Abstract
Bronchial asthma is a common chronic inflammatory disease of the airways. Although its pathogenic mechanism remains unknown, it is influenced by both genetic and environmental factors. The emergence and application of proteomic technologies can help to facilitate analysis of the changes in transcription factors, inflammatory mediators, chemokines, cytokines, and cell apoptosis-and proliferation-related proteins in the pathological processes of asthma. Proteomic technologies can unearth prospects and theoretical bases for improved understanding of the biological mechanism of asthma and effective identification of diagnostic and therapeutic targets.
Keywords: Proteomics, bronchial asthma, bioinformatics, mass spectrometry (MS)
Bronchial asthma is a common chronic inflammatory disease of the airways characterized by reversible bronchospasm, which is caused by bronchial hyperreactivity. Epidemiological studies have revealed that there has been a worldwide increase in the prevalence of bronchial asthma over the past four decades. In 2016, an estimated 300 million people worldwide had bronchial asthma; by 2025, this number is expected to reach 400 million (1,2).
Although the etiology of bronchial asthma remains unknown, its pathogenesis is well believed to be affected by both genetic and environmental factors. Environmental substances that may cause asthma include indoor allergens (e.g., dust mites, pets, and cockroaches), outdoor allergens (e.g., pollen and dust), sources of infection (e.g., bacteria, fungi, and parasites), occupational pollutants, or food additives (3). In terms of genetic factors, a genome-wide association (GWA) study identified more than 100 genes as being significantly associated with the onset of bronchial asthma. In addition, mutations in the ORM1 gene have been revealed in studies on single nucleotide polymorphisms (SNPs) to also be closely related to the onset of asthma (4,5).
The main pathophysiological characteristics of asthma are inflammation and airway remodeling, which include goblet cell hyperplasia, subepithelial fibrosis, collagen deposition, mucosal gland hyperplasia, smooth muscle hypertrophy, and changes in the extracellular matrix. These changes can result in immune system imbalance, eventually leading to airway hyperresponsiveness (6). Throughout the course of bronchial asthma, changes in the levels of transcription factors, inflammatory mediators, chemokines, cytokines, and cell apoptosis and proliferation-related proteins also take place (7). Therefore, studies on protein expression can enable better understanding of the pathological mechanism of bronchial asthma. The emergence of new genomic and proteomic technologies has facilitated high-throughput analysis of functional proteins in the body and has also promoted advances in bronchial asthma pathogenesis research. This article summarizes the results of proteomics research in relation to bronchial asthma carried out in recent years.
Proteomic technologies
A large amount of raw data has been provided for the analysis of biological indicators by modern bioinformatics including DNA microarrays, proteomics, peptide studies, and metabolomics. Through the application of computer-based statistical methods, bioinformatics can also help to build descriptive biological models from genomic and proteomic data (8). The ultimate goal of bioinformatics is to build predictive mathematical models that provide valuable information on the staging, diagnosis, and treatment of diseases. The Human Genome Project (HGP) has revolutionized life science but not all the information encoded in the genome will be expressed. Only a limited number of genes are activated, and large amounts of proteins and peptides are produced after the transcription, translation, and post-translational modifications of these genes. The body has a complex protein network, which ensures that every cell, tissue, and organ achieves its biological function by adhering to the physiological laws (9,10). Therefore, proteomic analysis is an important part of modern bioinformatics.
For more than 20 years, polyacrylamide gel electrophoresis (PAGE) has been used as the standard method for the isolation of complex proteome samples, with most commonly used technique being two-dimensional gel electrophoresis (2DG) (11). The molecular weights of the analyzed proteins usually range from 10–200 kDa, with a pH value of 4–10. First, proteins are isolated based on their pH values by using gels with fixed pH gradient. Subsequently, further separation is performed by SDS-PAGE according to relative quality. Different methods are then used to extract proteins from the gel, before mass spectrometry (MS) is carried out to quantify the proteins (12). Column chromatography can also be used to quantify proteins in complex mixtures and has the advantage of keeping the analytes in a liquid state at all times, thereby minimizing material loss. This method is especially suitable for analysis of plasma or serum containing a large amount of proteins (13). Since gene expression, protein synthesis, and post-translational modifications are dynamic processes, the identification and quantification of proteins are not adequate to draw functionally relevant conclusions. Hierarchical clustering analysis of the co-transcription, co-expression, and co-secretion of biomolecules assists with predicting the relevant proteins and the interaction networks. Therefore, it is necessary to dynamically analyze relevant complex interaction networks to construct functional models of the body and organ systems.
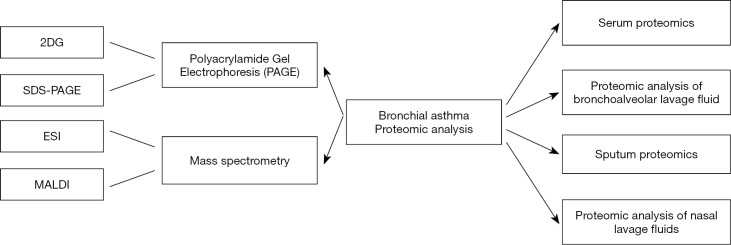
MS can accurately detect the total peptides in complex mixtures, determine the molecular weights of proteins, verify gene sequences, determine the number and location of disulfide bonds, and help clarify the structure-function relationships. Verification of the biological functions and interaction networks of newly discovered proteins has become increasingly important due to the rapid accumulation of data on gene and protein sequences (14). There are two techniques frequently used in MS: electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). Under the ESI mode, proteins are protonated multiple times and then transferred from solution to the gas phase, producing multiple charged ions (15). MALDI, on the other hand, uses a laser to transfer molecules directly from the solid phase to the gas phase, and transfers sufficient energy to the matrix molecules, generating single charged ions. The m/z (mass-to-charge ratio) of the peptide fragments obtained through HPLC or other chromatographic methods is then analyzed; if MALDI is applied, the time-of-flight (TOF) is usually obtained (16). The availability of tandem MS, which selectively isolates proteins in a mass spectrometer to read out part of a protein's amino acid sequence prior to MALDI-TOF analysis of each protein, further improves the analysis protocols (17). The combination of proteomic analysis and bioinformatic tools offers great prospects for the rapid discovery of the diagnostic and therapeutic targets of many diseases including asthma (As shown in Figure 1).
Figure 1.
Common techniques for proteomic analysis of bronchial asthma and their classifications.
Clinical application of proteomics in bronchial asthma
Body fluids including blood (serum), alveolar lavage fluids, sputum, and saliva make up the majority of the samples currently used to conduct proteomic analysis in bronchial asthma patients (As shown in Table 1). Serum is the most commonly used and is also the most important body fluid sample in asthma research. The development of fiberbronchoscopes has also facilitated the collection of cells and other soluble components from the airways and alveolar epithelium. Sputum, a substance excreted from the respiratory tract, provides a direct reflection of the pathophysiological conditions of the lungs, bronchi, or respiratory tract.
Table 1. Differently expressed proteins in various body fluids of patients with bronchial asthma.
| Categories | Proteins |
|---|---|
| Serum | C7, C3, C4, α-1-antitrypsin, PDE7, arginase, UK16 binding protein, phospholipase D, and cyclooxygenase |
| Bronchoalveolar lavage fluid | Lipocalin-1, cystatin S, phospholipase A2, α-1-antitrypsin, IgAb, IgMb, haptoglobin, α-1-acid glycoprotein 1, and PIGR airway protein |
| Sputum | S100A8, S100A9, SMR3B, SCGB1A110, hemopexin (HPX), and activated complement component 3 (C3a) |
| Nasal lavage fluid | β-2 globulin, galactin, and actin |
Serum proteomics
Serum, the preferred sample for proteomic analysis, has long been a valuable source of potential biomarkers for diseases. However, the complexity and dynamics of electrolytes and proteins in serum often become interfering factors in proteomic and metabolomic analyses, making serum protein characterization a challenging task. Nevertheless, serum proteomic analysis has been performed on bronchial asthma patients. Chung et al. developed a method for analyzing serum potential biomarkers in patients with bronchial asthma (18). Using functionalized magnetic beads, they enriched and analyzed serum proteins obtained from four asthmatic patients and four healthy subjects. After washing, the proteins were eluted, transferred to a pre-built sample holder, and then detected using a MALDI-TOF mass spectrometer. All obtained mass spectra were processed by ClinProTools software. Only two peaks (MW 1,846 and 6,444 Da) in serum proteins were found to be significantly different between the healthy and asthmatic groups. Terracciano et al. (19) carried out proteomic analysis of 16 children with bronchial asthma and 10 healthy children, and the results were verified by ELISA; they found that complement component C7 and alpha-1 antitrypsin were overexpressed in the serum samples of the bronchial asthma patients. Ghosal et al. (20) used LC-ESI MS to analyze serum proteins of bronchial asthma patients and found that when compared with healthy people, asthmatic patients had significantly increased levels of serum phosphodiesterase 7 (PDE7), arginase, UK16 binding protein, and phospholipase D, which are closely related to T cell-mediated immune response and hypersensitivity. Ruizcampillo et al. (21) carried out serum proteomic analysis of patients with aspirin-induced bronchial asthma (ASA) and found the serum levels of complements C3 and C4, and cyclooxygenase to be significantly increased in ASA patients and significantly associated with FEV1, which suggests that the complement pathway plays an important role in the pathogenesis of asthma.
Proteomic analysis of bronchoalveolar lavage fluid (BALF)
The airway mucosa and the alveolar surface constitute an interface for the exchange of air between the lungs and the external environment. As part of the host immune system, epithelial cells on this barrier are able to secrete fluids containing peptides and proteins. Since they are directly affected by external factors, these fluids are considered to reflect pathophysiological changes in the lungs (22). BALF contains cells and soluble components. Normally, the cells contained in BALF are mainly alveolar macrophages and a small number of T lymphocytes. Many lung diseases are characterized by changes in cell distribution. After these cells are removed, the resulting BALF contains only soluble proteins, which can be either endogenous (derived from alveolar macrophages, inflammatory cells, or epithelial cells) or exogenous (derived from serum and diffuse across the capillary-alveolar barrier) (23). Comparisons between serum and BALF proteomes have shown that several proteins have significantly higher levels in BALF than in serum, suggesting they are specifically generated in the airways. BALF analysis technology was introduced after the development of bronchoscopy in the 1970s. After a small amount of physiological saline is injected via a fiber bronchoscope into a specific area, BALF can be immediately obtained by aspiration. Proteomics of BALF has been widely used for the analysis of specific markers for a variety of respiratory diseases, including pulmonary fibrosis, acute lung injury, interstitial lung disease, and occupational lung disease (24). In 2012, the American Thoracic Association’s clinical guidelines on interstitial lung disease also recommended the use of BALF proteomic analysis for disease screening (25). Many studies have investigated the role of BALF proteomics in relation to the management of bronchial asthma.
Bogart et al. (26) first studied the BALF proteome of asthmatic patients and found that their levels of lipocalin-1, cystatin S, and phospholipase A2, which are related to the upper respiratory tract-stimulating pathways, were significantly increased. Haenen et al. (27) analyzed the BALF proteins of both asthmatic patients and healthy individuals by HPLC-MS and found that there were about 160 differentially expressed proteins, among which the upregulated proteins, including proteases, complement factors, acute phase proteins, and eosinophils, were closely associated with enhanced inflammatory response in asthmatic patients. Thus, the pathophysiological mechanism of asthma is quite complicated and involves processes such as inflammation, airway remodeling, repair of damaged tissues, and mucus production. Fujii et al. (28) reported that the BALF level of galectin-binding protein was significantly higher in asthmatic patients than in healthy controls, and galectin was an important factor involved in the regulation of inflammatory responses. Using enriched chromatography, they further verified the presence of galectin ligands in BALF specimens of asthmatic patients, identifying about 100 galectin-binding proteins, including complement C3, α1-antitrypsin, IgAb, IgMb, haptoglobin, α1-acid glycoprotein, and PIGR airway protein. These findings also revealed the important role of inflammatory response in the pathogenesis of asthma. Currently, diluted proteins and high electrolyte concentration are the main disadvantages of BALF proteomic analysis. In addition, issues surrounding the standardization of BALF sample preparation methods also affect the accuracy of analysis.
Sputum proteomics
Sputum is a complex secretion in the airway, containing information reflecting the inflammatory cells and mediators present. It holds great significance in the identification of the phenotypic characteristics of bronchial asthma. Atomized saline is often used clinically to induce sputum secretion, and the induced sputum may directly reflect pulmonary oxidative damage. Riccio et al. (29) extracted sputum specimens from 5 healthy subjects and 10 bronchial asthma patients and analyzed them by using MALDI-TOF/TOF MS after trypsin digestion. A total of 254 proteins were identified in the sputum specimens from all 15 subjects. The Gene Ontology database showed that these proteins were involved in defense responses, inflammation, and complement activation. In comparison with healthy subjects, the asthmatic patients had 7 significantly downregulated proteins, including S100A8, S100A9, SMR3B, and SCGB1A110, and 10 significantly upregulated proteins, including hemopexin (HPX) and activated complement component 3 (C3a). Wu et al. also reported (30) that the expression of S100A9 in the sputum of asthmatic patients was significantly increased and that this molecule was involved in the neutrophil-mediated inflammatory response. Despite the relatively non-invasive nature of proteomic analysis of sputum, some technical problems, such as contamination of the sputum specimens by saliva and difficulty in applying electrophoretic techniques in mucus specimen analysis, remain.
Proteomics of other body fluids
Nasal lavage fluid (NLF) is a fluid obtained by flushing the nasal mucosa with physiological saline. The use of NLF for the detection of lung disease biomarkers was first proposed by Erban et al. (31), who observed similarities between the protein components of NLF and serum. Mörtstedt et al. (32) isolated NLF from patients with bronchial asthma and performed LC/MS after trypsin digestion. It was found that β-2 globulin, galactin, and actin were highly expressed, although further investigation is required into their relationships with asthma. Proteomes analyses using other body fluids may also help to analyze the complex mechanisms of asthma at different stages, which provides a new perspective for finding new biomarkers for asthma.
Prospects and outlook
Unlike other diseases (e.g., cancer), the goal of proteomics in patients with bronchial asthma is not to develop a new method for asthma diagnosis through analysis of body fluids; instead, it aims to determine the causal relationship between changes of proteins in body fluids and clinical phenotypes, so as to identify target molecules that play key roles in the pathogenesis of asthma, eventually leading to the development of new treatment protocols. Although many proteomic analyses have been carried out on different body fluids obtained from asthmatic patients, the majority of studies were based on 2DE (33). At present, a new analytical method based on the 1D-LC-MS/MS system is the subject of research and development. This solution does not require the extraction of large amounts of proteins or the use of two-dimensional differential gel electrophoresis (2D-DIGE); it instead relies on chemical markers, or isotopically-encoded markers. However, its clinical feasibility requires further verification. In addition, post-translational modification is another research direction of proteomics. By introducing specifically modified proteins such as phosphorylated, acetylated, or nitrolated proteins, the amount of protein extracted for analysis can be reduced and accurate and detailed results can be obtained. The rapid advances seen in proteomics technology have led to the discovery of new proteins, which in turn has supported the progression of high-throughput bioinformatics research, which can quickly find a large number of target molecules with diagnostic and therapeutic implications and thus improve the prevention and treatment of relevant diseases.
Conclusions
Although the pathogenic mechanism of bronchial asthma remains unknown, it is affected by both genetic and environmental factors and may involve the endocrine system, neurologic system, and endocrine system. With the further developments in techniques including capillary electrophoresis, chromatography, and protein microarray, proteomic analysis can become more in-depth and accurate, providing a theoretical basis for a better understanding of the biological mechanism of bronchial asthma and the search for diagnostic and therapeutic targets for the disease.
Acknowledgments
Funding: This work was supported by the NSFC (No. 81770028), the Natural Science Foundation of Guangdong (2018A030310011), the Key Laboratory of Shenzhen Respiratory Disease (ZDSYS201504301616234), the Project of Shenzhen Basic Research Plan (JCYJ20170307095633450), and the Clinical research of Shenzhen Municipal Health and Family Planning Commission (SZLY2017024). This work also was supported by special funding for high-level disciplines from the Shenzhen Institute of Respiratory Diseases.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Fujii K, Nakamura H, Nishimura T. Recent mass spectrometry-based proteomics for biomarker discovery in lung cancer, COPD, and asthma. Expert Rev Proteomics 2017;14:373-86. 10.1080/14789450.2017.1304215 [DOI] [PubMed] [Google Scholar]
- 2.Cao C, Li W, Hua W, et al. Proteomic analysis of sputum reveals novel biomarkers for various presentations of asthma. J Transl Med 2017;15:171. 10.1186/s12967-017-1264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee CK. Nanotechnology as a savior in asthma management. Ann Transl Med 2019;7:517. 10.21037/atm.2019.09.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgstaller G, Oehrle B, Gerckens M, et al. The instructive extracellular matrix of the lung: basic composition and alterations in chronic lung disease. Eur Respir J 2017. doi: . 10.1183/13993003.01805-2016 [DOI] [PubMed] [Google Scholar]
- 5.Cox CA, Boudewijn IM, Vroegop SJ, et al. Associations of AMP and adenosine induced dyspnea sensation to large and small airways dysfunction in asthma. BMC Pulm Med 2019;19:23. 10.1186/s12890-019-0783-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valero A, Ribó P, Maíz L, et al. Asthma patient satisfaction with different dry powder inhalers. Expert Rev Respir Med 2019;13:133-8. 10.1080/17476348.2019.1567339 [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Hwang YH, Kim KJ, et al. Proteomic and transcriptomic analysis of lung tissue in OVA-challenged mice. Arch Pharm Res 2018;41:87-100. 10.1007/s12272-017-0972-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padding AM, Rutjes NW, Hashimoto S, et al. Young children experience little emotional burden during invasive procedures in asthma research. Eur J Pediatr 2019;178:207-11. 10.1007/s00431-018-3265-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu LH, Annex BH, Popel AS. Computational drug repositioning for peripheral arterial disease: prediction of anti-inflammatory and pro-angiogenic therapeutics. Front Pharmacol 2015;6:179. 10.3389/fphar.2015.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carr TF, Kraft M. Use of biomarkers to identify phenotypes and endotypes of severeasthma. Ann Allergy Asthma Immunol 2018;121:414-20. 10.1016/j.anai.2018.07.029 [DOI] [PubMed] [Google Scholar]
- 11.Cunsolo V, Muccilli V, Saletti R, et al. Mass spectrometry in food proteomics: a tutorial. J Mass Spectrom 2014;49:768-84. 10.1002/jms.3374 [DOI] [PubMed] [Google Scholar]
- 12.Toda M, Hellwig M, Henle T, et al. Influence of the Maillard Reaction on the Allergenicity of Food Proteins and the Development of Allergic Inflammation. Curr Allergy Asthma Rep 2019;19:4. 10.1007/s11882-019-0834-x [DOI] [PubMed] [Google Scholar]
- 13.Heyder T, Wiklundh E, Eklund A, et al. Altered Fc galactosylation in IgG4 is a potential serum marker for chronic lung disease. ERJ Open Res 2018. doi: . 10.1183/23120541.00033-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothe T, Spagnolo P, Bridevaux PO, et al. Diagnosis and Management of Asthma - The Swiss Guidelines. Respiration 2018;95:364-80. 10.1159/000486797 [DOI] [PubMed] [Google Scholar]
- 15.Clement CC, Aphkazava D, Nieves E, et al. Protein expression profiles of human lymph and plasma mapped by 2D-DIGE and 1D SDS-PAGE coupled with nanoLC-ESI-MS/MS bottom-up proteomics. J Proteomics 2013;78:172-87. 10.1016/j.jprot.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sircar G, Saha B, Bhattacharya SG, et al. Allergic asthma biomarkers using systems approaches. Front Genet 2014;4:308. 10.3389/fgene.2013.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulet LP. Airway remodeling in asthma: update on mechanisms and therapeutic approaches. Curr Opin Pulm Med 2018;24:56-62. 10.1097/MCP.0000000000000441 [DOI] [PubMed] [Google Scholar]
- 18.Chung KF, Adcock IM. Clinical phenotypes of asthma should link up with disease mechanisms. Curr Opin Allergy Clin Immunol 2015;15:56-62. 10.1097/ACI.0000000000000134 [DOI] [PubMed] [Google Scholar]
- 19.Terracciano R, Pelaia G, Preianò M, et al. Asthma and COPD proteomics: current approaches and future directions. Proteomics Clin Appl 2015;9:203-20. 10.1002/prca.201400099 [DOI] [PubMed] [Google Scholar]
- 20.Ghosal K, Saha B, Gupta Bhattacharya S. Clinical and immuno-proteomic approach on Lantana camara pollen allergy-a major health hazard. Allergy Asthma Clin Immunol 2016;12:33. 10.1186/s13223-016-0135-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz-Campillo MT, Molina Hernandez V, Escamilla A, et al. Immune signatures of pathogenesis in the peritoneal compartment during early infection of sheep with Fasciola hepatica. Sci Rep 2017;7:2782. 10.1038/s41598-017-03094-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miękus N, Olędzka I, Plenis A, et al. Gel electrophoretic separation of proteins from cultured neuroendocrine tumor cell lines. Mol Med Rep 2015;11:1407-15. 10.3892/mmr.2014.2864 [DOI] [PubMed] [Google Scholar]
- 23.Sulaiman I, Greene G, MacHale E, et al. A randomised clinical trial of feedback on inhaler adherence and technique in patients with severe uncontrolled asthma. Eur Respir J 2018. doi: . 10.1183/13993003.01126-2017 [DOI] [PubMed] [Google Scholar]
- 24.Chung KF, Adcock IM. Clinical phenotypes of asthma should link up with disease mechanisms. Curr Opin Allergy Clin Immunol 2015;15:56-62. 10.1097/ACI.0000000000000134 [DOI] [PubMed] [Google Scholar]
- 25.Qaseem A, Fihn SD, Williams S, et al. Diagnosis of stable ischemic heart disease: summary of a clinical practice guideline from the American College of Physicians/American College of Cardiology Foundation/American Heart Association/American Association for Thoracic Surgery/Preventive Cardiovascular Nurses Association/Society of Thoracic Surgeons. Ann Intern Med 2012;157:729-34. 10.7326/0003-4819-157-10-201211200-00010 [DOI] [PubMed] [Google Scholar]
- 26.Bogart M, Roberts A, Wheeler S. Cost-Effectiveness of Refractory Asthma Treatment Strategies: A Decision Tree Analysis. Value in Health 2015;18:a174 10.1016/j.jval.2015.03.1004 [DOI] [Google Scholar]
- 27.Haenen S, Clynen E, Nemery B, et al. Biomarker discovery in asthma and COPD: Application of proteomics techniques in human and mice. Eupa Open Proteomics 2014;4:101-12. 10.1016/j.euprot.2014.04.008 [DOI] [Google Scholar]
- 28.Fujii K, Nakamura H, Nishimura T. Recent mass spectrometry-based proteomics for biomarker discovery in lung cancer, COPD, and asthma. Expert Rev Proteomics 2017;14:373-86. 10.1080/14789450.2017.1304215 [DOI] [PubMed] [Google Scholar]
- 29.Riccio AM, Mauri P, De Ferrari L, et al. Galectin-3: an early predictive biomarker of modulation of airway remodeling in patients with severe asthma treated with omalizumab for 36 months. Clin Transl Allergy 2017;7:6. 10.1186/s13601-017-0143-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, Kobayashi M, Sousa EA, et al. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics 2005;4:1251-64. 10.1074/mcp.M500041-MCP200 [DOI] [PubMed] [Google Scholar]
- 31.Erban T, Harant K, Hubert J. Detailed two-dimensional gel proteomic mapping of the feces of the house dust mite Dermatophagoides pteronyssinus and comparison with D. farinae: Reduced trypsin protease content in D. pteronyssinus and different isoforms. J Proteomics 2017;162:11-19. 10.1016/j.jprot.2017.04.021 [DOI] [PubMed] [Google Scholar]
- 32.Mörtstedt H, Ali N, Kåredal M, et al. Targeted proteomic analyses of nasal lavage fluid in persulfate-challenged hairdressers with bleaching powder-associated rhinitis. J Proteome Res 2015;14:860-73. 10.1021/pr5009306 [DOI] [PubMed] [Google Scholar]
- 33.Sinha A, Sterk PJ. Proteomics in asthma: the clinicians were right after all, were not they? Clin Transl Med 2017;6:39. 10.1186/s40169-017-0170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]