were assembled using Taqman Fast Master Mix (Thermo Fisher, 4444556) according to the manufacturer’s instructions. qPCR amplifications were performed in the 7500 Real Time PCR System (AB Applied Biosystems). The thermal cycler conditions were as follows: step 150°C for 2:00 min, 95°C for 10:00 min, 95°C for 15 s, Step 2, 60°C for 1:00 min (40 cycles) followed by a melting curve analysis. All reactions were performed with 10 biological replicates with reference dye normalization. The median cycle threshold (CT) value was used for analysis, and all CT values were normalized to expression of three housekeeping genes: 18S ribosomal RNA (rRNA) (Thermo Fisher, Hs03928990_g1), GAPDH (Thermo Fisher, Hs03929097_g1) and HPRT1 (Thermo Fisher, Hs02800695_m1). 18S was chosen for final normalized expression analysis based upon stability. Differences in mRNA expression between pre- and post-drug CF groups and compared to controls were analyzed by pair-wise fixed reallocation randomization tests with REST 2009 software [17].
2.6. Gene validation by ratiometric calcium flux responses to lumacaftor/ivacaftor in human monocytes
Human peripheral blood mononuclear cells were isolated from whole blood layered in Ficoll Paque Plus (GE Healthcare; 17-1440-03). Then, monocytes were purified by negative selection using Easy Sep Direct Human Monocyte Isolation kit (Stem Cell Technology, 19669). Cells were re-suspended in RPMI 1640 (Life Technologies; 22400-089) plus 10 % FBS (HyClone, SH30071.03; Thermo Scientific), stained with the calcium indicators 1 μM Fluo-4 AM (Molecular Probes; F 23917) and 2 μM Fura-red AM (Life Technologies; F3021) and incubated at 37°C with 5% CO2 for 30 minutes. Monocytes were washed and treated with 5 μM VX-809(Lumacaftor; S1565, Selleck Chemicals) and 5 μM VX-770 (Ivacaftor; S1144, Selleck Chemicals) for 1h at 37°C and 5% CO2. Then cells were re-suspended in HBSS with Ca2+ and Mg2+ (Corning Cellgrow; 21-023-CV) plus 1% FBS. The accumulation of intracellular free calcium was assessed by FACS in duplicate with a LSR II flow cytometer using blue laser (488 nm) for excitation and the 505 LP 530/30 BP filter channel for Fluo-4 AM and 640 LP -660/32 for Fura-red AM. Baseline monocyte Ca2+ levels were collected before stimulation for 30 sec, following by the stimulation with 1 μM Thapsigargin (Sigma; T9033). The fluorescence emission of Fluo-4 and Fura-red was measured for 600 s. The calcium ratiometric analysis was calculated as the increasing signal in Fluo-4 over the decreasing signal in Fura-red using the kinetic platforms in Flow Jo Software version 10.1 (Ashland, OR).
2.7. Statistical analysis
Clinical data was reported as means ± the standard deviation. Changes in pre/post drug initiation parameters were analyzed using paired t-test. For transcriptomic data, a list of significant differentially expressed features between the two groups was generated using an absolute value of log2 fold change ≥1.5 and an adjusted p-value of ≤ 0.05 [5% false discovery rate (FDR)]. For pathway analysis, a 10% FDR was used for the pre/post drug comparison and are included in the deposited GEO data. For qRT-PCR analysis, statistical significance was determined using the integrated randomization and bootstrapping methods in the REST 2009 software package. Data analysis was performed in Stata/MP 13.1, R software, and GraphPad Prism 7.0, with a two-tail p<0.05 considered statistically significant.
3. Results
3.1. Patient demographics
The demographic characteristics of the overall CF cohort pre- and post-lumacaftor/ivacaftor have been previously published [11]. CF clinical characteristics before and after drug initiation were similar except for a decrease in the number of pulmonary exacerbations requiring hospitalization after initiating lumacaftor/ivacaftor compared to baseline (pre-drug) [11]. The characteristics of the 20 CF patients (at baseline) and 20 non-CF controls used in this study are shown in Table 1. CF and non-CF demographics were similar except the non-CF cohort was significantly older.
Table 1:
Cohort Demographics
| CF | Non-CF | P value | |
|---|---|---|---|
| Subjects (n) | 20 | 20 | |
| Female | 55% | 65% | 0.53 |
| Age (years) | 21.6 ± 9.5 | 37.9± 12.1 | 0.001 |
| Caucasian | 100% | 95% | 0.33 |
| Phe508del homozygous | 100% | ||
| BMI (kg/m2) | 21.2 ± 3.4 | ||
| FEV1 (% predicted) | 74.2 ± 20.5 | ||
| PEx hospitalization rate/6mos | 1.1 ± 1.7 |
Values as expressed as percentage or means ± standard deviation. BMI – Body mass index, FEV1 – Forced expiratory volume in 1 second, PEx – Pulmonary exacerbation
3.2. Gene expression signatures according to lumacaftor/ivacaftor use
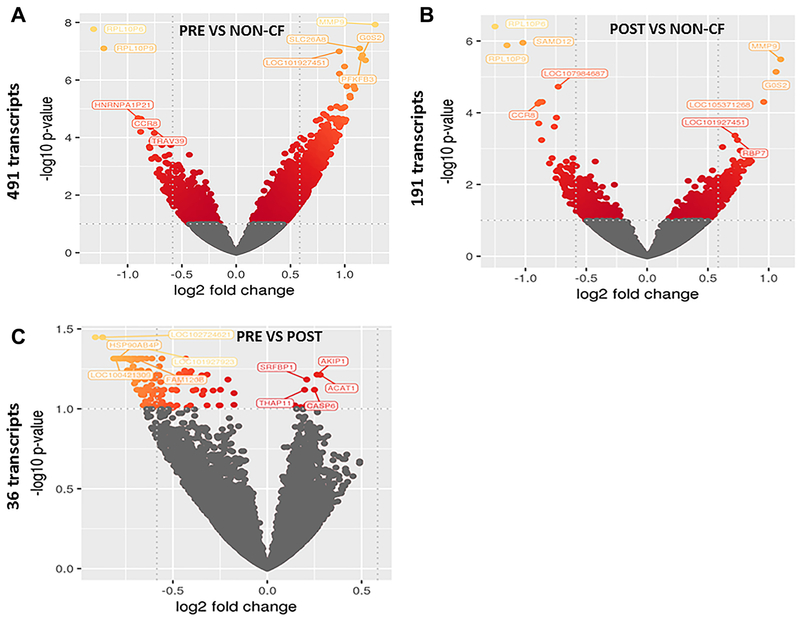
Whole blood samples from 20 patients pre- and six-month post-lumacaftor/ivacaftor initiation (total of 40 samples) and from 20 healthy, gender-matched controls were subjected to RNA-seq analysis. We identified 491 genes that were differentially expressed in CF patients at baseline, before initiating the drug, compared with controls (Fig. 1A, all genes listed in Supplemental Table 1). On the other hand, the number of significantly differentially expressed genes was lower when comparing CF patients that had received the drug for 6 months (post-drug) to controls, with only 191 differentially expressed genes detected (Fig. 1B, all genes listed in Supplemental Table 2). Last, when comparing pre-drug vs post-drug profiles in CF patients, only 36 genes were differentially expressed (Fig. 1C, all genes listed in Supplemental Table 3). For visualization of global transcriptomic profiles according to lumacaftor/ivacaftor status, we used PCA. This type of unsupervised clustering showed limited separation between CF patients pre or post drug use and healthy controls (Fig. 2A). However, when stratifying upon lumacaftor/ivacaftor use in CF patients, moderate clustering was observed (Fig. 2B).
Figure 1: Lumacaftor/ivacaftor alters gene expression profiles in CF.
Volcano plot of changes in RNA-Seq gene expression profiles for patients A) pre-lumacaftor/ivacaftor compared to non-CF (491 transcripts), B) post-lumacaftor/ivacaftor compared to non-CF (191 transcripts), and C) CF pre- vs post lumacaftor/ivacaftor (36 transcripts). Colored genes represent significant changes in expression, with yellow colors representing greater changes in comparison to red. Log fold changes are presented on the x axis and −log10 p values shown on the y axis.
Figure 2: Global transcriptomic profiling of whole blood.
Principal component analysis (PCA) of whole blood RNA-Seq profiles for A) CF patients pre- and post-lumacaftor/ivacaftor (drug) compared to non-CF persons and B) CF patients pre-lumacaftor/ivacaftor compared to post-lumacaftor/ivacaftor. Grouping are indicated by colors and shading.
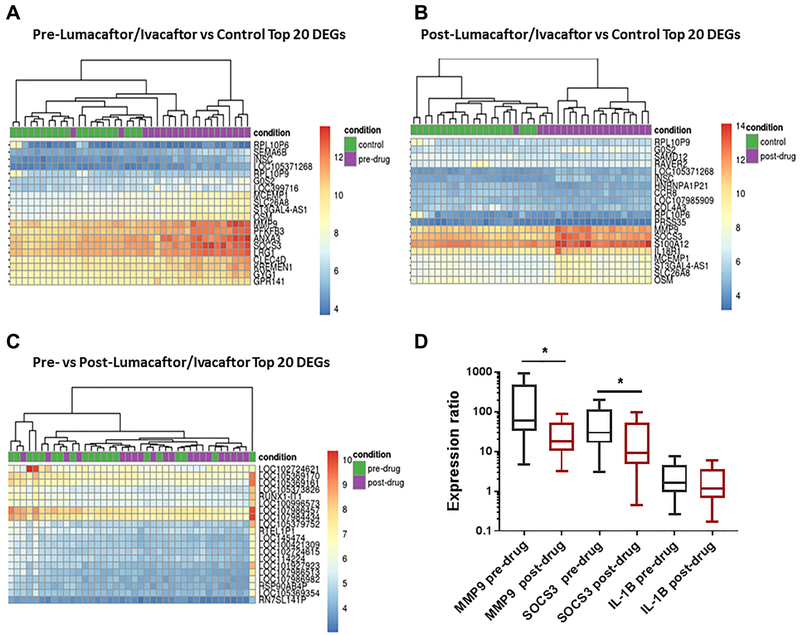
Next, we generated heat maps of the top 20 differentially expressed genes for each of the 3 comparison groups. Figure 3A demonstrates a clear separation between patients with CF at baseline (pre-lumacaftor/ivacaftor) and healthy controls. The top differentially expressed genes included matrix metallopeptidase 9 (MMP-9, involved in the breakdown of the extracellular matrix), suppressor of cytokine signaling 3 (SOCS3, negatively regulates cytokines that signal through the JAK/STAT pathway), and annexin A3 (ANXA3, regulates cellular growth and phospholipase A2 signal transduction pathways). We found a similar pattern of gene expression when comparing post-lumacaftor/ivacaftor CF patients to controls (Fig. 3B), with top differentially expressed genes including MMP-9 and SOCS3, but also other genes such as S100 calcium binding protein A12 (S100A12), a protein involved in cell cytoskeletal regulation. In contrast, there was less clear separation when comparing CF patients pre- and post-lumacaftor/ivacaftor administration (Fig. 3C). Within this comparison, the top genes included multiple pseudogenes, long non-coding RNAs, and genes encoding uncharacterized proteins.
Figure 3: Global transcriptomic profiling differentiates gene expression within CF.
Heat maps of top differentially expressed genes for A) CF patients pre-lumacaftor/lvacaftor compared to non-CF persons, B) CF patients post-lumacaftor/ivacaftor compared to non-CF persons, and C) CF patients pre-lumacaftor/ivacaftor compared to CF patients post-lumacaftor/ivacaftor. The top 20 differentially expressed genes are annotated. Each vertical row represents an individual patient with groupings indicated by color on the figure. D) qRT-PCR of MMP-9, SOCS3, and IL-1β gene expression in 10 CF patients pre- or post-lumacaftor/ivacaftor and compared to non-CF controls. Results are presented as gene expression ratios of CF patients relative to healthy controls and normalized to the housekeeping gene 18s. Statistical significance determined by REST mathematical modeling for pre-drug to post-drug measurements (“*” = p value <0.05).
We then validated by qRT-PCR two of the top differentially expressed genes (MMP-9 and SOCS3) in 10 CF patients at baseline and post-lumacaftor/ivacaftor and compared to 10 healthy controls (Fig. 3D). We used REST software to analyze the relative expression of these genes compared to the non-CF controls. This analysis confirmed a significant increase in MMP-9 and SOCS3 in CF patients compared to controls both at baseline and 6 months after drug administration as shown in the expression ratios in Figure 3D. Further, there was a significant reduction in both genes when comparing pre- to post-lumacaftor/ivacaftor treatment (Fig. 3D). We also analyzed the expression of a top upstream regulator IL-1β. However, there was no significant difference in IL-1β expression between drug treatments (Fig. 3D). Expression ratios, confidence intervals and p values for REST analyses can be found in Supplemental Table 4.
3.3. Gene pathway analyses in patients with CF before and after lumacaftor/ivacaftor use
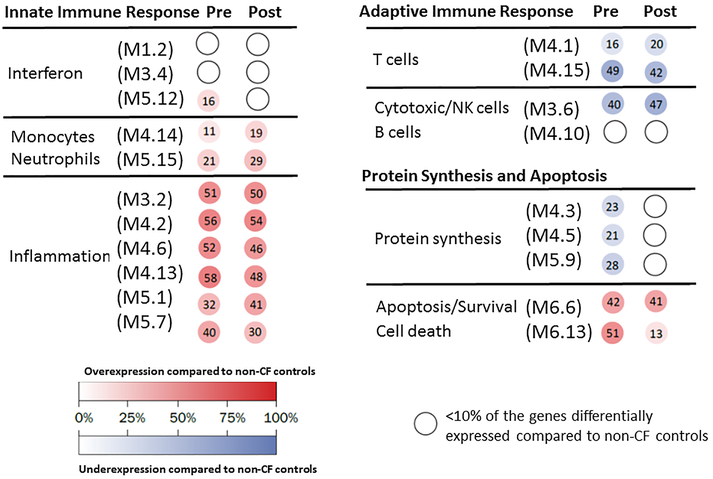
To define the biologic significance of the gene expression profiles, modular analyses were then performed in CF patients pre- versus post-lumacaftor/ivacaftor and compared to the non-CF cohort (Fig. 4). CF patients, independent of the drug administration, showed marked overexpression of inflammation and apoptosis related genes, and significant under-expression of T-cell and NK cell-related genes. Additionally, overexpression of cell death related genes, a subtle increase in the expression of one of the interferon modules, and decreased expression of genes related to protein synthesis were observed in CF patients pre-lumacaftor/ivacaftor. These latter changes in expression tended to normalize in these same patients after lumacaftor/ivacaftor use. Minimal changes in inflammation-related genes and B/T cell-related genes were observed in the post-lumacaftor/ivacaftor group compared to pre-lumacaftor/ivacaftor. Overall, these findings suggest that lumacaftor/ivacaftor has a mild impact on immunity-related genes that are altered in patients with CF.
Figure 4: Modular expression reveals responses to lumacaftor/ivacaftor.
Grouping of genes by modular expression in CF patients pre-lumacaftor/ivacaftor (pre) and post-lumacaftor/ivacaftor (post). Each group was compared to matched, healthy non-CF persons (n=20). Both CF group profiles demonstrated marked overexpression of inflammation-related genes and apoptosis genes, and significant under-expression of T cell and NK cell-related genes compared to non-CF persons. CF patients post-lumacaftor/ivacaftor demonstrated normalization of interferon-related and protein synthesis gene expression, and decreased cell-death related genes compared to CF patients pre-drug. The intensity of the modules (dots) indicates the proportion of overexpressed (in red) or under-expressed (in blue) transcripts within each module. Numeric values indicate the exact percentage of transcripts expressed in each specific module. A blank dot indicates that <10% of the genes in the module were differentially expressed.
Nevertheless, to further identify the relevant canonical pathways and regulators of gene function for each of the three comparisons (predrug vs control, post-drug vs control, pre and post drug), differentially expressed genes underwent pathway analysis via IPA. The top canonical pathways for persons with CF at baseline and after lumacaftor/ivacaftor use for 6 months compared to controls included: macrophage, fibroblast and endothelial cell pathways, as well as IL-6 signaling (Table 2). Shared top upstream regulators included lipopolysaccharide and IL-1β pathways. Fewer overlapping genes per pathway were present when we compared the pre- versus post-lumacaftor/ivacaftor profiles (Table 2). The top canonical pathways included RhoA, integrin, and actin cytoskeletal signaling, and the top upstream regulators myocyte enhancer factor 2C (MEF2C) and I-BET-151, an inhibitor of bromodomain and extra-terminal motif (BET) proteins such as BRD2.
Table 2:
Top canonical pathways and upstream regulators from pathway analysis
| Top Canonical Pathways | p-value | overlap |
|---|---|---|
| Pre-Luma/Iva vs Control | ||
| Role of macrophages, fibroblasts and endothelial cells | 9.30E-06 | 5.6% |
| IL-10 signaling | 1.24E-04 | 10.1% |
| Th1 and Th2 activation pathway | 1.74E-04 | 6.1% |
| Hepatic fibrosis/hepatic stellate cell activation | 1.82E-04 | 6.1% |
| II-6 signaling | 2.39E-04 | 7.0% |
| Post-Luma/Iva vs Control | ||
| Role of macrophages, fibroblasts and endothelial cells | 1.75E-04 | 3.0% |
| Role of pattern recognition receptors | 2.29E-04 | 4.6% |
| Role JAK family kinases in IL-6 type cytokine signaling | 5.73E-04 | 12.0% |
| Role of osteoblasts, osteoclasts and chrondrocytes | 7.83E-04 | 3.1% |
| Paxillin signaling | 7.99E-04 | 4.5% |
| Pre vs Post-Luma/lva | ||
| RhoA Signaling | 4.30E-02 | 0.8% |
| Integrin Signaling | 7.37E-02 | 0.5% |
| Actin cytoskeleton signaling | 7.68E-02 | 0.5% |
| Protein Kinase A signaling | 1.29E-01 | 0.3% |
| Axonal guidance signaling | 1.49E-01 | 0.2% |
| Responder vs non-responders | ||
| EIF2 Signaling | 4.46E-06 | 9.5 % |
| Oxidative Phosphorylation | 4.98E-04 | 10.1 % |
| Mitochondrial Dysfunction | 7.80E-04 | 8.2 % |
| IL-17A Signaling in Gastric Cells | 8.30E-04 | 20.0 % |
| Regulation of eIF4 and p70S6K Signaling | 1.07E-03 | 8.3 % |
| Top Upstream Regulators | p-value | predicted activation |
| Pre-Luma/Iva vs Control | ||
| Lipopolysaccharide | 2.96E-14 | activated |
| TNK | 1.78E-11 | activated |
| Immunoglobulin | 3.67E-11 | inhibited |
| TGM2 | 5.02E-09 | activated |
| IL-1β | 6.2E-08 | activated |
| Post-Luma/Iva vs Control | ||
| Lipopolysaccharide | 9.88E-11 | activated |
| OSM | 1.61E-09 | activated |
| Cg | 1.59E-08 | |
| PGR | 5.50E-07 | |
| IL-1β | 8.04E-07 | activated |
| Pre vs Post-Luma/Iva | ||
| I-BET-151 | 1.59E-03 | |
| HAND2 | 1.62E-02 | |
| TBX5 | 1.89E-02 | |
| MEF2C | 2.21 E-02 | |
| MYOCD | 2.60E-02 | |
| Responder vs non-responders | ||
| tomelukast | 3.05E-05 | |
| RICTOR | 3.23E-05 | Inhibited |
| HNF4A | 4.57E-05 | |
| Ifnar | 9.36E-05 | |
| STAT2 | 1.71E-04 | |
Luma/Iva – lumacaftor/ivacaftor
3.4. Changes in gene expression in clinical responders
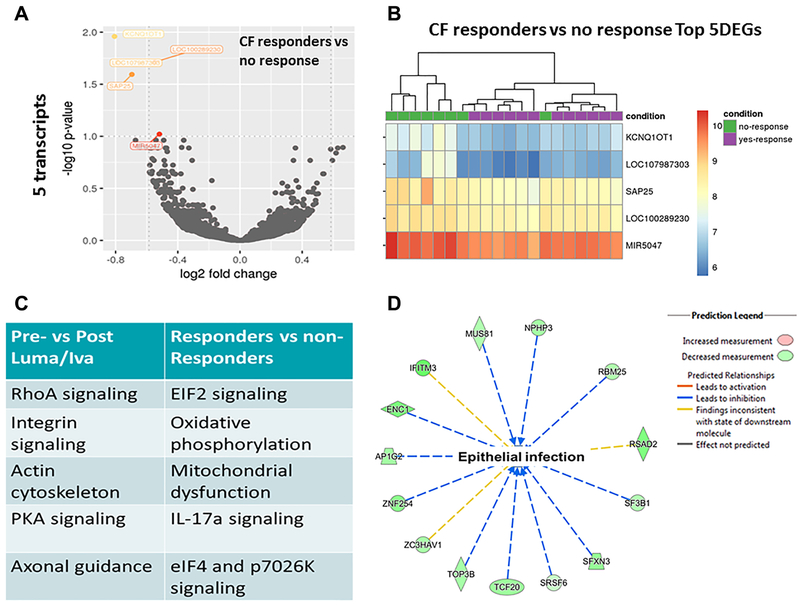
To define the profile of therapeutic response, we conducted further analyses in which CF patients were stratified according to clinical responders (n=12) and non-responders (n=8). Response to therapy was defined as the combined increase in BMI (≥ 0.3) and FEV1 (≥ 3%) as seen in previous clinical trials and our prior metabolomics analysis [6, 11]. A volcano plot of all transcripts and a corresponding heat map of the top 5 differentially expressed genes are presented in Figures 5A and 5B (genes listed in Supplemental Table 5). Of the top 5 differentially expressed genes, two were long non-coding RNAs and not annotated. The other three included KCNQ1OT, Sin3A associated protein 25kDa (SAP25), and microRNA 5047, which all decreased in expression after treatment. Notably, the prior top differential expressed genes identified, MMP-9, SOCS3, and ANXA3, were reduced by 1.8-fold, 1.3-fold, and 1.3-fold respectively in responders compared to non-responders. Pathway analysis of differentially expressed transcripts revealed changes in eIF2 signaling, oxidative phosphorylation, IL-17α signaling, and mitochondrial function for clinical responders in comparison to non-responders (Table 2 and Fig. 5C). Top upstream regulators for responders included RPTOR independent companion of mechanistic target of rapamycin (MTOR) complex 2 (RICTOR) and signal transducer and activator of transcription 2 (STAT2) (Table 2). Although RICTOR was noted to be an upstream regulator, there was no differential expression of MTOR or autophagy-related genes within the clinical responders compared to non-responders. Lastly, network analysis in responders compared to not responders revealed several downregulated genes predicted to be involved in the susceptibility to infection in epithelial cells (Fig. 5D). Combined, these data suggest that clinical responders to lumacaftor/ivacaftor demonstrated modest, but unique alterations in gene expression.
Figure 5: Clinical responders have altered gene expression profiles.
A) Volcano plot of changes in RNA-Seq gene expression profiles for CF patients post-lumacaftor/ivacaftor who were clinical responders (n=12) vs non-responders (n=8, defined by FEV1 and BMI change). B) Heat map of top differentially expressed genes for CF patients post-lumacaftor/ivacaftor clinical responders compared to non-responders. C) Top canonical pathways of gene expression analysis for pre/post lumacaftor/ivacaftor and clinical responder comparisons. D) Representative network analysis for predicted changes in susceptibility to epithelial infection in CF clinical responders compared to not responders. A colored prediction legend is listed.
3.5. Gene validation by functional assays
To correlate the gene expression changes in MMP-9 observed throughout all comparisons to a predicted functional change induced by lumacaftor/ivacaftor, we determined cytosolic calcium concentrations in CF and non-CF peripheral blood monocytes in response to thapsigargin. Thapsigargin is a non-competitive inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase [18]. CF peripheral blood mononuclear cells have been previously shown to constitutively express MMP9 due to alterations in their intracellular Ca2+ homeostasis [19]. CF monocytes from patients naive to lumacaftor/ivacaftor demonstrated increased cytosolic calcium concentrations compared to non-CF monocytes (Representative tracing in Fig. 6A with corresponding area under the curve analysis in Fig. 6B and summed end-point analysis for all experiments in Fig. 6C). Notably, CF monocytes treated with lumacaftor/ivacaftor in vitro restored cytosolic calcium responses to thapsigargin, as compared to untreated CF monocytes (Fig. 6C). In contrast, non-CF monocytes treated with lumacaftor/ivacaftor did not significantly decrease cytosolic calcium levels in response to thapsigargin (Fig. 6C). These data suggest that down-regulation of altered calcium homeostasis is one possible mechanism by which lumacaftor/ivacaftor may reduce the increased MMP9 expression in patients with CF.
Figure 6: Lumacaftor/Ivacaftor alters calcium concentrations in CF monocytes.
A) Graphical representation of cytosolic calcium concentrations over time in non-CF and CF monocytes with or without treatment with lumacaftor/ivacaftor. Monocytes were stimulated with thapsigargin. The signal ratio of Fluo-4 over Fura-red is presented. B) Area under the curve analysis for representative experiment in 6A. C) Summed end-point analysis of calcium ratios for all experiments, n=4 with statistical significance determined by ANOVA (“*” = p value <0.05).
4. Discussion
Therapeutic modulation of CFTR is continuing to advance at a rapid pace, with 3 drugs in current clinical use, and several more in pre-clinical or clinical trial evaluation [20]. As design and evaluation of these medications is still being optimized, it remains important to follow biochemical responses to existing CFTR modulators. To this end, we analyzed changes in gene expression profiles within patients with CF pre- and post-lumacaftor/ivacaftor initiation and found specific candidates in response to this drug that underwent further validation by PCR and functional studies. Our findings have implications for the continued evaluation of existing CFTR modulators in the newly initiated young CF population as well as ongoing development of novel agents. For young, nutritionally robust children with CF or persons with CF and mild clinical phenotypes it can be important to utilize blood biomarkers in the assessment of response to CFTR modulators when clinical markers (BMI, FEV1) may not be revealing. Further, gene expression gives insights into additional signaling pathways that are amenable to therapeutic manipulation in CF.
Overall, we found that whole blood RNA-Seq was a robust platform for differentiating CF and non-CF whole blood samples, with close to 500 differentially expressed transcripts between CF and non-CF controls. However, when comparing CF patient responses over time to lumacaftor-ivacaftor initiation, gene expression changes were less robust, with only 36 differentially expressed transcripts identified. We believe our findings reflect limited differences in CFTR expression induced by lumacaftor/ivacaftor in this population. RNA-Seq has been previously used in CF peripheral blood leukocytes to identify modifiers of disease severity [21], to identify biomarkers of pulmonary exacerbations in CF blood neutrophils [22], as well as to determine responses to a variety of individual pathogens in CF. Recently, transcriptional responses to treatment with the CFTR modulator ivacaftor were described in CF patients with at least one copy of the G551D variant [23]. The study by Sun and colleagues points out the utility of utilizing transcriptomic responses to predict clinical responsiveness to CFTR modulators. Together, these prior studies help validate the utility of RNA-Seq in our understanding of CF. Nevertheless, to our knowledge, we present the first RNA-Seq evaluation of whole blood gene expression changes in response to lumacaftor/ivacaftor in persons homozygous for the most common CF variant (F508del). This dataset is available to the CF community to serve as a resource for future transcriptomic studies comparing treatment responses in CF.
When comparing gene expression changes among patients with CF irrespective of CFTR modulator use, MMP-9 expression was consistently over-expressed in CF, but was reduced 1.8-fold post lumacaftor/ivacaftor initiation in CF patients that responded to therapy. MMP-9 has been implicated in several prior CF studies, and is thought to play a crucial role in CF disease progression [24-29]. In a separate study, we recently found that MMP-9 was within the top over-expressed transcripts in the blood of children with CF and exaggerated inflammation secondary to secondhand smoke exposure [30]. Studies have also shown that doxycycline reduced MMP-9 levels during acute CF exacerbations [24], and our collective data would support the investigation of doxycycline as an adjunctive therapy to CFTR modulators due to persisting elevations in MMP-9 expression during treatment. Further, our in vitro studies showed that CFTR modulators altered monocyte calcium homeostasis, a critical factor in initiating aberrant MMP-9 secretion in CF immune cells [19]. However, a recent study showed that CF monocytes have lower levels of MMP-9 mRNA compared to non-CF controls [31]. These differences could reflect differences in timing of mRNA extraction as MMP-9 mRNA has a biphasic release response [19], or result from technical differences between the studies. Regardless, MMP-9 and its relation to calcium homeostasis remains an important target for further studies in CF based on our overall findings.
Interestingly, activation of both SOCS3 and ANXA3 are also regulated by calcium signaling [32, 33]. These data are in agreement with our previous findings, where we demonstrated that CF neutrophils had increased cytosolic levels of calcium that were reduced by CFTR modulator treatment. This reduction in calcium also correlated with a recovery of antimicrobial mechanisms against B. cenocepacia [34]. Furthermore, SOCS3 induction from increased ceramide synthesis has been linked to leptin and insulin resistance in non-CF mice [35], which would indicate a potential significant role for altered SOCS3 signaling in CF, which is known to have alterations in ceramide accumulation and insulin resistance. Together, our data support an intriguing hypothesis that CFTR modulators can regulate exacerbated calcium responses in peripheral blood cells, and suggest that MMP-9 and genes regulated by calcium signaling such as SOCS3 are important therapeutic targets for CF patients. Persistently elevated MMP-9 or SOCS3 expression may also indicate a partial or incomplete response to CFTR modulator treatment that could be validated in future studies of personalized responses to treatment.
In addition to MMP-9, modular analysis revealed that altered gene expression in pathways of innate and adaptive immunity persisted in CF patients despite lumacaftor/ivacaftor administration. Although over-expression of genes related to cell-death decreased post-treatment, over-expression of inflammation-related transcripts as well as suppression of T cell and NK cell transcripts persisted. These findings highlight the importance of hyperactive innate inflammatory and suppressed adaptive immune responses in CF patients; and suggest that CFTR modulation with lumacaftor/ivacaftor may have a subtle impact on immune responses in older patients with established CF disease. In contrast, studies in patients on Ivacaftor alone for G551D variants have demonstrated reductions in inflammation [36] over time as well as improvement in neutrophil function [37]. Further studies are needed to confirm our findings in younger patient populations as well as measure responses to new CFTR modulators such as tezacaftor/ivacaftor or those in clinical development.
Although overall transcriptional profiles were minimally altered in clinically defined “responders” to lumacaftor/ivacaftor, several interesting pathways significantly altered in responders and non-responders were identified. These pathways included eIF2 signaling, oxidative phosphorylation, mitochondrial dysfunction, and IL-17 signaling. eIF2 is altered in newborn CFTR knockout ferrets [38] and it is integral to the dysfunctional unfolded protein response in CF [39]. eIF2 was also one of the top canonical pathways found to differentiate CF from non-CF persons in a recent microarray study of gene expression changes in plasma [40]. Similarly, dysfunctional mitochondrial responses have been demonstrated to play a role in inflammatory signaling and oxidative stress in CF [41-43]. IL-17 is markedly elevated in CF lungs and promotes inflammatory signaling [44]. Changes in these pathways in clinical responders suggests that further research in needed to better understand the role of these pathways in CF patients that do not respond to therapy, as well as why these particular pathways are impacted the greatest by CFTR modulation.
Other dysregulated pathways in CF patients that were independent of CFTR modulator status included macrophages, fibroblasts, and endothelial cells in inflammatory signaling. This was concordant with identification of the top upstream regulator IL-1β, as genes related to the IL-1β/IL-18 axis were highly upregulated in CF even after lumacaftor/ivacaftor initiation. We have previously shown that CF macrophage production of IL-1β is exaggerated in CF [45], especially in response to pathogens such as Burkholderia cenocepacia [46, 47]. IL-1 receptor antagonists have been proposed as a therapeutic blockade to abnormal IL-1β signaling in CF [48]. Combined with the prior findings, persistent exaggeration of macrophage-mediated inflammatory signaling provides additional evidence of a limited effect of lumacaftor/ivacaftor upon inflammation in CF as there was no significant difference in IL-1β expression after lumacaftor/ivacaftor treatment. New research is needed to determine how therapies that target IL-1β will interact with CFTR modulators and if there is an added benefit of IL-1β targeted treatment.
Our study was limited by a small, single-center cohort without an additional cohort available for confirmation. Furthermore, comparisons to initiation of ivacaftor alone or tezacaftor/lumacaftor were not available at the time this study commenced. Similarly, lumacaftor/ivacaftor was not available for children less than 12 years at study initiation. The use of blood transcriptional profiles may also limit interpretation of airway-specific gene responses, but is more generally applicable to all body systems in CF than bronchoalveolar lavage or sputum analysis. We also found non-coding genes to be the most differentially expressed during the pre- and post-lumacaftor/ivacaftor comparisons. Because of our strict filtering, we do not believe these represent a technical limitation, but rather potential future therapeutic targets. Despite these limitations, our study was strengthened by the comparison to a non-CF cohort, the ability of CF patients to serve as their own controls with repeated measurements, and comparisons within CF patients over a long time-period (6 months). It is noted that the six-month time point may limit interpretation of transcriptional responses that might occur shortly after treatment initiation, but may be milder at 6 months.
In summary, comprehensive blood RNA-seq transcriptome analyses revealed modest changes in gene expression post-lumacaftor/ivacaftor treatment, and help identify novel pathways in CF patients at baseline and in response to lumacaftor/ivacaftor.
Supplementary Material
Highlights.
CF gene expression profiles demonstrate marked changes in immunity-related genes
Chronic use of lumacaftor/ivacaftor minimally impacted gene expression profiles
Clinical response to lumacaftor/ivacaftor reflected changes in select gene pathways
Acknowledgements:
This work was supported in part by cystic fibrosis foundation grants (KOPP16I0 and PARTID18P0), the Sabatino Research Fund for Cystic Fibrosis, and the Cure CF Columbus Immune Core (C3IC) and Translational Core (C3TC). C3 Cores are supported by the Division of Pediatric Pulmonary Medicine, The Ohio State University Center for Clinical and Translational Science (National Center for Advancing Translational Sciences, Grant UL1TR002733) and by the Cystic Fibrosis Foundation (Research Development Program, Grant MCCOY19RO). Thank you to Terri Johnson, Laura Raterman, and Patti Olson for assistance in sample procurement, and all patients for their willing participation.
Abbreviations
- ANXA3
annexin A3
- BMI
body mass index
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- F508del
p.Phe508del
- FDR
false discovery rate
- FEV1
forced expiratory volume in one second
- MMP-9
matrix metallopeptidase 9
- PCA
Principal Components Analysis
- RIN
RNA Integrity Number
- rRNA
ribosomal RNA
- SOCS3
suppressor of cytokine signaling 3
Footnotes
Conflict of Interest Statement
BK has served on a Vertex Cystic Fibrosis Advisory Board. There are no other relevant conflicts of interest.
Portions of this manuscript were presented at the 2018 American Thoracic Society Conference.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Elborn JS. Cystic fibrosis. Lancet. 2016;388:2519–31. [DOI] [PubMed] [Google Scholar]
- [2].Accurso FJ, Rowe SM, Clancy JP, Boyle MP, Dunitz JM, Durie PR, et al. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. The New England journal of medicine. 2010;363:1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Elborn JS, Ramsey BW, Boyle MP, Konstan MW, Huang X, Marigowda G, et al. Efficacy and safety of lumacaftor/ivacaftor combination therapy in patients with cystic fibrosis homozygous for Phe508del CFTR by pulmonary function subgroup: a pooled analysis. Lancet Respir Med. 2016;4:617–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pettit RS, Fellner C. CFTR Modulators for the Treatment of Cystic Fibrosis. P T 2014;39:500–11. [PMC free article] [PubMed] [Google Scholar]
- [5].Ramsey BW, Davies J, McElvaney NG, Tullis E, Bell SC, Drevinek P, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. The New England journal of medicine. 2011;365:1663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. The New England journal of medicine. 2015;373:220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Taylor-Cousar JL, Munck A, McKone EF, van der Ent CK, Moeller A, Simard C, et al. Tezacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del. The New England journal of medicine. 2017;377:2013–23. [DOI] [PubMed] [Google Scholar]
- [8].Hubert D, Chiron R, Camara B, Grenet D, Prevotat A, Bassinet L, et al. Real-life initiation of lumacaftor/ivacaftor combination in adults with cystic fibrosis homozygous for the Phe508del CFTR mutation and severe lung disease. J Cyst Fibros. 2017;16:388–91. [DOI] [PubMed] [Google Scholar]
- [9].Ahmadi S, Bozoky Z, Di Paola M, Xia S, Li C, Wong AP, et al. Phenotypic profiling of CFTR modulators in patient-derived respiratory epithelia. NPJ Genom Med. 2017;2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jennings MT, Dezube R, Paranjape S, West NE, Hong G, Braun A, et al. An Observational Study of Outcomes and Tolerances in Patients with Cystic Fibrosis Initiated on Lumacaftor/ivacaftor. Ann Am Thorac Soc 2017;14:1662–6. [DOI] [PubMed] [Google Scholar]
- [11].Kopp BT, McCulloch S, Shrestha CL, Zhang S, Sarzynski L, Woodley FW, et al. Metabolomic responses to lumacaftor/ivacaftor in cystic fibrosis. Pediatr Pulmonol. 2018. [DOI] [PubMed] [Google Scholar]
- [12].Wainwright CE, Elborn JS, Ramsey BW. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. The New England journal of medicine. 2015;373:1783–4. [DOI] [PubMed] [Google Scholar]
- [13].Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Banchereau R, Jordan-Villegas A, Ardura M, Mejias A, Baldwin N, Xu H, et al. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 2012;7:e34390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rogers TB, Inesi G, Wade R, Lederer WJ. Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Biosci Rep. 1995;15:341–9. [DOI] [PubMed] [Google Scholar]
- [19].Averna M, Bavestrello M, Cresta F, Pedrazzi M, De Tullio R, Minicucci L, et al. Abnormal activation of calpain and protein kinase Calpha promotes a constitutive release of matrix metalloproteinase 9 in peripheral blood mononuclear cells from cystic fibrosis patients. Arch Biochem Biophys. 2016;604:103–12. [DOI] [PubMed] [Google Scholar]
- [20].Cystic Fibrosis Foundation. www.cff.org. 2017.
- [21].Kormann MSD, Dewerth A, Eichner F, Baskaran P, Hector A, Regamey N, et al. Transcriptomic profile of cystic fibrosis patients identifies type I interferon response and ribosomal stalk proteins as potential modifiers of disease severity. PLoS One. 2017;12:e0183526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jiang K, Poppenberg KE, Wong L, Chen Y, Borowitz D, Goetz D, et al. RNA sequencing data from neutrophils of patients with cystic fibrosis reveals potential for developing biomarkers for pulmonary exacerbations. J Cyst Fibros. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun T, Sun Z, Jiang Y, Ferguson AA, Pilewski JM, Kolls JK, et al. Transcriptomic responses to Ivacaftor and prediction of Ivacaftor clinical responsiveness. Am J Respir Cell Mol Biol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xu X, Abdalla T, Bratcher PE, Jackson PL, Sabbatini G, Wells JM, et al. Doxycycline improves clinical outcomes during cystic fibrosis exacerbations. Eur Respir J. 2017;49. [DOI] [PubMed] [Google Scholar]
- [25].Garratt LW, Sutanto EN, Ling KM, Looi K, Iosifidis T, Martinovich KM, et al. Matrix metalloproteinase activation by free neutrophil elastase contributes to bronchiectasis progression in early cystic fibrosis. Eur Respir J. 2015;46:384–94. [DOI] [PubMed] [Google Scholar]
- [26].Devereux G, Steele S, Jagelman T, Fielding S, Muirhead R, Brady J, et al. An observational study of matrix metalloproteinase (MMP)-9 in cystic fibrosis. J Cyst Fibros. 2014;13:557–63. [DOI] [PubMed] [Google Scholar]
- [27].Gaggar A, Li Y, Weathington N, Winkler M, Kong M, Jackson P, et al. Matrix metalloprotease-9 dysregulation in lower airway secretions of cystic fibrosis patients. Am J Physiol Lung Cell Mol Physiol. 2007;293:L96–L104. [DOI] [PubMed] [Google Scholar]
- [28].Ratjen F, Hartog CM, Paul K, Wermelt J, Braun J. Matrix metalloproteases in BAL fluid of patients with cystic fibrosis and their modulation by treatment with dornase alpha. Thorax. 2002;57:930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sagel SD, Kapsner RK, Osberg I. Induced sputum matrix metalloproteinase-9 correlates with lung function and airway inflammation in children with cystic fibrosis. Pediatr Pulmonol. 2005;39:224–32. [DOI] [PubMed] [Google Scholar]
- [30].Kopp BT TR, Kim J, Konstan R, Diaz A, Smith B, Shrestha C, Rogers LK, Hayes D, Tumin D, Woodley F, Ramilo O, Sanders DB, Groner J, Mejias A. Secondhand smoke alters arachidonic acid metabolism and inflammation in infants and children with cystic fibrosis. Thorax. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tarique AA, Sly PD, Cardenas DG, Luo L, Stow JL, Bell SC, et al. Differential expression of genes and receptors in monocytes from patients with cystic fibrosis. J Cyst Fibros. 2019;18:342–8. [DOI] [PubMed] [Google Scholar]
- [32].Zhang Y, Tao GJ, Hu L, Qu J, Han Y, Zhang G, et al. Lidocaine alleviates morphine tolerance via AMPK-SOCS3-dependent neuroinflammation suppression in the spinal cord. J Neuroinflammation. 2017;14:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meadows SM, Cleaver O. Annexin A3 Regulates Early Blood Vessel Formation. PLoS One. 2015;10:e0132580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Robledo-Avila FH, Ruiz-Rosado JD, Brockman KL, Kopp BT, Amer AO, McCoy K, et al. Dysregulated Calcium Homeostasis in Cystic Fibrosis Neutrophils Leads to Deficient Antimicrobial Responses. Journal of immunology. 2018;201:2016–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang G, Badeanlou L, Bielawski J, Roberts AJ, Hannun YA, Samad F. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. American journal of physiology Endocrinology and metabolism. 2009;297:E211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hisert KB, Heltshe SL, Pope C, Jorth P, Wu X, Edwards RM, et al. Restoring Cystic Fibrosis Transmembrane Conductance Regulator Function Reduces Airway Bacteria and Inflammation in People with Cystic Fibrosis and Chronic Lung Infections. Am J Respir Crit Care Med. 2017;195:1617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pohl K, Hayes E, Keenan J, Henry M, Meleady P, Molloy K, et al. A neutrophil intrinsic impairment affecting Rab27a and degranulation in cystic fibrosis is corrected by CFTR potentiator therapy. Blood. 2014;124:999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Keiser NW, Birket SE, Evans IA, Tyler SR, Crooke AK, Sun X, et al. Defective innate immunity and hyperinflammation in newborn cystic fibrosis transmembrane conductance regulator-knockout ferret lungs. Am J Respir Cell Mol Biol. 2015;52:683–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Blohmke CJ, Mayer ML, Tang AC, Hirschfeld AF, Fjell CD, Sze MA, et al. Atypical activation of the unfolded protein response in cystic fibrosis airway cells contributes to p38 MAPK-mediated innate immune responses. J Immunol. 2012;189:5467–75. [DOI] [PubMed] [Google Scholar]
- [40].Levy H, Jia S, Pan A, Zhang X, Kaldunski ML, Nugent ML, et al. Identification of molecular signatures of cystic fibrosis disease status using plasma-based functional genomics. Physiol Genomics. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Atlante A, Favia M, Bobba A, Guerra L, Casavola V, Reshkin SJ. Characterization of mitochondrial function in cells with impaired cystic fibrosis transmembrane conductance regulator (CFTR) function. J Bioenerg Biomembr. 2016;48:197–210. [DOI] [PubMed] [Google Scholar]
- [42].Rimessi A, Bezzerri V, Patergnani S, Marchi S, Cabrini G, Pinton P. Mitochondrial Ca2+-dependent NLRP3 activation exacerbates the Pseudomonas aeruginosa-driven inflammatory response in cystic fibrosis. Nature communications. 2015,’6:6201. [DOI] [PubMed] [Google Scholar]
- [43].Valdivieso AG, Santa-Coloma TA. CFTR activity and mitochondrial function. Redox Biol. 2013;1:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Roesch EA, Nichols DP, Chmiel JF. Inflammation in cystic fibrosis: An update. Pediatr Pulmonol. 2018. [DOI] [PubMed] [Google Scholar]
- [45].Kopp BT, Abdulrahman BA, Khweek AA, Kumar SB, Akhter A, Montione R, et al. Exaggerated inflammatory responses mediated by Burkholderia cenocepacia in human macrophages derived from Cystic fibrosis patients. Biochem Biophys Res Commun. 2012;424:221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kotrange S, Kopp B, Akhter A, Abdelaziz D, Abu Khweek A, Caution K, et al. Burkholderia cenocepacia O polysaccharide chain contributes to caspase-1-dependent IL-1beta production in macrophages. J Leukoc Biol. 2011;89:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Assani K, Tazi MF, Amer AO, Kopp BT. IFN-gamma stimulates autophagy-mediated clearance of Burkholderia cenocepacia in human cystic fibrosis macrophages. PLoS One. 2014;9:e96681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Iannitti RG, Napolioni V, Oikonomou V, De Luca A, Galosi C, Pariano M, et al. IL-1 receptor antagonist ameliorates inflammasome-dependent inflammation in murine and human cystic fibrosis. Nature communications. 2016;7:10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.