Abstract
BACKGROUND:
A significant proportion of patients with rectal cancer will present with synchronous metastasis at the time of diagnosis. Rates of OS for these patients are highly variable and previous attempts to build predictive models often have low predictive power, with concordance indexes (c-index) less than 0.70.
METHODS:
Using the National Cancer Database (2010–2014), we identified patients with synchronous metastatic rectal cancer. The data was split into a training dataset (diagnosis years 2010–2012), which was used to build the machine-learning model, and a testing dataset (diagnosis years 2013–2014), which was used to externally validate the model. A nomogram predicting 3-year overall survival was created using Cox proportional hazard regression with lasso-penalization. Predictors were selected based on clinical significance and availability in NCDB. Performance of the machine-learning model was assessed by c-index.
RESULTS:
A total of 4,098 and 3,107 patients were used to construct and validate the nomogram, respectively. Internally validated c-indexes at 1, 2, and 3 years were 0.816 (95% CI 0.813 – 0.818), 0.789 (95% CI 0.786 – 0.790), and 0.778 (95% CI 0.775 – 0.780), respectively. External validated c-indexes at 1, 2, and 3 years were 0.811, 0.779, and 0.778, respectively.
CONCLUSIONS:
There is wide variability in the OS for patients with metastatic rectal cancer, making accurate predictions difficult. However, using machine learning techniques, more accurate models can be built. This will aid patients and clinicians in setting expectations and making clinical decisions in this group of challenging patients.
Keywords: rectal cancer, machine learning, nomograms, lasso, NCDB
INTRODUCTION
Colon and rectal cancer is the fourth most common cancer diagnosis in the United States, with an estimated 140,000 new diagnoses and 50,000 deaths annually [1]. The most common metastatic site for patients with rectal cancer is the liver, and approximately 20–25% of patients will present with synchronous hepatic metastases [2, 3]. Other metastatic sites include lung, bone, peritoneum, and the nervous system [4]. Classically, metastatic disease has been considered incurable and is associated with poor overall survival [1]. However, with advances in chemotherapy and shifting paradigms in the definition of a resectable metastases, overall survival rates have improved in recent years [5–8].
In order to assist clinicians and patients with prognostic information and care-planning, predictive models have been built for this population of patients. When evaluating predictive models, the most commonly-used measure is the concordance index (c-index), which estimates the probability of concordance between predicted and observed outcomes [9]. A perfect concordance is 1.0, while a c-index of 0.50 is equatable to a coin-flip. Previous predictive models for metastatic colorectal cancer are often limited to a single metastatic organ (i.e. liver-only, lung-only) and/or have been hampered by c-indexes <0.70 [10–18]. This may be because outcomes in patients with metastatic disease are highly variable due to the wide variety of clinical conditions [19], making building accurate predictive models difficult. In addition, metastatic colon and rectal cancer are often grouped together as a single entity. However, there is mounting evidence that colon and rectal metastases are distinct entities, with different metastatic patterns and outcomes [4, 20]. Therefore, it may be prudent to build a rectum-specific predictive model for patients with metastatic disease.
Previously, models have been built on simple multivariable regression techniques, using either logistic regression or Cox proportional hazard modeling. However, advanced predictive modeling using machine learning can be used to build models that are more accurate, robust, and generalizable. Therefore, our aim was to utilize machine learning techniques to accurately predict 3-year overall survival in patients with metastatic rectal cancer. In addition, in order to make the model more accessible to clinicians and patients alike, we constructed a nomogram representing our predictive model in a graphical format.
MATERIALS AND METHODS
Patient Sample and Variables
The primary goal of this study is to construct accurate predictive nomograms for overall survival for patients with metastatic rectal cancer. Patients were identified from the National Cancer Database (NCDB), a national oncology outcomes database that is jointly sponsored by the American College of Surgeons’ Commissions on Cancer (CoC) and the American Cancer Society. The NCDB contains clinical oncology data sourced from over 1,500 CoC-accredited centers. Using the NCDB, all patients with metastatic rectal adenocarcinoma diagnosed from 2010 to 2014 were identified. Because the NCDB only contains de-identified patient information, this study was exempt from institution review board approval.
The primary tumor was identified as adenocarcinoma by International Classification of Disease for Oncology histology codes (8140–8145, 8210, 8211, 8220, 8221, 8255, 8261–8263, 8310, 8323, 8330–8332, 8480, 8481, 8490, 8525, 8530, 8570–8574). Survival time was defined as the number of months from diagnosis to an event (alive or dead). Because the model required non-zero survival times, survival times reported as 0 were transformed into 0.01 (equating to 0.3 days). Variables were selected due to clinical significance and availability within NCDB. Patient age was defined as the age of the patient at the time of diagnosis. In the NCDB, patient comorbidity is represented by the Charlson-Deyo Comorbidity Score (CDCS), which consists of 15 comorbidities that are assigned various point values [21]. Because of the small proportion of patients with CDCS of greater than 3, the CDCS variables is truncated to scores of 0, 1, 2, or ≥3 in the NCDB. Tumor grade was reported as well-differentiated, moderately-differentiated, poorly-differentiated, and undifferentiated/anaplastic. The CEA level, as defined in NCDB as the highest pre-treatment CEA level, was split into quartiles (≤6ng/mL, 6.1–28.9ng/mL, 29–97.9ng/mL, and ≥98ng/mL). Metastasis at the time of diagnosis to the liver, lung, brain, bone, and peritoneum were identified and reported as binary variables (i.e. yes/no). Resection of the primary tumor site and of a metastatic site (excluding resection of only distant lymph nodes) were also dichotomized. The number of positive lymph nodes was reported as a continuous variable, with aspiration of positive lymph nodes and unknown number of positive lymph nodes considered as one positive lymph node. Patients who had no lymph nodes examined was classified as having no positive lymph nodes. Lastly, any chemotherapy and/or radiation therapy received by the patient were included as separate binary variables in the model.
Nomogram Construction and Validation
Patients were split into a training set (diagnosis year 2010–2012) and testing set (2013–2014). Patients with any missing data were removed from analysis. Differences between the training and testing sets were evaluated using the chi-square test for categorical variables and the Mann-Whitney U test for continuous variables. Kaplan-Meier analysis with log-rank testing was used to compare the overall survival between the sets.
The predictive model was created using a 10-fold cross-validated Cox proportional hazard regression with lasso-penalization. The lasso (least absolute shrinkage and selection operator) is a machine-learning technique that can lead to superior performance over traditional multivariable regression because it performs both variable selection and penalization [22]. Variable selection reduces the number of predictors so that non-significant predictors are removed from the final model, while penalization (also known as regularization) decreases the predictors’ ability to affect the predicted outcome. By performing both variable selection and penalization, the lasso is able to build accurate models without under-fitting or over-fitting the training data. For our analysis, we combined properties of the lasso (i.e. variable selection and penalization) with Cox proportional hazard analysis, in which predictors for overall survival was subject to selection and penalization. The model was designed to predict the 3-year probability of overall survival for these patients. The machine-learning model formed the basis of the nomogram, with each predictor assigned points that can be summed to determine the 3-year probability of overall survival.
In evaluating the performance of our model, we employed both calibration (external) and validation (internal and external) assessments [23]. In external calibration, we determined how well our predictive model fits observed data from the testing dataset. We did this by stratifying patients in the testing dataset into 5 risk groups and reporting the predicted and actual probability of survival at 3 years for each risk group. In internal validation, we tested the congruence between model predictions and observed data in the training dataset by 100-repetition bootstrap resampling to determine the median time-dependent c-indexes at 1, 2, and 3 years with 95% confidence intervals. For external validation, we applied our predictive model to the observed data from the testing dataset to determine the time-dependent c-indexes at 1, 2, and 3 years.
All statistical and machine-learning analyses were performed using R (Version 3.3.2, R Foundation, Vienna, Austria) and R package hdnom [24]. The level of significance was set at 0.05 and all comparisons are two-tailed.
RESULTS
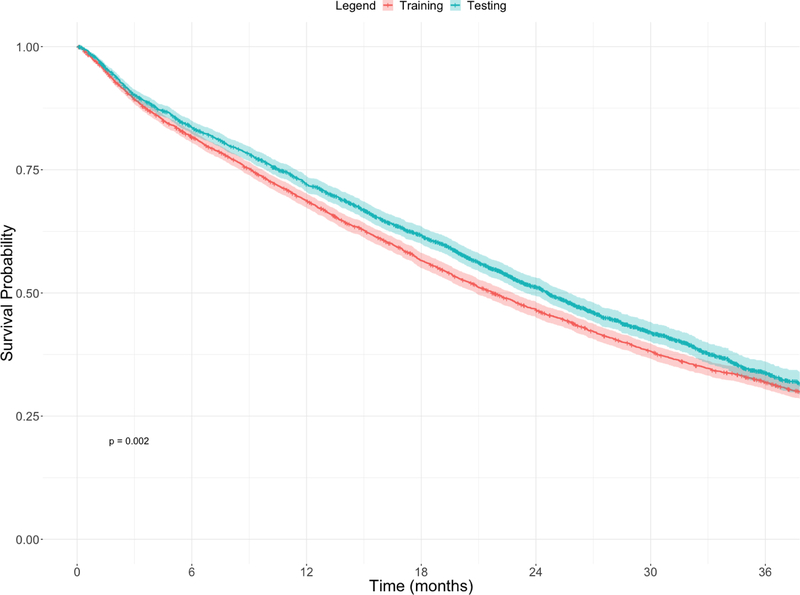
After exclusion, a total of 4,098 patients with rectal cancer were included in the training dataset, and a total of 3,107 patients with rectal cancer were included in the testing dataset. A comparison of the training and testing cohorts is shown in Table 1. The Kaplan-Meier analysis of the training and testing datasets is shown in Figure 1. The training group (21.7 months, 95% CI 20.9 – 22.8) had significantly shorter median OS than the testing group (24.6 months, 95% CI 23.5 – 25.7, p=0.002).
Table 1.
Comparison of the training and validation datasets for rectal cancer patients.
| Rectum | |||
|---|---|---|---|
| Training | Validation | p-value | |
| Total Number of Patients | 4059 | 3069 | - |
| Median Age (range) | 60 (18 – ≥90) | 60 (19 – ≥90) | 0.437 |
| Charlson-Deyo Score | 0.984 | ||
| 0 | 3176 (78.2%) | 2404 (78.3%) | |
| 1 | 681 (16.8%) | 511 (16.7%) | |
| 2 | 130 (3.2%) | 102 (3.3%) | |
| ≥3 | 72 (1.8%) | 52 (1.7%) | |
| Grade Differentiation | 0.172 | ||
| Well | 269 (6.6%) | 212 (6.9%) | |
| Moderate | 2949 (72.7%) | 2286 (74.5%) | |
| Poor | 776 (19.1%) | 525 (17.1%) | |
| Undifferentiated | 65 (1.6%) | 47 (1.5%) | |
| Highest CEA Level | 46 | 0.282 | |
| ≤6ng/mL | 1096 (27.0%) | 834 (27.2%) | |
| 6.1–28.9ng/mL | 1177 (29.0%) | 839 (27.3%) | |
| 29–97.9ng/mL | 768 (18.9%) | 573 (18.7%) | |
| ≥98ng/mL | 1018 (25.1%) | 823 (26.8%) | |
| Liver Metastasis | 3299 (81.3%) | 2503 (81.6%) | 0.763 |
| Lung Metastasis | 1506 (37.1%) | 1156 (37.7%) | 0.626 |
| Bone Metastasis | 344 (8.5%) | 229 (7.5%) | 0.119 |
| Brain Metastasis | 68 (1.7%) | 48 (1.6%) | 0.713 |
| Peritoneal Metastasis | 1050 (25.9%) | 844 (27.5%) | 0.122 |
| Primary Site Resected | 1565 (38.6%) | 1158 (37.7%) | 0.478 |
| Non-Primary Site Resected | 692 (17.0%) | 567 (18.5%) | 0.118 |
| Median # of Positive LNs (range) | 0 (0 – 50) | 0 (0 – 37) | 0.081 |
| Any Chemotherapy | 3372 (83.1%) | 2647 (86.2%) | <0.001 |
| Any Radiation Therapy | 1732 (42.7%) | 1248 (40.7%) | 0.089 |
Figure 1.
Kaplan-Meier curves comparing the training and testing datasets for patients with metastatic rectal cancer.
The nomogram for patients with metastatic rectal cancer is shown in Figures 2. For the nomogram, predictors are assigned a range of points that, when totaled, will equate to a given predicted probability of overall survival at 3 years. The higher the points that a patient receives, the worse their 3-year OS. The categorical variable receiving the most amount of points is omission of any type of chemotherapy (68 points). The categorical variable receiving the least amount of points is the presence of lung metastasis (4 points). In creating the machine-learning model, radiation therapy dropped out of the model as a predictor for 3-year overall survival.
Figure 2.
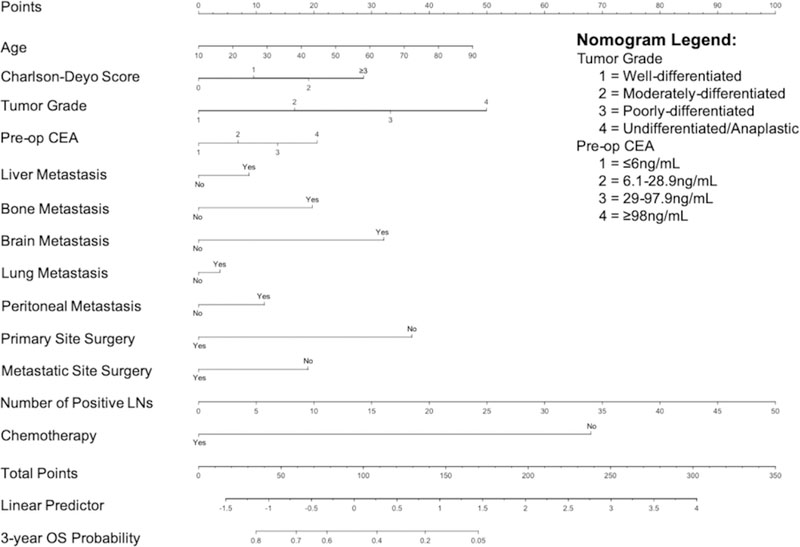
Nomograms predicting the 3-year overall survival for patients with metastatic rectal cancer.
External calibration, stratified into 5 risk-groups, is shown in Table 2. On external calibration, there was no significant difference in the predicted 3-year OS from the model compared to the actual 3-year OS in the testing dataset in 4 out of 5 risk groups. The predicted OS was outside of the 95% confidence interval of the actual 3-year OS for the highest risk group. The time-dependent internal and external validation at 1, 2, and 3 years are shown in Table 3. On internal validation, the c-indexes at 1, 2, and 3 years were 0.816 (95% CI 0.813 – 0.818), 0.789 (95% CI 0.786 – 0.790), and 0.778 (95% CI 0.775 – 0.780), respectively. On external validation, the c-indexes at 1, 2, and 3 years were 0.811, 0.779, and 0.778, respectively.
Table 2.
External calibration showing probability of 3-year OS for all cohorts
| Risk Group | Observed (95% CI) | Predicted |
|---|---|---|
| 1 (Highest Risk) | 0.096 (0.071 – 0.131) | 0.013* |
| 2 | 0.175 (0.138 – 0.220) | 0.179 |
| 3 | 0.289 (0.244 – 0.341) | 0.293 |
| 4 | 0.495 (0.445 – 0.550) | 0.447 |
| 5 (Lowest Risk) | 0.616 (0.565 – 0.670) | 0.644 |
Outside of 95% CI of observed probability
Table 3.
Time-dependent internal and external validation for all cohorts represented by area under the ROC
| Year | Internal Validation (95% CI) | External Validation |
|---|---|---|
| 1 | 0.816 (0.813 – 0.818) | 0.811 |
| 2 | 0.789 (0.786 – 0.790) | 0.779 |
| 3 | 0.778 (0.775 – 0.780) | 0.778 |
DISCUSSION
Patients with metastatic rectal cancer have high variability in overall survival due to their disease process, making accurate predictive models challenging in this patient population. However, by using a large nationwide oncology database and harnessing the predictive power of machine learning, we were able to construct nomograms for 3-year OS in metastatic rectal cancer patients with superior accuracy to those previously published [11, 12, 17, 18, 25].
Advances in the treatment of patients with metastatic rectal cancer is highlighted by the significant increase in median OS between the training and testing cohorts. The improvement in OS for these patients may be due to use of newer and more effective systemic agents and regimens [8, 26] and shifting paradigms in the definition of “resectable” metastases [27, 28]. This may have contributed to the significant increase in the proportion of patients who received any type of chemotherapy in the testing cohort. It was somewhat surprising that the proportion of patients who underwent resection of their rectal cancer was only 38.6% and 37.7% in the training and testing datasets, respectively. While there is equivocal evidence regarding benefits to resecting the primary site [29–31], it seems like this strategy is less commonly employed in metastatic rectal cancer compared to metastatic colon cancer [32]. This is likely due to the increased risk of morbidity associated with rectal resection. Likewise, the proportion of patients who had their metastatic site resected was only 17.0% and 18.5% in the training and testing datasets, respectively. While this difference did not reach statistical significance, it does suggest a small increase in the rate of metastasectomy in patients with metastatic rectal cancer.
In order to account for potential factors that are not captured in NCDB, models were built on patients diagnosed from 2010–2012, and were externally calibrated and validated on patients diagnosed from 2013–2014. This allowed us to incorporate time trends without explicitly including the year of diagnosis as a predictor (which would have limited use of these nomograms on future patients). This also prevented the training and testing data sets from being too similar, decreasing the risk of over-fitting the model to the training data. Because the testing dataset included patients diagnosed in 2013, we were not able to use this dataset to externally validate 5-year overall survival, limiting our analysis to 3-year overall survival. This is not uncommon in analysis of metastatic rectal cancer [11, 12, 33]. In the NCDB, 3-year OS for patients with metastatic disease is approximately 30%, highlighting the deadly nature of this disease despite advances in the field.
In our nomogram, the number of points assigned to each predictor is a measure of the predictor’s effect on 3-year OS. The number of points is determined by the lasso regression and produced some interesting results. For example, as seen in our study and in previous literature, the most common metastatic site for patients with rectal cancer is the liver [4]. However, bone metastasis, while much rarer, confers a far worse prognosis for patients [34]. Therefore, in our nomogram, the presence of bone metastasis is given more points than the presence of liver metastasis. The same phenomenon is true for the presence of peritoneal and brain metastasis, which are also associated with worse prognosis compared to liver metastases [35, 36]. However, metastasis to the lung was much less predictive of OS compared to metastasis to liver, peritoneum, bone, and/or brain in patients with metastatic rectal cancer. Though the lung represents the second most common metastatic site for patients with rectal cancer [4], overall survival after pulmonary metastasectomy has improved dramatically in the current era [37]. This may have led to a relative lack of predictive power of pulmonary metastasis, especially when compared to the effect of other metastatic sites on OS [4, 34–36]. This “crowding out” effect of pulmonary metastasis is seen in previous studies, in which the addition of lung metastasis to liver metastasis did not affect OS [38], and lung metastasis-associated variables were not prognostic for OS [18].
Though radiation therapy was included as a possible predictor, our lasso regression “selected” it out of the model. Therefore, because the nomogram is built from the lasso regression model, radiation therapy does not appear on the nomogram. This was unexpected, but may be because the role of radiation therapy in patients with metastatic rectal cancer is not well-defined, with previous studies yielding conflicting results [39–41]. Therefore, it may be possible that radiation therapy did not contribute significantly to the 3-year OS of these patients, leading the variable to be dropped by the lasso regression. In addition, radiation therapy may also have been “crowded out” by other predictors that contribute more to 3-year OS. For example, the predictor with the largest amount of points in the nomogram is treatment with any type of chemotherapy. However, of the 2,980 patients who received any radiation therapy, 2,760 (92.6%) also received chemotherapy. Therefore, it is possible that the effect of chemotherapy on OS simply “crowded out” the effects that radiation therapy had on OS.
Utilization of machine-learning techniques allowed us to build a predictive model with superior c-indexes compared to previous models [10–18]. However, accurate predictions for this cohort of patients remains challenging. When the testing dataset is split into 5 risk groups, our lasso model was able to accurately predict the 3-year OS in 4 out of 5 risk groups. The lasso model significantly under-predicted the 3-year OS for patients in the highest risk group. This may be related to the fact that there are predictors that contribute to survival in patients with high risk of death that are not captured in the NCDB, especially predictors that are not oncologic in nature (i.e. medical comorbidities, surgical complications).
Many consider machine learning as a “black box”, in which predictions are generated by a computer. Unfortunately, most clinicians have limited understanding of the machinations involved to generate these predictions. While medicine remains behind other disciplines in utilizing machine learning, its predictive power has been demonstrated with increasing frequency [42–44]. The lasso regression used in this study is a more approachable form of machine learning because it is based on multivariable regression. However, one advantage of the lasso over traditional multivariable regression is its ability to perform both variable selection and penalization. We also believe that techniques such as the lasso can also make nomograms easier to use. Because nomograms are graphical representations of complex algorithms, they can become too cumbersome and complex if too many predictors are included. The lasso is able to “select out” predictors that do not adequately contribute to predictive accuracy. In this study, radiation therapy was “selected out” by the lasso algorithm. This allows our nomogram to remain accurate but also user-friendly for both clinicians and patients alike, which makes it a useful tool in the shared decision-making process that is key in oncology care [45–47].
Limitations
These nomograms have certain limitations. First, the nomograms are constructed using a retrospective nationwide database. While this provides a large training and testing dataset, it limits the predictors that can be used to construct the dataset. There may be other predictors that are not included in NCDB that may be more predictive of OS, which could potentially make the models even more accurate. For example, the NCDB does not collect data on the specific chemotherapy regimen for each patient (e.g. FOLFOX versus FOLFIRI), nor whether a patient received targeted therapy (e.g. bevacizumab or erlotinib). In addition, emerging treatment options for patients with metastatic rectal cancer, such as immunotherapy, is not yet robustly recorded in the NCDB. However, our model can be updated with new data points as they are added to the NCDB, something that should be done regularly for all predictive models in order to provide accurate predictions. Our predictive outcome is limited to OS, as the NCDB does not capture disease-specific survival or recurrence data. The NCDB only includes metastases to the liver, lung, brain, bone, or peritoneum. While this covers the majority of metastatic sites for rectal tumors, rarer sites are not included in this analysis and the use of this nomogram in these patients may be limited. The NCDB does not collect data on the extent of metastases (e.g. the number of metastatic lesions), nor does it collect the location of lymph node metastases, though the number of positive lymph nodes is included in our analysis. Not all metastatic sites are biopsy-proven but are determined by clinical evidence. Inclusion of only biopsy-proven metastases could potentially limit our sample size and introduce additional bias. In addition, the NCDB only records metastatic sites at the time of diagnosis, so these nomograms are limited to patients who present with synchronous metastatic disease. Because the machine learning model can only be run with complete data, we excluded all patients with missing data. While imputation can be used to replace missing values, our large sample sizes ensured that sufficient power can be achieved with listwise deletion. Though we split our dataset into distinct training and testing datasets, the optimal method to externally validate our models is to use a separate dataset. However, because the NCDB covers such a high proportion of cancer diagnoses in the United States, it may be challenging to find patients that are not already represented in the NCDB. A potential solution is to prospectively collect validation data, though this is time-consuming and potentially unfeasible. Lastly, the NCDB contains data only from CoC-accredited centers. While that includes >1,500 centers across the U.S. and represents >70% of all cancer cases, the outcomes of patients at these institutions may be different than those treated at non-CoC-accredited centers, which may limit the generalizability of these models.
CONCLUSION
To our knowledge, this is the first application of machine learning to construct predictive models in metastatic rectal cancer. These models have superior performance compared to the currently available predictive models, showcasing the predictive power of machine learning. Nomograms created from these models can be of great assistance to both clinicians and patients in the treatment of metastatic rectal cancer.
ACKNOWLEDGEMENTS
Dr. Beiqun Zhao is funded by the National Library of Medicine Training Grant [NIH grant T15LM011271. The funding source had no role in the design and/or general conduct of this study; had no access to the data or role in data collection, management, analysis, or interpretation; had no role in the preparation, review, or approval of the manuscript; and had no role in the decision to submit the manuscript for publication.
Grant Support: Dr. Zhao is supported by the National Library of Medicine Training Grant [NIH Grant: T15LM011271]
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflicts of Interest Declaration: Dr. Zhao – None declared; Dr. Gabriel – None declared; Dr. Vaida – None declared; Dr. Lopez – None declared; Dr. Eisenstein – None declared; Dr. Clary – None declared
All authors have no conflicts to disclose.
REFERENCES
- 1. Cancer Facts & Figures 2018. Am Cancer Soc.
- 2.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM (2006) Epidemiology and management of liver metastases from colorectal cancer. Ann Surg 244:254–259. doi: 10.1097/01.sla.0000217629.94941.cf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackl C, Neumann P, Gerken M, Loss M, Klinkhammer-Shalke M, Schlitt HJ (2014) Treatment of colorectal liver metastases in Germany: a ten-year population-based analysis of 5772 cases of primary colorectal adenocarcinoma. BMC Cancer 14:810–820. doi: 10.1186/1471-2407-14-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riihimaki M, Hemminki A, Sundquist J, Hemminki K (2016) Patterns of metastasis in colon and rectal cancer. Sci Rep 6:1–9. doi: 10.1038/srep29765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlik TM, Scoggins CR, Zorzi D, Abdalla EK, Andres A, Eng C, Curley SA, Loyer EM, Muratore A, Mentha G, Capussotti L, Vauthey JN, Choti MA, Adams RB, Bolton JS, Hemming AW, Cofer JB, Smythe WR, Clary BM, Vauthey JN (2005) Effect of surgical margin status on survival and site of recurrence after hepatic resection for colorectal metastases. Ann Surg 241:715–724. doi: 10.1097/01.sla.0000160703.75808.7d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Portier G, Elias D, Bouche O, Rougier P, Bosset JF, Saric J, Belghiti J, Piedbois P, Guimbaud R, Nordlinger B, Bugat R, Lazorthes F, Bedenne L (2006) Multicenter randomized trial of adjuvant fluorouracil and folinic acid compared with surgery alone after resection of colorectal liver metastases: FFCD ACHBTH AURC 9002 trial. J Clin Oncol 24:4976–4982. doi: 10.1200/JCO.2006.06.8353 [DOI] [PubMed] [Google Scholar]
- 7.Aloia T, Sebagh M, Plasse M, Karam V, Lévi F, Giacchetti S, Azoulay D, Bismuth H, Castaing D, Adam R (2006) Liver Histology and Surgical Outcomes After Preoperative Chemotherapy With Fluorouracil Plus Oxaliplatin in Colorectal Cancer Liver Metastases. J Clin Oncol 24:4983–4990. doi: 10.1200/jco.2006.05.8156 [DOI] [PubMed] [Google Scholar]
- 8.Kopetz S, Chang GJ, Overman MJ, Eng C, Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM, McWilliams RR (2009) Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol 27:3677–3683. doi: 10.1200/JCO.2008.20.5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA (1982) Evaluating the Yield of Medical Tests. JAMA 247:2543–2546. doi: 10.1001/jama.1982.03320430047030 [DOI] [PubMed] [Google Scholar]
- 10.Beppu T, Sakamoto Y, Hasegawa K, Honda G, Tanaka K, Kotera Y, Nitta H, Yoshidome H, Hatano E, Ueno M, Takamura H, Baba H, Kosuge T, Kokudo N, Takahashi K, Endo I, Wakabayashi G, Miyazaki M, Uemoto S, Ohta T, Kikuchi K, Yamaue H, Yamamoto M, Takada T (2012) A nomogram predicting disease-free survival in patients with colorectal liver metastases treated with hepatic resection: Multicenter data collection as a project study for hepatic surgery of the Japanese Society of Hepato-Biliary-Pancreatic Surgery. J Hepatobiliary Pancreat Sci 19:72–84. doi: 10.1007/s00534-011-0460-z [DOI] [PubMed] [Google Scholar]
- 11.Kanemitsu Y, Kato T (2008) Prognostic models for predicting death after hepatectomy in individuals with hepatic metastases from colorectal cancer. World J Surg 32:1097–1107. doi: 10.1007/s00268-007-9348-0 [DOI] [PubMed] [Google Scholar]
- 12.Kanemitsu Y, Kato T, Hirai T, Yasui K (2004) Preoperative probability model for predicting overall survival after resection of pulmonary metastases from colorectal cancer. Br J Surg 91:112–120. doi: 10.1002/bjs.4370 [DOI] [PubMed] [Google Scholar]
- 13.Kattan MW, Gönen M, Jarnagin WR, DeMatteo R, D’Angelica M, Weiser M, Blumgart LH, Fong Y (2008) A nomogram for predicting disease-specific survival after hepatic resection for metastatic colorectal cancer. Ann Surg 247:282–287. doi: 10.1097/SLA.0b013e31815ed67b [DOI] [PubMed] [Google Scholar]
- 14.Nagashima I, Takada T, Matsuda K, Adachi M, Nagawa H, Muto T, Okinaga K (2004) A new scoring system to classify patients with colorectal liver metastases: Proposal of criteria to select candidates for hepatic resection. J Hepatobiliary Pancreat Surg 11:79–83. doi: 10.1007/s00534-002-0778-7 [DOI] [PubMed] [Google Scholar]
- 15.Nordlinger B, Guiguet M, Vaillant J, Balladur P, Boudjema K, Bachellier P, Jaeck D (1996) Surgical Resection of Colorectal Carcinoma Metastases to the Liver: A Prognostic Scoring System to Improve Case Selection, Based on 1568 Patients. Cancer 77:1254–62 [PubMed] [Google Scholar]
- 16.Sasaki K, Morioka D, Conci S, Margonis GA, Sawada Y, Ruzzenente A, Kumamoto T, Iacono C, Andreatos N, Guglielmi A, Endo I, Pawlik TM (2018) The Tumor Burden Score: A New “metro-ticket” Prognostic Tool for Colorectal Liver Metastases Based on Tumor Size and Number of Tumors. Ann Surg 267:132–141. doi: 10.1097/SLA.0000000000002064 [DOI] [PubMed] [Google Scholar]
- 17.Elias D, Faron M, Goéré D, Dumont F, Honoré C, Boige V, Malka D, Ducreux M (2014) A simple tumor load-based nomogram for surgery in patients with colorectal liver and peritoneal metastases. Ann Surg Oncol 21:2052–2058. doi: 10.1245/s10434-014-3506-z [DOI] [PubMed] [Google Scholar]
- 18.Kawai K, Ishihara S, Yamaguchi H, Sunami E, Kitayama J, Miyata H, Sugihara K, Watanabe T (2015) Nomograms for predicting the prognosis of stage IV colorectal cancer after curative resection: A multicenter retrospective study. Eur J Surg Oncol 41:457–465. doi: 10.1016/j.ejso.2015.01.026 [DOI] [PubMed] [Google Scholar]
- 19.Zacharakis M, Xynos ID, Lazaris A, Smaro T, Kosmas C, Dokou A, Felekouras E, Antoniou E, Polyzos A, Sarantonis J, Syrios J, Zografos G, Papalambros A, Tsavaris N (2010) Predictors of survival in stage IV metastatic colorectal cancer. Anticancer Res 30:653–660. doi: 10.1027/1614-0001.26.2.63 [DOI] [PubMed] [Google Scholar]
- 20.Qiu M, Hu J, Yang D, Cosgrove DP, Xu R (2015) Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget 6:38658–38666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deyo RA, Cherkin DC, Ciol MA (1992) Adapting a clinical comorbidity index for use with ICD-9-CM administrative databases. J Clin Epidemiol 45:613–619 [DOI] [PubMed] [Google Scholar]
- 22.Tibshirani R (1996) Regression Shrinkage and Selection via the Lasso. J R Stat Soc 58:267–288 [Google Scholar]
- 23.Dahabreh I, Chan J, Earley A, Moorthy D, Avendano E, Trikalinos T, Balk E, Wong J (2017) Modeling and Simulation in the Context of Health Technology Assessment: Review of Existing Guidance, Future Research Needs, and Validity Assessment [PubMed]
- 24.Xiao N (2016) hdnom: Building Nomograms for Penalized Cox Models with High-Dimensional Survival Data 1–21
- 25.Kawai K, Sunami E, Yamaguchi H, Ishihara S, Kazama S, Nozawa H, Hata K, Kiyomatsu T, Tanaka J, Tanaka T, Nishikawa T, Kitayama J, Watanabe T (2015) Nomograms for colorectal cancer: A systematic review. World J Gastroenterol 21:11877–11886. doi: 10.3748/wjg.v21.i41.11877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaunoit T, Alberts SR, Sergent DJ, Green E, Goldberg RM, Krook J, Fuchs C, Ramanathan RK, Williamson SK, Morton RF, Findlay BP (2005) Chemotherapy permits resection of metastatic colorectal cancer: Experience from intergroup N9741. Ann Oncol 16:425–429. doi: 10.1093/annonc/mdi092 [DOI] [PubMed] [Google Scholar]
- 27.Abdalla EK, Adam R, Bilchik AJ, Jaeck D, Vauthey J-N, Mahvi D (2006) Improving Resectability of Hepatic Colorectal Metastases: Expert Consensus Statement. Ann Surg Oncol 13:1271–1280. doi: 10.1245/s10434-006-9045-5 [DOI] [PubMed] [Google Scholar]
- 28.Rocha FG, Helton WS (2012) Resectability of colorectal liver metastases: An evolving definition. Hpb 14:283–284. doi: 10.1111/j.1477-2574.2012.00451.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulack B, Nussbaum D, Keenan J, Ganapathi A, Sun Z, Worni M, Migaly J, Mantyh C (2016) Surgical Resection of the Primary Tumor in Stage IV Colorectal without Metastasectomy is Associated with Improved Overall Survival Compared to Chemotherapy/Radiation Therapy Alone. Dis Colon Rectum 59:299–305. doi: 10.1097/DCR.0000000000000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nash GM, Saltz LB, Kemeny NE, Minsky B, Sharma S, Schwartz GK, Ilson DH, O’Reilly E, Kelsen DP, Nathanson DR, Weiser M, Guillem JG, Douglas Wong W, Cohen AM, Paty PB (2002) Radical resection of rectal cancer primary tumor provides effective local therapy in patients with stage IV disease. Ann Surg Oncol 9:954–960. doi: 10.1245/ASO.2002.03.068 [DOI] [PubMed] [Google Scholar]
- 31.Alawadi Z, Phatak UR, Hu CY, Bailey CE, You YN, Kao LS, Massarweh NN, Feig BW, Rodriguez-Bigas MA, Skibber JM, Chang GJ (2017) Comparative effectiveness of primary tumor resection in patients with stage IV colon cancer. Cancer 123:1124–1133. doi: 10.1002/cncr.30230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Temple LKF, Hsieh L, Wong WD, Saltz L, Schrag D (2004) Use of surgery among elderly patients with stage IV colorectal cancer. J Clin Oncol 22:3475–3484. doi: 10.1200/JCO.2004.10.218 [DOI] [PubMed] [Google Scholar]
- 33.Massacesi C, Norman A, Price T, Hill M, Ross P, Cunningham D (2000) A clinical nomogram for predicting long-term survival in advanced colorectal cancer. Eur J Cancer 36:2044–2052. doi: 10.1016/S0959-8049(00)00286-0 [DOI] [PubMed] [Google Scholar]
- 34.Baek SJ, Hur H, Min BS, Baik SH, Lee KY, Kim NK (2016) The Characteristics of Bone Metastasis in Patients with Colorectal Cancer: A Long-Term Report from a Single Institution. World J Surg 40:982–986. doi: 10.1007/s00268-015-3296-x [DOI] [PubMed] [Google Scholar]
- 35.Damiens K, Ayoub JM, Lemieux B, Aubin F, Saliba W, Campeau M, Tehfe MA (2012) Clinical features and course of brain metastases in colorectal cancer: Experience from a single institution. Curr Oncol 19:. doi: 10.1200/jco.2010.28.15_suppl.e14124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michl M, Thurmaier J, Schubert-Fritschle G, Wiedemann M, Laubender RP, Nüssler NC, Ruppert R, Kleeff J, Schepp W, Reuter C, Löhe F, Karthaus M, Neumann J, Kirchner T, Engel J, Heinemann V (2015) Brain Metastasis in Colorectal Cancer Patients: Survival and Analysis of Prognostic Factors. Clin Colorectal Cancer 14:281–290. doi: 10.1016/j.clcc.2015.05.009 [DOI] [PubMed] [Google Scholar]
- 37.Pfannschmidt J, Dienemann H, Hoffmann H (2007) Surgical Resection of Pulmonary Metastases From Colorectal Cancer: A Systematic Review of Published Series. Ann Thorac Surg 84:324–338. doi: 10.1016/j.athoracsur.2007.02.093 [DOI] [PubMed] [Google Scholar]
- 38.Engstrand J, Nilsson H, Strömberg C, Jonas E, Freedman J (2018) Colorectal cancer liver metastases - a population-based study on incidence, management and survival. BMC Cancer 18:1–11. doi: 10.1186/s12885-017-3925-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39., Manyam BV Mallick IH, Abdel-Wahab MM, Reddy CA, Remzi FH, Kalady MF, Lavery I, Koyfman SA (2015) The Impact of Preoperative Radiation Therapy on Locoregional Recurrence in Patients with Stage IV Rectal Cancer Treated with Definitive Surgical Resection and Contemporary Chemotherapy. J Gastrointest Surg 19:1676–1683. doi: 10.1007/s11605-015-2861-9 [DOI] [PubMed] [Google Scholar]
- 40.Repka MC, Aghdam N, Karlin AW, Unger KR (2017) Social determinants of stage IV anal cancer and the impact of pelvic radiotherapy in the metastatic setting. Cancer Med 6:2497–2506. doi: 10.1002/cam4.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renz P, Wegner RE, Hasan S, Brookover R, Finley G, Monga D, Raj M, McCormick J, Kirichenko A (2019) Survival Outcomes After Surgical Management of the Primary Tumor With and Without Radiotherapy for Metastatic Rectal Adenocarcinoma: A National Cancer Database (NCDB) Analysis. Clin Colorectal Cancer 1–7. doi: 10.1016/j.clcc.2018.12.005 [DOI] [PMC free article] [PubMed]
- 42.Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, Van Der Laak JAWM, Hermsen M, Manson QF, Balkenhol M, Geessink O, Stathonikos N, Van Dijk MCRF, Bult P, Beca F, Beck AH, Wang D, Khosla A, Gargeya R, Irshad H, Zhong A, Dou Q, Li Q, Chen H, Lin HJ, Heng PA, Haß C, Bruni E, Wong Q, Halici U, Öner MÜ, Cetin-Atalay R, Berseth M, Khvatkov V, Vylegzhanin A, Kraus O, Shaban M, Rajpoot N, Awan R, Sirinukunwattana K, Qaiser T, Tsang YW, Tellez D, Annuscheit J, Hufnagl P, Valkonen M, Kartasalo K, Latonen L, Ruusuvuori P, Liimatainen K, Albarqouni S, Mungal B, George A, Demirci S, Navab N, Watanabe S, Seno S, Takenaka Y, Matsuda H, Phoulady HA, Kovalev V, Kalinovsky A, Liauchuk V, Bueno G, Fernandez-Carrobles MM, Serrano I, Deniz O, Racoceanu D, Venâncio R (2017) Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318:2199–2210. doi: 10.1001/jama.2017.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upstill-Goddard R, Eccles D, Fliege J, Collins A (2013) Machine learning approaches for the discovery of gene-gene interactions in disease data. Brief Bioinform 14:251–260. doi: 10.1093/bib/bbs024 [DOI] [PubMed] [Google Scholar]
- 44.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. doi: 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 45.Kehl KL, Landrum MB, Arora NK, Ganz PA, Van Ryn M, Mack JW, Keating NL (2015) Association of actual and preferred decision roles with patient-reported quality of care: Shared decision making in cancer care. JAMA Oncol 1:50–58. doi: 10.1001/jamaoncol.2014.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP (2015) Nomograms in Oncology - More than Meets the Eye. Lancet Oncol 16:173–180. doi: 10.1586/14737175.2015.1028369.Focused [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caulfield S, Menezes G, Marignol L, Poole C (2018) Nomograms are key decision-making tools in prostate cancer radiation therapy. Urol Oncol Semin Orig Investig 36:283–292. doi: 10.1016/j.urolonc.2018.03.017 [DOI] [PubMed] [Google Scholar]