Abstract
Background and Objectives:
Notwithstanding the increased prevalence of Acinetobacter baumannii drug-resistant isolates, treatment options are progressively limiting. This study aims to provide a recent report on antibiotic susceptibility in burn wound isolates of A. baumannii, and the importance of OXA beta-lactamases in carbapenem resistance.
Materials and Methods:
The susceptibility levels to different antimicrobial categories were determined among 84 A. baumannii isolates from burn wound infection between 2016 and 2018. Multiplex PCR was used to detect OXA beta-lactamases genes, including blaOXA-51, blaOXA-23, blaOXA-24 and blaOXA-58. ISAba-1 association with blaOXA-51, blaOXA-23 and blaOXA-58 was detected by PCR mapping.
Results:
All the isolates were determined as multidrug-resistant (MDR) and 69% as extensively drug-resistant (XDR). Different carbapenems MIC ranges (MIC50 and MIC90) were observed among the isolates harboring blaOXA-like genes and isolates with the OXA-24-like enzyme showed higher carbapenems MIC ranges. The prevalence of blaOXA-51-like, blaOXA-23-like, blaOXA-24-like and blaOXA-58-like were 100%, 53.57%, 41.66% and 30.95%, respectively. ISAba-1 insertion sequence was found to be upstream to blaOXA-23-like and blaOXA-58-like genes in 23 out of 45 (71.1%) blaOXA-23-like-positive and 4 out of 23 (15.3) blaOXA-58-like-positive isolates, respectively.
Conclusion:
Resistance to carbapenems as the last resort for treatment of A. baumannii infections is growing. This study, for the first time in Iran, has observed the increased frequency of blaOXA-24-like and blaOXA-58-like genes and found an association between ISAba-1 and blaOXA-58-like gene, which signifies the possible risk of increased diversity in OXA beta-lactamases and growth in carbapenem resistance.
Keywords: Acinetobacter baumannii, Antibiotic susceptibility, Carbapenem, OXA beta-lactamases
INTRODUCTION
Acinetobacter baumannii, a ubiquitous and opportunistic Gram-negative pathogen, have shown insusceptibility to a wide range of antimicrobial agents, including β-lactamases, which are frequently used in clinical operations (1). Based on the molecular structure, beta-lactamases are subcategorized into four major classes, including A, B, C and D. Classes A to C are both chromosomally encoded and plasmid-encoded enzymes (2). Class D beta-lactamases, known as oxacillinases enzymes or OXA beta-lactamases, are relatively scarce and are identifiable only as plasmid-encoded beta-lactamases. These enzymes have a substrate profile limited to penicillin and oxacillin, though some of them confer resistance to cephalosporins. After considering by Bush et al. (3), the substrate profile of OXA beta-lactamases were designated as 2d, which OXA-1 to OXA-11 are representative enzymes of this class. Carbapenem-hydrolyzing class D beta-lactamases (CHDLs) are a subgroup of class D beta-lactamases that hydrolyze carbapenem antibiotics and have multidrug-resistant to A. baumannii.
An increased number of carbapenem-resistant A. baumannii isolates harboring blaOXA-23, blaOXA-24 (also named blaOXA-40), and blaOXA-58 genes have been reported since the 1980s of the last century (3, 4). It is also found that some intrinsic chromosomally encoded OXA-51-like enzymes can mediate resistance to carbapenems when their gene expression is promoted by the environment or mobile genetic elements (5, 6). OXA-23-like enzymes are pinpointed worldwide and are the most widespread OXA-like enzyme in A. baumannii (7). In Iran, blaOXA-23, as the most, and blaOXA-24, as the less, frequent CHDL-encoding genes are far been reported (8, 9, 10), and evidence has revealed that the blaOXA-58 gene distribution in Iran and the neighboring countries is less than other CHDL-encoding genes (9–15).
Expression and transformation of the OXA genes could be facilitated by insertion sequences (ISs) such as ISAba-1, ISAba-4, and ISAba1-25, which encode the transposases upstream of blaOXA genes and provide an effective promoter for the gene (16). ISAba-1, from IS4 family, has been found to be the upstream of blaOXA-51, blaOXA-23 and blaOXA-58 genes in Acinetobacter species (17, 18). It is well documented that resistance to carbapenems mediated by blaOXA-like genes can be regulated by the upstream presence of ISAba-1 sequence (12, 13, 18). Only a very limited studies in Iran have studied the association of the blaOXA-51 and blaOXA-23 genes with ISAba-1 (19, 20), and there is no report on the ISAba-1 up-regulation of blaOXA-58 gene.
The emergence of carbapenem resistance mediated by OXA enzymes in A. baumannii has demoted the clinical efficacy of this antibiotic, and few studies have unveiled the impact of these enzymes. Hence, the present study was aimed to evaluate the recent antibiotic susceptibility pattern of carbapenem in A. baumannii isolates recovered from burn wound infections at a general hospital of Tehran, Iran. This study also verified the presence of blaOXA-23-like, blaOXA-24-like and blaOXA-58-like genes and investigated their association with ISAba-1.
MATERIALS AND METHODS
Bacterial isolates.
The study included a total of 84 non-repetitive strains of A. baumannii isolated from patients with burn wounds in a general hospital of Tehran from 2016 to 2018. The enrolled population was adult patients, of both male and female genders. All patients had serious wound infections that developed to sepsis, uroinfection or pneumonia. The strains were identified by standard microbiological and biochemical techniques (21) and by PCR detection of the intrinsic carbapenemase gene blaOXA-51-like (22).
Ethical issues.
This study was conducted following the approved institutional guidelines of the Islamic Azad Medical University in Tehran (Code: IR.IAU.TMU.REC.1396.279) and volunteer’s data were anonymized before analysis.
Antibiotic susceptibility test.
Minimal inhibitory concentrations (MICs) for 15 antibiotics were determined by the broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) (23) and Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were used as references. Initial concentration of antibiotics (AppliChem, Germany) for MIC determination was (256 μg/ml). Antibiotics were selected based on antimicrobial categories proposed by Magiorakos et al. (24) as follows; gentamicin, amikacin, ceftriaxone, cefepime, ciprofloxacin, levofloxacin, ceftazidime, imipenem, meropenem, ploymyxin B, colistin, ampicillin, tetracycline, tigecycline, aztreoam. In this study MDR (multidrug-resistant) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, XDR (extensively drug-resistant) was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (i.e. bacterial isolates remain susceptible to only one or two categories) and PDR (pandrug-resistant) was defined as non-susceptibility to all agents in all antimicrobial categories (24).
blaOXA-like genes detection.
Genomic DNA was extracted using the DNA isolation kit (MBST, Iran) as recommended by the manufacturer. Detection of the intrinsic carbapenemase encoding gene blaOXA-51-like and three other OXA-carbapenemase genes including blaOXA-23-like, blaOXA-24-like and blaOXA-58-like was carried out by multiplex PCR using primers listed in Table 1 (22, 25).
Table 1.
Sequence of the primers used in this study.
| Target | Primer | Sequence (5′ to 3′) | Amplicon Size (bp) | Annealing Temperature (ºC) | Reference |
|---|---|---|---|---|---|
| blaOXA-51-like | OXA-51-F | TAATGCTTTGATCGGCCTTG | 353 | 58 | 22 |
| OXA-51-R | TGGATTGCACTTCATCTTGG | ||||
| blaOXA-23-like | OXA-23-F | GATGTGTCATAGTATTCGTCG | 1065 | 58 | 25 |
| OXA-23-R | TCACAACAACTAAAAGCACTG | ||||
| blaOXA-24-like | OXA-24-F | GGTTAGTTGGCCCCCTTAAA | 246 | 58 | 22 |
| OXA-24-R | AGTTGAGCGAAAAGGGGATT | ||||
| blaOXA-58-like | OXA-58-F | AAGTATTGGGGCTTGTGCTG | 599 | 58 | 22 |
| OXA-58-R | CCCCTCTGCGCTCTACATAC | ||||
| ISAba1- blaOXA-51-like | ISAba-1-F | CACGAATGCAGAAGTTG | 1200 | 60 | 17, 22 |
| OXA-51-R | TGGATTGCACTTCATCTTGG | ||||
| ISAba1- blaOXA-23-like | ISAba-1-F | CACGAATGCAGAAGTTG | 1600 | 60 | 17, 25 |
| OXA-23-R | TCACAACAACTAAAAGCACTG | ||||
| ISAba1- blaOXA-58-like | ISAba-1-F | CACGAATGCAGAAGTTG | 1259 | 60 | 17, 22 |
| OXA-58-R | CCCCTCTGCGCTCTACATAC |
The PCR reaction (25 μl) contained 3–5 μl of template DNA, 2.5 μl of 10× PCR buffer, 0.75 μl of 50 mM MgCl2, 0.5 μl of 10 mM dNTPs, 0.25 μl of 5 U/μl of Taq DNA polymerase, and 25 pmol of each primers. Primers used and annealing temperatures are given in Table 1. The PCR carried out in a thermocycler (Techne TC512, England) under the following conditions: initial denaturation at 94°C for 5 min followed by denaturation at 94°C for 1 min, annealing at 58°C for 1 min and extension at 72°C for 1 min (30 cycles), and a final extension at 72°C for 7 min. Identified isolates A. baumannii (harboring blaOXA-like genes), previously reported by Feizabadi et al. (8), were used as positive controls for studied genes. The PCR products were analyzed by electrophoresis on 1.5% agarose gel (Sigma- Aldrich, Germany) containing ethidium bromide (0.5 μg/ml).
ISAba-1 insertion gene detection.
The genetic association between ISAba1 sequence and blaOXA-51-like, blaOXA-23-like and blaOXA-58-like genes was investigated by PCR mapping using ISAba-1 forward (10) and bla genes reverse primers (22, 25) (Table 1). The PCR carried out as the blaOXA-like multiplex PCR, except that 20 pmol of each primer were used in PCR mixture, and an annealing temperature of 60°C for 45s and extension at 72°C for 3 min was used for 35 cycles of reaction. PCR products were sequenced in the absence of positive controls.
RESULTS
Antibiotic susceptibility.
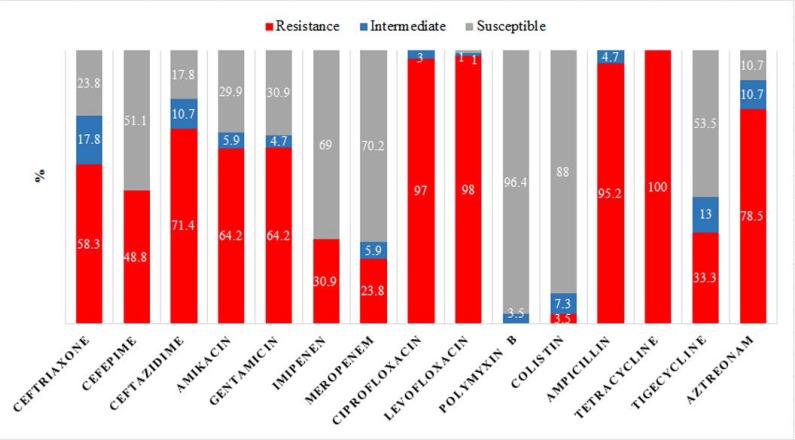
High percentage of A. baumannii isolates were resistant to tetracycline, ciprofloxacin, and levofloxacin, while polymyxin B and colistin were the most effective antibiotics. The results also showed a noticeable resistance rate to cephalosporins and tigecycline. The total antibiotic resistance results are presented in Fig. 1. According to the epidemiological definition, all 84 isolates (100%) were grouped as MDR, whereas 58 isolates (69%) were considered as XDR and none of the isolates were grouped as PDR. Different carbapenems MIC ranges (MIC50 and MIC90) were observed in the isolates harboring blaOXA-like genes. Among them, isolates with the OXA-24-like enzyme showed higher carbapenems MIC ranges (Table 2).
Fig. 1.
Susceptibility profile in 84 burn wound A. baumannii isolates
Table 2.
MIC values (μg/ml) for imipenem and meropenem in studied A. baumannii isolates harboring blaOXA-like genes.
| Gene | Number | MIC50 | MIC90 | MIC range | |||
|---|---|---|---|---|---|---|---|
| IM | MEM | IM | MEM | IM | MEM | ||
| blaOXA-23 | 45 | 0.5 | 0.5 | 16 | 8 | 0.062–32 | 0.032–32 |
| ISAba-1 –upstream blaOXA-23 | 32 out of 45 | 4 | 2 | 16 | 8 | 0.062–32 | 0.032–32 |
| blaOXA-24 | 35 | 1 | 1 | 32 | 16 | 0.032–32 | 0.032–16 |
| blaOXA-58 | 26 | 1 | 0.5 | 16 | 16 | 0.062–32 | 0.032–32 |
| blaOXA-23 only | 23 | 0.5 | 0.25 | 2 | 4 | 0.062–16 | 0.032–8 |
| blaOXA-24 only | 23 | 0.5 | 0.5 | 32 | 16 | 0.032–16 | 0.032–16 |
Notes: Breakpoints of IM and MEM, MIC ≤2 μg/mL: sensitive; MIC =4 μg/mL: intermediate; MIC ≥8 μg/mL: resistant (CLSI, 2017).
IM: Imipenem, MEM: Meropenem
Distribution of blaOXA-like genes.
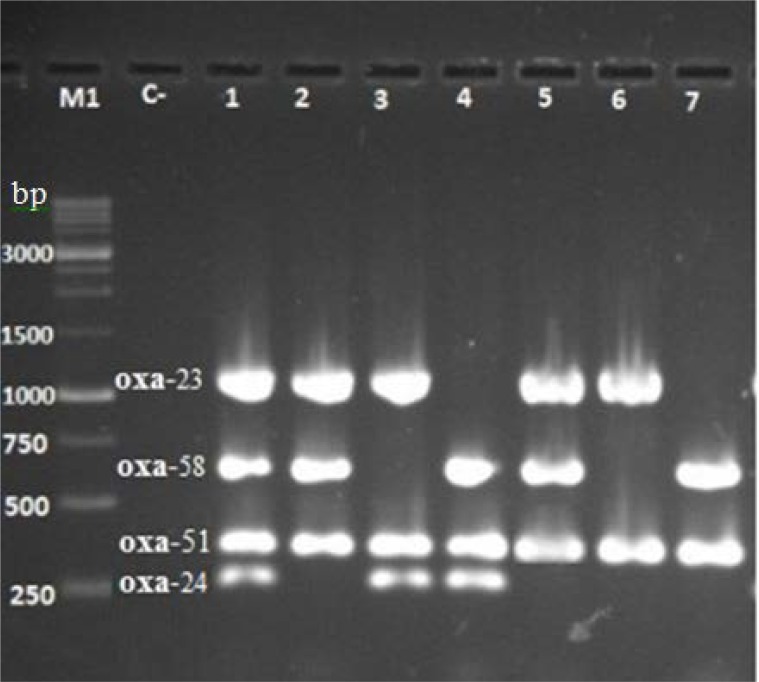
Multiple PCR reactions by blaOXA-like genes specific primers produced DNA fragments of 353, 1065, 246 and 599 bp, from blaOXA-51-like, blaOXA-23-like, blaOXA-24-like and blaOXA-58-like genes respectively (Fig. 2). The prevalence of blaOXA-51-like, blaOXA-23-like, blaOXA-24-like and blaOXA-58-like were 100%, 53.57%, 41.67% and 30.95%, respectively. The co-existence of blaOXA-51, blaOXA-23 and blaOXA-24 was observed in 3.57% (3/84) of isolates.
Fig. 2.
Detection of blaOXA-like genes from A. baumannii isolates by multiplex PCR amplification. Lanes 1 to 7: isolates harboring blaOXA-like genes. Lane C-: no chromosomal DNA (negative control). Lane M1: 1-kb DNA ladder (SINACLON, Iran).
Co-existence of blaOXA-51, blaOXA-23, blaOXA-58 and blaOXA-51, blaOXA-24, blaOXA-58 was seen in 20.23% (17/84) and 10.7% (9/84) of isolates respectively. There was an isolate (1.19%) that carried all four genes. Six isolates from 84 (7.14%) carried only the blaOXA-51 gene. The distribution of blaOXA-types among carbapenem-resistant A. baumannii isolates is shown in Table 3.
Table 3.
blaOXA-like genes and ISAba-1 insertion sequence distribution among imipenem and/or meropenem resistant A. baumannii isolates (n=28).
| Isolates* | blaOXA-like gene | ||||
|---|---|---|---|---|---|
| 51 | 23 | 24 | 58 | ISAba-1 upstream | |
| 1 | + | − | + | + | |
| 3 | + | − | + | − | |
| 4 | + | + | − | + | blaOXA-23 |
| 5 | + | + | − | − | blaOXA-23 |
| 6 | + | − | + | − | |
| 10 | + | + | − | − | blaOXA-23 |
| 14 | + | + | − | + | blaOXA-23 |
| 21 | + | − | + | − | |
| 25 | + | + | − | − | blaOXA-23 |
| 32 | + | − | + | − | |
| 33 | + | − | − | − | blaOXA-51 |
| 50 | + | + | − | + | blaOXA-23 |
| 53 | + | + | − | − | blaOXA-23 |
| 59 | + | + | − | − | blaOXA-23 |
| 65 | + | − | + | + | |
| 73 | + | − | + | − | |
| 74 | + | − | + | − | |
| 75 | + | + | − | + | blaOXA-23 |
| 76 | + | + | − | − | blaOXA-23 |
| 78 | + | + | − | + | blaOXA-23 |
| 79 | + | − | − | − | |
| 85 | + | − | + | − | |
| 90 | + | − | + | − | |
| 91 | + | − | + | − | |
| 92 | + | + | + | − | blaOXA-23 |
| 93 | + | + | − | − | blaOXA-23 |
| 97 | + | − | + | − | |
| 100 | + | − | − | + | |
Isolate numbering is not under the total number of studied isolates
ISAba-1 element association.
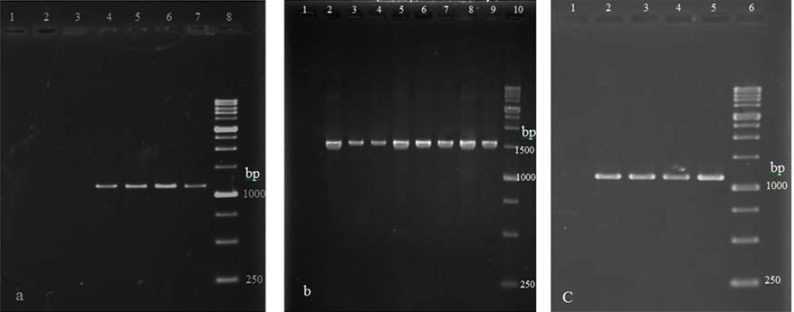
Among carbapenem-resistant isolates with only blaOXA-51-like, 4 isolates yielded a band of 1200 bp in a PCR reaction using ISAba-1 forward primer and the blaOXA-51-like reverse primer (Fig. 3). Isolates with positive PCR products for blaOXA-23-like and blaOXA-58-like genes showed a band of 1600 and 1259 bp respectively in PCR reactions using forward primer for ISAba-1 and the reverse primers for blaOXA-51-like and blaOXA-58-like genes (Fig. 3). The ISAba-1 element was found upstream blaOXA-23-like and blaOXA-58-like genes in 32 out of 45 (71.11%) and 4 out of 26 (15.38%) A. baumannii isolates respectively. ISAba-1 insertion sequence presence among 28 carbapenem-resistant is shown in Table 3.
Fig. 3.
PCR products obtained using the ISAba-1 forward primer (ISAba-1F) and blaOXA-like gene reverse primers. (a) Carbapenem-resistant isolates carrying only blaOXA-51-like after amplification with the ISAba-1F and OXA-51-R primer pair. Lane 1: no chromosomal DNA (negative control). Lanes 2 and 3: DNA of isolates failed to give a band. Lanes 4–7: DNA of isolates with ISAba-1 upstream of blaOXA-51-like gene with the 1200-bp PCR product. Lane 8: 1-kb DNA ladder. (b) Lane 1: no chromosomal DNA (negative control). Lanes 2–9: DNA of isolates with ISAba-1 upstream blaOXA-23-like gene with the 1600-bp PCR product. Lane 10: 1-kb DNA ladder. (c) Lane 1: no chromosomal DNA (negative control). Lanes 2–5: DNA of isolates with ISAba-1 upstream of blaOXA-58-like gene with the 1259-bp PCR product. Lane 6: 1-kb DNA ladder.
DISCUSSION
The present study showed a remarkable resistance of A. baumannii isolates to the tested antibiotics. The significance of these results becomes greater when using third and fourth-generation antibiotics, such as cephalosporins and carbapenems, for the treatment of A. baumannii infections. Almost all the isolates studied here indicated susceptibility to polymyxin B and colistin and similar result to that obtained by several numbers of previous studies conducted in Iran (26, 9, 27, 28). Our study found the susceptibility of 56 out of 58 XDR A. baumannii isolates to polymyxin B, meaning that polymyxin B has ability to re-emerge in medical practice for the treatment of infections caused by MDR and XDR A. baumannii strains. In a clinical study, it has been suggested that polymyxin B alone or in combination with other antibiotics can decrease the overall mortality and also can remove bacteria from patients. However, further investigations on the pharmacokinetics, pharmacodynamics, and toxicodynamics of polymyxin B are needed to find the appropriate doses of the drug (29). Aztreonam, a superior antibiotic to ceftazidime and more stable than carbapenemases (30), was tested in this study and indicated a high prevalence of resistance. The result is comparable with those of other studies (9, 26) and suggest to eliminate aztreonam from the list of therapeutic solutions for A. baumannii infections control.
The carbapenem-resistant isolates in the present work could be attributed to the frequent use of carbapenems, especially after explosive dissemination of ESBLs and CTX-M pandemic. Besides, this behavior may arise from multiple mechanisms of carbapenem resistance in A. baumannii and dissemination of carbapenemases by the acquisition of plasmid/chromosome-mediated resistance genes (4). It is noteworthy to say that, all imipenem- and meropenem-resistant A. baumannii strains of this study were identified as MDR and XDR.
To assay OXA beta-lactamases as a carbapenem resistance mechanism, the isolates were screened for the most prevalent blaOXA genes, including blaOXA-23-like, blaOXA-24-like and blaOXA-58-like (4). The products of these genes are consistently associated with resistance or at least reduced susceptibility to carbapenems (4). The isolates were also screened for blaOXA-51-like, a prevalent and an intrinsic gene in A. baumannii species with the chromosomal origin that has a relatively weak ability to hydrolyze carbapenems (4, 5). blaOXA-like genes are candidates for ISAba-1 acquisition, which is commonly associated with the expression of CHDL-encoding genes in A. baumannii and can contribute to carbapenemase genes spread among Acinetobacter species (4, 16). To survey this, the association of ISAba-1 with blaOXA-51-like (in blaOXA-23-like-, blaOXA-24-like- and blaOXA-58-like-negative isolates), blaOXA-23-like and blaOXA-58-like genes were also investigated in our isolates.
The blaOXA-23-like gene distribution frequency among all the studied A. baumannii isolates was the most, which coordinates with other studies in different regions of Iran and some neighboring countries (9–15, 19, 20, 28, 31). There were increased MIC values for imipenem and meropenem in A. baumannii isolates harboring blaOXA-23 gene, especially in ones with upstream ISAba-1 element (Table 2). Meanwhile, from 13 carbapenem-resistant A. baumannii harboring blaOXA-23-like all but one showed upstream ISAba-1 element, (Table 3). These observations emphasize the blaOXA-23-like gene role and its up-regulation by ISA-ba-1 element as a major mechanism for carbapenem resistance phenotype.
OXA-24-like carbapenemase is widely disseminated, but its prevalence is less than OXA-23-like (16). OXA-24-like frequency among A. baumannii isolates has formerly been reported in Iran and some neighboring countries. Having looked at previous studies, we found that over time, blaOXA-24-like gene distribution has increased (9–15, 19, 20, 28, 31). In the present study, the spread of the blaOXA-24-like gene (41.66%) was noticeably higher than the rates reported previously (9–15, 19, 20, 28, 31). A possible explanation for such elevation could be the cephalosporin antibiotics overuse, especially because almost all the A. baumannii isolates harboring blaOXA-24-like genes were resistant to ceftriaxone, cefepime, and ciprofloxacin. According to the results summarized in Table 2, A. baumannii isolates harboring the blaOXA-24-like gene showed higher MIC values than those with blaOXA-23-like. This evidence reflects the higher contribution of the blaOXA-24-like gene in carbapenem resistance than blaOXA-23-like, which is in accordance with a recent study (32).
blaOXA-58-like gene was identified in 30.95% of the studied isolates, which was remarkably higher than other reports in Iran (9–11, 20, 28) and neighboring countries (12–15, 31). Association of ISAba-1 gene with the blaOXA-58-like in A. baumannii isolates of the present study, which was observed for the first time in Iran, is a justification for its growth in dissemination and suggests the possibility of wider dissemination of blaOXA-58-like gene.
In this study, from 28 carbapenem-resistant A. baumannii isolates, 27 harbored blaOXA-23-like and/or blaOXA-24-like and/or blaOXA-58-like genes which signifies the responsibility of these genes in carbapenem resistance. However, examining the higher number of carbapenem-resistant A. baumannii isolates could assist to come to a stronger conclusion regarding the importance of OXA β-lactamases in carbapenem resistance.
In conclusion, the high frequency of MDR and XDR A. baumannii strains, in the present study, represents the wide dissemination of antibiotic-resistant A. baumannii strains in the healthcare centers of Iran and only polymyxin B, colistin, imipenem, and meropenem can be considered as effective drugs for the treatment of A. baumannii infection. This problem can be managed by Iran’s annual reports on drug resistance to the World Health Organization (WHO) and Central Asian and Eastern European Surveillance of Antimicrobial Resistance (CAESAR). Our results provided evidence for higher prevalence of blaOXA-24-like and blaOXA-58-like genes than past and the association of the blaOXA-58-like gene with the ISAba-1 insertion sequence can speculate a shift in blaOXA-like genes distribution in Iran. More studies are required to strengthen the proposed hypothesis. As carbapenems are not as toxic as colistin, it is essential to preserve their efficacy for clinical success against A. baumannii. This goal may achieve by accurate monitoring of resistance mechanisms to carbapenems.
REFERENCES
- 1.Wieland K, Chhatwal P, Vonberg RP. Nosocomial outbreaks caused by Acinetobacter baumannii and Pseudomonas aeruginosa: Results of a systematic review. Am J Infect Control 2018; 46:643–648. [DOI] [PubMed] [Google Scholar]
- 2.Gutkind GO, Di Conza J, Power P, Radice M. β-Lactamase-mediated resistance: a biochemical, epidemiological and genetic overview. Curr Pharm Des 2013; 19:164–208. [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2010; 54: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 2006; 12: 826–836. [DOI] [PubMed] [Google Scholar]
- 5.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol 2006; 44: 2974–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figueiredo S, Poirel L, Papa A, Koulourida V, Nordmann P. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob Agents Chemother 2009; 53: 4045–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mugnier PD, Poirel L, Naas T, Nordmann P. World wide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis 2010; 16: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, et al. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis 2008; 61: 274–278. [PubMed] [Google Scholar]
- 9.Kooti S, Motamedifar M, Sarvari J. Antibiotic resistance profile and distribution of oxacillinase genes among clinical isolates of Acinetobacter baumannii in Shiraz teaching hospitals, 2012–2013. Jundishapur J Microbiol 2015; 8(8):e20215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rezaei A, Fazeli H, Moghadampour M, Halaji M, Faghri J. Determination of antibiotic resistance pattern and prevalence of OXA-type carbapenemases among Acinetobacter baumannii clinical isolates from inpatients in Isfahan, central Iran. Infez Med 2018; 26: 61–66. [PubMed] [Google Scholar]
- 11.Taherikalani M, Fatolahzadeh B, Emaneini M, Soroush S, Feizabadi MM. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran hospitals. New Microbiol 2009; 32: 265–271. [PubMed] [Google Scholar]
- 12.Ganjo Aryann R, Maghdid Delshad M, Mansoor Isam Y, Kok Dik J, Severin Juliette A, Verbrugh Henri A, et al. OXA-carbapenemases present in cClinical Acinetobacter baumannii-calcoaceticus complex isolates from patients in Kurdistan region, Iraq. Microb Drug Resist 2016; 22: 627–637. [DOI] [PubMed] [Google Scholar]
- 13.Hasan B, Perveen K, Olsen B, Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol 2014; 63: 50–55. [DOI] [PubMed] [Google Scholar]
- 14.Zanganeh Z, Eftekhar F. Correlation of oxacillinase gene carriage with the genetic fingerprints of imipenem-resistant clinical isolates of Acinetobacter baumannii. Jundishapur J Microbiol 2015; 8(9): e26545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed SS, Alp E, Ulu-Kilic A, Dinc G, Aktas Z, Ada B, et al. Spread of carbapenem-resistant international clones of Acinetobacter baumannii in Turkey and Azerbaijan: a collaborative study. Eur J Clin Microbiol Infect Dis 2016; 35: 1463–1468. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother 2010; 54: 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segal H, Garny S, Elisha BG. Is ISAba-1 customized for Acinetobacter? FEMS Microbiol Lett 2005;243: 425–429. [DOI] [PubMed] [Google Scholar]
- 18.Turton JF1, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 2006; 258: 72–77. [DOI] [PubMed] [Google Scholar]
- 19.Peymani A, Higgins PG, Nahaei MR, Farajnia S, Seifert H. Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, northwest Iran. Int J Antimicrob Agents 2012; 39: 526–528. [DOI] [PubMed] [Google Scholar]
- 20.Farshadzadeh Z, Hashemi FB, Rahimi S, Pourakbari B, Esmaeili D, Haghighi MA, et al. Wide distribution of carbapenem resistant Acinetobacter baumannii in burns patients in Iran. Front Microbiol 2015; 6: 1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forbes B.A., Sahm D.F., and Weissfeld A. Bailey and Scott’s Diagnostic Microbiology. 12th Edition, Mosby Elsevier, St Louis, Missouri; 2007. 334–339. [Google Scholar]
- 22.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006; 27: 351–353. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards. Institute Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement 2017; M100-S27.CLSI.
- 24.Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–281. [DOI] [PubMed] [Google Scholar]
- 25.Afzal-Shah M, Woodford N, Livermore DM. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 2001; 45: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karmostaji A, Peerayeh SN, Salmanian AH. Distribution of OXA-type class D β-lactamase genes among nosocomial multi drug resistant Acinetobacter baumannii isolated in Tehran hospitals. Jundishapur J Microbiol 2013; 6(5): e8219. [Google Scholar]
- 27.Shoja S, Moosavian M, Peymani A, Tabatabaiefar MA, Rostami S, Ebrahimi N. Genotyping of carbapenem resistant Acinetobacter baumannii isolated from tracheal tube discharge of hospitalized patients in intensive care units, Ahvaz, Iran. Iran J Microbiol 2013; 5: 315–322. [PMC free article] [PubMed] [Google Scholar]
- 28.Bagheri Josheghani S, Moniri R, Firoozeh F, Sehat M, Dasteh Goli Y. Susceptibility pattern and distribution of oxacillinases and blaPER-1 genes among multidrug resistant Acinetobacter baumannii in a teaching hospital in Iran. J Pathog 2015; 2015: 957259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zavascki AP, Goldani LZ, Li J, Nation RL. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 2007; 60: 1206–1215. [DOI] [PubMed] [Google Scholar]
- 30.Livermore DM, Mushtaq S, Warner M, Zhang J, Maharjan S, Doumith M, et al. Activities of NXL104 combinations with ceftazidime and aztreonam against carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 2011; 55: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the gulf cooperation Council States: Dominance of OXA-23-type producers. J Clin Microbiol 2015; 53: 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang TH, Leu YS, Wang NY, Liu CP, Yan TR. Prevalence of different carbapenemase genes among carbapenem-resistant Acinetobacter baumannii blood isolates in Taiwan. Antimicrob Resist Infect Control 2018; 7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]