Abstract
Epilepsy is a widespread neurological disease characterized by abnormal neuronal activity resulting in recurrent seizures. There is mounting evidence that a circadian system disruption, involving clock genes and their downstream transcriptional regulators, is associated with epilepsy. In this study, we characterized the hippocampal expression of clock genes and PAR bZIP transcription factors (TFs) in a mouse model of temporal lobe epilepsy induced by intrahippocampal injection of kainic acid (KA). The expression of PAR bZIP TFs was significantly altered following KA injection as well as in other rodent models of acquired epilepsy. Although the PAR bZIP TFs are regulated by proinflammatory cytokines in peripheral tissues, we discovered that the regulation of their expression is inflammation-independent in hippocampal tissue and rather mediated by clock genes and hyperexcitability. Furthermore, we report that hepatic leukemia factor (Hlf), a member of PAR bZIP TFs family, is invariably downregulated in animal models of acquired epilepsy, regulates neuronal activity in vitro and its overexpression in dentate gyrus neurons in vivo leads to altered expression of genes associated with seizures and epilepsy. Overall, our study provides further evidence of PAR bZIP TFs involvement in epileptogenesis and points to Hlf as the key player.
Subject terms: Circadian rhythms and sleep, Diseases of the nervous system, Neuroimmunology, Neurology, Epilepsy
Introduction
Epilepsy is a chronic brain disease characterized by the occurrence of recurrent seizures, which are the result of an excessive electrical discharge or hypersynchronization of a group of neurons in distinct areas of the brain. Temporal lobe epilepsy (TLE) is the most common subtype of acquired epilepsy1. TLE, affecting the hippocampal formation and surrounding cortices of the temporal lobe, is associated with pathological changes including neuroinflammation and neurodegeneration resulting in impaired cognition2.
Recent studies provided converging evidence that the circadian system might be disrupted in epilepsy3,4. At the molecular level, core clock genes form a transcriptional-translational feedback loop that drives their diurnal oscillations. The positive loop members CLOCK (or NPAS2) and BMAL1 form heterodimers that activate the expression of negative feedback loop members, CRY and PER, by binding to E-box motives in their promoters. CRY and PER consequently inhibit the transcriptional activity of CLOCK/NPAS2:BMAL1 complex and thereby their own expression. The inhibition of BMAL1 expression is additionally mediated in a second negative feedback loop by REV-erb-α and RORC5,6. Moreover, core clock components interact with the expression of the proline and acidic amino acid-rich basic leucine zipper (PAR bZIP) transcription factors (TFs). This family is composed of three activators, DBP (albumin D-site-binding protein), HLF (hepatic leukemia factor), TEF (thyrotrophic embryonic factor) and one suppressor of transcription E4 Promoter-Binding Protein 4 (E4BP4), known also as NFIL37–10. It has been shown that E4BP4 has an opposite rhythm of oscillation compared to positive members of PAR bZIP TFs and compete with them for the same binding sites11. Importantly, the triple knockout mouse deficient for the main three PAR bZIP transcription factors (Hlf, Dbp and Tef) developed generalized spontaneous seizures12. Additionally, a significant reduction in seizure threshold was reported in Bmal1 knockout mouse13 and decreased levels of CLOCK, CRY1, PER1 and DBP protein were found in epileptic tissue obtained from surgical resection3.
Proinflammatory cytokines, which may be produced following seizures and contribute to their generation and susceptibility14, have been shown to be key regulators of clock gene expression. Decreased expression of clock genes as well as PAR bZIP TFs was observed in NIH-3T3 cells or human synovial fibroblasts exposed to tumor necrosis factor α (TNF-α) or interleukin 1-β (IL-1β), and in the liver of mice injected with TNF-α or with CD40 agonistic antibodies15–18. The functional significance of cytokine-mediated suppression of clock genes remains elusive.
In this study, our objective was to characterize the hippocampal expression of clock genes and PAR bZIP TFs in a kainic acid (KA) model of TLE and to dissect the interaction between them and neuroinflammation. The intrahippocampal KA injection in adult mice induces an acute status epilepticus, followed by 14 days of a latent period without ictal activity. Afterwards, animals develop spontaneous recurrent seizures19. In this model, seizures are accompanied by progressive neurodegeneration and neuroinflammation in hippocampus20.
We found significant changes in PAR bZIP TFs expression characterized by downregulation of Hlf, Dbp, Tef and upregulation of E4bp4. Surprisingly, we were not able to mimic these changes by simulating only neuroinflammation. This suggests a tissue-specific and inflammation-independent pathway. The changes in Clock, Bmal1, Npas2 and Period genes expression at an early stage of KA-induced epilepsy might be driving the subsequent changes in PAR bZIP TFs expression. Additionally, we demonstrated that Hlf over-expression in primary hippocampal neurons leads to altered neuronal excitability in vitro and differential expression of genes involved in neuronal excitability, seizures and epilepsy in vivo.
Methods
Animals
All methods were carried out in accordance with guidelines and regulations of University of Zurich, Switzerland and University of Los Andes, Santiago, Chile. All experimental protocols were approved by the Cantonal Veterinary Office of Zurich, Switzerland and the Bioethical Committee of University of Los Andes, Santiago, Chile. Adult C57Bl6/J male mice (12–14 weeks old, from Charles River) and Sprague Dawley female rats (from the breeding colony of Universidad de los Andes, Chile) were used in the study. Animals were housed at standard conditions (20–24 °C, minimum 40% relative humidity) under a 12-hour light/dark cycle (lights on at 7 a.m., lights off at 7 p.m), with access to food and water ad libitum.
Stereotaxic injections into hippocampus
Mice were anaesthetized with isoflurane and placed into a motorized stereotaxic robot (Neurostar) equipped with the drill and microinjector Nanoinject II (Drummond). The concentration of isoflurane was adjusted until all pain reactions disappeared. After fixation, a small opening was drilled in the skull to allow for the injection needle to access the hippocampal formation. Afterwards, sterile solutions of PBS, KA (5 nM, 70 nl, Sigma Aldrich), N-methyl-D-aspartic-acid (NMDA, 70 mM, 140 nl, Sigma Aldrich), (-)-Bicuculline methiodide (1 μM, 0.5 μl, Tocris), mouse recombinant cytokines TNF-α (15 pmol, 0.5 μl, Peprotech) and IL-1β (5 pmol, 0.5 μl, Peprotech), lipopolysaccharide (LPS, 10 μg/1 μl, 1 μl, Sigma Aldrich) or AAV vectors (described below) were stereotaxically injected into the right hippocampus (−1.8 AP, +1.6 ML, +1.9 DV). The injection capillary was left in the brain for additional 5 min after the injection. Control animals received a matching volume of sterile PBS or control AAV vector. All injections were performed between 7 a.m. and noon. The concentration and volume injected is summarized in Table 1. Additionally, before and immediately after surgery, mice received an injection of Temgesic (buprenorphine, 0.06 mg/kg, i.p.).
Table 1.
Primers used in the RT-qPCR.
| Gene Name | Forward primer | Reverse primer |
|---|---|---|
| Hlf | TGCTTCGTCGTGCGTCTC | CAAGAGGGAAATGGAGAAAGTGAA |
| Tef | TCCCCTTAGTCCCCGTTCT | CTCCAAGAAACAAGCAGACAGT |
| Dbp | TAGAAGGAGCGCCTTGAGTC | GCAACCCTCCAGTATCCAGA |
| E4bp4 | ATGGGAAGCTCTTTCTCCACT | TACCCGAGGTTCCATGTTTC |
| Npas2 | TCGGGACCAGTTCAATGTTCT | GATTTCTGTTTGTGCTGAGACTTC |
| Clock | TTCCTTCCTTAGAGACGAGACT | CTAAATGCTACCCTGAGGATAGAG |
| Bmal1 | AACTACAAGCCAACATTTCTATCAG | TTCCCTCGGTCACATCCTAC |
| Per1 | CAGGCTAACCAGGAATATTACCAGC | CACAGCCACAGAGAAGGTGTCCTGG |
| Per2 | CTCAGCCTCCCACTCTATATGT | AGGACCAGCAGCACAGATAG |
| Per3 | TCAGAAGAAGCCAAGCCAATC | GTTGTTTCCTGTTTCCGTATGTC |
| Cry1 | GATCCACCATTTAGCCAGACAC | ACAGCCACATCCAACTTCCA |
| Cry2 | CAAGCACTTGGAACGGAAGG | GAAGAGGCGGCAGGAGAG |
| Rev-erb-α | GTCTCTCCGTTGGCATGTCT | CCAAGTTCATGGCGCTCT |
| Rorc | ACTACGGGGTTATCACCTGTGAG | GTGCAGGAGTAGGCCACATTAC |
| Hprt | TCCTCCTCAGACCGCTTTT | AGGTATACAAAACAAATCTAGGTCAT |
| Eef1a1 | AAGCCCATGTGTGTTGAGAG | CTCCAGCAGCCTTCTTGTC |
| TNF-α | GAAGATGTGCCTGTCCTGTGT | CGCTCAGGTCAGTGATGTTAA |
| Il-1β | AAACAGATGAAGTGTAGG | TGGAGAACACCACTTGTT |
| Il-10 | CAGGGATCTTAGCTAACGGAAA | GCTCAGTGAATAAATAGAATGGGAAC |
In KA model experiments (n = 125, 4–7 mice per group), animals were sacrificed at either 3 hours, 1-day post injection (dpi), 6 dpi, 14 dpi or 28 dpi. Additionally, at 1 and 28 dpi, mice were sacrificed at four different Zeitgeber time-points (ZT = Zeitgeber time, ZT0 lights on, ZT12 lights off): ZT0, ZT6, ZT12 and ZT18. At 6 and 14 dpi, mice were sacrificed at two ZT: ZT0 and ZT12. In experiments investigating acute effect of recombinant murine cytokines TNF-α and IL-1β (n = 15, 5 mice per group), animals were sacrificed at 6 hours after their injection. In experiments with LPS (n = 10, 5 mice per group), NMDA and (-)-Bicuculline methiodide (n = 15, 5 mice per group), animals were sacrificed at 24 h after the injection to resemble the 1 dpi time-point in KA experiment.
AAV-vectors
Self-complementary (sc) adenovirus-associated virus (AAV) vector TNF (AAV-TNF) scAAV-2/8-hCMV-chI-floxedmTNFalpha-SV40p(A) and its corresponding control AAV vector (AAV-control) scAAV-2/8-hCMV-chI-loxP-SV40p(A) were kindly provided by the laboratory of Christopher Pryce (University of Zurich, Switzerland). The AAV vectors were characterized in their recent publication [21]. AAV-control and AAV-TNF were injected as described above at a final titer of 1.1 × 1011 vg/ml (280 μl, n = 10, 5 mice per group). Animals were sacrificed 14 days after AAV injection and their brain processed for the gene expression analysis.
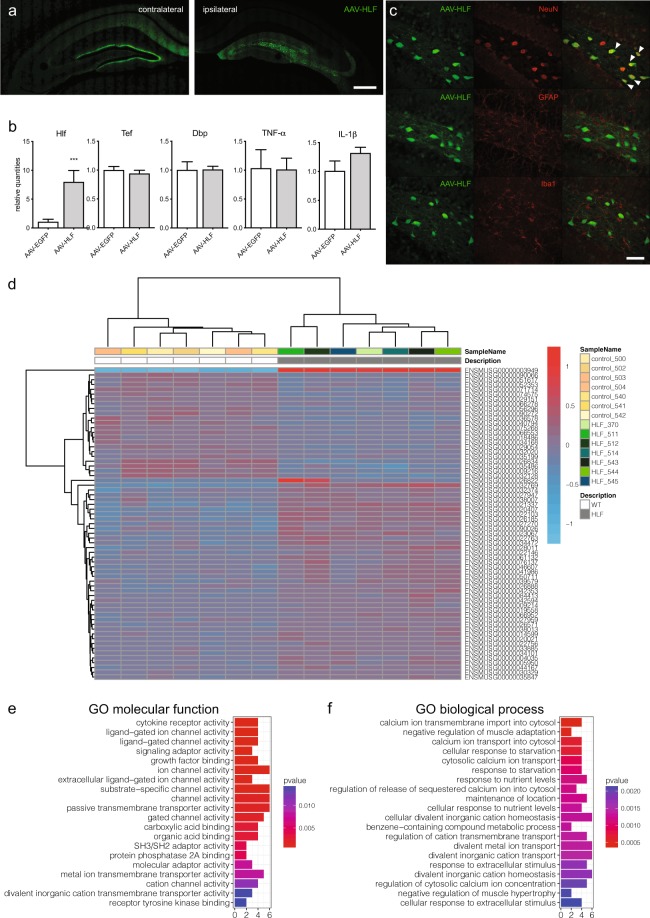
Single-stranded (ss) AAV vector HLF (AAV-HLF) ssAAV-2/8-hSyn1-chI-EGFP_2A_mHLF-WPRE-SV40p(A) and its corresponding control AAV vector (AAV-EGFP) ssAAV-2/8-hSyn1-chI-EGFP-WPRE-SV40p(A) were produced by the Viral Vector Facility (VVF, University of Zurich) as described before21. AAV-HLF was constructed by inserting the PCR-amplified mHLF ORF (Origene, MR204073, NM_172563) downstream and in-frame of an EGFP_2A sequence. AAV-EGFP and AAV-HLF vectors were injected at a final titer of 1.2 × 1012 vg/ml (280 μl, n = 26, 7 mice per group for transcriptomics; 12 mice were used in the pilot study to determine the transgene expression levels, localization and subcellular specificity in the hippocampus, Fig. 1a–c). After 14 days of expression, animals were sacrificed, and their brain processed for the gene expression and fluorescent microscopy analyses. Histology and immunohistochemistry were performed as described in our previous work22,23. Brain sections were stained with the following antibodies: GFAP (DAKO Schweiz, Z334, 1:20000), Iba-1 (Wako, 019–19741, 1:3000) and NeuN (Chemicon, MAB377, 1:1000).
Figure 1.
Identification of genes associated with Hlf over-expression in dentate gyrus neurons of adult male mice. (a) The representative expression pattern of EGFP reporter expression after AAV-HLF injection in the ipsilateral hippocampus. Scalebar = 400 μm (b) Relative mean expression (±SD) of PAR bZIP TFs and inflammatory cytokines after AAV-HLF injection. The t-test was performed to evaluate significance. ***p < 0.001. (c) Co-localization of EGFP reporter expression with immunohistological staining specific for neurons (NeuN), astrocytes (GFAP) and microglia (Iba1). White arrows show co-localization of HLF-EGFP reporter with neuronal NeuN staining. Scalebar = 50 μm. (d) Heatmap shows the z-score from the transformed counts of each individual sample replicate, for all differentially expressed genes. (e) Summary of enrichment for GO molecular function and (f) Biological process. Figures were created using Adobe Illustrator version 23.1.1. www.adobe.com/Illustrator, R version 3.4.3.; https://www.R-project.org and GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
RNA isolation and gene expression analysis
In all experiments, the dorsal parts of ipsilateral and contralateral hippocampi were dissected on ice and were immediately frozen in RNAPure PeqGOLD solution (VWR). At the day of RNA isolation, brain tissue samples were homogenized (Qiagen TissueLyser) and whole cell RNA extracted using NucleoSpin-RNA II kit (Macherey-Nagel). 500 μg RNA was subsequently reverse-transcribed using random hexamer primers (Thermo Scientific) and M-MuLV reverse transcriptase (Life Technologies). 20 ng of cDNA was amplified in a CFX384 detection system (Biorad) using PrecisionPLUS qPCR Mastermix (Primerdesign). The relative levels of each RNA sample were calculated by the 2-□ΔCT method24 using qbase software (Biogazelle). Hprt1 and eEF1a1 were used as housekeeping genes. Each CT value used for these calculations was the mean of triplicates of the same reaction. All primers used were synthetized by Eurofins genomics. Sequences of all used primers are listed in Table 1. The t-test with multiple-testing correction was performed to evaluate statistical significance in the gene expression using the function “t-test” on R statistical package25.
Analysis of GSE47752 dataset
The Affymetrix microarray data described by Dingledine et al.26 was obtained from the Gene expression omnibus (GEO), under accession number GSE47752. This data is composed of 172 samples and 31099 features and was analyzed using the R statistical software25. The following samples were included in the analysis: kainic acid (Wadman-day 1-rat 1–6, Wadman-control-rat 1–6, Nadler-day 1-rat 1–6, Nadler-control-rat 1–6; n = 12 control, n = 12 kainic acid), pilocarpine (Coulter-day 1-rat 1–6, Coulter-control-rat 1–6, Dingledine-day 1-rat 1–6, Dingledine-control-rat 1–6; n = 12 control, n = 12 pilocarpine), kindling (McNamara-stage 2-rat 1–6, McNamara-control-rat 1–6, Rogawski-stage 2-rat 1–6, Rogawski-control-rat 1–6; n = 12 control, n = 12 kindling) and self-sustained status epilepticus (Wasterlain-day 1-rat 1–6, Wasterlain-control-rat 1–5; n = 5 control, n = 6 self-sustained status epilepticus).
Normalization was performed using the RMA normalization using all probes regardless of Affymetrix ‘Present’ (P) or ‘Absent’ (A) calls. Only probes with the “at” suffix were retained to ensure that only probes specific to a single gene were carried forward for further analysis. Differential expression of the genes Hlf, Tef, Dbp and E4bp4 was performed using the Limma (Linear models for microarray) package27.
Primary neuronal cultures & patch clamp
Primary hippocampal cultures were prepared from E18 Sprague Dawley rats as previously described28. This protocol was approved by the Universidad de los Andes Animal Care and Use Committee. Cells were cultured on cover slips and transduced with at 2 DIV with AAV-HLF or AAV-EGFP (MOI = 50000, 1.2 × 1013 vg/ml). Medium was changed every 3 days. At ~12–16 DIV, the coverslip was transferred to a 35 mm plate containing ACSF which contained (in mM) 120 NaCl, 3 KCl, 2.5 CaCl2, 2 MgSO4, 1 NaH2PO4, 25 NaHCO3, and 20 glucose (pH 7.4) for electrophysiology experiments. The intracellular solution consisted of (in mM) 146 KGluconate (KGluc), 1 NaCl, 1 MgSO4, 0.2 CaCl2, 2 EGTA, 10 HEPES, 4 MgATP, 0.3 NaGTP, (pH 7.3, 290 mOsM/Kg). Patch clamp experiments were performed under an inverted NIKON TE-2000U microscope to visualize transduced primary hippocampal neurons expressing EGFP. Whole-cell pipettes were pulled from thin wall glass capillaries (1.5 O.D. × 1.17 I.D., Harvard Apparatus, Massachusetts, USA) using a Sutter puller9 (model P-97). Electrode resistances in the bath were 5–7 MΩ. Series resistance was monitored by a 5 mV step, and cells were discarded if this changed significantly (~20%). Cells were clamped at a holding potential of −70 mV with an axopatch 200B (Molecular Devices, Pennsylvania, USA), filtered a 2 KHz and digitized at 5 kHz using a Digidata 1550 (Molecular Devices) and Clampex 10.0 (Molecular Devices). Stimulation of neurons was achieved by adding bicuculline (20 μM) to the recording bath, while delivering glutamate (2.5 mM) with a patch pipette with a resistance of ~2MΩ placed near the recorded cell (~50μm away). Glutamate concentration in the pipette was 2.5 mM, and delivery of glutamate was obtained by a 50 μl Hamilton syringe mounted to a SP101i syringe pump (flow rate of 1 μl/min, World Precision Instruments, Fl, USA) and connected to the pipette by a tight silicon tubing. Action potentials were obtained by applying a series of current steps (50 pA each) under current clamp mode. Traces were recorded with pClamp 10 and analysis of intrinsic properties and sPSCs were performed with MiniAnalysis (Synaptosoft). Average traces were obtained by selecting events in MiniAnalysis, aligning the peak of the events and averaging those traces under the “group analysis” function. Average traces from each cell were averaged to obtain one trace per group. Decay fit was calculated in Mini Analysis by using a 2 exponential function approach (Amplitude1 exp(−time/τ1) + Amplitude2 exp(−time/τ2).
Two-way ANOVAs and two-tailed t-tests were performed with GraphPad Prism version 8 (GraphPad Software). The amplitude and interevent interval of PSCs were analyzed according to the highest level of activity shown, in one-minute long traces. To make the KS tests more stringent we applied a bootstrap estimation of the p-value by extracting random samples of size 100 (either currents or amplitudes) using the function “sample” on R statistical package25. After obtaining a p-value from these samples the procedure was repeated 1000 times. The percentage of times the random samples gave a p-value < 5% was used to establish if cumulative distributions are different. We considered distributions to be significantly different when more than 50% of random samples had a p-value < 5%This ensures that the KS test is not as sensitive to small differences between the associated functions. Density plot of sPSCs amplitudes and interevent intervals were used to visually confirm our statistical results.
Transcriptomics analysis
Next-generation sequencing was performed by Functional Genomics Center Zurich, University of Zurich, Switzerland. Consequent data analysis was than performed by the Swiss Institute of Bioinformatics, Switzerland. Total RNA was processed using the TruSeq RNA stranded protocol (Illumina) in order to produce sequencing libraries. Libraries were then sequenced on the Illumina HiSeq. 4000, single-end, 125 cycles. Reads were aligned against Mus Musculus.GRCm38.86 genome using STAR (v.2.5.2b)29. The number of read counts per gene locus was summarized with htseq-count (v0.6.1)30 using Mus Musculus.GRCm38.86 gene annotations. Quality of the RNA-seq data alignment was assessed using RSeQC (v. 2.3.7)31. Reads were also aligned to the Mus Musculus.GRCm38.86 transcriptome using STAR (v. 2.5.2b)29 and the estimation of the isoforms abundance was computed using RSEM (v. 1.2.31)32. To assess differential expression, we used the R Bioconductor package DESeq. 2 (version 1.14.1)33. Differentially expressed Genes were identified at the Benjamini-Hochberg (BH) adjusted P < 0.05 level, using the Wald test. For gene set enrichment analysis, no direction criterion on fold change was applied. Enriched GO (Gene Onthology) categories were identified using the enrichment analysis package in R/Bioconductor, clusterProfiler34 with significant terms at BH P adjusted <0.05 reported.
Results
Status epilepticus induces changes in the expression of PAR bZIP transcription factors and does not affect expression of clock core genes
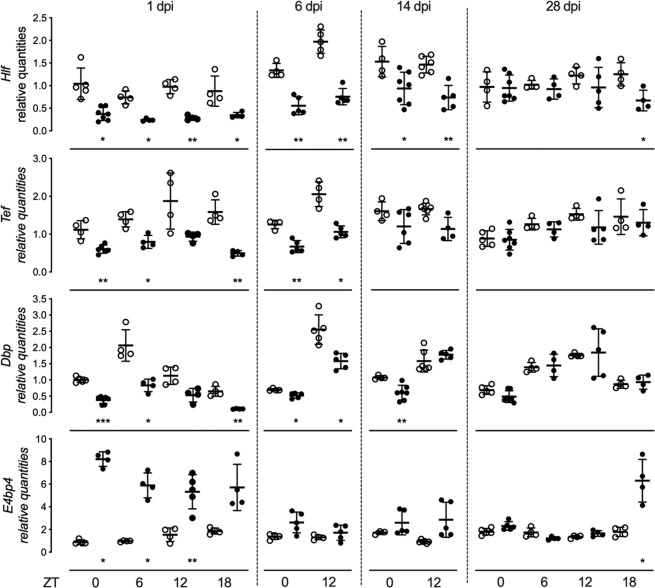
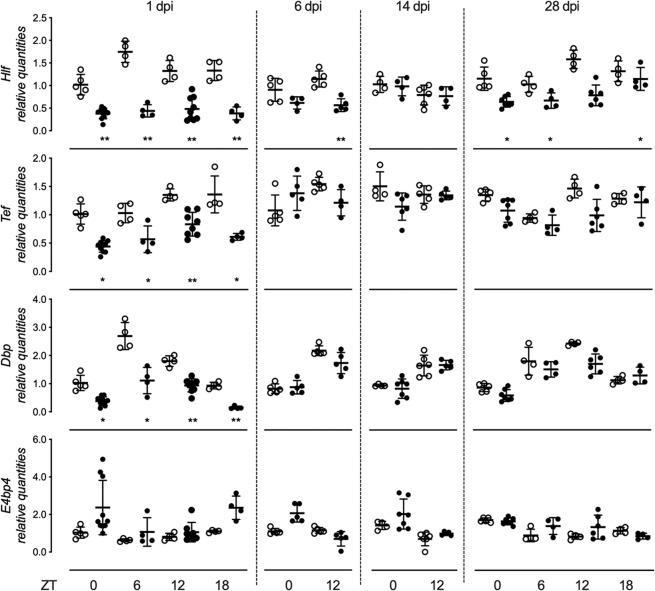
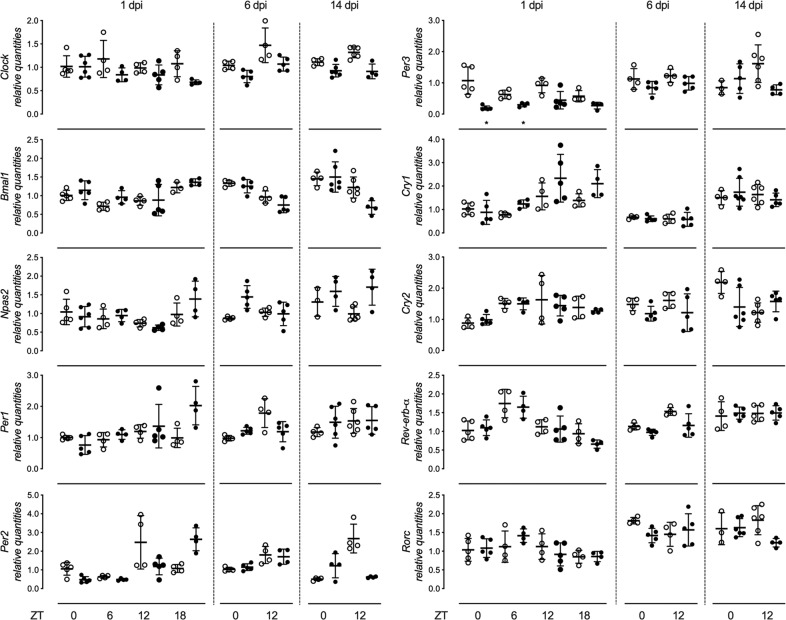
We evaluated the hippocampal expression of core clock genes and PAR bZIP TFs at different stages during epileptogenesis by injecting KA into the right dorsal hippocampus, a well-established model of TLE35. The pathogenesis of this model follows a stereotypic pattern23,35, characterized by an initial status epilepticus lasting up to 1 dpi, followed by a silent latent period and the occurrence of spontaneous recurrent seizures in the chronic period starting at about 14 dpi. The expression of clock genes and PAR bZIP TFs was evaluated at two (6 and 14 dpi) or four ZT time-points (1 and 28 dpi) over a day to exclude possible phase shift in clock gene expression amplitude. We found significant alteration in the expression of PAR bZIP TFs (Fig. 2). The transcriptional activators Hlf, Dbp and Tef were significantly down-regulated while the transcriptional repressor E4bp4 was up-regulated. Changes were most evident during the acute and epileptogenesis phases (1 and 6 dpi, respectively) of the status-epilepticus and tend to normalize thereafter. The expression of Hlf remained significantly down-regulated also at the beginning of the chronic phase, 14 days after the induction of status epilepticus. At 28 dpi, Hlf was significantly suppressed only at ZT18. Interestingly, similar changes in expression of PAR bZIP TFs were found in the contralateral (not-injected) hippocampus at 24 h post injection. Additionally, Hlf remained significantly downregulated during the later chronic period (28 dpi) in the contralateral hippocampus (Fig. 3). Surprisingly, we did not observe any significant changes in the expression of the following clock genes: Clock, Bmal1, Npas2, Per1, Per2, Cry1, Cry2, Rev-erb-α and Rorc (Fig. 4). Only the expression of Per3 was downregulated 24 h after the induction of status epilepticus (Fig. 4). In the contralateral hippocampus, there were no significant changes in the expression of core clock genes except downregulation of Per3 at 1 dpi, ZT0 and 6 (see Supplementary Fig. 1). To ensure that the observed changes are due to the local KA injection, we analyzed in addition the expression of PAR bZIP TFs and core clock genes in the cortical tissue above the injected hippocampus. We did not detect any significant changes in this tissue (see Supplementary Fig. 2).
Figure 2.
Relative mean expression (±SD) of PAR bZIP TFs (Hlf, Tef, Dbp and E4bp4) in ipsilateral dorsal hippocampus at 1, 6, 14 and 28 days post injection (dpi) of KA in adult male mice. ZT0, ZT6, ZT12 and ZT18 (ZT = Zeitgeber time, ZT0 lights on, ZT12 lights off). White and black circles represent individual values for control and KA groups, respectively. The t-test with multiple-testing correction was performed to evaluate significance between control and KA group for each gene. *p < 0.05, **p < 0.005, ***p < 0.00. Figures were created using GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
Figure 3.
Relative mean expression (±SD) of PAR bZIP TFs (Hlf, Tef, Dbp and E4bp4) in contralateral (non-injected) dorsal hippocampus at 1, 6, 14 and 28 days post injection (dpi) of KA in adult male mice. ZT0, ZT6, ZT12 and ZT18 (ZT = Zeitgeber time, ZT0 lights on, ZT12 lights off). White and black circles represent individual values for control and KA groups, respectively. The t-test with multiple-testing correction was performed to evaluate significance between control and KA groups. *p < 0.05, **p < 0.005. Figures were created using GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
Figure 4.
Relative mean expression (±SD) of clock genes (Clock, Bmal1, Npas2, Per1, Per2, Per3, Cry1, Cry2, Rev-erb-α and Rorc) in ipsilateral dorsal hippocampus at 1, 6 and 14 days post injection (dpi) of KA in adult male mice. ZT0, ZT6, ZT12 and ZT18 (ZT = Zeitgeber time, ZT0 lights on, ZT12 lights off). White and black circles represent individual values for control and KA groups, respectively. The t-test with multiple-testing correction was performed to evaluate significance between control and KA groups. *p < 0.05. Figures were created using GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
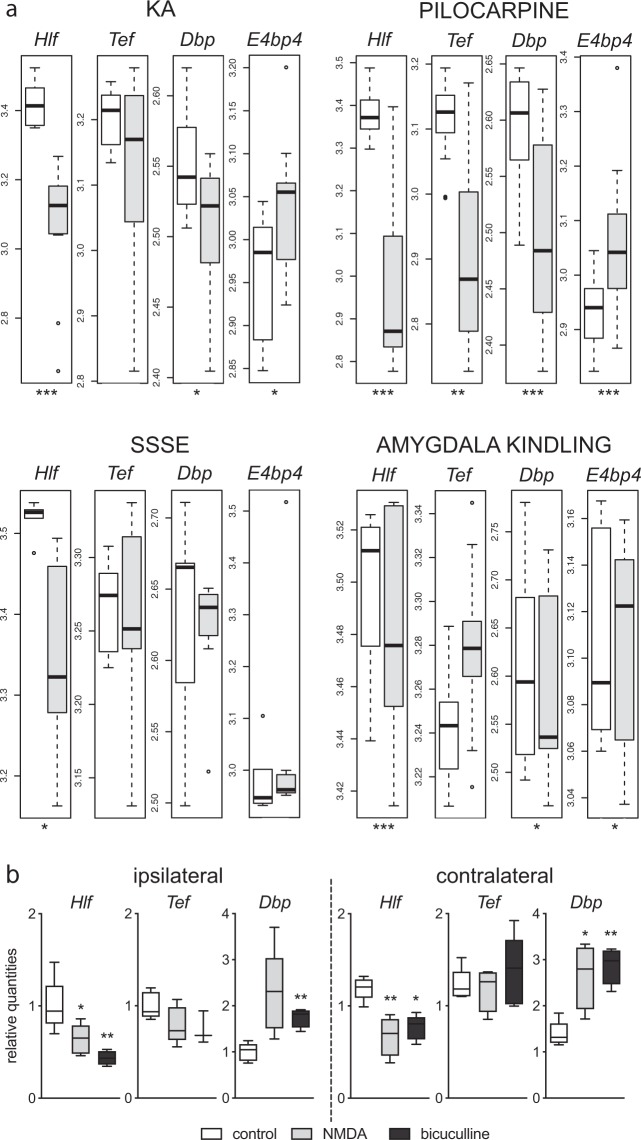
To evaluate whether changes in PAR bZIP TFs can be observed in other rodent models of TLE, we performed analysis on previously published26 microarray data (NCBI Gene Expression Omnibus. GSE47752) profiling the hippocampal expression of genes in four different rat models of acquired epilepsy: kainate (intraperitoneal, i.p.), pilocarpine, self-sustained status epilepticus (SSSE) and amygdala kindling. We focused our analysis on the differential expression of PAR bZIP factors in these models at 24 h after induction of status epilepticus. Our analysis revealed signification downregulation of Hlf in all four models (See Fig. 5a). In addition, Tef was significantly downregulated and the transcriptional repressor E4bp4 was significantly up-regulated in all models except for SSSE. Dbp was significantly downregulated in the pilocarpine model. Taken together the microarray data of four rat models of epilepsy support our finding of altered expression of PAR bZIP TFs in KA induced epilepsy.
Figure 5.
The expression of PAR bZIP TFs in other models of acquired epilepsy. (a) Hippocampal expression of PAR bZIP transcription factors (Hlf, Dbp, Tef and E4bp4) in four different rat models of status epilepticus: systemic KA (n = 12 in each group) and pilocarpine administration (n = 12 in each group), self-sustained status epilepticus (SSSE, n = 5 in control group, n = 6 in SSSE group) and amygdala kindling (n = 12 in each group)26. All rats involved in the study were male. White and grey boxes represent control and epilepsy model groups, respectively. Differential expression of the genes Hlf, Tef, Dbp and E4bp4 was performed using the Linear models for microarray (Limma) (b) Relative expression of PAR bZIP TFs after intrahippocampal injection of NMDA or bicuculline in adult male mice (n = 5 in each group). The t-test was performed to evaluate significance. *p < 0.05, **p < 0.005, ***p < 0.001. Figures were created using R version 3.4.3., https://www.R-project.org and GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
Excitotoxicity and seizure activity induce the downregulation of Hlf
Next, we investigated whether acute hyperexcitability in general is responsible for the altered behavior of PAR bZIP TFs. To explore this possibility, we injected either N-methyl-D-aspartic acid (NMDA) or bicuculline into the right dorsal hippocampus. Hippocampal injection of NMDA stimulates ionotropic NMDA receptors, induces acute excitotoxicity and status epilepticus; however, unlike KA-induced status epilepticus, does not cause an early loss of hippocampal interneurons22,36. Bicuculline is a competitive antagonist of GABAA receptors and it is known to produce seizures in the absence of neurodegenerative events37. In both models, Hlf was significantly downregulated 24 h after status epilepticus (Fig. 5b), whereas Dbp was upregulated only after NMDA injection. These effects were mirrored also in the contralateral hippocampus (Fig. 5b).
Changes in the expression of PAR bZIP transcription factors are independent of inflammation
Both clinical38–40 and experimental20,41,42 evidence suggest that inflammatory processes are involved in the pathogenesis of TLE. Hence, we characterized the expression of proinflammatory cytokines TNF-α and IL-1β, as well as the anti-inflammatory cytokine interleukin 10 (IL-10) in the intrahippocampal KA model of TLE (Fig. 6). TNF-α expression was significantly upregulated 1, 6 and 14 days after the KA infusion (15-fold, 10-fold and 5-fold respectively). IL-1β beta remained upregulated at all timepoints studied, the effect being about 5-fold. The expression of IL-10 was increased at later time points, at 14 and 28 dpi. These cytokines were also significantly upregulated in the contralateral hemisphere, albeit the increase being less pronounced (Fig. 6).
Figure 6.
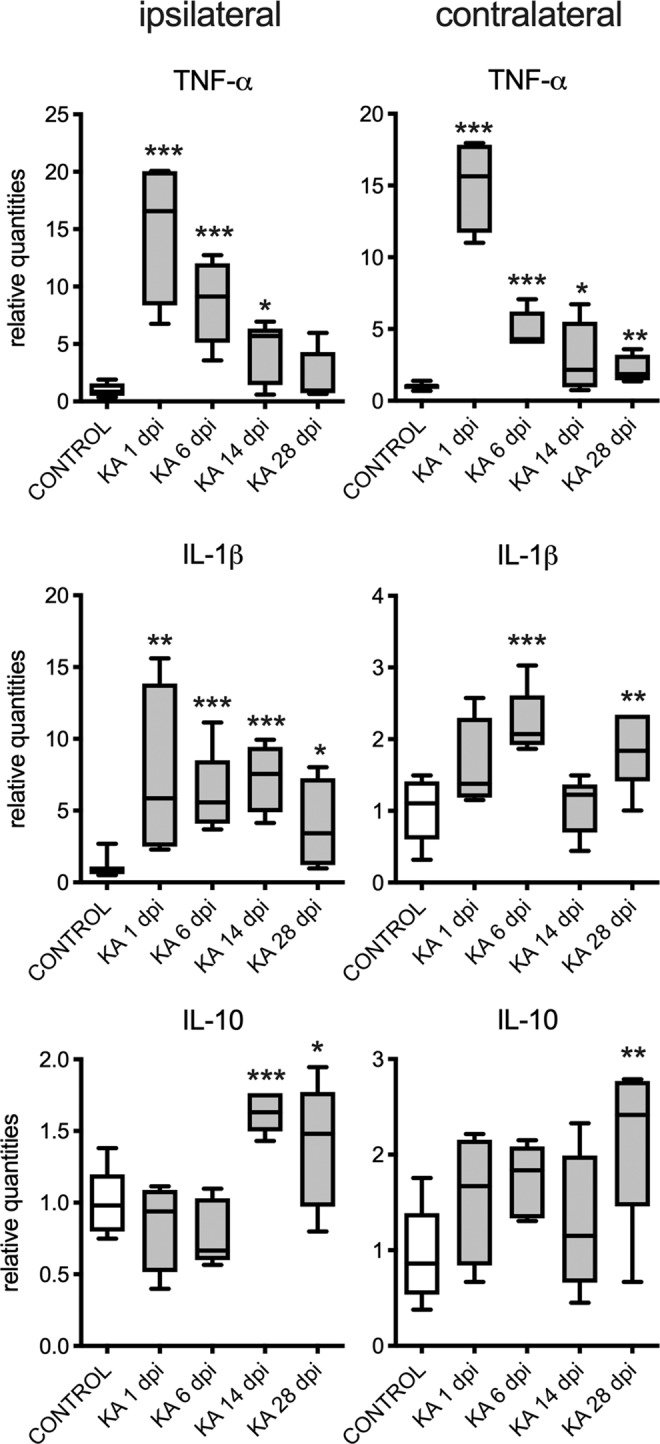
The relative expression of proinflammatory (TNF-α and IL-1β) and anti-inflammatory cytokines (IL-10) in ipsilateral and contralateral hippocampi at 1, 6, 14 and 28 days post injection (dpi) of KA in adult male mice (n = 5 in each group). The left panel shows changes in ipsilateral hippocampus. The right panel shows changes in contralateral hippocampus. The t-test with multiple-testing correction was performed to evaluate significance between control and KA groups *p < 0.05, **p < 0.005, ***p < 0.001. Figures were created using GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
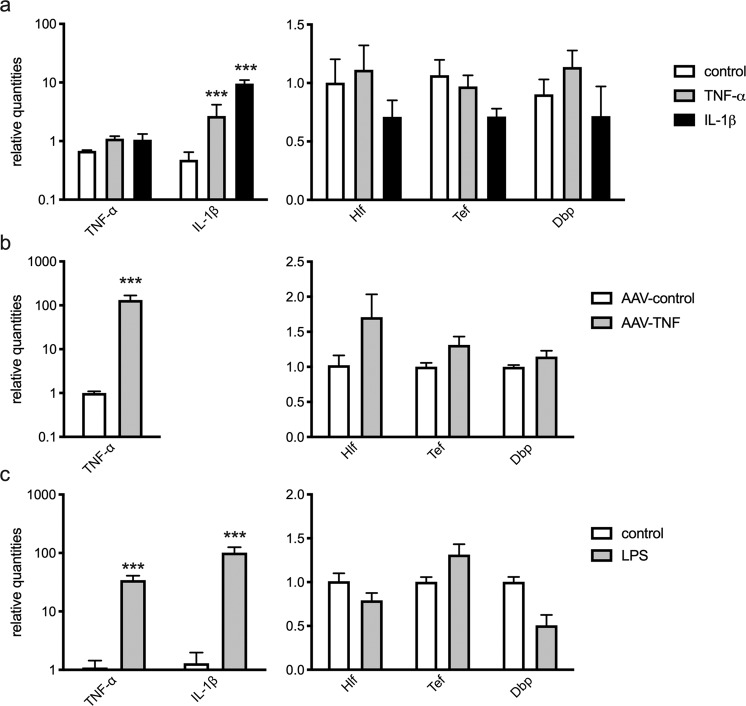
As outlined above, inflammation regulates the expression of PAR bZIP TFs. Thus, we hypothesized that the KA induced inflammatory response drives the observed changes of PAR bZIP TFs expression. To test this hypothesis, we infused either mouse cytokine TNF-α or IL-1β into the hippocampus. Whereas TNF-α mRNA expression did not increase upon TNF-α or IL-1β injection, the expression of IL-1β mRNA was augmented about 5- and 10-fold respectively (Fig. 7a). However, we observed no changes in the expression of PAR bZIP factors (Fig. 7a). As cytokine levels reached by this treatment may have been insufficient to inhibit PAR bZIP TFs, we decided to simulate sub-chronic TNF-α exposure by infusion of an adenovirus-associated viral TNF vector (AAV-TNF). The AAV-TNF induced robust TNF-α expression (100- fold) in the hippocampus; however, it did not affect the expression of PAR bZIP TFs (Fig. 7b). We corroborated this result by inducing neuroinflammation by infusion of lipopolysaccharide (LPS). LPS activates microglia interacting with their Toll-like receptor-4 and induces production of inflammatory cytokines. The intrahippocampal infusion of LPS induced significant overexpression of TNF-α (50-fold) and IL-1β (100-fold); however, the expression of PAR bZIP TFs remained unchanged (Fig. 7c). Surprisingly, the hippocampal expression of PAR bZIP TFs was not affected by a local increase in the expression of pro-inflammatory cytokines such as TNF-α and IL-1β, as has been described for other murine tissues15. This evidence is in line with our previous finding that the expression of PAR bZIP TFs is unchanged in brains of mice with experimental autoimmune encephalomyelitis, a model of multiple sclerosis (unpublished data). Hence, this evidence suggests that early changes in hippocampal expression of PAR bZIP TFs in rodent models of TLE are not a direct response to inflammatory processes that occur during the initial stages of epileptogenesis. Thus, PAR bZIP TFs in the hippocampal tissues are regulated in a different way than in the periphery.
Figure 7.
The effect of proinflammatory environment on the expression of PAR bZIP TFs in dorsal hippocampus of adult male mice (n = 5 in each group). Relative mean expression (±SD) of inflammatory cytokines and PAR bZIP TFs after (a) acute injection of mouse recombinant cytokines (TNF-α or IL1-β, 6 h after injection) (b) chronic overexpression of TNF-α (c) acute inflammation induced by injection of LPS (24 h after injection). The t-test with multiple-testing correction was performed to evaluate significance. ***p < 0.001. Figures were created using GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
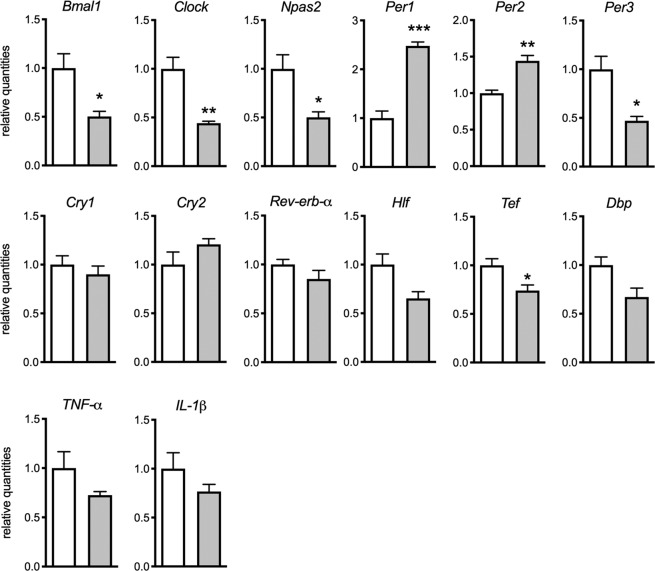
KA induces early response of clock genes
To further explore the mechanism responsible for the downregulation of PAR bZIP TFs during epileptogenesis, we decided to characterize the hippocampal expression of PAR bZIP TFs and core clock genes, at 3 hours after the induction of status epilepticus by intrahippocampal KA (Fig. 8). At this early time point, we observed significant reduction of Clock, Bmal1, Npas2 and Per3 expression. Additionally, the expression of Per1 and Per2 was significantly upregulated. The expression of PAR bZIP TFs Hlf and Dbp did not show statistically significant decrease, while the expression of Tef was significantly downregulated. This expression pattern suggests that early changes in core clock genes might be involved in the initiation of changes in PAR bZIP TFs levels. The positive regulators of their expression (Clock, Bmal1, Npas2) were downregulated and their negative regulators (Per1 and Per2) were upregulated. Additionally, the expression of TNF-α and IL-1β was not altered yet at this early time point. These results might explain the changes in PAR bZIP TFs expression at 1 dpi after the KA lesion.
Figure 8.
Relative mean expression (±SD) of clock genes, PAR bZIP TFs and proinflammatory cytokines at three hours after the injection of KA in adult male mice (n = 5 in each group). White and grey boxes represent control and KA groups, respectively. The t-test with multiple-testing correction was performed to evaluate significance. *p < 0.05, **p < 0.005, ***p < 0.001. Figures were created using GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
Neurons over-expressing Hlf showed a significant decrease in the frequency and increase in the amplitude of spontaneous events in the presence of bicuculline and glutamate
As we observed consistent downregulation of Hlf expression in various models of epilepsy, likely as a result of acute hyperexcitability, we examined the effects of Hlf expression in neurons under basal and hyperexcitable conditions. Hence, we overexpressed Hlf by transducing primary hippocampal neurons at 2 DIV with either a control vector (AAV-EGFP) or a vector overexpressing Hlf (AAV-HLF) and recorded spontaneous currents at ~12–16 DIV by whole-cell voltage clamp. Under basal activity levels (Fig. 9a,b,c), overexpression of Hlf resulted in a significant reduction in the amplitude of sPSCs, with no differences in the frequency or decay kinetics (Fig. 9d, AAV-EGGP: n = 10, 8.72 ± 0.89 msec; AAV-HLF: n = 11, 8.01 ± 1.06 msec, p = 0.6204), of events compared to neurons transduced with an AAV-EGFP. In the presence of bicuculline and pressure-ejected glutamate (Fig. 9e,f,g), neurons overexpressing Hlf showed a significant decrease in the frequency and an increase in the amplitude of sEPSCs compared to neurons carrying the EV control. We observed no differences between groups in the number of action potentials [AAV-EGFP: n = 10, 5.1 ± 0.86, AAV-HLF: n = 11, 4.64 ± 0.92, AAV-EGFP (BIC + GLU): n = 12, 5 ± 0.75, AAV-HLF/(BIC + GLU): n = 6, 5.67 ± 0.56. ANOVA: F (3, 35) = 0.2016, P = 0.8946)] or amplitude [AAV-EGFP: n = 10, 71.49 ± 4.45 mV AAV-HLF: n = 11, 64.57 ± 4.92 mV, AAV-EGFP/(BIC + GLU): n = 12, 61.83 ± 3.51 mV, AAV-HLF/(BIC+GLU): n = 6, 57.08 ± 5.03 mV ANOVA: F (3, 35) = 1.545, P = 0.2200)]; Fig. 9i,j.
Figure 9.
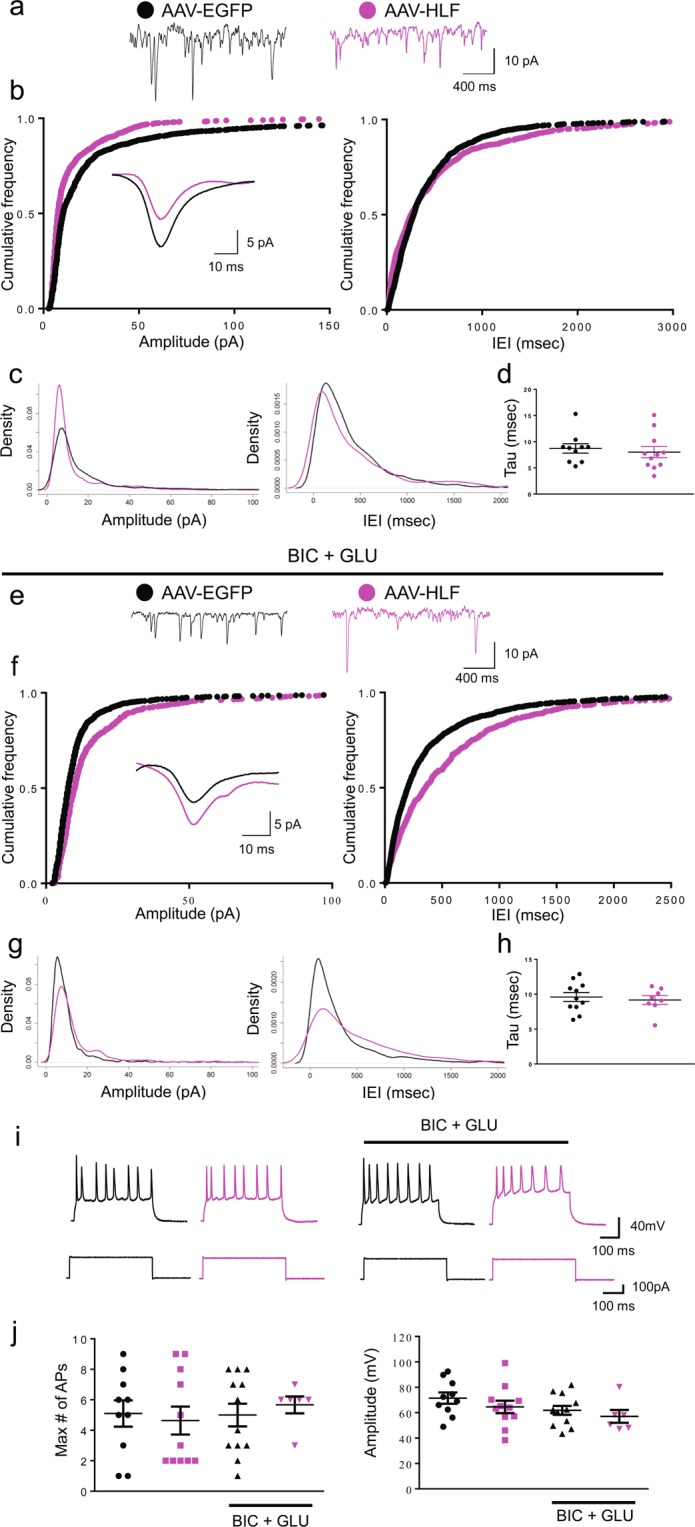
(a–c) Hlf overexpression decreases the amplitude but does not affect the frequency of spontaneous currents under basal conditions. (a) Representative traces (sPSCs). (b) (left) Cumulative amplitude distributions for sPSCs. 64.3% of simulated samples had a p-value < 5%. Inset: Mean spontaneous currents. (right) Cumulative interevent interval distributions for sPSCs. 20.7% of simulated samples had a p-value < 0.05. (c) Density plot of sPSCs (left) amplitudes and (right) interevent intervals. (d) Decay kinetics of sPSCs. (e,f) Hlf overexpression reduces the frequency of spontaneous excitatory currents in the presence of bicuculline and glutamate (BIC+GLU). (e) Representative sPSCs traces. (f) (left) Cumulative amplitude distributions for sPSCs. 67.6% of simulated samples had a p-value < 0.05. (right) Cumulative interevent interval distributions for sPSCs. 72.6% of the simulated samples had a p-value < 0.05. Inset: Mean spontaneous currents. (g) Density plot of sPSCs (left) amplitudes and (right) interevent intervals. (h) No significant differences were observed in decay kinetics between groups (i,j) There are no differences in action potential number between neurons transduced with AAV-EGFP or a AAV-HLF in basal conditions or in the presence of bicuculline and glutamate. (i) representative action potential (AP) traces fired by a current step of 250 nA in (left) basal conditions and (right) in the presence of BIC+GLU. (j) Left panel: AP number. Right panel: AP amplitude. AAV-EGFP: black traces and dots. AAV-HLF: magenta traces and dots. Figures were created using Adobe Illustrator version 23.1.1., www.adobe.com/Illustrator; R version 3.4.3., https://www.R-project.org; Mini Analysis version 6.0.7., http://www.synaptosoft.com/MiniAnalysis/; Pclamp suite version 10, https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite#gref and GraphPad Prism version 8.3.1 for macOS, www.graphpad.com.
Gene expression analysis
To determine which genes are regulated by hepatic leukemia factor, we performed transcriptome analysis of the dorsal hippocampal tissue from mice over-expressing mouse Hlf restricted to the dentate gyrus (Fig. 1a). It has been shown that that mouse endogenous Hlf is expressed mostly in the dentate gyrus, while in CA1 and CA3 subregions of hippocampus the expression is limited43, (Experiment: 565 and 566, Probe Name: Rp_Baylor_103366)44. These samples were compared with samples from mice injected with a control vector (AAV-EGFP). Hlf was over-expressed 7-fold in the dorsal hippocampal tissue while there were no changes in the expression of Tef and Dbp (Fig. 1b). In addition, the AAV-HLF injection did not induce local neuroinflammation detected as expression of TNF-α and IL-1β (Fig. 1b). The expression of AAV-HLF vector was restricted to neurons and was detected in neither astrocytes nor microglia (Fig. 1c). Only samples with confirmed overexpression of Hlf using qPCR were sequenced (AAV-HLF n = 7, AAV-EGFP n = 7). In total, the expression of 65 genes was significantly altered (Fig. 1d). The list of all differentially expressed genes is included in the Supplementary Table S1. Raw data are available at NCBI GEO database under accession number GSE140046. Additionally, the gene ontology classification for molecular function and biological process were performed using all 65 significantly differentially expressed genes (Fig. 1e,f).
We identified genes whose expression is influenced by neuronal Hlf. We found changes in expression of genes coding for ion channels such as Trpa1, P2rx5, Grin3a, Kcng1 and Gabrd. Next, the expression of Synpr, Gfra2, Lcn2, Slc30a3, Rasd2, Igfbp5, Fxyd7, Cdkn1a and Slc6a8 was altered. Synaptoporin, the protein coded by Synpr gene, is a marker of mossy fiber sprouting, a phenomenon associated with epileptogenesis after KA injection45. Gfra2 was reported to modulate threshold of kindling evoked seizures46, Lcn2 was identified as a chemokine inducer in KA model47 and Slc30a3 modulates transport of zinc into synaptic vesicles that is co-released with glutamate and regulates excitability and its loss is associated with febrile seizures48. Rasd2 and Igfbp5 were identified by a recent study as candidate genes associated with epileptogenicity in KA model of TLE49 and Fxyd7 to be altered after seizure preconditioning using pilocarpine model50. Human studies reported Cdkn1a to be upregulated in patients with epilepsy51 and deficiency in Slc6a8 was reported to result in intractable epilepsy and cognitive impairment52. Although, with the given dataset, we are not able to deduce any direct involvement in seizure modulation, many of those genes were previously described to be associated with neuronal excitability, seizures or epilepsy.
Discussion
Even though the occurrence of epileptic seizures in circadian patterns has been documented in animal models as well as humans53 the relationship between epileptic seizures and the circadian system at the molecular level is still unclear. A recent study reported decreased Clock expression in human epileptogenic tissue and decreased seizure threshold in mice with Clock deletion in excitatory cortical neurons suggesting that alterations in Clock expression are epileptogenic3. Furthermore, mice deficient for Bmal1 exhibited reduced seizure thresholds13.
In this study, we utilized the KA model of TLE to study the expression of core clock genes and their downstream transcription factors, the PAR bZIP TFs family. We showed that the expression of Hlf, Dbp, Tef is suppressed while the expression of E4bp4 is significantly upregulated during epileptogenesis. This effect was most evident during the acute phase (1 dpi); however, it was significant also during epileptogenesis (6 dpi) and at the beginning of the chronic phase (14 dpi). Additionally, our analysis of existing microarray data set (Dingledine R. 2013. NCBI Gene Expression Omnibus. GSE47752) revealed identical trends in the expression of PAR bZIP TFs in rat models of acquired epilepsy. Surprisingly, the expression of core clock-genes was not altered at 1, 6, 14 or 28 dpi after KA injection. These data are in accordance with another study performing gene profiling in dorsal hippocampus after 6 hours, 15 days and 6 months post KA injection54. They did not find significant changes in core-clock gene expression while they identified Hlf to be an important transcription factor associated with changes in gene expression at 15 dpi. The functional importance of PAR bZIP TFs in regulating neuronal excitability was proven by a study showing that triple-knockout mice deficient for Hlf, Dbp and Tef develop audiogenic and spontaneous seizures12. The authors pointed at Tef as the gene associated with seizures by regulating expression of pyridoxal kinase, an enzyme involved in the metabolism of neurotransmitters. In another study, the deletion of Hlf in mouse exacerbated seizures and reduced survival in the Snc2aQ54 mouse model of epilepsy55. Likewise, DBP was found downregulated together with CLOCK, CRY1 and PER1 at protein levels in patients with focal cortical dysplasia3. Furthermore, neuroprotective effects were described for E4bp4, which we found here to be upregulated after induction of status epilepticus. Besides modulating circadian rhythms, E4bp4 serves as a survival factor in motoneurons during their development56. By limiting neuronal injury, the upregulation of E4bp4 in experimental epilepsy may be a part of the response to neurotoxic insults.
Interestingly, KA induced changes in the expression of PAR bZIP TFs as well as neuroinflammation were mirrored in the contralateral (non-injected) hippocampus. Hlf remained downregulated there even at the late chronic phase (28dpi). At the acute phase, this effect is most likely attributed to hyperactivity spreading from the injected hippocampus via commissural projections57. However, during the chronic phase the epileptic activity does not spread into the contralateral hippocampus58. Additionally, it has been shown that contralateral hippocampus can provide important regulatory input and modulate ongoing seizures59. Thus, it is not clear whether the downregulation of Hlf in contralateral hippocampus during the chronic phase is caused by ongoing changes in the ipsilateral hippocampus or it is entrained by intrinsic changes in the contralateral hippocampus induced during the status epilepticus.
What induces the changes in PAR bZIP TFs expression? We have shown in our previous research that the expression of PAR bZIP TFs is suppressed by the proinflammatory cytokines TNF-α and IL-1β by interfering with their E-boxes in mouse liver and fibroblasts15. Besides suppressing Hlf, Dbp and Tef, TNF-α was found to enhance the expression of E4bp4 in synovial fibroblasts18. This picture is identical to the one detected here in our KA induced model of epilepsy. Since we observed an elevated expression of TNF-α and IL-1β in the KA model, we thought that the cytokine induced neuroinflammatory effect might be responsible for the modulation of PAR bZIP TFs. However, we were not able to suppress the expression of PAR bZIP TFs in hippocampus by simulating the proinflammatory environment. This suggests that their expression in the hippocampus is regulated by mechanisms than differ from those in periphery. Distinct circadian oscillations of PAR bZIP TFs in different tissues supports this hypothesis. In liver and kidney, PAR bZIP TFs oscillate diurnally with a high amplitude60,61 while in brain tissue they oscillate with a low amplitude or not at all11,12.
Early changes in clock core-genes expression might be driving the initial downregulation of PAR bZIP TFs. At an early time-point (3 h) after KA injection, when the inflammatory cytokines are not yet elevated, the expression of Clock, Npas2 and Bmal1 were significantly downregulated while Per genes 1 and 2 were upregulated. The expression of Dbp and presumably also Hlf and Tef is directly activated by CLOCK-BMAL162,63 and inhibited by PER and CRY64, thus their deficient expression results in altered expression of PAR bZIP TFs. In addition, it has been shown that Hlf and Tef expression in neurons is reduced in mouse with neuronal Clock deletion3. However, it is unclear whether these early changes in core clock gene expression contribute to alterations in PAR bZIP TFs at later stages of KA model, since their expression is already normalized at this stage. Alternatively, the neuroinflammation, excitotoxicity or seizure activity itself might play a role in sustaining the altered levels of PAR bZIP TFs.
To further investigate the role of PAR bZIP TFs in the regulation of neuronal excitability, we induced Hlf over-expression in primary neurons under hyperexcitable conditions induced by glutamate and bicuculline exposure. It resulted in decrease in the frequency and an increase in the amplitude of sEPSCs. In another study, Hlf deletion has been shown to increase the frequency of spontaneous seizures in two mouse models involving mutations on voltage-gated sodium channels; the gain-of-function Scn2aQ54 (Q54) mouse model and the heterozygous loss-of-function Scn1a mouse model55. Interestingly, these models seem to induce seizures from different mechanisms; a reduction in GABAergic inhibition in the Scn1a model and an increase in excitability in CA1 glutamatergic neurons in the Q54 mouse model65. This posits the possibility that Hlf is involved in regulation of both pathways, which may explain the lower excitability we observed when exposing hippocampal primary neurons to hyperexcitable conditions. This decrease in excitability does not appear to involve changes in intrinsic properties of neurons nor in dendritic filtering, as we did not observe changes in the number or amplitude of action potentials, nor in the decay time of spontaneous currents. This result suggests that Hlf, as a member of PAR bZIP TFs, is an intrinsic regulatory element modulating synaptic activity by a yet unknown mechanism.
Taken together, Hlf seems to be a promising transcription factor associated with epileptogenesis. We found consistent deficiency of Hlf in animal models of acquired epilepsy, demonstrated that Hlf regulates neuronal activity and its overexpression in neurons leads to altered expression of genes associated with epilepsy. Upon resolving the molecular mechanism and causality between PAR bZIP TFs alterations and seizures, these findings should initiate further studies into PAR bZIP TFs as a target to prevent epileptogenesis or to modulate seizure activity.
Supplementary information
Acknowledgements
The investigators are grateful for the continuous support of the study by Peter Fuchs, M.D.
Author contributions
L.R., T.G., A.F., C.L. and J.F. designed the study. L.R., T.G. and C.L performed the experiments. J.P. created the AAV vectors. L.D. performed bioinformatics analyses. L.R., T.G. and C.L. wrote the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60638-7.
References
- 1.Wiebe S. Epidemiology of Temporal Lobe Epilepsy. Can. J. Neurol. Sci. 2000;27:S6–S10. doi: 10.1017/S0317167100000561. [DOI] [PubMed] [Google Scholar]
- 2.Alexander A, Maroso M, Soltesz I. Organization and control of epileptic circuits in temporal lobe epilepsy. in. Progress in brain research. 2016;226:127–154. doi: 10.1016/bs.pbr.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li P, et al. Loss of CLOCK Results in Dysfunction of Brain Circuits Underlying Focal Epilepsy. Neuron. 2017;96:387–401.e6. doi: 10.1016/j.neuron.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karoly PJ, et al. Circadian and circaseptan rhythms in human epilepsy: a retrospective cohort study. Lancet Neurol. 2018;17:977–985. doi: 10.1016/S1474-4422(18)30274-6. [DOI] [PubMed] [Google Scholar]
- 5.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/S0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 6.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 7.Mueller CR, Maire P, Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990;61:279–291. doi: 10.1016/0092-8674(90)90808-R. [DOI] [PubMed] [Google Scholar]
- 8.Drolet DW, et al. TEF, a transcription factor expressed specifically in the anterior pituitary during embryogenesis, defines a new class of leucine zipper proteins. Genes Dev. 1991;5:1739–1753. doi: 10.1101/gad.5.10.1739. [DOI] [PubMed] [Google Scholar]
- 9.Inaba T, et al. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science (80-.). 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- 10.Cowell IG, Skinner A, Hurst HC. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol. Cell. Biol. 1992;12:3070–3077. doi: 10.1128/MCB.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gachon F, et al. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerstner JR, et al. BMAL1 controls the diurnal rhythm and set point for electrical seizure threshold in mice. Front. Syst. Neurosci. 2014;8:121. doi: 10.3389/fnsys.2014.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, et al. Cytokines and epilepsy. Seizure. 2011;20:249–256. doi: 10.1016/j.seizure.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Cavadini G, et al. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc. Natl. Acad. Sci. USA. 2007;104:12843–8. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taraborrelli C, et al. TNFR1 is essential for CD40, but not for lipopolysaccharide-induced sickness behavior and clock gene dysregulation. Brain. Behav. Immun. 2011;25:434–442. doi: 10.1016/j.bbi.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Hashiramoto A, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J. Immunol. 2010;184:1560–5. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida K, et al. TNF-α modulates expression of the circadian clock gene Per2 in rheumatoid synovial cells. Scand. J. Rheumatol. 2013;42:276–280. doi: 10.3109/03009742.2013.765031. [DOI] [PubMed] [Google Scholar]
- 19.Bouilleret V, et al. Recurrent seizures and hippocampal sclerosis following intrahippocampal kainate injection in adult mice: Electroencephalography, histopathology and synaptic reorganization similar to mesial temporal lobe epilepsy. Neuroscience. 1999;89:717–729. doi: 10.1016/S0306-4522(98)00401-1. [DOI] [PubMed] [Google Scholar]
- 20.Zattoni M, et al. Brain infiltration of leukocytes contributes to the pathophysiology of temporal lobe epilepsy. J. Neurosci. 2011;31:4037–4050. doi: 10.1523/JNEUROSCI.6210-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaus F, et al. Differential effects of peripheral and brain tumor necrosis factor on inflammation, sickness, emotional behavior and memory in mice. Brain. Behav. Immun. 2016;58:310–326. doi: 10.1016/j.bbi.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Rambousek L, et al. Rat intra-hippocampal NMDA infusion induces cell-specific damage and changes in expression of NMDA and GABAA receptor subunits. Neuropharmacology. 2016;105:594–606. doi: 10.1016/j.neuropharm.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 23.Gschwind T, et al. Contribution of early Alzheimer’s disease-related pathophysiology to the development of acquired epilepsy. Eur. J. Neurosci. 2018;47:1534–1562. doi: 10.1111/ejn.13983. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0 URL, http://www.R-project.org/
- 26.Dingledine R, et al. Data Descriptor: Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models. Sci. Data. 2017;4:170061. doi: 10.1038/sdata.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval R, et al. Homeostatic NMDA receptor down-regulation via brain derived neurotrophic factor and nitric oxide-dependent signalling in cortical but not in hippocampal neurons. J. Neurochem. 2011;118:760–772. doi: 10.1111/j.1471-4159.2011.07365.x. [DOI] [PubMed] [Google Scholar]
- 29.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Wang S, Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 32.Li, B. & Dewey, C. N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome in Bioinformatics: The Impact of Accurate Quantification on Proteomic and Genetic Analysis and Research (ed. Liu, Y.) 41–74 (Apple Academic Press, 2014). doi:10.1201/b16589 [DOI] [PMC free article] [PubMed]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riban V, et al. Evolution of hippocampal epileptic activity during the development of hippocampal sclerosis in a mouse model of temporal lobe epilepsy. Neuroscience. 2002;112:101–111. doi: 10.1016/S0306-4522(02)00064-7. [DOI] [PubMed] [Google Scholar]
- 36.Bouilleret V, Loup F, Kiener T, Marescaux C, Fritschy JM. Early loss of interneurons and delayed subunit-specific changes in GABA(A)-receptor expression in a mouse model of mesial temporal lobe epilepsy. Hippocampus. 2000;10:305–324. doi: 10.1002/1098-1063(2000)10:3<305::AID-HIPO11>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 37.Vezzani A, et al. Powerful anticonvulsant action of IL-1 receptor antagonist on intracerebral injection and astrocytic overexpression in mice. Proc. Natl. Acad. Sci. USA. 2000;97:11534–9. doi: 10.1073/pnas.190206797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crespel A, et al. Inflammatory reactions in human medial temporal lobe epilepsy with hippocampal sclerosis. Brain Res. 2002;952:159–169. doi: 10.1016/S0006-8993(02)03050-0. [DOI] [PubMed] [Google Scholar]
- 39.Choi J, et al. Cellular injury and neuroinflammation in children with chronic intractable epilepsy. J. Neuroinflammation. 2009;6:38. doi: 10.1186/1742-2094-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varella PPV, et al. Relationship between fluid-attenuated inversion-recovery (FLAIR) signal intensity and inflammatory mediator’s levels in the hippocampus of patients with temporal lobe epilepsy and mesial temporal sclerosis. Arq. Neuropsiquiatr. 2011;69:91–99. doi: 10.1590/S0004-282X2011000100018. [DOI] [PubMed] [Google Scholar]
- 41.Ravizza T, et al. Innate and adaptive immunity during epileptogenesis and spontaneous seizures: Evidence from experimental models and human temporal lobe epilepsy. Neurobiol. Dis. 2008;29:142–160. doi: 10.1016/j.nbd.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Maroso M, et al. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat. Med. 2010;16:413–419. doi: 10.1038/nm.2127. [DOI] [PubMed] [Google Scholar]
- 43.Hitzler JK, et al. Expression patterns of the hepatic leukemia factor gene in the nervous system of developing and adult mice. Brain Res. 1999;820:1–11. doi: 10.1016/S0006-8993(98)00999-8. [DOI] [PubMed] [Google Scholar]
- 44.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 45.Häussler U, Rinas K, Kilias A, Egert U, Haas CA. Mossy fiber sprouting and pyramidal cell dispersion in the hippocampal CA2 region in a mouse model of temporal lobe epilepsy. Hippocampus. 2016;26:577–588. doi: 10.1002/hipo.22543. [DOI] [PubMed] [Google Scholar]
- 46.Nanobashvili A, et al. Development and persistence of kindling epilepsy are impaired in mice lacking glial cell line-derived neurotrophic factor family receptor α2. Proc. Natl. Acad. Sci. USA. 2000;97:12312–12317. doi: 10.1073/pnas.97.22.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chia WJ, Dawe GS, Ong WY. Expression and localization of the iron-siderophore binding protein lipocalin 2 in the normal rat brain and after kainate-induced excitotoxicity. Neurochem. Int. 2011;59:591–599. doi: 10.1016/j.neuint.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 48.Hildebrand MS, et al. Loss of synaptic Zn2+ transporter function increases risk of febrile seizures. Sci. Rep. 2015;5:17816. doi: 10.1038/srep17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winden KD, et al. A systems level, functional genomics analysis of chronic epilepsy. PLoS One. 2011;6:6. doi: 10.1371/journal.pone.0020763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinjo ÉR, Arida RM, de Oliveira DM, da Silva Fernandes MJ. The Na+/K+ATPase activity is increased in the hippocampus after multiple status epilepticus induced by pilocarpine in developing rats. Brain Res. 2007;1138:203–207. doi: 10.1016/j.brainres.2006.12.068. [DOI] [PubMed] [Google Scholar]
- 51.Chen H, Xu G, Du H, Yi M, Li C. Integrative analysis of gene expression associated with epilepsy in human epilepsy and animal models. Mol. Med. Rep. 2016;13:4920–4926. doi: 10.3892/mmr.2016.5122. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Q. et al. A novel SLC6A8 mutation associated with intellectual disabilities in a Chinese family exhibiting creatine transporter deficiency: Case report 06 Biological Sciences 0604 Genetics. BMC Med. Genet. 19, (2018). [DOI] [PMC free article] [PubMed]
- 53.Khan S, et al. Circadian rhythm and epilepsy. The Lancet Neurology. 2018;17:1098–1108. doi: 10.1016/S1474-4422(18)30335-1. [DOI] [PubMed] [Google Scholar]
- 54.Motti D, et al. Gene expression analysis of the emergence of epileptiform activity after focal injection of kainic acid into mouse hippocampus. Eur. J. Neurosci. 2010;32:1364–1379. doi: 10.1111/j.1460-9568.2010.07403.x. [DOI] [PubMed] [Google Scholar]
- 55.Hawkins NA, Kearney JA. Hlf is a genetic modifier of epilepsy caused by voltage-gated sodium channel mutations. Epilepsy Res. 2016;119:20–3. doi: 10.1016/j.eplepsyres.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Junghans D, et al. The CES-2-related transcription factor E4BP4 is an intrinsic regulator of motoneuron growth and survival. Development. 2004;131:4425–4434. doi: 10.1242/dev.01313. [DOI] [PubMed] [Google Scholar]
- 57.Yasuda H, Fujii M, Fujisawa H, Ito H, Suzuki M. Changes in Nitric Oxide Synthesis and Epileptic Activity in the Contralateral Hippocampus of Rats Following Intrahippocampal Kainate Injection. Epilepsia. 2008;42:13–20. doi: 10.1046/j.1528-1157.2001.083032.x. [DOI] [PubMed] [Google Scholar]
- 58.Arabadzisz D, Antal K, Parpan F, Emri Z, Fritschy J-M. Epileptogenesis and chronic seizures in a mouse model of temporal lobe epilepsy are associated with distinct EEG patterns and selective neurochemical alterations in the contralateral hippocampus. Exp. Neurol. 2005;194:76–90. doi: 10.1016/j.expneurol.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 59.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nat. Commun. 2013;4:1376. doi: 10.1038/ncomms2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–62. doi: 10.1002/j.1460-2075.1996.tb00365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wuarin J, Schibler U. Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell. 1990;63:1257–66. doi: 10.1016/0092-8674(90)90421-A. [DOI] [PubMed] [Google Scholar]
- 62.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 63.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 64.Yamaguchi S, et al. Role of DBP in the Circadian Oscillatory Mechanism. Mol. Cell. Biol. 2000;20:4773–4781. doi: 10.1128/MCB.20.13.4773-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kearney JA, et al. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102:307–317. doi: 10.1016/S0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.