Abstract
Background
The further progression of credible expression profiling analysis of genes continues to expand our understanding of the biological characteristics in lung cancer. In this study, RNA sequencing (RNA-Seq) was used to contrast the transcriptomics profiling of small cell lung cancer (SCLC) that acquired partial response (PR) and stable disease (SD)/progressive disease (PD) after first-line chemotherapy. We aimed to illuminate the underlying mechanisms of long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) in the efficacy of SCLC first-line chemotherapy.
Methods
Six male patients (mean age, 64.2 years) with SCLC were enrolled in this study. RNA-Seq was executed on the tumor tissues from 3 patients with PR outcome and 3 patients with SD or PD therapeutic effect after first-line chemotherapy.
Results
RNA-Seq generated 26.67×106 (±8.7×106) reads in SCLC tissues [mean (±standard deviation)]. Analysis revealed that 64 lncRNAs had higher expression and 194 had lower expression in the PR group ≥2-fold (P<0.05). Three downregulated genes in the PR group [HOXA-AS3, cancer susceptibility 9 (CASC9), and KEGG] could have a role in the insensitivity of SCLC. A total of 1,303 differential miRNAs were defined between PR and the SD or PD SCLC group, while 520 miRNAs had higher expression, and 783 had lower expression in the PR group. Two lower expressed miRNAs in the PR group (miRNA 601 and miRNA 596) might be the key genes in SCLC chemotherapy insensitivity.
Conclusions
The expression of 3 gene (HOXA-AS3, CASC9, and KEGG) and 2 miRNAs (miRNA 601 and miRNA 596) were markedly decreased in SCLC patients who achieved PR. They thus might be the promising candidate genes in SCLC chemotherapy insensitivity.
Keywords: Small cell lung cancer (SCLC), long noncoding RNAs (lncRNAs), microRNAs (miRNAs), chemotherapy insensitivity
Introduction
Small cell lung cancer (SCLC) accounts for 10–15% of lung cancer with about 75% patients being extensive-stage disease (ED). Despite objective response rates (ORRs) of chemotherapy for SCLC being 60–65%, median overall survival (OS) is only approximately 10 months, with the outcomes of SCLC remaining stagnant (1,2). The standard treatment of SCLC is chemotherapy, but most patients undergoing this treatment eventually still suffer disease progression as a result of chemotherapy insensitivity (3). Therefore, further research is needed to identify molecular biomarkers and drug targets to provide new strategies for SCLC therapy.
Long noncoding RNAs (lncRNAs) are involved in the pathogenesis and development of various diseases. In terms of cell function, they can be divided into tumor-suppressive and carcinogenic lncRNA types (4,5). One function of lncRNAs has been shown to be as gene regulators in complex mechanisms of oncogenic pathways (6-8). It was also reported that lncRNAs are correlated with cancer diagnosis, insensitivity to target therapy, and prognosis. Consequently, it is critical to identify those lncRNAs that correspond to SCLC and explore their cross functions with protein-encoding genes to enhance our comprehension of SCLC.
MicroRNAs (miRNAs) are small noncoding RNAs with a length of approximately 21–23 nucleotides (nt). Single-stranded miRNA usually leads to mRNA degradation or protein translation as a common means of post-transcriptional regulation (9). Recent studies have reported that an abnormal level of miRNAs can cause tumorigenesis via the accelerated production of oncogenes or the repression of the tumor suppressor genes (10). Various miRNAs are indicated to have aberrant expression in non-small cell lung cancer (NSCLC) (11), and theya similarly productivefunction However, no study has identified which specific miRNAs are involved in lung cancer or the related targets of these miRNAs (11).
Therefore, this study used RNA sequencing (RNA-Seq) to identify novel the gene expression profiles involved in SCLC treatment efficacy.
Methods
Patients
We obtained the data of 6 male smokers who received needle biopsy for diagnosis between October 2018 and February 2019 from the Shanghai Pulmonary Hospital (Shanghai). The study was approved by Shanghai Pulmonary Hospital ethics committee. All participants gave their informed consent.
RNA isolation and RNA-Seq
Fine-needle aspiration specimens were assessed by routine procedures of frozen-section and stored at −80°C. SCLC was diagnosed by 2 pathologists.
For RNA-Seq, we used RNeasy 96 Universal Tissue Kit (Qiagen, Gaithersburg, MD, USA) to extract RNA from tissue. Spectrophotometric and electrophoretical verification (Nano-Drop 1000 spectrometer; Thermo Scientific, Bioanalyzer 2100; Agilent Technologies, USA) was used to identify the quality and quantity of total RNA. In accordance with the manufacturer’s instructions, TruSeq RNA library preparation kit (Illumina, San Diego, CA, USA) was used for construct Illumina-compatible libraries. A TruSeq library of double-stranded (ds) cDNA was constructed. The short ds-cDNA fragment was ligated to the sequencing adaptors, and the appropriate fragment was separated by agarose gel electrophoresis. We used quantitative polymerase chain reaction (qPCR) to quantify the TruSeq RNA library constructed by PCR amplification and evaluated its quality by electrophoresis. A HiSeqTM2000 platform (Illumina) was used for performing RNA-Seq.
Differentially expressed gene (DEG) selection
TopHat was used for mapping the human genome by RNA-Seq reads (12), and Cufflinks software (version1.2.1) (12) was used to identify the DEG profiles. Genome reference sequence and data annotations were downloaded from the website, http://genome.uscs.edu. Gene levels were calculated by using the transcript number, and we used the Cufflinks software (version 1.2.2) to confirm the relative transcript abundance as the number of fragments plotted per thousand base pairs per million base exons (FPKM) (12). Using this method, the expression level of 47,362 Ref-Seq genes based on a unique alignment of RNA-Seq reads could be measured. The extracted raw data were the FPKM values in all samples, and we excluded zero-value samples in more than 50% of the genes.
We used the paired t-test to determine statistical significance of the changed fold in expression. The null hypothesis was that there was no difference between the 2 values. The Benjamini-Hochberg algorithm was used to align the p values to limit the false discovery rate. R2.15.1 (www.r-project.org) was employed to analyze all data and visualize the DEGs.
Signal transduction pathway analysis
The Gene Ontology (GO) database is a collection of gene annotation terms for large-scale genomic or transcriptomic data. An online tool, Database for Annotation Visualization and Integrated Discovery (DAVID), was used for systematically relating the functional terms with large gene or protein lists. We measure the degree of association through the enrichment score (ES) and input maximum (MES) of all the gene sets in the true data set of the SCLC patient sample. About 1,000 random arrangements for phenotypes was designed as the number of the MES score significance. We acquired the nominal P value, normalized enrichment score (NES), and numerical statement between gene sets and the actual data. Finally, a P<0.01 and q<0.20 gene set was deemed to be a significant enrichment.
Results
Patient characteristics
The clinical information of all 6 Chinese patients with SCLC is summarized in Table 1 with details in Table S1. All the patients were males who smoked (mean age 64.2 years). All patients were diagnosed with SCLC. Limit-stage disease (LD) was identified in 2 patients, and extended disease (ED) SCLC was identified in 4 patients.
Table 1. Clinical characteristics of SCLC patients.
| Characteristic | Value |
|---|---|
| Age, year (mean ± SD) | 64.2±6.14 |
| Male, n [%] | 6 [100] |
| Smoking, PY (mean ± SD)a | |
| Histological cell type, n [%] | |
| SCLC | 6 [100] |
| Stage, n [%] | |
| LD | 2 [33] |
| ED | 4 [67] |
| Metastasis, n [%] | |
| Y | 4 [67] |
| N | 2 [33] |
| Treatment, n [%] | |
| EP | 1 [17] |
| EC | 5 [83] |
| Assessment, n [%] | |
| PR | 3 [50] |
| SD | 2 [34] |
| PD | 1[16] |
a, PY, pack year. SCLC, small cell lung cancer; SD, standard deviation; LD, limited disease; ED, extensive disease; EP, etoposide plus cisplatin; EC, etoposide plus carboplatin.
Table S1. All of the patients were male, smokers, and SCLC.
| ID | Gender | Age | Smoking history | Tumor type | T | N | M | Stagea | Stageb | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LU-01-1120 | Male | 67 | 1 | SCLC | 4 | 0 | 1 | 4 | ED | EP | SD |
| LU-01-1142 | Male | 68 | 1 | SCLC | 3 | 2 | 0 | 3 | LD | EC | PR |
| LU-01-1147 | Male | 54 | 1 | SCLC | 4 | 1 | 1 | 4 | ED | EC | PD |
| LU-01-1192 | Male | 66 | 1 | SCLC | 2 | 3 | 1 | 4 | ED | EC | PR |
| TYQ | Male | 69 | 1 | SCLC | 4 | 3 | 1 | 4 | ED | EC | SD |
| PXG | Male | 63 | 1 | SCLC | 4 | 3 | 0 | 3 | LD | EC | PR |
a, TNM stage, tumor node metastasis; b, Veterans Administration Lung Study Group (VALSG), limited disease and extensive disease. SCLC, small cell lung cancer; ED, extensive disease; EP, etoposide plus cisplatin; LD, limited disease; EC, etoposide plus carboplatin; PR, partial response. SD, stable disease; PD, progressive disease.
RNA-Seq analysis results
RNA-Seq analysis produced 26.67×106 (±8.7×106) base pairs (bp) from the SCLC tissues. In total, 25.23×106 (±9.3×106) reads in partial response (PR) tissues and 29.12×106 (±12.73×106) reads in SD or PD were proposed. There was no significant difference between the 2 groups (Table 2).
Table 2. RNA sequencing results.
| Features | PR group | SD or PD group | P |
|---|---|---|---|
| Total throughput, M bp (mean ± SD) | 5,352±1,263 | 5,024±1,024 | NS |
| Counted reads, M (mean ± SD) | 23.52±8.10 | 23.17±6.12 | NS |
| Left processed, M (mean ± SD) | 23.61±6.93 | 26.78±6.21 | NS |
| Right processed, M (mean ± SD) | 23.72±9.03 | 24.35±5.60 | NS |
| Total mapped, M (mean ± SD) | 39.16±10.19 | 36.15±16.63 | NS |
Reads counts are expressed in the millions (M); P values were calculated by the Student’s t-test. bp, base pair; NS, non-significant; SD, standard deviation; PR, partial response; PD, progressive disease; SD, stable disease.
Differentially expressed lncRNAs
The PR treatment group and SD or PD tissue transcriptomes all received the high-through put sequence which identified several differentially expressed transcripts. In total, there were 258 genes with fold changes ≥2 and P values <0.05 that were differentially expressed. Compared with the SD or PD tissue, there were included 64 higher expressed genes and 194 lower expressed genes with ≥2-fold changes in the PR tissue. The 10 most upregulated genes in the SD or PD tissues and highest expressed genes in the PR tissues are respectively summarized in Tables 3,4. Three downregulated genes, HOXA-AS3, cancer susceptibility 9 (CASC9), and KEGG, have been reported in various tumors and lung cancers. The genes with ≥4-fold alterations are presented in Tables S2,S3.
Table 3. The top 10 downregulated genes differentially expressed in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (PR/SD PD, ± SD) | P valuea | |
|---|---|---|---|---|
| SD of PD | PR | |||
| AC090692.1 | 2.17±1.17 | 3.62±0.35 | 34.14±1.22 | 2.26×10−18 |
| LINC02500 | 0.89±0.72 | 1.99±0.36 | 12.67±0.81 | 2.37×10−21 |
| LINC02525 | 1.06±0.61 | 2.16±0.39 | 11.17±0.20 | 2.90×10−22 |
| AC087260.1 | 2.23±1.26 | 3.55±0.24 | 11.14±1.19 | 4.16×10−14 |
| LINC01515 | 2.13±0.93 | 3.23±0.23 | 11.08±1.02 | 1.83×10−14 |
| LINC02506 | 0.84±0.33 | 1.83±0.29 | 10.00±0.70 | 1.24×10−23 |
| LINC02128 | 1.74±0.75 | 2.22±0.21 | 9.24±0.78 | 6.16×10−19 |
| LINC01896 | 0.27±0.23 | 1.94±0.76 | 9.14±0.82 | 2.32×10−18 |
| AL354993.2 | 0.97±0.22 | 1.86±0.37 | 8.74±0.67 | 3.71×10−22 |
| AC090692.1 | 0.61±0.52 | 1.56±0.38 | 8.63±0.61 | 9.85×10−23 |
a, P values were calculated by the paired t-test. FPKM, fragments per kilobase of exon per million fragments mapped; SD, standard deviation; PR, partial response; PD, progressive disease; SD, stable disease.
Table 4. The top 20 genes upregulated in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (SD or PD/PR, ± SD) | P valuea | |
|---|---|---|---|---|
| SD or PD | PR | |||
| AL359475.1 | 1.77±0.61 | 0.61±0.44 | 18.26±0.94 | 3.95×10−28 |
| HOXA-AS3 | 1.13±1.32 | 0.19±0.16 | 8.88±1.33 | 2.57× 10−10 |
| AC005307.1 | 1.24±0.65 | 0.47±0.65 | 8.36±0.91 | 4.02×10−13 |
| CASC9 | 1.34±1.07 | 0.41±0.38 | 7.01±1.14 | 3.32×10−11 |
| AC010776.2 | 1.28±1.38 | 0.45±0.37 | 6.42±1.44 | 2.03×10−7 |
| KCNQ5-IT1 | 0.91±0.90 | 0.12±0.16 | 6.07±0.91 | 7.32×10−13 |
| AC104463.2 | 1.02±0.80 | 0.38±0.32 | 5.78±0.36 | 1.19×10−13 |
| KEGG | 0.86±0.23 | 0.20±0.35 | 5.65±0.94 | 6.49× 10−12 |
| AC010776.2 | 0.91±0.82 | 0.18±0.61 | 5.18±0.95 | 5.07×10−17 |
| AC005609.1 | 0.70±0.21 | 0.09±0.73 | 5.16±0.63 | 4.22×10−20 |
| HIF1A-AS2 | 1.13±0.19 | 0.32±0.31 | 5.03±0.85 | 2.83×10−12 |
| AC012414.5 | 0.73±0.93 | 0.03±0.13 | 5.21±0.63 | 4.41×10−17 |
| AP001993.1 | 1.10±0.81 | 0.30±0.25 | 4.88±0.75 | 2.89×10−12 |
| AL031772.1 | 1.86±0.47 | 0.37±0.25 | 4.94±0.55 | 6.72×10−20 |
| ERVMER61-1 | 1.11±0.66 | 0.52±0.36 | 4.93±0.71 | 1.13×10−13 |
| AL162464.2 | 0.81±0.43 | 0.24±0.17 | 4.72±0.27 | 1.20×10−21 |
| AC078789.1 | 0.84±0.67 | 0.18±0.23 | 4.55±0.50 | 3.99×10−15 |
| AC005062.1 | 1.18±0.61 | 0.45±0.21 | 4.22±0.66 | 1.47×10−16 |
| AC005609.3 | 0.94±0.93 | 0.28±0.15 | 4.67±0.93 | 3.00×10−9 |
| PSPC1-AS2 | 0.83±0.59 | 0.18±0.21 | 4.45±0.62 | 2.49×10−15 |
a, P values were calculated by the paired t-test. FPKM, fragments per kilobase of exon per million fragments mapped; SD, standard deviation; SD, stable disease; PR, partial response; PD, progressive disease.
Table S2. Genes with >4-fold upregulated expression in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (SD or PD/PR, ± SD) | P valuea | |
|---|---|---|---|---|
| SD or PD | PR | |||
| AL359475.1 | 1.87±0.61 | 0.63±0.44 | 17.26±0.94 | 3.95×10−28 |
| HOXA-AS3 | 1.14±1.32 | 0.15±0.16 | 9.88±1.33 | 2.56×10−10 |
| AC005307.1 | 1.34±0.65 | 0.42±0.65 | 8.26±0.92 | 4.06×10−13 |
| CASC9 | 1.33±1.07 | 0.49±0.38 | 7.02±1.14 | 3.37×10−11 |
| AC010776.2 | 1.29±1.38 | 0.49±0.37 | 6.32±1.43 | 2.13×10−7 |
| KCNQ5-IT1 | 0.92±0.90 | 0.14±0.16 | 6.05±0.91 | 7.34×10−13 |
| AC104463.2 | 1.12±0.80 | 0.36±0.32 | 5.79±0.86 | 1.19×10−13 |
| KEGG | 0.86±0.93 | 0.10±0.15 | 5.75±0.94 | 6.41×10−12 |
| AC010776.2 | 0.90±0.62 | 0.18±0.21 | 5.28±0.65 | 5.03×10−17 |
| AC005609.1 | 0.80±0.51 | 0.09±0.13 | 5.06±0.53 | 4.24×10−20 |
| HIF1A-AS2 | 1.03±0.79 | 0.32±0.31 | 5.01±0.85 | 2.83×10−12 |
| AC012414.5 | 0.83±0.63 | 0.13±0.10 | 5.01±0.63 | 4.43×10−17 |
| AP001993.1 | 1.00±0.81 | 0.30±0.25 | 4.98±0.85 | 2.79×10−12 |
| AL031772.1 | 1.06±0.47 | 0.37±0.28 | 4.94±0.55 | 6.62×10−20 |
| ERVMER61-1 | 1.21±0.66 | 0.52±0.36 | 4.90±0.76 | 1.03×10−13 |
| AL162464.2 | 0.91±0.43 | 0.24±0.18 | 4.78±0.47 | 1.10×10−21 |
| AC078789.1 | 0.86±0.67 | 0.19±0.22 | 4.75±0.70 | 3.99×10−15 |
| AC005062.1 | 1.17±0.61 | 0.49±0.21 | 4.72±0.64 | 1.47×10−16 |
| AC005609.3 | 0.84±0.93 | 0.18±0.15 | 4.57±0.94 | 3.00×10−9 |
| PSPC1-AS2 | 0.73±0.59 | 0.08±0.26 | 4.46±0.64 | 2.49×10−15 |
| AC007223.1 | 0.68±0.86 | 0.03±0.08 | 4.44±0.87 | 1.54×10−10 |
| AC108734.4 | 0.74±0.56 | 0.10±0.17 | 4.43±0.58 | 4.49×10−17 |
| AC121338.2 | 0.82±0.88 | 0.18±0.14 | 4.28±0.89 | 9.16×10−10 |
| LINC01305 | 1.75±0.57 | 1.12±0.35 | 4.27±0.66 | 6.39×10−16 |
| AC011825.2 | 1.08±0.58 | 0.45±0.17 | 4.23±0.60 | 2.35×10−15 |
a, P values were calculated by the paired t-test. FPKM, fragments per kilobase of exon per million fragments mapped; SD, standard deviation; PR, partial response, PD, progressive disease; SD, stable disease.
Table S3. Genes with >4-fold downregulated expression in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (PR/SD or PD), ± SD) |
P valuea | |
|---|---|---|---|---|
| PR | SD or PD | |||
| AC090692.1 | 2.07±1.17 | 3.61±0.35 | 35.14±1.22 | 2.25×10−18 |
| LINC02500 | 0.88±0.72 | 1.98±0.36 | 12.67±0.81 | 2.27×10−21 |
| LINC02525 | 1.00±0.69 | 2.06±0.39 | 11.57±0.80 | 2.49×10−22 |
| AC087260.1 | 2.29±1.06 | 3.35±0.24 | 11.04±1.09 | 4.64×10−14 |
| LINC01515 | 2.23±0.99 | 3.28±0.23 | 11.06±1.02 | 1.17×10−14 |
| LINC02506 | 0.89±0.63 | 1.93±0.29 | 11.00±0.70 | 1.20×10−23 |
| LINC02128 | 1.24±0.75 | 2.22±0.24 | 9.44±0.78 | 6.38×10−19 |
| LINC01896 | 0.97±0.73 | 1.94±0.36 | 9.34±0.82 | 2.30×10−18 |
| AL354993.2 | 0.91±0.62 | 1.86±0.27 | 8.94±0.67 | 3.74×10−22 |
| AC090692.1 | 0.99±0.55 | 1.88±0.25 | 7.80±0.60 | 4.57×10−24 |
| AC020659.1 | 1.04±0.54 | 0.83±0.59 | 6.21±0.79 | 1.72×10−15 |
| AC024610.2 | 0.93±0.61 | 1.72±0.44 | 6.18±0.75 | 4.39×10−16 |
| AP005263.1 | 1.35±0.95 | 2.11±1.02 | 5.70±1.40 | 3.94×10−6 |
| AL450311.1 | 1.43±0.63 | 2.19±0.30 | 5.70±0.70 | 2.28×10−17 |
| AC023794.1 | 0.70±0.48 | 1.36±0.33 | 4.51±0.59 | 5.49×10−16 |
| AL359317.1 | 1.63±0.52 | 2.29±0.34 | 4.48±0.62 | 6.78×10−18 |
| AC079610.2 | 0.32±0.34 | 0.97±0.30 | 4.47±0.45 | 9.94×10−23 |
| AC105206.1 | 1.79±0.55 | 2.44±0.41 | 4.46±0.69 | 2.29×10−12 |
| AC027117.1 | 0.92±0.46 | 1.57±0.24 | 4.44±0.52 | 1.74×10−19 |
| AP003500.1 | 1.22±0.66 | 1.87±0.48 | 4.42±0.81 | 2.82×10−11 |
| AC020659.1 | 1.33±0.52 | 1.87±0.48 | 4.42±0.69 | 9.49×10−15 |
| AC024610.2 | 1.08±0.58 | 1.72±0.22 | 4.41±0.62 | 1.53×10−15 |
a, P values were calculated by the paired t-test; b, P values were adjusted with the Benjamini-Hochberg algorithm. FPKM, fragments per kilobase of exon per million fragments mapped; SD, standard deviation; PR, partial response; SD, stable disease; PD, progressive disease.
According to the tumor stage, we also identified differential gene expression. The data are reported in the Table S4. Keratin family members were overexpressed only in LD cases.
Table S4. The top 10 genes upregulated in ED stage tissue compared to LD stage tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (ED/LD, ± SD) | P valuea | ||
|---|---|---|---|---|---|
| ED | LD | ||||
| KRT6A | 2.28±1.29 | 0.18±0.16 | 127.76±1.30 | 5.905×10−11 | |
| KRT5 | 2.28±0.95 | 0.23±0.41 | 57.09±1.03 | 3.40×10−12 | |
| KRT17 | 1.63±0.96 | 0.12±0.18 | 32.56±0.98 | 5.00×10−11 | |
| AC008870.1 | 1.61±0.93 | 0.19±0.14 | 26.60±0.84 | 4.79×10−12 | |
| KRT16 | 1.61±0.93 | 0.31±0.24 | 20.28±0.96 | 8.58×10−10 | |
| AC091564.5 | 1.96±0.65 | 0.71±0.46 | 18.06±0.80 | 1.08×10−13 | |
| LINC00498 | 1.28±0.65 | 0.05±0.13 | 17.22±0.95 | 4.15×10−9 | |
| AC009137.2 | 1.21±0.79 | 0.02±0.02 | 15.60±0.79 | 3.20×10−10 | |
| AC121247.1 | 1.57±0.80 | 0.39±0.35 | 15.25±0.88 | 2.88×10−9 | |
| DAPK1-IT1 | 1.52±0.89 | 0.35±0.34 | 14.67±0.95 | 1.20×10−8 | |
a, P values were calculated by the paired t-test. ED, extensive disease; LD, limited disease; FPKM, fragments per kilobase of exon per million fragments mapped; SD, standard deviation.
An additional analysis for the associated genes of metastasis status is also presented. No significant correlation was found between metastasis and non-metastasis.
miRNA differential expression
Overall, 1,303 genes were identified as differentially expressed with ≥2-fold changes and P<0.05, including 520 higher expressed genes and 783 lower expressed genes with ≥2-fold changes in PR compared to SD or PD. The top 10 most changed genes (upregulated and downregulated) in SD or PD tissue are displayed in Tables 5,6, respectively. Two downregulated genes, miRNA 601 and miRNA 596, exhibited the maximum difference between the PR and SD or PD group. Genes with ≥4-fold changes are shown in Tables S5,S6.
Table 5. The top 10 miRNAs upregulated in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (SD or PD/PR, ± SD) | P valuea | |
|---|---|---|---|---|
| SD or PD | PR | |||
| hsa-miRNA 601 | 1.57±0.32 | 1.64±0.50 | 6.24±0.61 | 1.18×10−19 |
| hsa-miRNA 596 | 0.93±0.56 | 1.05±0.54 | 6.21±0.79 | 1.62×10−15 |
| hsa-miR-615-5p | 1.78±0.46 | 0.95±0.61 | 6.08±0.75 | 4.29×10−16 |
| hsa-miR-224-5p | 1.75±0.24 | 0.91±0.56 | 6.07±0.61 | 1.44×10−20 |
| hsa-miR-6718-5p | 1.37±0.48 | 0.57±0.55 | 6.15±0.72 | 2.87×10−19 |
| hsa-miR-3616-3p | 2.11±0.64 | 1.32±0.78 | 6.11±0.95 | 8.07×10−13 |
| hsa-miR-504-5p | 1.32±0.29 | 0.54±0.49 | 6.01±0.57 | 2.29×10−23 |
| hsa-miR-3180-5p | 1.72±0.30 | 0.94±0.65 | 5.96±0.71 | 2.68×10−16 |
| hsa-miR-4802-3p | 1.64±0.23 | 1.65±0.63 | 5.73±0.58 | 3.01×10−20 |
| hsa-miR-1260a | 2.13±1.02 | 1.05±0.95 | 5.10±1.40 | 3.34×10−6 |
a, P values were calculated by the paired t-test. SD, standard deviation; PR, partial response; SD, stable disease; PD, progressive disease; FPKM, fragments per kilobase of exon per million fragments mapped.
Table 6. The top 10 miRNAs downregulated in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (PR/SD or PD, ± SD) | P valuea | |
|---|---|---|---|---|
| SD or PD | PR | |||
| hsa-miR-6073 | 2.43±0.63 | 1.19±0.30 | 5.73±0.70 | 3.28×10−17 |
| hsa-miR-3152-5p | 2.95±0.87 | 1.70±0.13 | 5.59±0.88 | 8.10×10−12 |
| hsa-miR-4652-5p | 2.04±0.72 | 1.37±0.67 | 5.56±0.97 | 1.79×10−10 |
| hsa-miR-3162-3p | 1.02±0.53 | 1.86±0.28 | 5.51±0.57 | 2.44×10−22 |
| hsa-miR-3607-5p | 0.94±0.61 | 1.64±0.31 | 5.47±0.67 | 6.87×10−17 |
| hsa-miR-135a-3p | 0.74±0.45 | 1.53±0.24 | 5.36±0.51 | 1.23×10−21 |
| hsa-miR-3617-5p | 1.39±0.78 | 2.41±0.44 | 5.27±0.99 | 8.01×10−11 |
| hsa-miR-758-5p | 0.62±0.60 | 1.74±0.30 | 5.21±0.66 | 2.71×10−17 |
| hsa-miR-4301 | 0.44±0.42 | 1.25±0.36 | 5.20±0.55 | 1.60×10−19 |
| hsa-miR-3653-5p | 0.39±0.42 | 1.50±0.28 | 5.01±0.50 | 6.31×10−24 |
a, P values were calculated by the paired t-test. SD, standard deviation; PR, partial response; SD, stable disease; PD, progressive disease; FPKM, fragments per kilobase of exon per million fragments mapped.
Table S5. miRNAs with >4-fold upregulated expression in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (SD or PD/PR, ± SD) | P valuea | ||
|---|---|---|---|---|---|
| SD or PD | PR | ||||
| hsa-miRNA 601 | 1.47±0.35 | 0.67±0.50 | 6.28±0.61 | 1.08×10−19 | |
| hsa-miRNA 596 | 0.83±0.59 | 1.04±0.54 | 6.21±0.79 | 1.72×10−15 | |
| hsa-miR-615-5p | 1.72±0.44 | 0.93±0.61 | 6.18±0.75 | 4.39×10−16 | |
| hsa-miR-224-5p | 1.71±0.24 | 0.92±0.56 | 6.17±0.61 | 1.14×10−20 | |
| hsa-miR-6718-5p | 1.36±0.47 | 0.58±0.55 | 6.14±0.72 | 2.07×10−19 | |
| hsa-miR-3616-3p | 2.12±0.54 | 1.33±0.78 | 6.10±0.95 | 8.67×10−13 | |
| hsa-miR-504-5p | 1.31±0.29 | 0.53±0.49 | 6.02±0.57 | 2.39×10−23 | |
| hsa-miR-3180-5p | 1.71±0.30 | 0.93±0.65 | 5.95±0.71 | 2.48×10−16 | |
| hsa-miR-4802-3p | 1.91±0.23 | 1.15±0.53 | 5.73±0.58 | 3.11×10−20 | |
| hsa-miR-1260a | 2.11±1.02 | 1.35±0.95 | 5.70±1.40 | 3.94×10−6 | |
| hsa-miR-1260b | 0.83±0.75 | 0.20±0.37 | 4.35±0.83 | 1.85×10−10 | |
| hsa-miR-7975 | 1.61±0.22 | 0.97±0.43 | 4.30±0.49 | 1.09×10−20 | |
| hsa-miR-1294 | 0.96±0.23 | 0.33±0.34 | 4.30±0.41 | 3.12×10−23 | |
| hsa-miR-561-3p | 1.24±0.53 | 0.61±0.53 | 4.30±0.75 | 4.15×10−13 | |
| hsa-miR-20b-3p | 0.98±0.30 | 0.35±0.40 | 4.24±0.50 | 5.68×10−19 | |
| hsa-miR-204-5p | 0.91±0.30 | 0.28±0.33 | 4.23±0.45 | 7.43×10−21 | |
| hsa-miR-5584-5p | 1.24±0.24 | 0.62±0.41 | 4.22±0.48 | 3.26×10−24 | |
| hsa-miR-452-5p | 1.37±0.36 | 0.75±0.42 | 4.20±0.55 | 6.36×10−18 | |
| hsa-miR-4501 | 1.05±0.36 | 0.42±0.38 | 4.20±0.52 | 1.60×10−18 | |
a, P values were calculated by the paired t-test. SD, standard deviation; PR, partial response; PD, progressive disease; FPKM, fragments per kilobase of exon per million fragments mapped.
Table S6. miRNAs with >4-fold downregulated expression in SD or PD tissue compared to PR tissue.
| Gene symbol | FPKM (mean ± SD) | Mean fold change (PR/SD or PD, ± SD) | P valuea | ||
|---|---|---|---|---|---|
| SD or PD | PR | ||||
| hsa-miR-6073 | 1.43±0.63 | 2.19±0.30 | 5.70±0.70 | 2.28×10−17 | |
| hsa-miR-3152-5p | 1.95±0.87 | 2.70±0.13 | 5.58±0.88 | 8.00×10−12 | |
| hsa-miR-4652-5p | 1.04±0.70 | 1.77±0.67 | 5.46±0.97 | 1.89×10−10 | |
| hsa-miR-3162-3p | 1.02±0.50 | 1.76±0.28 | 5.41±0.57 | 2.54×10−22 | |
| hsa-miR-3607-5p | 0.91±0.60 | 1.64±0.31 | 5.37±0.67 | 6.77×10−17 | |
| hsa-miR-135a-3p | 0.70±0.45 | 1.43±0.24 | 5.35±0.51 | 1.26×10−21 | |
| hsa-miR-3617-5p | 1.59±0.88 | 2.31±0.44 | 5.24±0.99 | 8.02×10−11 | |
| hsa-miR-758-5p | 0.92±0.60 | 1.64±0.30 | 5.23±0.66 | 2.76×10−17 | |
| hsa-miR-4301 | 0.43±0.42 | 1.15±0.36 | 5.22±0.55 | 1.30×10−19 | |
| hsa-miR-3653-5p | 0.59±0.42 | 1.30±0.28 | 5.21±0.50 | 6.41×10−24 | |
| hsa-miR-1251-5p | 0.78±0.53 | 1.38±0.24 | 4.06±0.58 | 3.62×10−15 | |
| hsa-miR-2681-5p | 0.43±0.40 | 1.04±0.40 | 4.06±0.57 | 1.09×10−19 | |
| hsa-miR-509-3-5p | 1.25±0.46 | 1.85±0.17 | 4.05±0.49 | 3.45×10−18 | |
| hsa-miR-1246 | 1.66±0.66 | 2.26±0.17 | 4.00±0.68 | 1.08×10−12 | |
| hsa-miR-1271-3p | 2.00±0.92 | 2.60±0.19 | 4.00±0.94 | 2.90×10−8 | |
a, P values were calculated by the paired t-test. SD, standard deviation; PR, partial response; SD, stable disease; PD, progressive disease; FPKM, fragments per kilobase of exon per million fragments mapped.
We also compared genes with differential expression on the basis of tumor stage. There was no significant correlation between LD and ED stage.
Analysis of GO and pathway enrichment
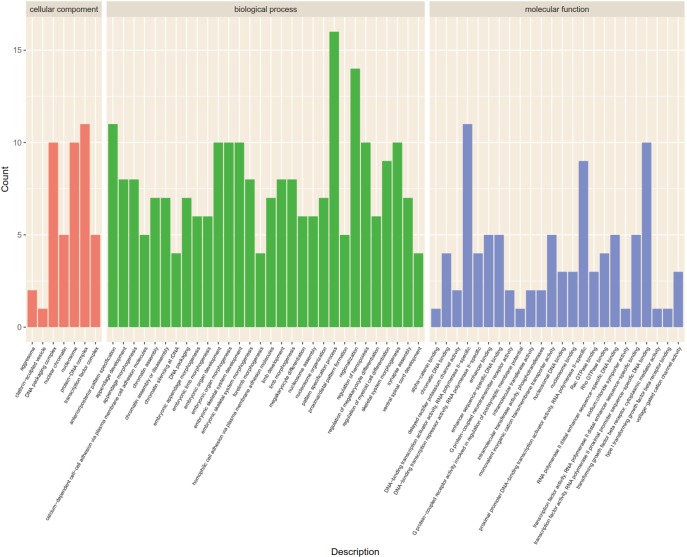
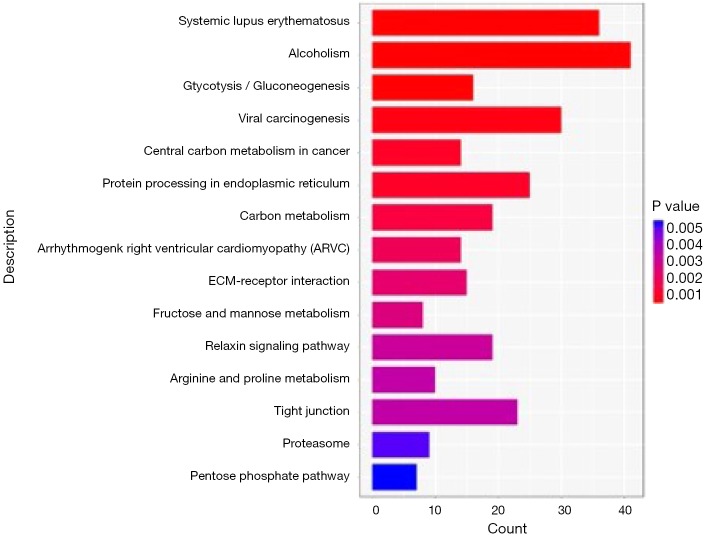
The Gene Ontology biological processes (GO-BP) and pathway enrichment studies for differential mRNAs in the SCLC PR and SD or PD groups were performed. The lower expressed PR gene GO-BP terms were mainly related to nucleosome, DNA packaging complex, protein-DNA complex, and other functions (Table 7 and Figure 1). Systemic lupus erythematosus and alcoholism event were enriched in the SD or PD group (Table 8 and Figure 2). In particular, LIME1, LAT, SLA2, GBP1, DEFB4A, and GBP2, which were overexpressed in the PR group, were significantly involved in immunological function.
Table 7. The enriched CO-BP terms of differentially co-expressed mRNAs.
| Term | Site | P |
|---|---|---|
| GO:0000786 | Nucleosome | 2.31E-21 |
| GO:0044815 | DNA packaging complex | 4.31E-20 |
| GO:0032993 | Protein-DNA complex | 4.01E-16 |
| GO:0000788 | Nuclear nucleosome | 2.18E-09 |
| GO:1904813 | Ficolin-1-rich granule lumen | 3.39E-09 |
| GO:0005788 | Endoplasmic reticulum lumen | 4.83E-07 |
| GO:0000786 | Nucleosome | 2.31E-21 |
GO-BP, Gene Ontology biological processes.
Figure 1.
GO blot of differentially expressed mRNAs between the PR and SD/PD group. GO, Gene Ontology; PR, partial response; SD, stable disease; PD, progressive disease.
Table 8. The KEGG enriched pathways of differentially expressed mRNAs.
| Pathway Name | rawP |
|---|---|
| Systemic lupus erythematosus | 3.41E-12 |
| Alcoholism | 9.30E-11 |
| Glycolysis/gluconeogenesis | 2.66E-05 |
| Viral carcinogenesis | 0.000166396 |
| Central carbon metabolism in cancer | 0.000445778 |
| Systemic lupus erythematosus | 3.41E-12 |
P values were calculated by hypergeometric test (rawP).
Figure 2.
KEGG pathway bar-blot of differentially expressed mRNAs between the PR and SD or PD group. PR, partial response; SD, standard deviation; PD, progressive disease.
Discussion
Previous research has discovered that aberrant lncRNA and miRNA levels are related to the metastasis, invasion, and chemo-insensitivity of cancer (13,14). Furthermore, lncRNAs and miRNAs have been found to generate in multiple abnormal pathways, contributing to the development of chemo-insensitivity. Through RNA-Seq, we confirmed the genes that were differentially expressed between SCLC cases gaining PR and cases that were SD or PD after first-line chemotherapy.
We identified the lncRNA difference between the SCLC cases with SD or PD and PR. HIF1A-AS2 was shown to be upregulated in the SD or PD group, while CYP4F26P, a biomarker of cancer, which is involved in the proliferation and invasiveness in SCLC, was also upregulated in the SD or PD group in our study. The expression levels of PSPC1-AS2, DLGAP1-AS4, DAPK1-IT1, AQP4-AS1, and KIRREL3-AS3 were upregulated in resistant samples. The RNA-Seq data were partially related to previous reports in lung cancer (15-17).
It is widely acknowledged that microarray analyses can identify changes in gene networks or those genes responsible for physiological and pathological cases. However, RNA-Seq has many capabilities beyond microarray analysis. By using a method called cross-hybridization, microarray analyses may cause hybridization artifacts that do not uniformly affect expression measures. Hundreds of millions or even billions of RNA fragments can be analyzed by RNA-Seq. Thus, this method possesses not only a higher sensitivity but also greater dynamic reading range than microarray analyses. These characteristics may amplify the likelihood of discovering novel biomarkers.
This study found that HOXA-AS3 RNA expression levels were increased in SD or PD tissues. HOXA-AS3 is one of HOX family genes. HOX is a cluster that regulates embryological development with highly homologous transcription factors. Also, this group can regulate differentiation and hematopoietic lineage. There are only 2 studies that have reported on the function of HOXA-AS3, as it is a relatively novel lncRNA. In glioma, the high-expression of HOXA-AS3 predicted poor prognosis (18), while other family members of the HOXA were correlated with lung cancer cell proliferation (19). However, there is no report on the role of HOXA-AS3 in SCLC, and the mechanism behind HOXA-AS3’s function as an antisense transcript is poorly understood. It has been speculated that HOXA-AS3 may promote cancer insensitivity by regulating cell proliferation, but more research is needed to clarify the biological functions of HOXA-AS3.
CASC9 was identified as another insensitivity biomarker in our study. CASC9 has a length of 1,316 bp and is located on chromosome 8. Recently, next generation sequencing (NGS) analysis has shown that CASC9 is a noncoding proto-oncogene, and is related to lung cancer. Specifically, Shang et al. found that the expression of CASC9 was significantly up-regulated in gastric cancer, while the higher expression of CASC9 was associated with poor prognosis in nasopharyngeal carcinoma (20). Functional experimentation demonstrated that CASC9 could promote nasopharyngeal carcinoma cell growth via stabilizing HIF1α. Wu et al. reported that the higher expression of CASC9 was associated with poor prognosis in esophageal squamous cell carcinoma (ESCC) patients (21). Meanwhile, CASC9 was found to serve as an oncogene by negatively regulating PDCD4 expression through recruiting EZH2 (18). In short, CASC9 induces cancer development. Although functional assays have shown that CASC9 contributes to the development and progression of cancer, its mechanisms remain unclear, and the function of CASC9 in SCLC needs to be explored further.
The cell cycle, DNA, and nucleotide metabolism-associated pathways that are generally upregulated in SCLC patients with SD or PD were analyzed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. GO identified the upregulation in the metabolic process of macromolecules, but the immunologic pathway investigated by KEGG analysis has not been studied previously for SCLC.
The nucleosome signaling pathways (GO:0000786), were revealed by GO analysis. Nuclear nucleosome signaling pathways (GO:0000788) and nucleosome signaling pathways (GO:0000786) were downregulated in the PR group. Downregulated systemic lupus erythematosus pathways were also found in the PR group by KEGG analysis. In particular, alcoholism event developmental processes were downregulated in the SCLC PR group, implying a relation between SCLC chemotherapy insensitivity and neuron-associated developmental processes. Semaphorin-3B (SEMA3B) and Kallman syndrome 1 sequence (KAL1) were significantly downregulated in the PR group in the above-mentioned pathways. Further studies are required to clarify the roles of these genes in SCLC chemotherapy insensitivity.
Conclusions
This study used RNA-Seq to identify the lncRNAs and miRNAs which are differentially regulated in SCLC PR patients compared with SD or PD patients after first-line chemotherapy. The main roles of these genes and miRNAs in SCLC first-line chemotherapy insensitivity should be illuminated in further studies.
Acknowledgments
Funding: This study was supported in part by grants from National Natural Science Foundation of China (81802255), the Shanghai Pujiang Program (17PJD036), the Shanghai Municipal Commission of Health and Family Planning Program (20174Y0131), the National Key Research & Development Project (2016YFC0902300), the Major Disease Clinical Skills Enhancement Program of Three-year Action Plan for Promoting Clinical Skills and Clinical Innovation in Municipal Hospitals, the Shanghai Shen Kang Hospital Development Center Clinical Research Plan of SHDC (16CR1001A), the “Dream Tutor” Outstanding Young Talents Program (fkyq1901), the Key disciplines of Shanghai Pulmonary Hospital (2017ZZ02012), and the Shanghai Science and Technology Commission (16JC1405900).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by Shanghai Pulmonary Hospital ethics committee (No. 18151). All participants gave their informed consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Farago AF, Keane FK. Current standards for clinical management of small cell lung cancer. Transl Lung Cancer Res 2018;7:69-79. 10.21037/tlcr.2018.01.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park CK, Oh IJ, Kim YC. Is transformed small cell lung cancer (SCLC) different from de novo SCLC? Transl Cancer Res 2019;8:346-9. 10.21037/tcr.2019.03.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarvi S, Mackinnon AC, Avlonitis N, et al. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res 2014;74:1554-65. 10.1158/0008-5472.CAN-13-1541 [DOI] [PubMed] [Google Scholar]
- 4.Tsoukalas N, Aravantinou-Fatorou E, Baxevanos P, et al. Advanced small cell lung cancer (SCLC): new challenges and new expectations. Ann Transl Med 2018;6:145. 10.21037/atm.2018.03.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Su W, Chen X, et al. Validation of a serum 4-microRNA signature for the detection of lung cancer. Transl Lung Cancer Res 2019;8:636-48. 10.21037/tlcr.2019.09.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 7.Zhang A, Xu M, Mo YY. Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol 2014;6:181-91. 10.1093/jmcb/mju013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao H, Yang JJ, Zhou X, et al. Emerging role of long noncoding RNAs in lung cancer: Current status and future prospects. Respir Med 2016;110:12-9. 10.1016/j.rmed.2015.10.006 [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 10.Bandyopadhyay S, Mitra R, Maulik U, et al. Development of the human cancer microRNA network. Silence 2010;1:6. 10.1186/1758-907X-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, Yang Y, Kuang P, et al. Seven-microRNA panel for lung adenocarcinoma early diagnosis in patients presenting with ground-glass nodules. Onco Targets Ther 2017;10:5915-26. 10.2147/OTT.S151432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu G, Li J, Chu J, et al. 1,25(OH)2D3 deficiency increases TM40D tumor growth in bone and accelerates tumor-induced bone destruction in a breast cancer bone metastasis model. Biomed Pharmacother 2017;95:1033-9. 10.1016/j.biopha.2017.09.018 [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Yang B, Zhang M, et al. lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer. Cancer Cell 2018;33:706-20.e9. 10.1016/j.ccell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu HS, Somvanshi S, Patel E, et al. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep 2018;23:297-312.e12. 10.1016/j.celrep.2018.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeh HW, Hsu EC, Lee SS, et al. PSPC1 mediates TGF-beta1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat Cell Biol 2018;20:479-91. 10.1038/s41556-018-0062-y [DOI] [PubMed] [Google Scholar]
- 16.Lin Q, Geng J, Ma K, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol 2009;135:1675-84. 10.1007/s00432-009-0614-4 [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, Wen X, Jiang Z, et al. Aquaporin 1 and aquaporin 4 are involved in invasion of lung cancer cells. Clin Lab 2012;58:75-80. [PubMed] [Google Scholar]
- 18.Wu Y, Hu L, Liang Y, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer 2017;16:150. 10.1186/s12943-017-0715-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Liu Y, Yan L, et al. Increased levels of the long noncoding RNA, HOXA-AS3, promote proliferation of A549 cells. Cell Death Dis 2018;9:707. 10.1038/s41419-018-0725-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su X, Li G, Liu W. The Long Noncoding RNA Cancer Susceptibility Candidate 9 Promotes Nasopharyngeal Carcinogenesis via Stabilizing HIF1alpha. DNA Cell Biol 2017;36:394-400. 10.1089/dna.2016.3615 [DOI] [PubMed] [Google Scholar]
- 21.Gao GD, Liu XY, Lin Y, et al. LncRNA CASC9 promotes tumorigenesis by affecting EMT and predicts poor prognosis in esophageal squamous cell cancer. Eur Rev Med Pharmacol Sci 2018;22:422-9. [DOI] [PubMed] [Google Scholar]