Immune checkpoint inhibitors (ICIs) that target programmed cell death protein 1 (PD-1) and its ligand (PD-L1), or cytotoxic T lymphocyte antigen-4 (CTLA-4) elicit durable antitumor responses in multiple cancer types. Yet, only a minority of patients could derive clinical benefit (1). Understanding genomic correlates of response to ICIs could lay a foundation for the development of novel biomarkers and treatment to further enhance the therapeutic benefit (2,3). Previous studies demonstrated that genetic alterations of KEAP1 would dysregulate oxidative stress pathway, resulting in oncogenesis and drug- and radio-resistance in different cancers (4,5). Recently, some exploratory analyses with limited samples showed the association between KEAP1 mutations and clinical benefit to ICI. However, a comprehensive analysis of KEAP1 mutation frequency and their predictive significance for ICI treatment outcome in diverse cancers has not yet been investigated. Therefore, we conducted this pan-cancer analysis by using online database to systematically characterize the prevalence and predictive value of KEAP1 mutations across multiple cancer types.
All included patients and sequencing data were identified from the cBioPortal online database (https://www.cbioportal.org) (6). KEAP1 mutations were defined as all kinds of nonsynonymous mutations including missense, frame-shift, splice site, nonstop, nonsense, and translation start site changes. To evaluate the difference of tumor mutation burden (TMB) level between KEAP1 mutant and wild type groups, a subset generated from MSK-IMPACT cohort was selected to avoid the selection bias and ensure the TMB could be comparable (7). The six immune infiltrates abundances including B cells, CD4+ T cells, CD8+ T cells, dendritic cells, macrophages and neutrophils were estimated by using a web server for comprehensive analysis of tumor-infiltrating immune cells, named TIMER (Tumor Immune Estimation Resource, https://cistrome.shinyapps.io/timer/) (8). Kaplan-Meier curves with log-rank tests were used to determine the survival difference.
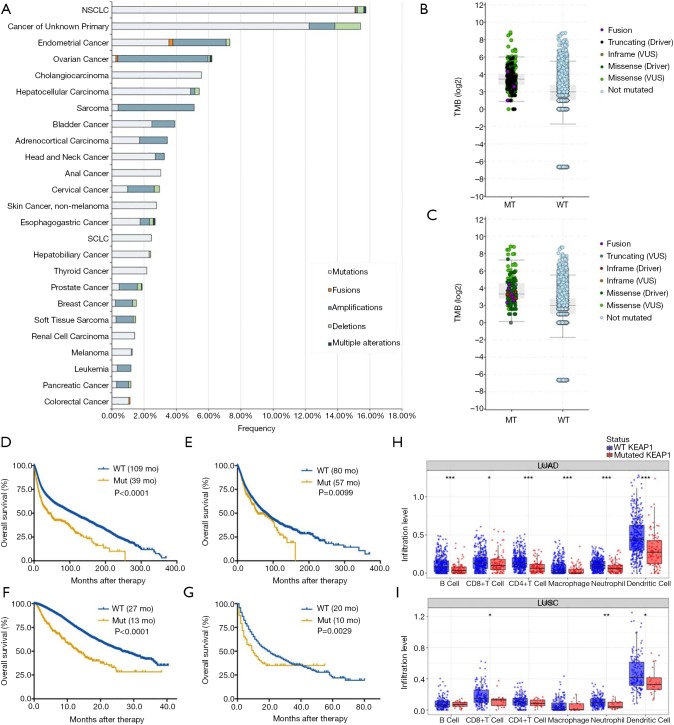
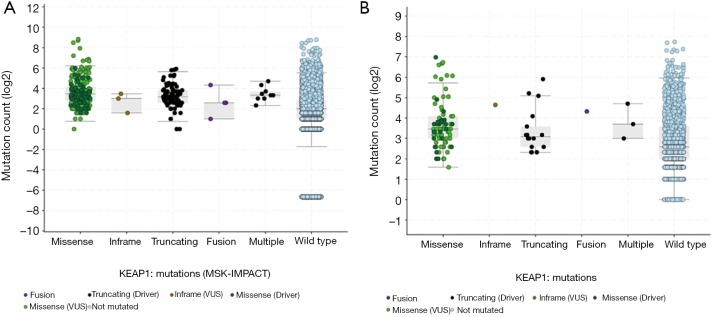
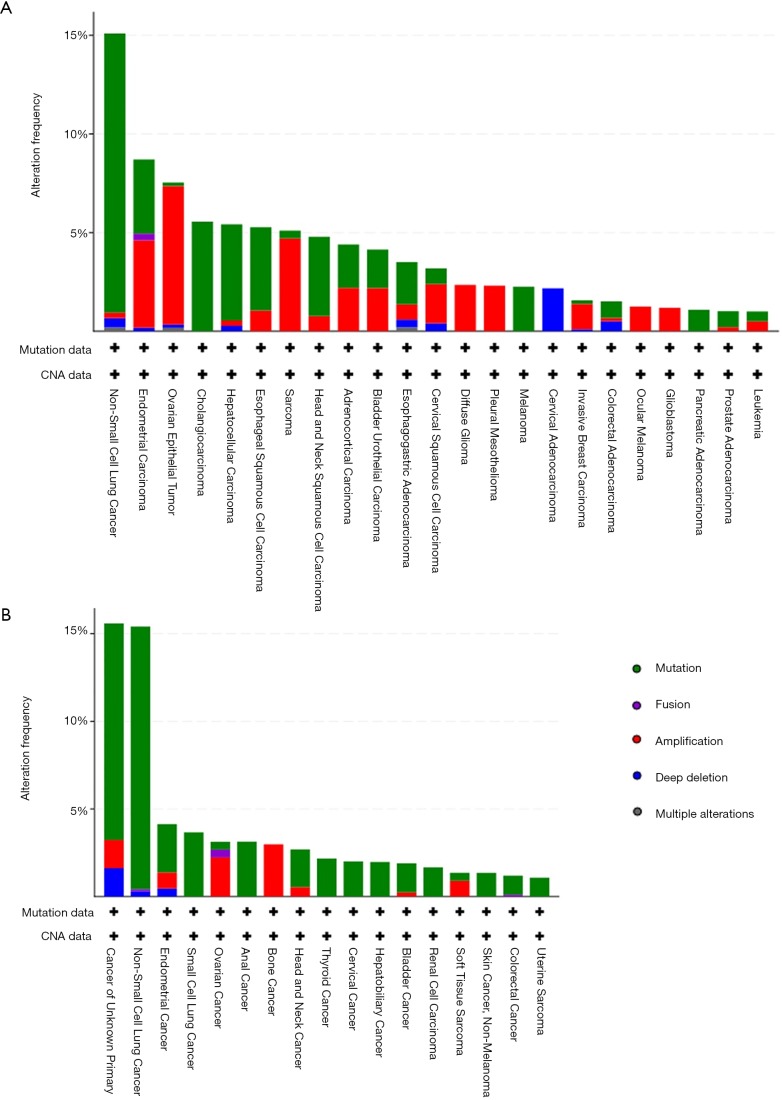
We summarized all the relevant data in Figure 1. The prevalence of KEAP1 mutations in 40,167 patients with distinct cancer types was 2.7% (Figure 1A), with patients with non-small cell lung cancer (NSCLC) having the highest levels of KEAP1 mutations (15.8%). Most of the alterations were missense mutations. The prevalence and spectrum of KEAP1 mutations were slightly different in early-stage (441/10,967, TCGA cohort; Figure S1A) versus advanced-stage cancers (388/10,945, MSK-IMPACT cohort; Figure S1B). In the MSK-IMPACT cohort (7), TMB of patients with KEAP1 mutations was significantly higher than in those without the mutations (10 vs. 4 mutations/Mb, P<0.0001; Figure 1B). This was validated in the ICI-treated cohort (Figure 1C) (9). Notably, cancers with KEAP1 missense mutations had the highest TMB level (Figure S2). Together, these findings reveal a high prevalence of KEAP1 mutations and its close relationship with TMB level across cancer types, suggesting that KEAP1 mutations should be considered as biomarkers when conducting ICI treatment.
Figure 1.
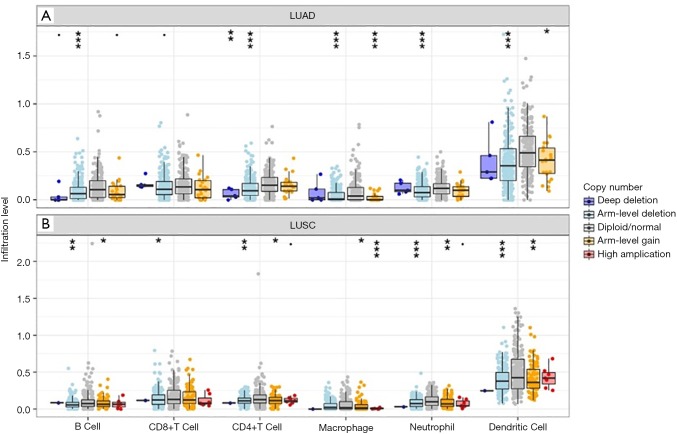
Pan-cancer analysis of KEAP1 mutations as biomarkers for immunotherapy outcomes. (A) Prevalence of KEAP1 mutations in different cancer types; (B) the association between TMB and KEAP1 mutations in MSK-IMPACT cohort; (C) the association between TMB and KEAP1 alterations in immune checkpoint inhibitors treatment cohort; (D) prognostic value of KEAP1 mutations in all cancers; (E) prognostic value of KEAP1 mutations in early-stage cancers (TCGA cohort); (F) prognostic value of KEAP1 mutations in advanced-stage cancers (MSK-IMPACT cohort); (G) predictive value of KEAP1 mutations in patients received ICI therapy; (H) the association between KEAP1 mutations and six immune infiltrates in lung adenocarcinoma; (I) the association between KEAP1 mutations and six immune infiltrates in lung squamous cell carcinoma. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; Mut, mutation; WT, wild type; TMB, tumor mutation burden; ICI, immune checkpoint inhibitor.
Figure S2.
The association between TMB and KEAP1 mutations subtypes in MSK-IMPACT cohort (A) and immune checkpoint inhibitors treatment cohort (B). TMB, tumor mutation burden.
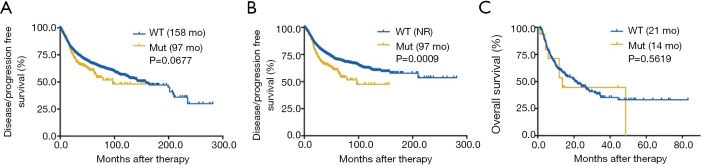
Next, we surveyed the relationship between KEAP1 mutations and overall survival (OS) in both whole group and ICI-treated cohort. We firstly found that patients with KEAP1 mutations showed a significantly shorter OS (39 vs. 109 months, P<0.0001; Figure 1D) than those without in whole populations. The prognostic value of KEAP1 mutations was also found in early-stage (P=0.0099; Figure 1E) and advanced-stage cancers (P<0.0001; Figure 1F). Although KEAP1 mutations were associated with marginally significantly shorter disease-free survival (DFS, 97 vs. 158 months, P=0.0677; Figure S3A), it was associated with markedly inferior DFS in early-stage cancers (P=0.0009; Figure S3B). In the ICI treatment cohort (9), we identified 1,661 patients with different cancers receiving ICI therapy and 99 of them with KEAP1 mutations. Patients with KEAP1 mutations also had a substantially inferior OS of 10 vs. 20 months in the wild-type group (P=0.0029; Figure 1G). Further investigation showed that only KEAP1 mutations could not predict OS in patients with microsatellite-stable (MSS) solid tumors (14 vs. 21 months, P=0.5619; Figure S3C).
To unravel the potential mechanism of the predictive value of KEAP1 mutations for ICI treatment, we then investigated the association between KEAP1 mutations and immune landscape across multiple cancer types. We observed that these mutations were associated with significantly lower CD8+ T cells infiltrations in most of the cancer types including endometrial cancer, breast cancer, bladder cancer, colorectal cancer, lung adenocarcinoma (Figure 1H), lung squamous cell carcinoma (Figure 1I) and so on. Notably, patients with KEAP1-mutant lung adenocarcinoma had dramatically lower CD8+ T cells, neutrophils and dendritic cells infiltrations than those without, which was consistent with our recent publication (10). Of note, copy number variations (especially deep deletion or arm-level deletion) of KEAP1 were associated with substantially lower immune infiltrates in most cancer types including lung adenocarcinoma (Figure S4A) and lung squamous cell carcinoma (Figure S4B).
To our knowledge, this study firstly reported a high frequency of KEAP1 mutations in diverse cancers including lung cancer, endometrial cancer, hepatocellular carcinoma, head and neck cancer, bladder cancer, colorectal cancer, esophagogastric cancer, etc. and negative prognostic value of KEAP1 mutations for patients with different types of cancer. We also observed that KEAP1 mutations were a negative predictive biomarker and might be utilized to predict a survival benefit from ICI treatment across multiple cancers. Although KEAP1 mutations were correlated with significantly higher TMB level, they were also associated with significantly lower immune infiltrates especially CD8+ T cells, suggesting that tumor with these mutations could promote establishment of a cold-tumor immune microenvironment. Considering the high prevalence of KEAP1 mutations, there is an urgent need for the development of rational and novel therapeutic. We are planning to initiate a prospective study to investigate the efficacy of PD-1 antibody plus vascular endothelial growth factor receptor tyrosine kinase inhibitors for patients with solid cancer and KEAP1 mutations. Collectively, our findings highlight the important value of KEAP1 alterations as pan-cancer predictive biomarkers for ICI treatment.
Figure S1.
The frequency of KEAP1 mutations in early-stage cancer (A) and advanced-stage cancer (B).
Figure S3.
Predictive value of KEAP1 mutations in all cancers (A), in TCGA cohort (B) and in patients with MSS solid tumors (C). MSS, microsatellite-stable.
Figure S4.
The association between KEAP1 copy number variations and six immune infiltrates in lung adenocarcinoma (A) and lung squamous cell carcinoma (B).
Acknowledgment
Funding: This study was supported in part by grants from the National Natural Science Foundation of China (No. 81672286, 81772467 and 81874036) and Medical Guidance Project of Shanghai Science and Technology Commission (No. 17411969200).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Sacher AG, Gandhi L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol 2016;2:1217-22. 10.1001/jamaoncol.2016.0639 [DOI] [PubMed] [Google Scholar]
- 2.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med 2019;25:389-402. 10.1038/s41591-019-0382-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeong Y, Hoang NT, Lovejoy A, et al. Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov 2017;7:86-101. 10.1158/2159-8290.CD-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Y, Wu K, Liu Q, et al. Modification of platinum sensitivity by KEAP1/NRF2 signals in non-small cell lung cancer. J Hematol Oncol 2016;9:83. 10.1186/s13045-016-0311-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202-6. 10.1038/s41588-018-0312-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang T, Shi J, Dong Z, et al. Genomic landscape and its correlations with tumor mutational burden, PD-L1 expression, and immune cells infiltration in Chinese lung squamous cell carcinoma. J Hematol Oncol 2019;12:75. 10.1186/s13045-019-0762-1 [DOI] [PMC free article] [PubMed] [Google Scholar]