Abstract
Here we present a blood-brain barrier (BBB) model that enables high-resolution imaging of nanoparticle (NP) interactions with endothelial cells and the capture of rare NP translocation events. The enabling technology is an ultrathin silicon nitride (SiN) membrane (0.5 μm pore size, 20% porosity, 400 nm thickness) integrated into a dual-chamber platform that facilitates imaging at low working distances (~50 μm). The platform, the μSiM-BBB (microfluidic Silicon Membrane-BBB), features human brain endothelial cells and primary astrocytes grown on opposite sides of the membrane. The human brain endothelial cells form tight junctions on the ultrathin membranes and exhibit a significantly higher resistance to FITC-dextran diffusion than commercial membranes. The enhanced optical properties of the SiN membrane allow for high-resolution live-cell imaging of three types of NPs, namely 40 nm PS-COOH, 100 nm PS-COOH, and apolipoprotein E (ApoE)-conjugated 100 nm SiO2, interacting with the BBB. Despite the excellent barrier properties of the endothelial layer, we are able to document rare NP translocation events of NPs localized to lysosomal compartments of astrocytes on the ‘brain side’ of the device. Although the translocation is always low, our data suggest that size and targeting ligand are important parameters for NP translocation across the BBB. As a platform that enables the detection of rare transmission across tight-BBB layers, the μSiM-BBB is as an important tool for the design of nanoparticle-based delivery of drugs to the CNS.
Keywords: blood-brain barrier, co-culture, live-cell imaging, nanoparticle, ultrathin silicon nitride membrane
Graphical Abstract
The size and controllable surface properties of nanosized materials potentially present special opportunities to overcome physiological barriers by active biological trafficking processes.1,2 For the blood-brain barrier (BBB), where clinical successes have been frustratingly limited for decades, the potential of nanoscale therapies to actively traffic and deliver therapies would attract particular attention.3-6 While a wide variety of efforts have been reported,7-24 progress towards a clinical outcome has been slower than hoped for. Thus, increasingly researchers investigating the potential of nanoscale BBB-related therapies have become convinced that real progress in understanding the detailed mechanisms of interaction between nanoparticles (NPs) and barriers may be a prerequisite for critical advances to occur in the field. However, progress there also has been hampered because the research tools (both in vitro and in vivo) have generally not been tailored for NPs and restrict our ability to probe detailed mechanisms. Indeed, the unmodified use of some existing tools even has the potential to confound understanding, and there is now a need for rethinking of the strategy. It is worthy noting that to this day disagreement remains between some scientists as to the extent of truly active (transcytosis driven) barrier crossing even in cell models, let alone in vivo. Here we describe a methodological approach and model that addresses these problems.
The key problem is that for most NP systems, the efficiency of barrier crossing is so low that active crossings typically appear as ‘rare events’.25 Furthermore, many of the cellular barrier models upon which such studies rely possess quite high densities of defects ranging from incomplete cell-cell junction to cell-sized holes, and NP crossing via those represent a significant background to genuine active crossings.25,26 Thus while, in our opinion, there is little doubt that a very small fraction of true active barrier crossings do occur, it is difficult to identify and study them. Certainly, imaging studies represent a credible strategy of picking out such key events from the background and unraveling the specific processes that lead to those events,10,25 but there the methods currently available have significant limitations.
For instance, endothelial cells and astrocyte in 2D contact BBB co-cultures growing on multiwell plates with microporous membrane filters (e.g. Transwell® or ThinCert™ inserts) may be at a distance of 10 - 50 μm depending on the thickness of membranes and collagen coating layer, several orders of magnitude larger than their spacing in vivo (basement membranes that separate endothelial cells and astrocytes are ~40 - 80 nm27). Not only does this exclude many near-cell contacts and paracrine effects28 (which we consider may be highly significant mechanistically), but the materials used also typically lead to non-specific particle-pore associations affecting the transmission of particles across the model barrier.25,26,29
Conventional polymeric membranes are also incompatible with in situ high-resolution imaging using optical microscopy because of light scattering and autofluorescence from the support material.30 By contrast, ultrathin silicon-based ‘nanomembranes’ provide optical transparency and high permeability to enhance endogenous intercellular communications while enabling direct in situ image analysis.28,30-35 Studies have shown that many types of cells can grow on nanomembranes with normal proliferation and viability and appropriate markers of cell differentiation,31,36,37 as both monocultures31,33,38,39 and co-cultures,31,32,34,40 including an early attempt to make a BBB co-culture model.37
We recently reported the use of ultrathin (50 nm thick) nanoporous silicon nitride (NPN) membranes integrated into a dual-chamber “Transwell® mimetic” for the visualization of T-cell migration across a monoculture model of the BBB.41 The device, which we called the μSiM-CVB (microfluidic silicon membrane cerebrovascular barrier), represents an important advance because it provides a live-cell imaging platform for the study of a fundamental mechanism in multiple sclerosis. In a similar fashion, here we hypothesized that ultrathin silicon nanomembrane technologies would enable the visualization of rare NP translocation events across a live BBB model. To distinguish the current work from that of Mossu et al.,41 we refer to the model used here as the μSiM-BBB.
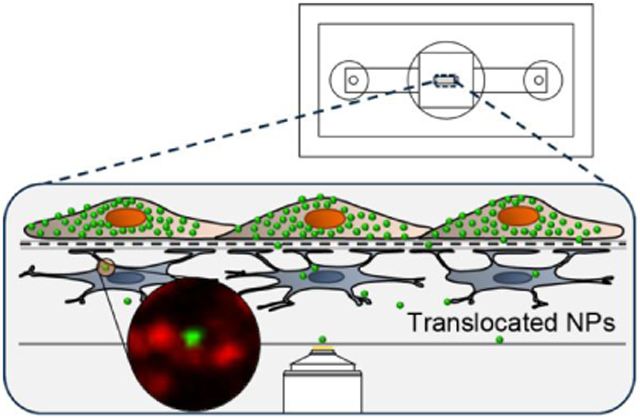
The μSiM-BBB employs photolithographically patterned 0.5 μm diameter pores in ultrathin (400 nm thick), highly porous (20% porosity), silicon nitride (SiN) and a co-culture of immortalized human brain capillary endothelial hCMEC/D3 cells42 and primary human normal astrocytes (NHA) grown in juxtaposition on the opposite side of the SiN membrane. In comparison to an equivalent Transwell® model, the optical visualization by transmitted light and fluorescence of both endothelial cells and astrocytes was significantly enhanced in the μSiM-BBB. In addition, the μSiM-BBB demonstrated an elevated barrier compared to Transwell® system in a permeability assay. Results confirmed that the hCMEC/D3 cells express claudin-5 tight junction protein formed a confluent monolayer, and NHAs expressed astrocytic marker glial fibrillary acidic protein (GFAP) on collagen/fibronectin-coated SiN membranes. Furthermore, we employed high-resolution three-dimensional live-cell imaging to analyze NP translocation across the barrier in situ. While the overwhelming number of NPs become trapped in the endothelial cells, the platform allows us to identify rare translocation events across well-formed BBBs. We found that NPs with different size and surface targeting moiety exhibited significant differences in their ability to translocate across the endothelial cells and reach the ‘brain side’ astrocyte layer. Given the challenges of designing NPs that can successfully deliver drugs into the CNS, these results suggest that the nanomembrane platform will be useful for identifying and amplifying those characteristics of NPs that can enable successful translocation of drug carriers into the BBB.
RESULTS AND DISCUSSION
Co-culture device prototype
The μSiM co-culture device used in this paper is a custom-designed prototype that is fabricated in-house, and it consists of two main components: a silicone-based housing and a highly porous SiN membrane chip (Figure 1). The SiN membrane (2 mm x 0.7 mm) has 0.5 μm pore size and 400 nm thickness and is supported on a 5.4 mm square silicon chip with one or three freestanding membrane windows. The silicone-housing consists of precision-cut silicone gaskets and a hole-punched polydimethylsiloxane (PDMS) block. The height of the silicone gasket is 50 μm (i.e. the membrane is in 50 μm distance from the bottom cover glass) that allows the use of high magnification water and oil immersion objectives. The PDMS block consists of two smaller, hole-punched PDMS blocks (Figure 1a). The top PDMS block contains one 7 mm hole and two 5 mm holes. The bottom PDMS block contains one 7 mm hole and two 840 μm holes. The 7 mm holes of the top and the bottom PDMS block forms a continuous volume to host cells on top of the SiN membrane. The 840 μm holes allow the insertion of a p200 pipette tip to introduce cells into the bottom channel. The 5 mm holes on top of the 840 μm holes host additional media volume for long-term culture. All components of the housing, the membrane chip, and the #1.5 cover glass that forms the floor of the bottom channel are covalently-bonded together using ozone plasma treatment.43
Figure 1.
Nanomembrane cell culture device. (a) Layer-by-layer schematic of the co-culture device. The device consisted mainly of two components: the silicone-based housing that hosts the membrane chip and the membrane chip itself. The larger well (diameter: 7 mm) of the top PDMS block hosts the media for long-term cell culture and allows direct seeding of the human brain endothelial cells onto the top side of the membrane. The two smaller wells (diameter: 5 mm) are connected to the bottom channel where the astrocytes were grown and provide the additional media volume. (b) The top view highlighting the chip in the center of the larger well, and the two smaller wells on the side with access-holes to the bottom channels. (c) Inset shows scanning electron micrographs that highlight the feature of the microporous SiN membrane. The scale bars represent 10 μm.
Enhanced imaging properties of the μSiM-BBB
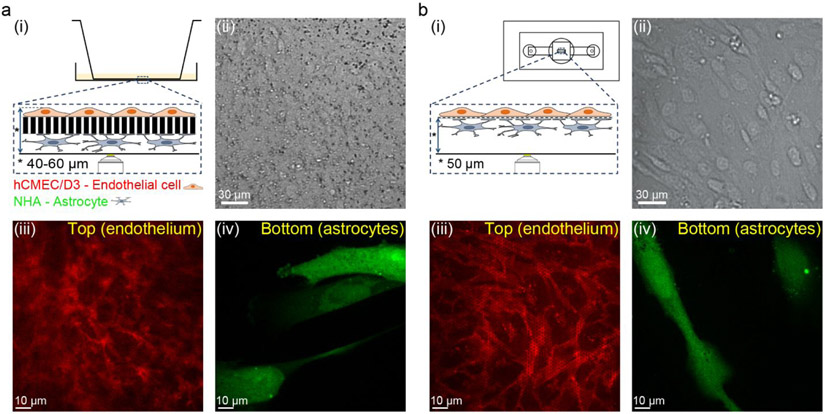
Immortalized human brain capillary endothelial hCMEC/D3 cells and primary human NHA astrocytes were cultured on either side of a collagen/fibronectin-coated SiN membrane in the μSiM device to create the μSiM-BBB model. For comparison, the equivalent in vitro co-culture model was set up on a Transwell® system, where the hCMEC/D3 and NHA cells were cultured on a 10 μm thick polyester membrane with a pore size of 0.4 μm (Figure S1). We compared live-cell imaging of the BBB co-culture based on the SiN or polyester track-etched membranes using transmitted light microscopy and high-resolution fluorescence microscopy. For the imaging analysis, the Transwell® insert was directly placed into a glass bottom imaging dish filled with cell culture medium and imaged using an inverted microscope (Figure 2a (i)). The μSiM-BBB has a built-in #1.5 cover glass at the bottom of the device that allows the direct mounting of the device to the stage of an inverted microscope, i.e. co-culture can be imaged in situ (Figure 2b (i)). The images were obtained using a 20x magnification objective. It is apparent that under transmitted light, the cells can only be clearly visualized through the transparent SiN (Figure 2a (ii) and Figure 2b (ii)). For the fluorescence live-cell imaging, the hCMEC/D3 and NHA cells were stained with CellMask™ Orange plasma membrane dye (red) and CellTracker™ Green cell body dye (green), respectively. Images were obtained using spinning disk confocal microscopy with a 60x magnification objective. In the Transwell® model, the astrocytes were readily visualized, as they were directly placed on the cover glass (Figure 2a (iv)). However, the imaging quality of the endothelial cells on the Transwell® membrane was significantly compromised due to the thickness and poor optical properties (e.g. autofluorescence and light scattering) of the membrane Figure 2a (iii)). In contrast, it was straightforward to obtain sharp and focused images of both astrocytes (Figure 2b (iv)) and endothelial cells (Figure 2a (iii)) in the μSiM-BBB. This finding is in good agreement with other studies comparing ultrathin silicon-based membranes to commercial track-etched membranes for cell microscopy.30,31,34,39 Taken together, the transparent SiN membrane provides enhanced optical properties to allow cell visualization in situ by both transmitted light and high-resolution fluorescence microscopy. This greatly facilitates researchers to directly monitor cell growth, morphology, and spatial organization of the BBB co-culture in real time.
Figure 2.
Enhanced optical properties of the μSiM-BBB co-culture model in comparison to the Transwell® BBB model. Schematic illustration of the imaging approach of the in vitro BBB co-culture model grown on Transwell® insert and on the μSiM-BBB (a (i) and b (i)). Transmitted light micrographs of in vitro BBB co-culture model grown on 10 μm thick polyester membrane (0.4 μm pore size) and on 400 nm thick SiN membrane (0.5 μm pore size) (a (ii) and b (ii), respectively). 2D confocal micrographs of the apical (top, endothelial cells) and basal (bottom, astrocytes) side of the in vitro BBB co-culture model grown on traditional 0.4 μm pore size, 10 μm thick polyester membrane (a (iii) and b (iv)) and on 0.5 μm pore size, 400 nm thick SiN membrane (b (iii) and b (iv)). The hCMEC/D3 cells are labeled by 7.5 μg/mL CellMask™ Orange (red) and NHA cells are labeled by 25 μM CellTracker™ Green CMFDA (green). Image acquisition: 20x air objective (transmitted light micrographs) and 60x oil objective (fluorescent images).
Enhanced barrier characteristics of the μSiM-BBB
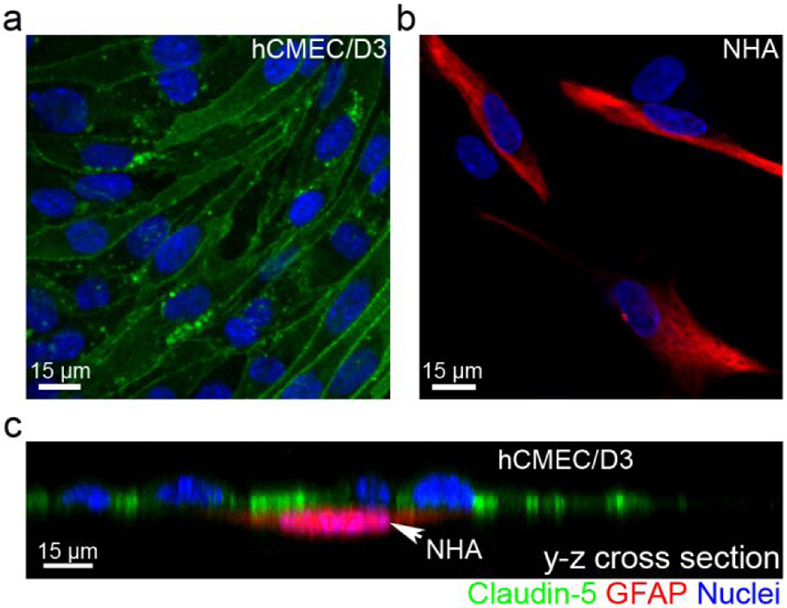
Next, we sought to examine the barrier property of the μSiM-BBB. In a functional BBB, endothelial cells express tight junction, adherens junction proteins, and junctional adhesion molecules, such as claudins, occludins, and zonula occludens,44 to form a tight physical barrier that only allows small water-soluble compounds to pass. To analyze the formation of cell adhesion by endothelial cells in the BBB co-culture, cells were directly fixed, stained with anti-claudin-5 tight junction protein antibody, and imaged in the μSiM-BBB (Figure 3a). One of the best-known astrocyte markers, GFAP, was immunostained to identify the astrocytes (Figure 3b). By labeling the cells with their distinctive markers, the two adjacent layers of cells became clear (Figure 3c). Our data showed that claudin-5 tight junction protein was uniformly abundant in cells that were lacking GFAP expression (i.e. hCMEC/D3 cells). The high expression of claudin-5 tight junction protein suggests the formation of a tight and functional barrier that has very low permeability and high resistance.
Figure 3.
Immunocytochemical analysis of the μSiM-BBB. (a-b) Expression of tight junction protein claudin-5 (green) in hCMEC/D3 cells and astrocytic marker GFAP (red) in NHA cells. (c) The y-z cross-sectional view of the co-culture model showing the distinctive two layers of cells. The nuclei were stained by DAPI (blue). Image acquisition: 40x oil objective.
It was important to confirm the integrity of the barrier, i.e. measure the transendothelial electrical resistance (TEER) and/or paracellular permeability of the SiN membrane BBB model. Due to the configuration of the device, the measurement of TEER is challenging, therefore the integrity of the barrier was further confirmed by paracellular permeability assays of a commonly used marker, fluorescently labeled 4 kDa dextran (FITC-dextran). The paracellular permeability of the μSiM-BBB is 1.51 ± 0.50E-06 cm/sec, 3.5-9.2 times smaller than the permeability of hCMEC/D3 monoculture models,42,45,46 and approx. 3.3 times smaller than the permeability of the equivalent BBB co-culture grown on Transwell® inserts (4.93 ± 0.10E-06 cm/sec) (Table 1, Figure S1), indicating enhanced barrier characteristics of the μSiM-BBB in comparison to the same cells grown in a Transwell® model. The tighter barrier properties might be explained by the high permeability of the ultrathin membrane enabling the facile crosstalk of soluble factors between human astrocytes and human brain endothelial cells.47,48 It is also noted that the permeability presented here was around an order of magnitude higher than the permeabilities reported in vivo for 10 times larger dextran (40 kDa),49 suggesting that the in vitro BBB model may be leakier than in vivo BBB. This could be due to the oversimplification of some features (e.g. glycocalyx on the luminal side and extravascular structure) in the in vitro model. In order to improve the barrier properties and tightness of our current BBB model and to better mimic the in vivo physiological conditions, shear stress50,51 and endothelial cells derived from human induced pluripotent stem cells52-56 are key parameters for future developments.
Table 1.
Paracellular permeability of 4 kDa FITC-dextran. The first two rows present the permeability value (mean ± SEM, n ≥ 3) of the μSiM-BBB.
| Permeability (x10−6 cm/sec) | Membrane specifications | BBB model |
|---|---|---|
| 1.51 ± 0.50 | μSiM-BBB (SiN, 0.5 μm) | hCMEC/D3 - NHA |
| 4.93 ± 0.10 | TW (0.4 μm, polyester) | hCMEC/D3 - NHA |
| 5.42 | TW (3 μm, PC) | hCMEC/D3 42 |
| 13.8 ± 0.50 | TW (0.4 μm, PET) | hCMEC/D3 45 |
| 8.83 ± 0.33 | TW (0.4 μm, PET), suppl. with HC | hCMEC/D3 45 |
| 12.0 ± 1.17 | N.I. | hCMEC/D3 46 |
hCMEC/D3: immortalized human brain capillary endothelial cells, HC: hydrocortisone, NHA: primary human astrocytes, N.I.: not indicated, PC: polycarbonate, PET: polyethylene terephthalate TW: Transwell® insert
Nanoparticle transport in the μSiM-BBB
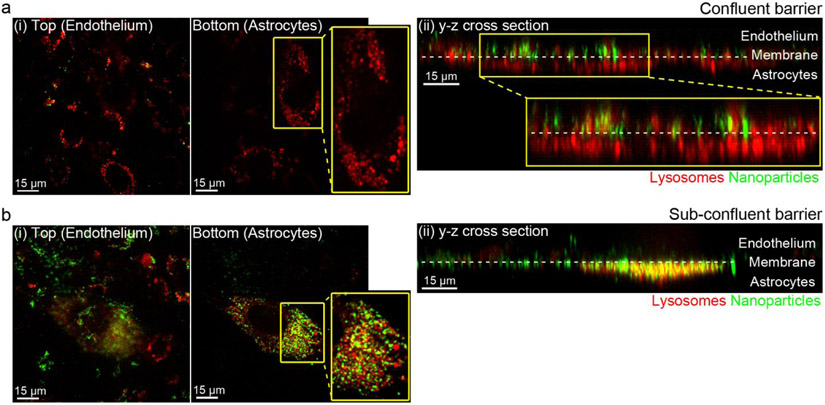
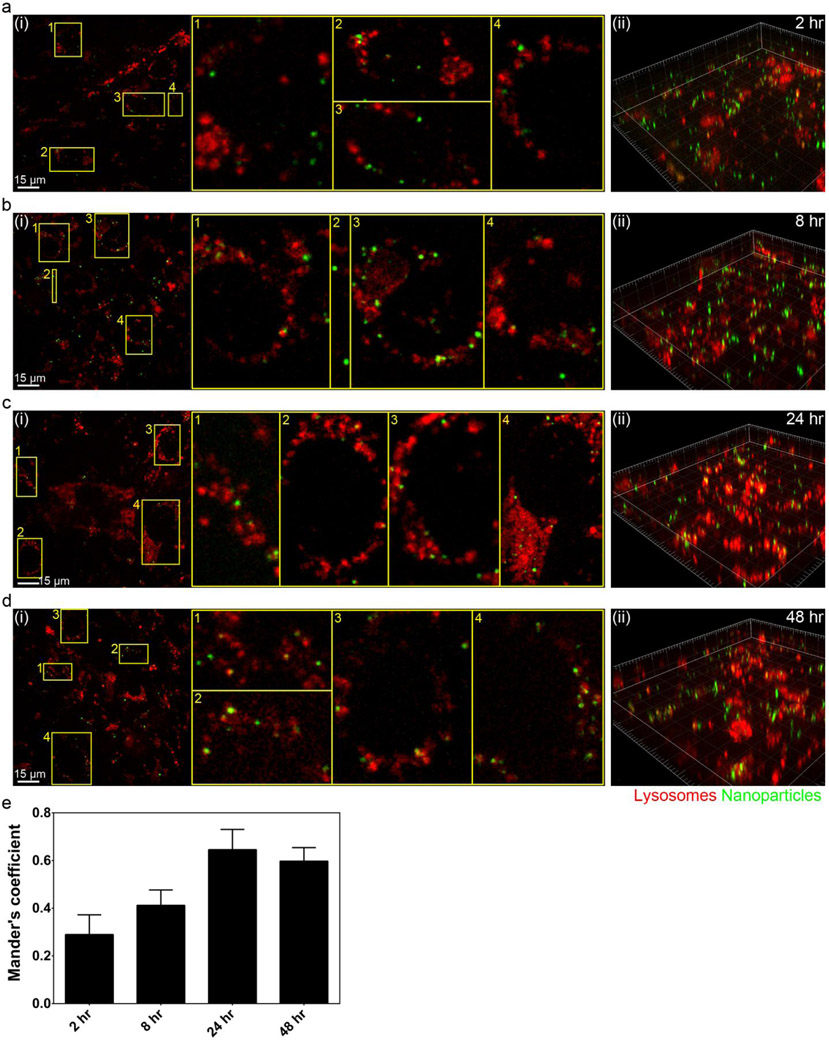
We first chose to confirm the integrity of the μSiM-BBB model using commercial fluorescently labeled 100 nm PS-COOH NPs that have been shown to rarely translocate across the BBB.25 The physicochemical characterization of the PS-COOH NPs used in this study was shown in Table S1. The apical side of the in vitro BBB co-culture model was exposed with PS-COOH NPs (100 μg/mL in cell culture medium supplemented with 10% fetal bovine serum (FBS)) for 10 min followed by removal of the NPs in excess and further culturing for up to 48 h (chase time). First, we determined the optimal magnification for high-resolution fluorescence imaging for tracking NPs. Our data have shown that 60x oil objective gave the best quality of imaging, as any higher magnification and/or shorter working distance resulted in images out of focus (Figure S2). To outline the two layers of cells in the co-culture, the acidic organelles of both endothelial cells and astrocytes were fluorescently labelled (LysoTracker™ Deep Red). Due to the distinctively different nature of cell functions, the lysosomes in astrocytes exhibited much stronger staining, allowing clear identification of the two layers (Figure 4). By scanning the entire apical and basal compartments (including the bottom cover glass located 50 μm from the membrane), we found that virtually all NPs were retained at the endothelial barrier (Figure 4a). In contrast, when the endothelial layer was not fully confluent, NPs were observed in both apical and basal compartments including a significant population in astrocytes (Figure 4b). Then we examined the subcellular location of NPs in the endothelial cells. LysoTracker™ Deep Red was used to stain the lysosomes of endothelial cells. NPs distribution was examined at different chase time points (i.e. 2, 8, 24, and 48 hours) (Figure 5a-d). Mander’s coefficient was used to quantify the co-localization between NPs and lysosomes. Our data showed an increasing lysosomal accumulation of NPs in the endothelial cells from 2 to 48 h chase time periods (Figure 5e). This result is consistent with previous reports showing NPs become trapped in the lysosomes of endothelial monocultures visualized by transmission electron microscopy and high-resolution fluorescence microscopy.25,26
Figure 4.
Subcellular location of 100 nm PS-COOH NPs in the μSiM-BBB with confluent (a) and sub-confluent (b) endothelial cell layer 24 hours post-exposure. (a) Representative 2D micrographs of the apical (top) and basolateral (bottom) side (i) and y-z cross-sectional view (ii) of the intact co-culture model showing that no NPs (green) were found inside the astrocytes (red) or in the basal compartment. (b) Representative 2D micrograph of the basolateral side (i) and y-z compressed cross-sectional view (ii) of a co-culture model with sub-confluent endothelial cell barrier showing that significant number of NPs (green) accumulated inside astrocytes where they co-localized with or located closed to (yellow) the lysosomes (red) 24 h post-exposure. Both endothelial cells and astrocytes were labelled by LysoTracker® Deep Red lysosomal dye (red). The fluorescence intensity of lysosomal staining in astrocyte was much stronger than in endothelial cells, therefore, endothelial cells are not visible on the three-dimensional reconstructed image (b(ii)). Image acquisition: 60x oil objective.
Figure 5.
Subcellular localization of 100 nm PS-COOH NPs in the μSiM-BBB over time. (a-d) 2D images (i) from the top (apical, endothelial) side of the membrane and three-dimensional rendering of the BBB co-culture model (ii) showing the lysosomes (LysoTracker® Deep Red; red) and PS-COOH NPs (green) 2, 8, 24, and 48 h after a 10 min NP exposure. Yellow spots correspond to NPs co-localized with/close to lysosomes. Image acquisition: 60x oil objective. (e) Mander's overlap coefficient represents the co-localization of NPs with lysosomes over time. Results represent mean ± standard deviation of at least four random regions where both endothelial cells and astrocytes were found.
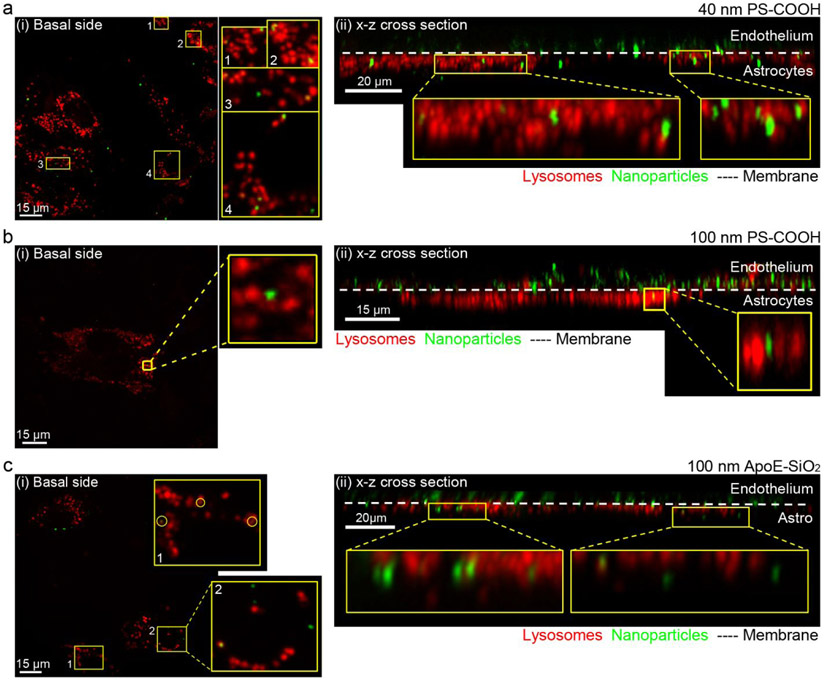
To challenge the sensitivity of high-resolution fluorescence imaging using the μSiM-BBB, we then sought to detect the translocation events of three types NPs (40 nm PS-COOH NPs, 100 nm PS-COOH, 100 nm ApoE-conjugated SiO2). Briefly, the apical side of the in vitro BBB co-culture model was exposed to NPs for 10 min, then the translocation was assessed at different time intervals (2, 4, 8, 24, and 48 h). We scanned the apical and basal compartments with three-dimensional (‘z-stack’) images (the actual imaging area is estimated in Supporting Information). Using the built-in spot algorithm of Imaris software, we were able to identify all NPs immobilized by the cells in the BBB model, and then distinguish the ones translocated to the astrocytes or basolateral compartments. Both the translocation of 40 nm PS-COOH NPs and 100 nm ApoE-SiO2 was apparent at the 2 h chase time point, whereas the first translocation event of 100 nm PS-COOH NPs was identified at chase time of 8 h (Figure 6, Table 2, Figure S3-S8). Furthermore, combining three independent experiments, only five out of more than 11,000 identified 100 nm PS-COOH NPs were found in the basolateral compartment with a chase phase up to 48 h (Table 2). By contrast, over 300 and 450 translocation events were detected for 40 nm PS-COOH and 100 nm ApoE-SiO2NPs, respectively (Table 2).
Figure 6.
Translocation of 40 nm and 100 nm PS-COOH and 100 nm ApoE-SiO2 NPs across the μSiM-BBB. (a, c) Representative maximum intensity projection images of the astrocytes located on the basal side of the membrane (i) and its corresponding x-z cross-sectional view (ii) showing that both 40 nm PS-COOH and 100 nm ApoE-SiO2 NPs (green) were able to cross the intact BBB. (b) 2D slice from the basal side (i) and x-z cross-sectional view of the μSiM-BBB showing a rare case, when 100 nm PS-COOH NPs (green) were able to cross an intact BBB co-culture model. Both endothelial cells and astrocytes were labeled by LysoTracker® Deep Red lysosomal dye (red). Chase times: 24 h (a, b) and 4 h (c). Image acquisition: 60x oil or water immersion objectives.
Table 2.
The number of observed and translocated NPs across the μSiM-BBB after pulse for 10 min
| Chase Time |
PS-COOH, 40 nm (6.62 x 108 NPs*) |
PS-COOH, 100 nm (6.62 x 108 NPs*) |
ApoE-SiO2, 100 nm (2.61 x 108 NPs*) |
|||
|---|---|---|---|---|---|---|
| Observed | Translocated | Observed | Translocated | Observed | Translocated | |
| NPs** | NPs | NPs** | NPs | NPs** | NPs | |
| 2 h | 185 | 18 | 1000 | 0 | 2729 | 161 |
| 4 h | 114 | 10 | 1502 | 0 | 2205 | 89 |
| 8 h | 160 | 49 | 1797 | 1 | 1829 | 70 |
| 24 h | 358 | 165 | 5481 | 2 | 2458 | 108 |
| 48 h | 177 | 49 | 1262 | 1 | N/A | N/A |
| Coverslip | 22 | 1 | 46 | |||
| SUM | 994 | 313 | 11042 | 5 | 9221 | 474 |
Total number of NPs exposed to all the cells grown on the membrane
Number of NPs identified by microscopy
It is worth noting that the translocation analysis was carried out at 100 μg/ml for 100 nm PS-COOH, and 6.4 μg/ml for 40 nm PS-COOH (which results in the same concentration by particle number). Therefore, our data suggest that the translocation is size-dependent. Although translocation increased with decreasing particle size, the translocation rate should still be interpreted as low given the number of delivered NPs was six orders of magnitude higher than the number of translocated NPs. This observation is in agreement with previous studies, which have shown that the translocation of NPs across the BBB is rare for a wide range of NP sizes,15,25,26 and that larger sized NPs exhibit a lower efficiency to translocate the BBB.25 Hence, the infrequency of 100 nm PS-COOH NP translocation events observed in this study underscores the exceptional sensitivity of our system.
ApoE is known to be able to cross the BBB via receptor-mediated transcytosis (RMT)57 and is often used to functionalize the surface of NPs to trigger RMT pathway and facilitate BBB crossing.19,20,22,23,58 In our experiment, even though a lower concentration by particle number for ApoE-SiO2 NPs was used compared to PS-COOH NPs with similar size (Table 2), the number of translocation events at all time points were significantly increased for the ApoE-SiO2 NPs, consistent with the notion that ApoE plays an important role in the NP-mediated transport across the BBB.
It is also worth mentioning that the translocation rate is often estimated based on the administrated dose of NPs for in vivo studies, whereas it is calculated based on the numbers of NPs that are associated with the BBB model in in vitro studies. With these definitions, it is assured that the in vivo rates of translocation will be far smaller than in vitro measurements. The in vitro studies can be thought of as ‘zooming in’ on those particles that directly interact with the BBB in the in vivo studies and thus have the greatest chance to transmigrate. Although the portion of translocated NPs is still very low in both types of studies, the ability to see and count rare translocations in vitro represents a useful tool for designing safe and effective nanocarriers of CNS-targeted drugs.
CONCLUSION
While imaging-based analysis, such as point scanning or spinning disk confocal microscopy and total internal reflection fluorescence microscopy, can provide validation of translocation,10,25 these studies would ideally be done on permeable substrates that support a valid BBB model with the cell types of the neurovascular unit. Imaging on permeable substrates, however, presents a different set of challenges, which were met here by the use of ultrathin silicon-based membranes. Our in vitro BBB co-culture model, the μSiM-BBB, is customized for high-resolution imaging of NP translocation. Both hCMEC/D3 endothelial cells and NHA astrocytes preserved their morphology and phenotype on the collagen/fibronectin pre-treated membrane and presented induced barrier properties in comparison with Transwell® model presumably because of better cell-cell communications. One of the clear advantages of the μSiM-BBB is that it is compatible with both in situ fixed-cell and live-cell high-resolution imagings that enable the monitoring of different cellular and spatiotemporal events, including rare events of NP translocation across the BBB. By using 100 nm PS-COOH NPs as a model delivery system, we have illustrated a high degree of lysosomal co-localization of NPs in the endothelial layer. With these particles we were also able to identify 5 translocation events after surveying over 10,000 NPs associated with the BBB. Reducing the size of PS-COOH NPs, or using functionalized NPs of the same size, increased these translocation rates significantly. Our results are a testament to the sensitivity of nanomembrane-enabled high-resolution imaging for the quantification of rare NP translocations. Given that most CNS-targeted NPs are captured by the BBB but do not translocate, the μSiM-BBB platform may be a key tool for identifying and amplifying those characteristics of drug carriers that facilitate translocation.
METHODS
Nanomembrane Fabrication
Microporous SiN membranes were commercially manufactured by and purchased from SiMPore Inc. (West Henrietta, NY) as previously described by Nehilla et al.33 Briefly, 150mm, <100> oriented, 300 μm thick, double-side polished silicon wafers (WaferPro, San Jose, CA) were deposited with low-stress (250 MPa) SiN (400 nm) using low-pressure chemical vapor deposition (LPCVD). Front and backside of wafers were patterned independently with OiR 620 Positive photoresist (FujiFilm) and exposed on an ASML PAS 5500/200 5x reduction stepper with 6 mm thick quartz-chrome masks (HTA Photomask, San Jose, CA). The backside pattern formed chip boundaries and defined window sizes of 2 x 0.7 mm2. On the front side, the pores were patterned as 0.5 circles on a rectangular coordinate system with a 1 or 6 μm center-center spacing respectively. This resulted in membranes with a 20% porosity. The backside pattern and micropores were then etched into the SiN using a DryTek 482 Quad Etcher. After nitride etching, a custom one-sided etch cell was used to mount the silicon wafer. The backside was exposed to heated ethylenediamine pyrocatechol (EDP) until the microporous membrane on the front side was reached. The backside pattern resulted in individual chips of size 5.4 x 5.4 mm2 connected by thin (perforated) bridges of silicon. The chips were manually cleaved from the wafer and inspected optically and by scanning electron microscopy and transmission electron microscopy.
Device assembly
The μSiM co-culture device consisted of two main components: the silicone-based housing and the microporous SiN membrane chip. The silicone-housing consisted of precision-cut silicone gaskets and hole-punched polydimethylsiloxane (PDMS) block. Patterns in the silicone gaskets were optimized as shown in Figure 1a and cut using the Silhouette CAMEO cutter (Silhouette America, Oren, UT). The ‘sealing layer’ isolated the top and the bottom chambers and prevented any fluid leakage within the compartments, while the ‘membrane’ layer acted as a spacer to accommodate the membrane-chip. These two layers were cut from 300 μm gasket sheets, while the bottom layer was cut from 50 μm gasket. The thinness of the bottom layer reduced the working distances for high-resolution optical microscopy. PDMS blocks were made by mixing Sylgard 184 elastomer and curing agent (both from Dow Corning, USA) in a 10:1 ratio and cured at 70°C for 2 hours. The PDMS block contained one 7 mm hole and two 5 mm holes. The 7 mm holes of the PDMS block formed a continuous volume to host cells on top of the SiN membrane. The 840 μm holes in the ‘sealing layer’ allow the insertion of a p200 pipette tip to introduce cells into the bottom channel (Figure 1a). The 5 mm holes on top of the 840 μm holes hosted additional media volume for long-term culture. All components of the housing, the membrane chip, and the #1.5 cover glass that forms the floor of the bottom channel were covalently-bonded together using UV-Ozone treatment (Novascan technologies, Boone, IA). The surfaces to be bonded were exposed to UV-Ozone atmosphere for 10 min and brought in the immediate contact of each other to establish the covalent bond. The treated surfaces were then cured in the oven at 70°C for another hour for irreversible bonding. The coverslips were pre-treated with piranha solution (1:3 solution of hydrogen peroxide and concentrated sulfuric acid) and cleaned with distilled water and dried using isopropanol. Piranha treatment cleaned the glass surfaces and enhanced the surface charge to improve the bonding between 50 μm gaskets and the glass coverslips. The entire device assembly were sterilized using standard autoclaving procedures or UV light.
Routine cell culturing
Immortalized human brain capillary endothelial hCMEC/D3 cells (provided by P.-O. Couraud, INSERM, France) were used between passages 28 and 35. For routine cell culture, cells were seeded in collagen-coated flask (Corning® BioCoat™ Collagen I flasks or rat tail type I collagen (Sigma Aldrich) pre-coated flasks from Greiner Bio-One GmbH) and supplemented with EBM-2 basal medium containing vascular endothelial growth factor (VEGF), insulin-like growth factor-1 (IL-1), epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), ascorbic acid, gentamicin sulfate/amphotericin B (GA-1000), hydrocortisone (all from Lonza Biosciences) and 2.5% fetal bovine serum (FBS) (Gibco®). For functionality assays and other experiments, all cultureware was coated with rat tail type I collagen (Sigma Aldrich) at a concentration of 100 μg/ml or with the mixture of 100 μg/ml rat tail type I collagen and 50 μg/ml bovine plasma fibronectin (Thermo Fisher Scientific) (collagen/fibronectin) and incubated for 1 hour at 37°C; the cells were supplemented with growth factor depleted EBM-2 medium containing bFGF, hydrocortisone, GA-1000 (all from Lonza Biosciences), 2% FBS (Gibco®), and 10 mM HEPES (Sigma Aldrich), referred as assay medium.
Clonetics™ Normal Human Astrocytes (NHA) (Lonza Biosciences) were used between passage 1 and 6. For routine cell culture, cells were seeded in Nunclon™ Delta treated flasks (Thermo Scientific™ Nunc™ Cell Culture Treated EasYFlasks™) and supplemented with ABM basal medium containing rhEGF, insulin, L-Glutamine, ascorbic acid, GA-1000 (all from Lonza Biosciences), and 3% FBS (Gibco®), referred as astrocyte medium.
Cells were cultured in an incubator at 37°C with 5% CO2/95% air and saturated humidity. The cell culture medium was changed every 2 days, and the assay medium twice weekly.
BBB co-culture model on Transwell® membrane
Twenty thousand hCMEC/D3 cells were seeded on the upper side of collagen pre-coated polyester Transwell® inserts (12-well format, 1.12 cm2, 0.4 μm pore size, 4x106 pores/cm2, i.e. approx. 0.5%, porosity, Corning® #3460) and maintained in assay medium (0.5 mL of assay medium in the apical compartment and 1.5 mL of assay medium in the basolateral compartment) at 37°C with 5% CO2/95% air and saturated humidity. The medium was changed twice a week. Ten thousand unstained or pre-labeled NHA cells (in 150 μL astrocyte medium) were seeded on the bottom side of the flipped membrane inserts on day 5 or 6. After 1.5-2 hours of incubation at 37°C, the membrane inserts were flipped back to the original orientation and fresh assay medium was added to both apical and basolateral compartments. The experiments were conducted 7 days post-Transwell® seeding of hCMEC/D3 cells.
BBB co-culture model on cell culture device with SiN membrane
Prior to cell seeding, the μSiM-BBB devices were sterilized by autoclaving or by UV light (1 hour), and both sides of the membrane were coated with collagen/fibronectin. The top reservoir was seeded with 10,500 hCMEC/D3 cells and the bottom reservoir was filled with assay medium. The device was then placed inside a 6 mm cell culture dish (Greiner Bio-One GmbH) with sterile MilliQ water wetted Kimcare wipes (Kimberly Clark) and kept in an incubator at 37°C with 5% CO2/95% air and saturated humidity. The cells were grown under static condition, and the assay medium (100 μl medium/top reservoir) was replaced 2-3 hours after plating the cells and every second day afterwards. Unstained or pre-stained NHA cell suspension (12 μl of 300,000-400,000 cells/ml cell suspension) was seeded to the bottom reservoir on day 4 (or once hCMEC/D3 cells reached confluency). After the two loading channels were filled with astrocyte medium, the device was covered with parafilm and was flipped upside down to allow the settlement of astrocytes onto the bottom side of the membrane. After 1.5-2 hours of incubation at 37°C, the device was flipped back to its original orientation and fresh astrocyte medium (approx. 25 μl) was added to the two 5 mm reservoirs to allow long-term cell culture. On the next day, the astrocyte medium was replaced with assay medium. Due to the small volume (2.4 μl) of the bottom reservoir, the medium was replaced every day in the lower channel. The BBB model was used for experiments 5-7 days after plating the hCMEC/D3 cells.
Immunocytochemistry
Endothelial cells and astrocytes were cultured on the opposite side of the Transwell® or the μSiM-BBB as described above. On the day of the experiment, the cells were washed with preheated medium and were fixed with 4% paraformaldehyde (Sigma Aldrich) for 20 min at room temperature (RT), washed three times with phosphate buffer saline (PBS) (Sigma Aldrich), permeabilized with 1 x saponin (Life Technologies) for 10 min, and blocked in 1% bovine serum albumin (BSA) (Sigma Aldrich) (in 0.05% PBS-Tween 20 (Sigma Aldrich) (PBS-T)) for 30 min. Cells were then incubated for 2 hours at RT or overnight at 4°C with relevant primary antibody (Ab) (mouse monoclonal Ab to claudin-5 [4C3C2] (35-2500) (Invitrogen™, Thermo Fisher Scientific); 2.5 μg/ml concentration in 1% BSA-PBS-T, rabbit monoclonal [EP672Y] (ab33922) or polyclonal (ab7260) Ab to GFAP (Abcam); 2 μg/ml concentration in 1% BSA-PBS-T). After that, the cells were washed three times with PBS and incubated with Alexa Fluor® 488 or Alexa Fluor® 546 -mouse or anti-rabbit IgG secondary Ab (Invitrogen™, Thermo Fisher Scientific) (the concentration of secondary Ab was half of the primary Ab) for 1 hour in dark at RT, followed by rinsing three times with PBS. In addition, the nuclei of both hCMEC/D3 NHA cells were stained with 4’,6-diamidino-2-phenylindole (DAPI) (1 μg/ml concentration in PBS) (Sigma Aldrich). In the case of Transwell® co-culture, the polyester membrane was excised using scalpel and scissors from the plastic insert and was mounted on a glass coverslip using MOWIOL (Polysciences, Inc.). The cells were observed and photographed using a spinning disk confocal microscopy system consisting of a CSU22 spinning disk unit (Yokogawa Electric Corporation, Japan) and an Andor iXon3 897 EMCCD camera (Andor, UK), mounted on an inverted fully motorized Nikon Ti microscope body (Nikon Instruments, US). The raw images were processed using Imaris imaging software version 7.4.2 (Bitplane AG, Switzerland).
Paracellular permeability assay
To determine the tightness of BBB models, we measured the paracellular permeability of fluorescein isothiocyanate (FITC) labeled 4 kDa dextran (Sigma Aldrich).
Paracellular permeability of Transwell® co-culture model
The hCMEC/D3 and NHA cells were cultured on the opposite side of the Transwell® inserts as described above. The permeability was measured 7 days after seeding the hCMCE/D3 cells. The apical side (upper chamber) was filled with 500 μl 100 μg/ml FITC-dextran in assay medium, and 1500 μl assay medium was added to the basolateral side (bottom compartment). In every 20 min for a period of 120 min, 100 μl medium was removed from the basolateral side. The fluorescence intensity of the sample was read by a Varioskan® Flash spectral scanning multimode reader (Thermo Scientific) (λExc = 490 nm, λEm = 515 nm, Bandwidth = 12 nm), and the paracellular permeability was calculated according to Czupalla et al. (see equation (1)).59
| (1) |
Where Papp is the apparent permeability, B is the relative fluorescence unit (RFU) at time t, T is the top chamber RFU at time 0 (constant), Vb is the volume of the bottom channel [ml], A is the cross-section area of the membrane [cm2], and t is the time [min].
The final permeability of the co-culture (Pco-culture) was calculated based equation (2):
| (2) |
Where Papp, co-culture is the permeability of the apparent permeability of the co-culture model (cells + insert), and Pinsert is the permeability of the collagen-coated blank porous membrane.
Measurement was performed in technical triplicates at least three times, and the data are represented as mean ± standard error of the mean (SEM).
Paracellular permeability in the μSiM-BBB
The hCMEC/D3 and NHA cells were cultured on the opposite side of the SiN membrane device as described previously. The permeability was measured 5-6 days after seeding the hCMEC/D3 cells. The apical side of the membrane was filled with 100 μl 250 μg/ml 4 kDa FITC-dextran and the permeability was determined by measuring the fluorescence intensity by plate reader. Due to the small volume of the basal side (2.4 μl volume in total), 10 μl sample was taken through one of the inlet/outlet points of the bottom channel (cell culture medium from the bottom channel and the other bottom reservoirs) and from the apical compartment at 120 min time point. The samples were diluted in assay medium and the permeability was determined and calculated as it was described above. Collagen/fibronectin-coated cell-free devices were used as negative controls.
Cell labeling and live-cell imaging
Unstained hCMEC/D3 cells were seeded and grown on the apical side of Transwell® insert or the μSiM-BBB, and CellTracker™ Green CMFDA (CTG) (Molecular Probes™) pre-stained NHA cells were cultured on the opposite side of the membrane. For CTG labeling, NHA cells in suspension were incubated with 25 μM CTG (100 μl /1000 cells) for 30 min at 37°C, and then the excess dye was washed by incubating the cells in a large volume of cell culture medium for 20 min at 37°C. The pre-stained cells were diluted in astrocyte medium at required seeding densities. See seeding conditions and growth times of the different cell types and device formats above. Prior to image acquisition, hCMEC/D3 cells were stained with CellMask™ Orange (Molecular Probes™) at 7.5 μg/ml for 10 min at 37°C and washed twice with PBS and once with assay medium. Live visualization of the BBB co-culture was performed on a spinning disk confocal microscopy system consisting of a CSU22 spinning disk unit (Yokogawa Electric Corporation, Japan) and an Andor iXon3 897 EMCCD camera (Andor, UK), mounted on an inverted fully motorized Nikon Ti microscope body (Nikon Instruments, US) with climate control chamber (37°C, controlled CO2 and humidity). In the case of Transwell® models, the inserts were directly placed into a glass bottom imaging dish filled with cell culture medium that allowed direct imaging of cells on both sides of the membrane. CTG labelled NHA cells were excited with a 488 nm laser line (λEm=505/530 nm bandpass filter), whereas CellMask™ Orange labelled hCMEC/D3 cells were excited using a 561 nm laser line (λEm=610 nm long pass filter). Plan Apo 10x/0.45 (WD = 4.0 mm) and Plan Apo 20x/0.75 (WD =1.0 mm) air objectives, and Plan Apo 40x/1.0 (WD = 0.16 mm), Plan Apo VC 60x/1.40 (WD = 0.13 mm), and Plan Apo VC 100x/1.40 (WD = 0.13 mm) oil immersion objectives were used (Nikon). Three-dimensional (‘z-stack’) images were acquired using Andor iQ software and processed using Imaris imaging software version 7.4.2 (Bitplane AG, Switzerland).
Nanoparticle synthesis and characterization
FluoSpheres® carboxylate-modified polystyrene (PS-COOH) (YG: λExc/ λEm = 505/515 nm, Red: λExc/ λEm = 580/605 nm) (Life Technologies, Thermo Fisher Scientific) with 40 nm and 100 nm nominal size were purchased from Life Technologies, Thermo Fisher Scientific. Silica NPs of 110 nm were synthesized by a sol-gel approach including the incorporation of fluorescein isothiocyanate (FITC) followed by their surface amination with the organofunctional silane (3-aminopropyl)trimethoxysilane according to a method previously described.60 To these particles dispersed in HEPES buffer (50 mM, pH 7.4), the recombinant ApoE protein expressing an N-terminal Cysteine was conjugated with a short heterobifunctional PEG of 8 ethylene μlycol units bearing an N-hydroxysuccinimide ester and a maleimide group. After several washing steps with HEPES buffer (10 mM, pH 7.4) by centrifugation between, the protein excess was removed. The concentration of the particles was measured by relative fluorescence according to a calibration curve and the protein concentration was measured by bicinchoninic acid assay (Micro BCA™ Protein Assay Kit, Thermo Fisher Scientific).The size distribution was determined in PBS, cell culture medium supplemented with 10% FBS, or 10 mM HEPES buffer by dynamic light scattering using a Malvern Zetasizer 3000HSa and a Zetasizer Nano ZS (Malvern Instruments Ltd., UK) under experimental conditions. The results are the average of minimum three separate runs, the errors represent the standard deviation over measurement and indicates the reproducibility of the measurement. The effective surface charge (zeta potential) of the NPs was also measured using Malvern Zetasizer 3000HSa. The results are the average of minimum three separate runs, the errors represent the standard deviation over measurement.
Subcellular localization and translocation studies of nanoparticle in the μSiM-BBB using live-cell imaging
The BBB co-cultures on μSiM devices were_achieved described above using unstained or CTG pre-labelled NHA cells. 40 nm and 100 nm PS-COOH NPs (FluoSpheres™ Carboxylate-Modified Microspheres, 0.04 μm (actual size: 47 nm, therefore) yellow/green (505/515) fluorescent, 5% solids, and FluoSpheres™ Carboxylate-Modified Microspheres, 0.1 μm, red (580/605) or yellow/green (505/515) fluorescent, 2% solids) were dispersed in assay medium supplemented with 10% FBS at 37°C prior to cellular exposure. ApoE-SiO2 NPs were found to be aggregated when they were directly dispered in assay medium (both serum-free or medium supplemented with 10% FBS). Thus, these NPs were firstly diluted in HBSS supplemented with 1mM Ca2+ and 0.1 %w/v BSA, and the colloidal stability was confirmed by DLS (Table S1). The NP dispersion was then added to the cells and imaged. Endothelial cells on the apical side of the SiN membrane were exposed to 1.82 x 1010 PS-COOH NPs (e.g. 100 μL of 100 nm PS-COOH NP dispersions at 100 μg/ml concentration) and 7.18x109 ApoE-SiO2 NPs (i.e. 100 μL of 100 μg/ml concentration). After 10 min NP exposure, the medium was removed, and the samples were washed three times with NP-free assay medium supplemented with 10% FBS or HBSS supplemented with Ca2+ and BSA, and finally 100 μl fresh assay medium was added to the cells. The devices were kept at 37°C until imaging. Prior to imaging, the acidic organelles of the cells were stained with 100 nM LysoTracker™ Deep Red (Molecular Probes™) for 1 hour and washed three times with cell culture medium (the dye was only added to and washed away from the apical side). Three-dimensional images (‘z-stacks’) were obtained up to 48 h (2, 4, 8, 24, and 48 h timepoints, imaging for up to 1 hour/time point) by using a spinning disk confocal microscopy system consisting of a CSU22 or CSU-X1 spinning disk unit (Yokogawa Electric Corporation, Japan) and an Andor iXon3 897 or Andor iXon3 EMCCD camera (Andor, UK), mounted on an inverted fully motorized Nikon Ti (Nikon Instruments, US) or Olympus IX83 (Olympus Corporation, Japan) microscope body and Plan Apo VC 60x/1.40 (WD = 0.13 mm) oil immersion objective (Nikon) or a UPlanSApo 60x/NA1.20 (WD = 0.28 mm) water immersion objective (Olympus Corporation, Japan). The following excitation laser lines and emission filters were used: CTG, Yellow/Green PS-COOH NPs, FITC labelled ApoE-SiO2 NPs: λexc = 488 nm, λem = 505/530 nm bandpass filter, LysoTracker™Deep Red and Red PS-COOH NPs: λexc = 560 nm, λem = 610 nm long pass filter or λexc = 640 nm, λem = 440/521/607/700 nm quad-band bandpass filter. Images were acquired using Andor iQ software and processed using Imaris imaging software version 7.4.2 or 8.2.1 (Bitplane AG, Switzerland).
Image analyzes
Imaris imaging software version 7.4.2 and 8.2.1 (Bitplane AG, Switzerland) was used for all image analyzes. Both NPs and lysosomes were identified using the ‘spot detection’ algorithm with a Mexican hat filter for dynamic background subtraction. The identified spots were classified and filtered using a quality filter and a z-position filter. The parameters for the 'quality intensity threshold' were adjusted manually for each image to account for differences in background intensity (the threshold was adjusted until the majority of identifiable NPs and vesicles were labeled). The value range for this parameter are always determined empirically for each data set. Since both astrocytes and endothelial cells were labeled with the same dye, the use of z-position filter enabled us to determine the position of the membrane and distinguish between astrocytic and endothelial lysosomes (the number of identified vesicles dropped around the position of the membrane). NPs detected below the determined membrane position were considered as translocated NPs.
The lysosomal co-localization of NPs was determined using ImarisColoc using Mander’s overlap coefficient and manual thresholding.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by EU FP7 Marie-Curie Initial Training Network PathChooser (PITN-GA-2013-608373, K.A.D. and D.H.), the Science Foundation Ireland (SFI) Starting Investigator Researcher Grant (Agreement No. 15/SIRG/3423, Y.Y.), and EuroNanoMed III (Grant No. 16/ENM- ERA/3457, Y.Y.). L.A. was supported by Science Foundation Ireland (SFI) and the National Natural Science Foundation Of China (NSFC) under the SFI-NSFC Partnership Programme Grant Number 17/NSFC/4898. J.L.M., T.K., and H.L.C were supported by NIH R01 HL125265. D.G. and E.L. were supported by NIH P50AR07200. M.S.N. was supported by Research Initiative on Brain Barriers and Drug Delivery funded by the Lundbeck Foundation (Grant no. 2013-14113). The authors acknowledge the Conway Imaging Core Facility, the imaging facilities at the School of Biology, UCD, and the AU Health Bioimaging Core Facility. P.-O. Couraud, I. A. Romero, and B. Weksler are also acknowledged for providing endothelial hCMEC/D3 cells. Xiaoliang Yang is acknowledged for the purification of recombinant ApoE protein.
Footnotes
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: J.L.M. declares a competing financial interest as a co-founder and equity holder of SiMPore Inc., a commercial manufacturer of NPN and other silicon-based membrane materials.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. Calculation of number concentration of nanoparticles, area of the apical side, membrane, and imaging field of view. Tables showing the number of NPs the apical side of the μSiM-BBB device, the membrane, and the individual imaging fields were exposed to, the physicochemical characterization of the NPs, and the total number of observed and translocated NPs across the μSiM-BBB 2 to 48 h after a 10 min exposure. Figures showing the validation of the in vitro BBB co-culture model grown on Transwell® insert, demonstration of the in vitro BBB co-culture model grown on 0.5 μm pore size SiN membrane and the application of different objectives for live-cell imaging of the model, and quantification of the translocation of 40 and 100 nm PS-COOH and 100 nm ApoE-SiO2 nanoparticles in the μsiM-BBB model.
REFERENCES
- 1.Chou LY; Ming K; Chan WC Strategies for the Intracellular Delivery of Nanoparticles. Chem. Soc. Rev 2011, 40, 233–245. [DOI] [PubMed] [Google Scholar]
- 2.Blanco E; Shen H; Ferrari M Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat. Biotechnol 2015, 33, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masserini M Nanoparticles for Brain Drug Delivery. ISRN Biochem. 2013, 2013, 238428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreuter J Drug Delivery to the Central Nervous System by Polymeric Nanoparticles: What Do We Know? Adv. Drug Delivery Rev 2014, 71, 2–14. [DOI] [PubMed] [Google Scholar]
- 5.Saraiva C; Praca C; Ferreira R; Santos T; Ferreira L; Bernardino L Nanoparticle-Mediated Brain Drug Delivery: Overcoming Blood-Brain Barrier to Treat Neurodegenerative Diseases. J. Controlled Release 2016, 235, 34–47. [DOI] [PubMed] [Google Scholar]
- 6.Furtado D; Bjornmalm M; Ayton S; Bush AI; Kempe K; Caruso F Overcoming the Blood-Brain Barrier: The Role of Nanomaterials in Treating Neurological Diseases. Adv. Mater. (Weinheim, Ger.) 2018, 30, 1801362. [DOI] [PubMed] [Google Scholar]
- 7.Ulbrich K; Knobloch T; Kreuter J Targeting the Insulin Receptor: Nanoparticles for Drug Delivery across the Blood-Brain Barrier (BBB). J. Drug Targeting 2011, 19, 125–132. [DOI] [PubMed] [Google Scholar]
- 8.Huile G; Shuaiqi P; Zhi Y; Shijie C; Chen C; Xinguo J; Shun S; Zhiqing P; Yu H A Cascade Targeting Strategy for Brain Neuroglial Cells Employing Nanoparticles Modified with Angiopep-2 Peptide and EGFP-EGF1 Protein. Biomaterials 2011, 32, 8669–8675. [DOI] [PubMed] [Google Scholar]
- 9.Gao H; Zhang S; Cao S; Yang Z; Pang Z; Jiang X Angiopep-2 and Activatable Cell-Penetrating Peptide Dual-Functionalized Nanoparticles for Systemic Glioma-Targeting Delivery. Mol. Pharmaceutics 2014, 11, 2755–2763. [DOI] [PubMed] [Google Scholar]
- 10.Tian X; Nyberg S; P SS; Madsen J; Daneshpour N; Armes SP; Berwick J; Azzouz M; Shaw P; Abbott NJ; Battaglia G LRP-1-Mediated Intracellular Antibody Delivery to the Central Nervous System. Sci Rep 2015, 5, 11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aktaş Y; Yemisci M; Andrieux K; Gürsoy RN; Alonso MJ; Fernandez-Megia E; Novoa-Carballal R; Quiñoá E; Riguera R; Sargon MF; Çelik HH; Demir AS; Hincal AA; Dalkara T; Çapan Y; Couvreur P Development and Brain Delivery of Chitosan-PEG Nanoparticles Functionalized with the Monoclonal Antibody 0X26. Bioconjngate Chem. 2005, 16, 1503–1511. [DOI] [PubMed] [Google Scholar]
- 12.Ulbrich K; Hekmatara T; Herbert E; Kreuter J Transferrin- and Transferrin- Receptor-Antibody-Modified Nanoparticles Enable Drug Delivery across the Blood-Brain Barrier (BBB). Eur. J. Pharm. Biopharm 2009, 77, 251–256. [DOI] [PubMed] [Google Scholar]
- 13.Salvati E; Re F; Sesana S; Cambianica I; Sancini G; Masserini M; Gregori M Liposomes Functionalized to Overcome the Blood-Brain Barrier and to Target Amyloid-Beta Peptide: The Chemical Design Affects the Permeability across an In Vitro Model. Int. J. Nanomed 2013, 8, 1749–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang J; Jallouli Y; Kroubi M; Yuan XB; Feng W; Kang CS; Pu PY; Betbeder D Characterization of Endocytosis of Transferrin-Coated PLGA Nanoparticles by the Blood-Brain Barrier. Int. J. Pharm. (Amsterdam, Neth.) 2009, 379, 285–292. [DOI] [PubMed] [Google Scholar]
- 15.Clark AJ; Davis ME Increased Brain Uptake of Targeted Nanoparticles by Adding an Acid-Cleavable Linkage between Transferrin and the Nanoparticle Core. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 12486–12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiley DT; Webster P; Gale A; Davis ME Transcytosis and Brain Uptake of Transferrin-Containing Nanoparticles by Tuning Avidity to Transferrin Receptor. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 8662–8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kratzer I; Wernig K; Panzenboeck U; Bernhart E; Reicher H; Wronski R; Windisch M; Hammer A; Malle E; Zimmer A; Sattler W Apolipoprotein A-I Coating of Protamine-Oligonucleotide Nanoparticles Increases Particle Uptake and Transcytosis in an In Vitro Model of the Blood-Brain Barrier. J. Controlled Release 2007, 117, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreuter J; Hekmatara T; Dreis S; Vogel T; Gelperina S; Langer K Covalent Attachment of Apolipoprotein A-I and Apolipoprotein B-100 to Albumin Nanoparticles Enables Drug Transport into the Brain. J. Controlled Release 2007, 118, 54–58. [DOI] [PubMed] [Google Scholar]
- 19.Kreuter J; Shamenkov D; Petrov V; Ramge P; Cychutek K; Koch-Brandt C; Alyautdin R Apolipoprotein-Mediated Transport of Nanoparticle-Bound Drugs Across the Blood-Brain Barrier. J. Drug Targeting 2002, 10, 317–325. [DOI] [PubMed] [Google Scholar]
- 20.Michaelis K; Hoffmann MM; Dreis S; Herbert E; Alyautdin RN; Michaelis M; Kreuter J; Langer K Covalent Linkage of Apolipoprotein E to Albumin Nanoparticles Strongly Enhances Drug Transport into the Brain. J. Pharmacol. Exp. Ther 2006, 317, 1246–1253. [DOI] [PubMed] [Google Scholar]
- 21.Petri B; Bootz A; Khalansky A; Hekmatara T; Muller R; Uhl R; Kreuter J; Gelperina S Chemotherapy of Brain Tumour Using Doxorubicin Bound to Surfactant-Coated Poly(Butyl Cyanoacrylate) Nanoparticles: Revisiting the Role of Surfactants. J. Controlled Release 2007, 117, 51–58. [DOI] [PubMed] [Google Scholar]
- 22.Wagner S; Zensi A; Wien SL; Tschickardt SE; Maier W; Vogel T; Worek F; Pietrzik CU; Kreuter J; von Briesen H Uptake Mechanism of ApoE-Modified Nanoparticles on Brain Capillary Endothelial Cells as a Blood-Brain Barrier Model. PLoS ONE 2012, 7, e32568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zensi A; Begley D; Pontikis C; Legros C; Mihoreanu L; Wagner S; Buchel C; von Briesen H; Kreuter J Albumin Nanoparticles Targeted with Apo E Enter the CNS by Transcytosis and Are Delivered to Neurones. J. Controlled Release 2009, 137, 78–86. [DOI] [PubMed] [Google Scholar]
- 24.Zensi A; Begley D; Pontikis C; Legros C; Mihoreanu L; Buchel C; Kreuter J Human Serum Albumin Nanoparticles Modified with Apolipoprotein A-I Cross the Blood-Brain Barrier and Enter the Rodent Brain. J. Drug Targeting 2010, 18, 842–848. [DOI] [PubMed] [Google Scholar]
- 25.Bramini M; Ye D; Hallerbach A; Nic Raghnaill M; Salvati A; Aberg C; Dawson KA Imaging Approach to Mechanistic Study of Nanoparticle Interactions with the Blood-Brain Barrier. ACS Nano 2014, 8, 4304–4312. [DOI] [PubMed] [Google Scholar]
- 26.Ye D; Raghnaill MN; Bramini M; Mahon E; Aberg C; Salvati A; Dawson KA Nanoparticle Accumulation and Transcytosis in Brain Endothelial Cell Layers. Nanoscale 2013, 5, 11153–11165. [DOI] [PubMed] [Google Scholar]
- 27.Banks WA From Blood-Brain Barrier to Blood-Brain Interface: New Opportunities for CNS Drug Delivery. Nat. Rev. Drug Discovery 2016, 15, 275–292. [DOI] [PubMed] [Google Scholar]
- 28.Ryu S; Yoo J; Jang Y; Han J; Yu SJ; Park J; Jung SY; Ahn KH; Im SG; Char K; Kim BS Nanothin Coculture Membranes with Tunable Pore Architecture and Thermoresponsive Functionality for Transfer-Printable Stem Cell-Derived Cardiac Sheets. ACS Nano 2015, 9, 10186–10202. [DOI] [PubMed] [Google Scholar]
- 29.Raghnaill MN; Bramini M; Ye D; Couraud PO; Romero IA; Weksler B; Aberg C; Salvati A; Lynch I; Dawson KA Paracrine Signalling of Inflammatory Cytokines from an In Vitro Blood Brain Barrier Model upon Exposure to Polymeric Nanoparticles. Analyst (Cambridge, U. K.) 2014, 139, 923–930. [DOI] [PubMed] [Google Scholar]
- 30.Kim MY; Li DJ; Pham LK; Wong BG; Hui EE Microfabrication of High-Resolution Porous Membranes for Cell Culture. J. Memb. Sci 2014, 452, 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal AA; Nehilla BJ; Reisig KV; Gaborski TR; Fang DZ; Striemer CC; Fauchet PM; McGrath JL Porous Nanocrystalline Silicon Membranes as Highly Permeable and Molecularly Thin Substrates for Cell Culture. Biomaterials 2010, 31, 5408–5417. [DOI] [PubMed] [Google Scholar]
- 32.Mazzocchi AR; Man AJ; DesOrmeaux J-PS; Gaborski TR Porous Membranes Promote Endothelial Differentiation of Adipose-Derived Stem Cells and Perivascular Interactions. Cell. Mol. Bioeng 2014, 7, 369–378. [Google Scholar]
- 33.Nehilla BJ; Nataraj N; Gaborski TR; McGrath JL Endothelial Vacuolization Induced by Highly Permeable Silicon Membranes. Acta Biomater. 2014, 10, 4670–4677. [DOI] [PubMed] [Google Scholar]
- 34.Carter RN; Casillo SM; Mazzocchi AR; DesOrmeaux JS; Roussie JA; Gaborski TR Ultrathin Transparent Membranes for Cellular Barrier and Co-Culture Models. Biofabrication 2017, 9, 015019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casillo SM; Peredo AP; Perry SJ; Chung HH; Gaborski TR Membrane Pore Spacing Can Modulate Endothelial Cell-Substrate and Cell-Cell Interactions. ACS Biomater. Sci. Eng 2017, 3, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DesOrmeaux JP; Winans JD; Wayson SE; Gaborski TR; Khire TS; Striemer CC; McGrath JL Nanoporous Silicon Nitride Membranes Fabricated from Porous Nanocrystalline Silicon Templates. Nanoscale 2014, 6, 10798–10805. [DOI] [PubMed] [Google Scholar]
- 37.Ma SH; Lepak LA; Hussain RJ; Shain W; Shuler ML An Endothelial and Astrocyte Co-Culture Model of the Blood–Brain Barrier Utilizing an Ultra-Thin, Nanofabricated Silicon Nitride Membrane. Lab Chip 2005, 5, 74–85. [DOI] [PubMed] [Google Scholar]
- 38.Harris SG; Shuler ML Growth of Endothelial Cells on Microfabricated Silicon Nitride Membranes for an In Vitro Model of the Blood-Brain Barrier. Biotechnol. Bioprocess Eng 2003, 8, 246–251. [Google Scholar]
- 39.Miller JJ; Carter RN; McNabb KB; DesOrmeaux JPS; Striemer CC; Winans JD; Gaborski TR Lift-Off of Large-Scale Ultrathin Nanomembranes. J. Micromech. Microeng 2015, 25, 015011. [Google Scholar]
- 40.Jud C; Ahmed S; Müller L; Kinnear C; Vanhecke D; Umehara Y; Frey S; Liley M; Angeloni S; Petri-Fink A; Rothen-Rutishauser B Ultrathin Ceramic Membranes as Scaffolds for Functional Cell Coculture Models on a Biomimetic Scale. BioRes. Open Access 2015, 4, 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mossu A; Rosito M; Khire T; Li Chung H; Nishihara H; Gruber I; Luke E; Dehouck L; Sallusto F; Gosselet F; McGrath JL; Engelhardt B A Silicon Nanomembrane Platform for the Visualization of Immune Cell Trafficking across the Human Blood-Brain Barrier under Flow. J. Cereb. Blood Flow Metab 2019, 39, 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weksler BB; Subileau EA; Perriere N; Charneau P; Holloway K; Leveque M; Tricoire-Leignel H; Nicotra A; Bourdoulous S; Turowski P; Male DK; Roux F; Greenwood J; Romero IA; Couraud PO Blood-Brain Barrier-Specific Properties of a Human Adult Brain Endothelial Cell Line. FASEB J. 2005, 19, 1872–1874. [DOI] [PubMed] [Google Scholar]
- 43.Chung HH; Chan CK; Khire TS; Marsh GA; Clark A Jr.; Waugh RE; McGrath JL Highly Permeable Silicon Membranes for Shear Free Chemotaxis and Rapid Cell Labeling. Lab Chip 2014, 14, 2456–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tietz S; Engelhardt B Brain Barriers: Crosstalk between Complex Tight Junctions and Adherens Junctions. J. Cell Biol 2015, 209, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Förster C; Burek M; Romero IA; Weksler B; Couraud PO; Drenckhahn D Differential Effects of Hydrocortisone and TNFα on Tight Junction Proteins in an In Vitro Model of the Human Blood-Brain Barrier. J. Physiol. (Oxford, U. K.) 2008, 586, 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weksler B; Romero IA; Couraud PO The hCMEC/D3 Cell Line as a Model of the Human Blood Brain Barrier. Fluids Barriers CNS 2013, 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abbott NJ; Ronnback L; Hansson E Astrocyte-Endothelial Interactions at the Blood-Brain Barrier. Nat. Rev. Neurosci 2006, 7, 41–53. [DOI] [PubMed] [Google Scholar]
- 48.Cheslow L; Alvarez JI Glial-Endothelial Crosstalk Regulates Blood-Brain Barrier Function. Curr. Opin. Pharmacol 2016, 26, 39–46. [DOI] [PubMed] [Google Scholar]
- 49.Kutuzov N; Flyvbjerg H; Lauritzen M Contributions of the Glycocalyx, Endothelium, and Extravascular Compartment to the Blood–Brain Barrier. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E9429–E9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santaguida S; Janigro D; Hossain M; Oby E; Rapp E; Cucullo L Side by Side Comparison Between Dynamic versus Static Models of Blood-Brain Barrier In Vitro: A Permeability Study. Brain Res. 2006, 1109, 1–13. [DOI] [PubMed] [Google Scholar]
- 51.Cucullo L; Couraud P-O; Weksler B; Romero I-A; Hossain M; Rapp E; Janigro D Immortalized Human Brain Endothelial Cells and Flow-Based Vascular Modeling: A Marriage of Convenience for Rational Neurovascular Studies. J. Cereb. Blood Flow Metab 2008, 28, 312–328. [DOI] [PubMed] [Google Scholar]
- 52.Lippmann ES; Azarin SM; Kay JE; Nessler RA; Wilson HK; Al-Ahmad A; Palecek SP; Shusta EV Derivation of Blood-Brain Barrier Endothelial Cells from Human Pluripotent Stem Cells. Nat. Biotechnol 2012, 30, 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lippmann ES; Al-Ahmad A; Palecek SP; Shusta EV Modeling the Blood-Brain Barrier Using Stem Cell Sources. Fluids Barriers CNS 2013, 10, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lippmann ES; Al-Ahmad A; Azarin SM; Palecek SP; Shusta EV A Retinoic Acid-Enhanced, Multicellular Human Blood-Brain Barrier Model Derived from Stem Cell Sources. Sci. Rep 2014, 4, 4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Appelt-Menzel A; Cubukova A; Gunther K; Edenhofer F; Piontek J; Krause G; Stüber T; Walles H; Neuhaus W; Metzger M Establishment of a Human Blood-Brain Barrier Co-Culture Model Mimicking the Neurovascular Unit Using Induced Pluri- and Multipotent Stem Cells. Stem Cell Rep. 2017, 8, 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neal EH; Marinelli NA; Shi Y; McClatchey PM; Balotin KM; Gullett DR; Hagerla KA; Bowman AB; Ess KC; Wikswo JP; Lippmann ES A Simplified, Fully Defined Differentiation Scheme for Producing Blood-Brain Barrier Endothelial Cells from Human iPSCs. Stem Cell Rep. 2019, 12, 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dehouck B; Fenart L; Dehouck MP; Pierce A; Torpier G; Cecchelli R A New Function for the LDL Receptor: Transcytosis of LDL Across the Blood-Brain Barrier. J. Cell Biol 1997, 138, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dal Magro R; Ornaghi F; Cambianica I; Beretta S; Re F; Musicanti C; Rigolio R; Donzelli E; Canta A; Ballarini E; Cavaletti G; Gasco P; Sancini G ApoE-Modified Solid Lipid Nanoparticles: A Feasible Strategy to Cross the Blood-Brain Barrier. J. Controlled Release 2017, 249, 103–110. [DOI] [PubMed] [Google Scholar]
- 59.Czupalla CJ; Liebner S; Devraj K In Vitro Models of the Blood-Brain Barrier In Cerebral Angiogenesis: Methods and Protocols, Milner R, Ed.; Humana Press: New York, 2014; pp 415–437. [DOI] [PubMed] [Google Scholar]
- 60.Reinhardt N; Adumeau L; Lambert O; Ravaine S; Mornet S Quaternary Ammonium Groups Exposed at the Surface of Silica Nanoparticles Suitable for DNA Complexation in the Presence of Cationic Lipids. J. Phys. Chem. B 2015, 119, 6401–6411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.