Abstract
The processes that lead to lung adenocarcinoma (LUAD) metastasis are poorly characterized. Spindle and kinetochore associated complex subunit 3 (SKA3) plays a key role in cervical cancer development, but its contribution to LUAD is unknown. Here, we found that SKA3 is overexpressed in LUAD and its expression correlates with lymph node metastasis and poor prognosis. SKA3 silencing experiments identified SKA3 as an oncogene that promotes the metastasis of LUAD cell lines and tissues. SKA3 was found to induce the expression of matrix metalloproteinase (MMP)-2, -7, and -9, which activate PI3K–AKT. SKA3 was also found to bind and activate EGFR to activate PI3K–AKT. In summary, we identify a role for SKA3 in LUAD metastasis through its ability to bind EFGR and activate PI3K–AKT signaling.
Keywords: cancer, EGFR, LUAD, metastasis, PI3K-AKT, SKA3
Introduction
Lung cancer (LCa) remains the major cause of cancer mediated fatalities globally, with 5-year survival rates less than 20% [1]. Approximately 234,030 new LCa cases were diagnosed in the US alone in 2018, of which 154,050 are expected to die [2]. Non-small cell lung cancer (NSCLC) is the major subtype of LCa, with lung adenocarcinoma (LUAD) the major NSCLC subtype [3]. Despite progress in LUAD therapeutics, the molecular mechanisms governing LUAD progression and development are poorly understood [4,5]. Further mechanistic studies to fully understand LUAD progression are therefore urgently required.
SKA3 is present in the kinetochore layer of the SKA complex, and is a key mediator of mitotic exit in the cooperation with NDC80 [6–8]. SKA3 is also a key to the migration of meiotic spindles and the stability of the spindles during anaphase [9]. It has been shown that SKA3 mediates cancer development and progression. SKA3 mutations are common in breast cancer, and were regulated cell growth [10]. Studies have shown that SKA3 mediates the metastatic profile and outcome of several cancers [11,12]. Using the GEPIA database to analyze LUAD tissue, SKA3 mRNA expression was elevated and correlated to poor survival outcomes in LUAD patients. The molecular mechanisms underlying the oncogenic effects of SKA3 in LUAD were not, however, characterized.
Here, we confirm that SKA3 is overexpressed in LUAD and shows a negative association with the survival of LUAD patients. Using SKA3 silencing approaches, we highlight that SKA3 acts as an oncogene through direct binding to EGFR, and the subsequent activation of pro-cancer signaling pathways to promote LUAD metastasis.
Methods
Patients and sample collection
We analyzed 26 LUAD tissues and their corresponding healthy tissue in samples collected from our local hospital. Our local ethics committee approved the study protocols and all participants provided informed consent. Patient information that could lead to patient identification remained confidential throughout the study.
qRT-PCR analysis
Trizol was added to cell/tissues for RNA extraction that was quantified on a nanodrop. cDNA synthesis was then performed and gene expression was quantified through qRT-PCR analysis [13]. Primer sequences are shown in Table 1.
Table 1. Sequence of primers for qRT-PCR.
| Primer | Sequence (5′ to 3′) |
|---|---|
| SKA3 forward primer | CAGATCCCTCTTCACCTACGA |
| SKA3 reverse primer | TCAACGTTTAAAGGGGGACA |
| MMP2 forward primer | TCTTGACCAGAATACCATCG |
| MMP2 reverse primer | TACTTCACACGGACCACTTG |
| MMP7 forward primer | AGTGGTCACCTACAGGATCGTA |
| MMP7 reverse primer | ATCTCCTCCGAGACCTGTCC |
| MMP9 forward primer | CAACATCACCTATTGGATCC |
| MMP9 reverse primer | GGGTGTAGAGTCTCTCGCTG |
| GAPDH forward primer | TGTGGGCATCAATGGATTTGG |
| GAPDH reverse primer | ACACCATGTATTCCGGGTCAAT |
Cell-based assays
MRC-5 cells (non-cancer) and H226 and SK-MES-1 cells (LUAD) were grown in complete culture media (DMEM plus 10% FBS at 37°C, 5% CO2). Cells were stimulated with recombinant human EGF (Abcam). siRNAs targeting SKA3 were synthesized by Gene Pharma Co. Overexpression plasmids for HA-tagged EGFR, pmyr-AKT, and shSKA3 were synthesized by Genechem. Cells were transfected with the indicated plasmids and packaged lentivirus was collected. Cells were then infected with each lentivirus and PI3K activity was assayed as previously described [14].
Cell cycle assessments
LUAD cells were transfected with siRNA-SKA3 or scrambled controls. Ten hours post-transfection, cells seeded into 96-well plates (∼1 × 104 cells/well). Cells were BrdU labeled (CST) to assess cell cycle progression. Experiments were repeated on a minimum of three independent occasions.
Assessment of the invasiveness and migratory ability of LUAD cells
Assays were performed in Boyden chambers with pore sizes of 8 mm in PET membranes. LUAD cells were counted and assessed via transwell assays. After 48 h, non-invasive cells in the upper Matrigel membrane were removed with cotton-tipped swabs and cells in the bottom wells were fixed in methanol and toluidine blue stained. Cells were invasive when they passed through the lower membrane surface. All cells were imaged on an inverted light microscope (200x magnifications) and quantified from three widefield images. Data shown are the averages of three independent experiments.
Co-immunoprecipitation assays
Post-thawing, LUAD cells were harvested in lysis buffer containing NaF, 50 mM; NaCI, 100 mM; Tris-HCl pH 7.5, 50 mM; Na3VO4, 1 mM; Na4P2O7, 30 mM; 0.5% NP-40 and PMSF, 0.5 mM supplemented with protease inhibitors. Antibodies were added to the lysates as previously described [15] and Co-IPs were assessed by Western blot analysis to confirm interactions. Antibodies used in the study are listed in Table 2.
Table 2. The primary antibodies used for WBs and the antibodies used for IP were as follows:
| Antibody | Supplier |
|---|---|
| SKA3 | ab186003, Abcam, 1:1,000 |
| GAPDH | ab181602, Abcam, 1:1,000 |
| Vimentin | ab193555, Abcam,1:1,000 |
| E-cadherin | ab194982, Abcam,1:1,000 |
| N-cadherin | ab202030, Abcam,1:1,000 |
| PTEN | ab32119, Abcam, 1:1,000 |
| STAT3 | ab119352, Abcam,1:1,000 |
| JAK2 | ab108596, Abcam,1:1,000 |
| Total p38(T-p38) | ab31828, Abcam,1:1,000 |
| Total Akt (T-Akt) | ab179463, Abcam,1:1,000 |
| Phosphorylated EGFR(p-EGFR, at Y992) | ab81440, Abcam, 1:1,000 |
| Phosphorylated EGFR(p-EGFR, at Y1045) | ab24928, Abcam, 1:1,000 |
| Phosphorylated EGFR(p-EGFR, at Y1068) | ab40815, Abcam, 1:1,000 |
| Phosphorylated p38(p-p38) | ab4822, Abcam, 1:1,000 |
| Phosphorylated STAT3(p-STAT3) | ab76315, Abcam, 1:1,000 |
| Phosphorylated JAK2(p-JAK2) | Ab32101, Abcam,1:000 |
| Phosphorylated Akt (p-Akt) | ab38449, Abcam, 1:1,000 |
| HA-probe | sc-2362, Santa Cruz Biotechnology; 1:800 |
| FLAG-probe | F1804, Sigma-Aldrich; 1:2,000 |
| EGFR | ab52894, Abcam, 1:1,000 |
| FLAG M2-affinity gel | A2220, Sigma-Aldrich; 20 ml per reaction |
In vivo metastasis assays
All animal experiments were carried out in Shanghai Kaixue Biological Technology Development Co., Ltd., Shanghai, China (Experiment code: 2018-03-07). In vivo assessments were performed in male nude mice aged 6 weeks (Beijing Vitonlihua Experimental Animal Technology Co. Ltd, Beijing, China). Animals were housed in specified cages that were approved by the national animal guidelines of our institute. Mice were injected with either H226-shSKA3 (Group 1) or H226-shNT (Group 2) cells (4 × 105 cells, 5 mice per group) in the tail-vein to produce the pulmonary metastasis model. Ten weeks following injection, mice were humanely killed in accordance with ethical study requirements and H&E stained to identify the presence of metastatic foci in the lungs. None anaesthetics were used during animal experiments.
Statistical analysis
SPSS19.0 was used foe data analysis. Student’s t-tests were used for repeated variance assessments. Wilcoxon signed-rank tests and Mann–Whitney U tests were performed for group comparisons. Kaplan–Meier curves were constructed to assess patient survival. Log rank tests were employed for subgroup comparisons. Unless otherwise stated, data are the mean ± SE. P < 0.05 were deemed statistical significance.
Results
SKA3 is up-regulated in LUADs
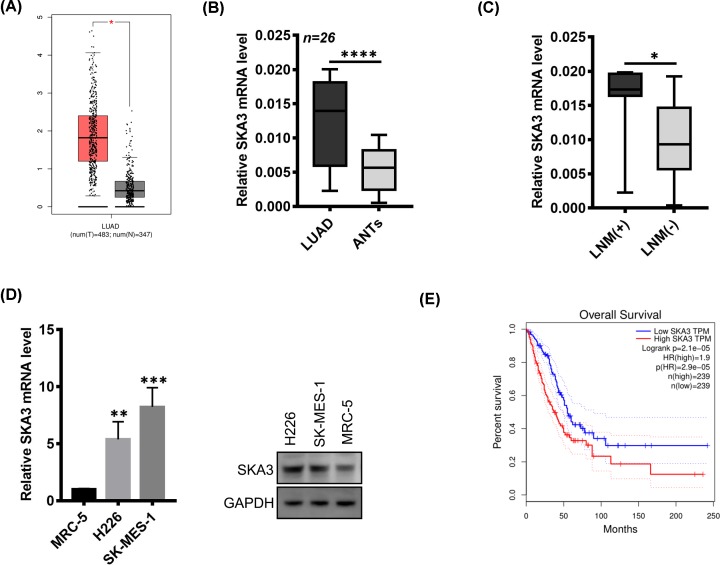
Analysis of the GEPIA suggested that SKA3 is up-regulated in LUAD versus normal tissue (fold-change > 2, P-value < 0.05, Figure 1A). To confirm this observation, SKA3 mRNA was quantified by qRT-PCR in 26 human LUAD and adjacent normal tissues (ANTs). Significantly higher expression of SKA3 mRNA was observed in LUAD versus ANT tissue (Figure 1B). SKA3 mRNA expression across LUAD tissue positively correlated with the occurrence of lymph node metastasis (Figure 1C). We further noted that SKA3 expression was elevated in vitro LUAD cell lines (H226 and SK-MED-1 cells) compared with non-lung cancer cells (MRC-5, Figure 1D). To confirm the prognostic value of SKA3 LUAD, the GEPIA database was analyzed which indicated that elevated SKA3 expression leads to a reduced OS versus tumors with low expression levels of SKA3 (Figure 1E). An intimate relationship therefore was observed between SKA3 overexpression, LUAD metastasis and poor patient prognosis.
Figure 1. SKA3 is up-regulated in LUAD.
(A) Gene Expression Profiling Interactive Analysis (GEPIA) indicated that the SKA3 expression is enhanced in LUAD tissues compared with normal tissue (fold-change > 2, P-value < 0.05). (B) SKA3 mRNA in 26 paired LUADs and ANTs examined by qRT-PCR. Data were compared via a Student’s t test. (C) SKA3 mRNA in 19 LUADs lacking lymph node metastasis and 7 with lymph node metastasis. (D) SKA3 expression in MRC-5, H226 and SK-MES-1 cells. (E) GEPIA analysis revealing the association of high SKA3 expression with a poor OS. Data were compared via two-sided log-rank tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
SKA3 promotes LUAD metastasis
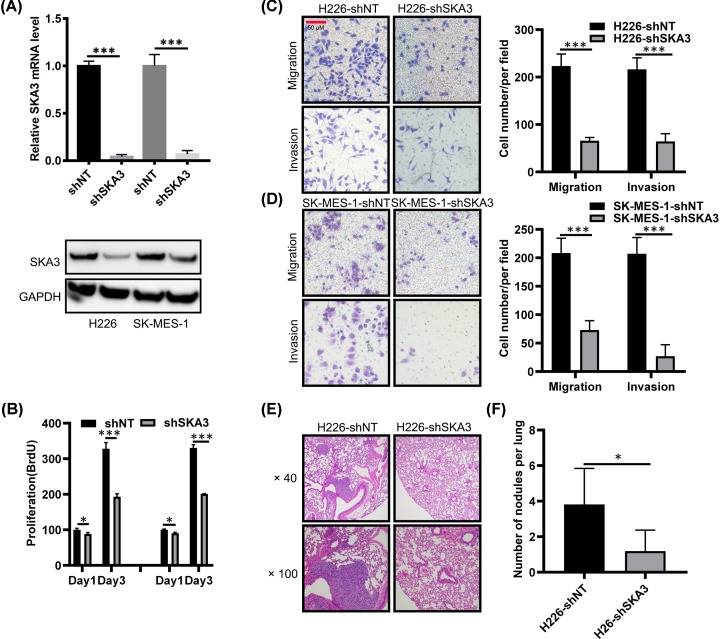
The data to this point inferred a role for SKA3 during LUAD metastasis. To fully define the role of SKA3 in LUAD tumorigenesis, we performed silencing experiments in in vitro and in vivo models of LUAD. To this end, we designed shRNAs targeting SKA3 to silence its expression in the H226 and SK-MES-1 LUAD cells (Figure 2A). SKA3-silencing strikingly inhibited the in vitro proliferation of H226 and SK-MES-1 cells (Figure 2B). Similarly, SKA3-silencing reduced the metastatic phenotypes of these LUAD lines, as decreased motility was observed in silenced versus shNT (shRNA non-target control) cells (Figure 2C,D). To confirm these findings, in vivo assessments of SKA3 expression in situations of LUAD metastasis were performed. In these experiments, SKA3 was silenced in H226-shSKA3 that was subcutaneously injected into the tail veins of nude mice to assess metastatic growth. Ten weeks post-injection, lungs were H&E stained and micro-metastases assessed (Figure 2E). Mice injected with H226-shSKA3 cells showed fewer numbers of metastatic foci that upon examination were of smaller size versus the H226-shNT group (Figure 2F). This suggested that SKA3 mediates the metastasis of LUAD cells.
Figure 2. SKA3 enhances the metastatic phenotypes of LUAD.
(A) SKA3 silencing (KD, shSKA3) in the indicated cell lines. (B–D) Effects of SKA3 silencing on cell proliferation (B). A one way ANOVA was used for data comparisons. (C and D) Migration and invasion assays of H226 and SK-MES-1 cells, respectively. Data were compared via a Student’s t test. (E) H & E staining of mouse lung tissues from H226-shNT and H226-shSKA3 groups (40×, metastatic nodules are indicated by arrows). (F) Numbers of metastatic foci observed in each group (n = 5). Data were analyzed through a Student’s t-test. *P<0.05; **P<0.01.
SKA3 controls matrix metalloproteinase-2, -7 and -9 expression through PI3K–AKT activation
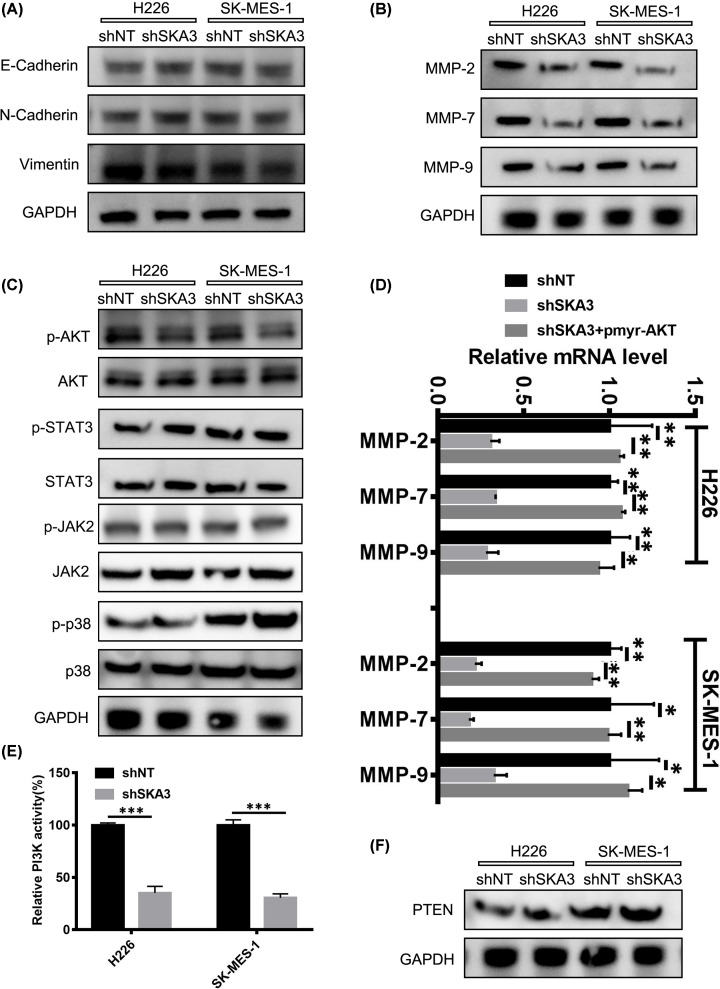
To study the regulation of LUAD via SKA3, we examined its role(s) in EMT, a key process during cancer metastasis. The data revealed that SKA3 did not influence the expression of vimentin, N-cadherin or E-cadherin (Figure 3A) suggestive that the metastasis induced by SKA3 occurs independently of EMT processes. A key process during tumor invasion is the enhanced proteolytic activity of cancer cell expressed MMPs that mediate the degradation of the stroma of neighboring cells and enhance the spread of tumor cells. In LUAD cells overexpressing SKA3, a marked increase in MMP-2, -7 and -9 proteins was observed, which decreased in response to SKA3-silencing (Figure 3B). In terms of other known oncogenic pathways, we observed low levels of active p-AKT in H226-shSKA3 and SK-MES-1 -shSKA3 cell lines, whilst levels of JAK, STAT3 and p38 did not differ between SKA3 silenced cells or shNT cells (Figure 3C). Upon the exogenous transfection of H226 and SK-MES-1 cells with shSKA3, shNT or pmyr-AKT for overexpression studies, the overexpression of AKT enhanced the expression of MMP-2, -7 and -9 mRNA in cells silenced for SKA3 (Figure 3D). No such effects on PTEN expression were observed (Figure 3E,F). These findings suggested that SKA3 mediated its effects on tumor metastasis through MMPs, mediated via the PI3K–AKT signaling axis.
Figure 3. SKA3 modulates MMP-2, -7 and -9 expressions through PI3K–AKT activation.
(A) Expression of the indicated EMT markers. (B) Expression of MMP-2, -7 and -9. GAPDH was probed as a loading control. (C) AKT (p-AKT), JAK2 (p-JAK2), STAT3 (STAT3) and p38 (p-p38) were evaluated by Western blot analysis. GAPDH was probed as a loading control. (D) Relative mRNA expression of MMP-2, -7 and -9. (E) PI3K kinase activity in the specified cell lines. (F) Western blot analysis of PTEN expression *P<0.05; **P<0.01; ***P<0.001.
Enhanced SKA3-mediated PI3K–AKT signaling requires SKA3 binding to EGFR
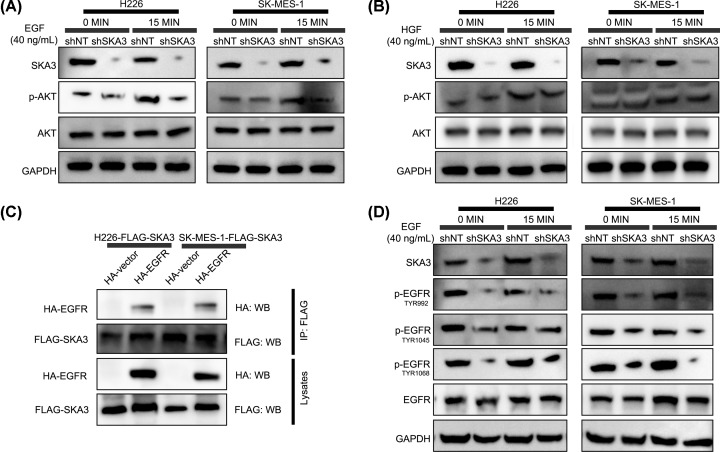
PI3K–AKT signaling occurs via RTKs and their downstream interactions with cognate ligands. We further explored the role of SKA3 in response to HGF and EGF. As shown in (Figure 4A,B) SKA3-silencing strongly inhibited EGF-mediated AKT activation in LUAD-shSKA3 cells versus shNT cells, whilst HGF stimulation led to no such observable changes. To assess a possible interaction of SKA3 with EGFR, FLAG-tagged SKA3 was co-transfected into LUAD cells with or without HA-EGFR or empty vector controls. Co-IPs were performed to investigate potential interactions (Figure 4C). We found that EGFR enhances AKT activation via SKA3, evidenced through the enhanced phosphorylation on TYR992, TYR1045 and TYR1068 that decreased following SKA3-silencing in LUAD cells (Figure 4D). These data confirmed that the oncogenic effects of SKA3 were mediated through its binding to EGFR, which would promote PI3K–AKT activation and enhance EGFR-phosphorylation.
Figure 4. SKA3 and EGFR interact to enhance PI3K–AKT signaling.
(A and B) Cells were stimulated with EGF (A) or HGF (B) for 0, 5 or 15 min and assessed for the expression of the indicated proteins. GAPDH was probed as a loading control. (C) Co-Ips using anti-FLAG antibodies in LUAD cells co-transfected with FLAG-SKA3 along with HA-EGFR or empty vector controls, as specified. (D) Cells were EGF treated for 0 or 5 min.
Discussion
Despite improvements in LUAD diagnostics, treatment and subsequent patient prognosis, LUAD frequently metastasizes and tumor recurrence is common [16–21]. Despite this, the molecular basis of LUAD metastasis is poorly characterized. Biomolecular markers that can predict both tumor recurrence and patient outcomes are urgently required for efficient patient management and the exploration of new therapeutic targets. Here, we investigated the role of SKA3 in LUAD and highlighted its critical requirement for LUAD metastasis. At a mechanistic level, we found that SKA3 increased MMP-2, -7 and -9 expressions in a PI3K–AKT dependent manner. SKA3 also interacted with EGFR to enhance its activation and protein pro-oncogenic PI3K-AKT signaling. These data highlight SKA3 as a novel therapeutic target to inhibit the metastasis of LUAD.
We investigated human LUAD samples and in vitro cell lines for SKA3 expression to fully define its role during LUAD metastasis. Analysis of the online database showed that SKA3 expressed was enhanced in clinical LUAD samples, and higher levels of lymph node metastasis were observed in LUAD cell lines. SKA3 expression positively correlated with survival time’s post-curative resection. Moreover, SKA3 silencing impaired the motility and invasion of LUAD cells both in vivo and in vitro. This implicated SKA3 in the pro-metastatic phenotypes of LUAD, and suggested that SKA3 acts as a biomarker to predict LUAD prognosis. SKA3 therefore represents a putative therapeutic target for LUAD treatment.
We further assessed the role of SKA3 in EMT processes [22–25] but no associations were observed. Upon further analysis, we found that AKT activation strongly correlated with SKA3 expression, and that the expression of MMP following SKA3 silencing decreased in a manner that could be rescued by exogenous AKT overexpression, suggestive that SKA3 influences MMP-2, -7 and -9 to promote of LUAD occurrence and development [26–32]. These data further suggested that the selective modulation of SKA3 during EGFR–PI3K–AKT signaling in LUADs leads to EGF modulation, with no effects on HGF-signaling. Co-Ips indicated an interaction between SKA3 and EGFR, which influenced EGFR phosphorylation and activation. Similar to the present study, recent findings indicated that SKA3 promotes cell proliferation and migration in cervical cancer tissue through its ability to activate PI3K/Akt signaling [11]. However, we found that SKA3 interacts with EGFR to enhance PI3K–AKT activity, which was a novel finding. The mechanisms governing this EGFR interaction now require further exploration.
In summary, we report for the first time that LUAD metastasis is enhanced by SKA3/EGFR mediated PI3K–AKT signaling. This reveals new information on mechanisms of metastasis in LUAD and highlights SKA3 as an exciting anti-LUAD therapeutic.
Abbreviations
- ANT
adjacent normal tissue
- GEPIA
Gene Expression Profiling Interactive Analysis
- LCa
lung cancer
- LUAD
lung adenocarcinoma
- MMP
matrix metalloproteinase
- NSCLC
non-small cell lung cancer
- SKA3
Spindle and kinetochore associated complex subunit 3
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The study was funded by Zhejiang Natural Science Foundation Youth Fund Project [grant number LQ18H270002].
Author Contribution
Guo-liang Yang performed conception and design, and was responsible for experiments performance. Dan-dan Hu and Li-ming Lou contributed to experiments performance. Hai-ling Chen performed data analysis. Hong Zhang contributed to conception and design, data analysis and manuscript writing. Guo-liang Yang contributed to financial support and final approval of manuscript.
References
- 1.Siegel R.L., Miller K.D. and Jemal A. (2017) Cancer Statistics, 2017. CA Cancer J. Clin. 67, 7–30 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D. and Jemal A. (2018) Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30, 0000-0002-0000-4111 AO 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 3.Blandin K.S., Crosbie P.A., Balata H., Chudziak J., Hussell T. and Dive C. (2017) Progress and prospects of early detection in lung cancer. Open Biol. 7, 9 10.1098/rsob.170070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh M. and Jadhav H.R. (2018) Targeting non-small cell lung cancer with small-molecule EGFR tyrosine kinase inhibitors. Drug Discov. Today 23, 745–753 10.1016/j.drudis.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 5.Hill A., Gupta R., Zhao D., Vankina R., Amanam I. and Salgia R. (2019) Targeted Therapies in Non-small-Cell Lung Cancer. Cancer Treat. Res. 178, 3–43 10.1007/978-3-030-16391-4_1 [DOI] [PubMed] [Google Scholar]
- 6.Jeyaprakash A.A., Santamaria A., Jayachandran U. et al. (2012) Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol. Cell 46, 274–286 10.1016/j.molcel.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 7.Sivakumar S., Daum J.R., Tipton A.R., Rankin S. and Gorbsky G.J. (2014) The spindle and kinetochore-associated (Ska) complex enhances binding of the anaphase-promoting complex/cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol. Biol. Cell 25, 594–605 10.1091/mbc.e13-07-0421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daum J.R., Wren J.D., Daniel J.J. et al. (2009) Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr. Biol. 19, 1467–1472 10.1016/j.cub.2009.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q.H., Qi S.T., Wang Z.B. et al. (2012) Localization and function of the Ska complex during mouse oocyte meiotic maturation. Cell Cycle 11, 909–916 10.4161/cc.11.5.19384 [DOI] [PubMed] [Google Scholar]
- 10.Jiao X., Hooper S.D., Djureinovic T. et al. (2013) Gene rearrangements in hormone receptor negative breast cancers revealed by mate pair sequencing. BMC Genomics 14, 165 10.1186/1471-2164-14-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu R., Wang M.Q., Niu W.B. et al. (2018) SKA3 promotes cell proliferation and migration in cervical cancer by activating the PI3K/Akt signaling pathway. Cancer Cell Int. 18, 183 10.1186/s12935-018-0670-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M., Williams K.A., Hu Y. et al. (2015) GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin. Exp. Metastasis 32, 769–782 10.1007/s10585-015-9745-y [DOI] [PubMed] [Google Scholar]
- 13.Yuan J.H., Yang F., Wang F. et al. (2014) A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681 10.1016/j.ccr.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 14.Wen W., Ding J., Sun W. et al. (2012) Cyclin G1-mediated epithelial-mesenchymal transition via phosphoinositide 3-kinase/Akt signaling facilitates liver cancer progression. Hepatology 55, 1787–1798 10.1002/hep.25596 [DOI] [PubMed] [Google Scholar]
- 15.Zheng H., Yang Y., AUID- Oho et al. (2019) Tropomodulin 3 modulates EGFR-PI3K-AKT signaling to drive hepatocellular carcinoma metastasis. Mol. Carcinog. 58, 1897–1907 10.1002/mc.23083 [DOI] [PubMed] [Google Scholar]
- 16.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J. and Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 17.Tsiambas E., Lefas A.Y., Georgiannos S.N. et al. (2016) EGFR gene deregulation mechanisms in lung adenocarcinoma: A molecular review. Pathol. Res. Pract. 212, 672–677 10.1016/j.prp.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 18.Han L., Lee C.K., Pang H. et al. (2017) Genetic predisposition to lung adenocarcinoma among never-smoking Chinese with different epidermal growth factor receptor mutation status. Lung Cancer 114, 79–89 10.1016/j.lungcan.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 19.Zhou W. and Christiani D.C. (2011) East meets West: ethnic differences in epidemiology and clinical behaviors of lung cancer between East Asians and Caucasians. Chin. J. Cancer 30, 287–292 10.5732/cjc.011.10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavin Y., Kobayashi S., Leader A. et al. (2017) Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 169, 750–765 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y., Wang R., Shen X. et al. (2016) Minor Components of Micropapillary and Solid Subtypes in Lung Adenocarcinoma are Predictors of Lymph Node Metastasis and Poor Prognosis. Ann. Surg. Oncol. 23, 2099–2105 10.1245/s10434-015-5043-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A., Greninger P., Rhodes D. et al. (2009) A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15, 489–500 10.1016/j.ccr.2009.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak P., Leav I., Pursell B. et al. (2010) ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell 17, 319–332 10.1016/j.ccr.2010.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng H., Shen M., Zha Y.L. et al. (2014) PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Cancer Cell 26, 358–373 10.1016/j.ccr.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brabletz T. (2012) EMT and MET in metastasis: where are the cancer stem cells. Cancer Cell 22, 699–701 10.1016/j.ccr.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 26.Zhu H.E., Yin J.Y., Chen D.X., He S. and Chen H. (2019) Agmatinase promotes the lung adenocarcinoma tumorigenesis by activating the NO-MAPKs-PI3K/Akt pathway. Cell Death. Dis. 10, 854 10.1038/s41419-019-2082-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun T., Du B., Diao Y., Li X., Chen S. and Li Y. (2019) ATAD2 expression increases [18F]Fluorodeoxyglucose uptake value in lung adenocarcinoma via AKT-GLUT1/HK2 pathway. BMB Rep. 52, 457–462 10.5483/BMBRep.2019.52.7.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Mao Y., Mao Q. et al. (2019) FLOT1 promotes tumor development, induces epithelial-mesenchymal transition, and modulates the cell cycle by regulating the Erk/Akt signaling pathway in lung adenocarcinoma. Thorac. Cancer 10, 909–917 10.1111/1759-7714.13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei C., Dong X., Lu H. et al. (2019) LPCAT1 promotes brain metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC pathway. J. Exp. Clin. Cancer Res. 38, 95 10.1186/s13046-019-1092-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu G.S., Li M., Xu C.X. and Wang D. (2018) APE1 stimulates EGFR-TKI resistance by activating Akt signaling through a redox-dependent mechanism in lung adenocarcinoma. Cell Death. Dis. 9, 1111 10.1038/s41419-018-1162-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Ning Z., Zhou X. et al. (2018) Neuregulin1 acts as a suppressor in human lung adenocarcinoma via AKT and ERK1/2 pathway. J. Thorac. Dis. 10, 3166–3179 10.21037/jtd.2018.05.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sui J., Yang R.S., Xu S.Y. et al. (2017) Comprehensive analysis of aberrantly expressed microRNA profiles reveals potential biomarkers of human lung adenocarcinoma progression. Oncol. Rep. 38, 2453–2463 10.3892/or.2017.5880 [DOI] [PubMed] [Google Scholar]