Significance
This study applies coalescence modeling to a “slowly evolving” bacterial pathogen, Bacillus anthracis, to derive estimates of infection durations and founding population sizes from natural anthrax mortalities. Although coalescence modeling has been applied to highly mutable chronic pathogens (i.e., HIV), to date, methodological hurdles have prevented its wider application. Our findings show it is possible to obtain pathological data from infections, post hoc, which may be applicable to other pathogens and settings, including clinical. Given their higher resolution, microsatellites will remain useful in shorter evolutionary timeframe studies.
Keywords: pathology, population dynamics, bacterial pathogens
Abstract
Bacillus anthracis, the etiological agent of anthrax, is a well-established model organism. For B. anthracis and most other infectious diseases, knowledge regarding transmission and infection parameters in natural systems, in large part, comprises data gathered from closely controlled laboratory experiments. Fatal, natural anthrax infections transmit the bacterium through new host−pathogen contacts at carcass sites, which can occur years after death of the previous host. For the period between contact and death, all of our knowledge is based upon experimental data from domestic livestock and laboratory animals. Here we use a noninvasive method to explore the dynamics of anthrax infections, by evaluating the terminal diversity of B. anthracis in anthrax carcasses. We present an application of population genetics theory, specifically, coalescence modeling, to intrainfection populations of B. anthracis to derive estimates for the duration of the acute phase of the infection and effective population size converted to the number of colony-forming units establishing infection in wild plains zebra (Equus quagga). Founding populations are small, a few colony-forming units, and infections are rapid, lasting roughly between 1 d and 3 d in the wild. Our results closely reflect experimental data, showing that small founding populations progress acutely, killing the host within days. We believe this method is amendable to other bacterial diseases from wild, domestic, and human systems.
Questions regarding pathology of microorganisms are often addressed using animal models. Since the validation of germ theory (using Bacillus anthracis) (1), animal models have been used to elucidate various parameters of infection, such as infectious dose, strain lethality, disease pathology, and host immune response (2, 3). In most studies, inbred, small-animal lines are used where age, sex, diet, and other variables are controlled to reduce immune response variation among individuals. Yet, it is difficult to assess to what degree these controlled studies reflect how these infectious agents behave in natural hosts. This is due to variation in immune response within heterogeneous host populations where genetic and life history variation can affect the outcome of an infection (4). Furthermore, use of natural hosts in pathological studies can be, in practice, impossible, due to necessary permissions, facilities, and ethical considerations. As a result, disease pathology data are lacking in most large, wild hosts and leave general pathological questions, regarding these species, open.
The Gram-positive, spore-forming bacterium B. anthracis causes anthrax. An acute infection, anthrax can start via several routes of infection: inhalational, cutaneous, ingestional, and injection. The pathogen occurs globally where its main hosts are large ungulates, yet most mammals and even birds can be susceptible (5, 6). B. anthracis is an “obligately lethal pathogen,” where the host must die for transmission to occur. In some anthrax endemic areas, transmission may be enhanced with the involvement of biting flies and blowflies (7). Yet, regardless of these other types of transmission, anthrax associated with grazing at carcass sites by new hosts is the backbone of its epidemiology across systems (5, 8).
According to Glomski et al. (9), ingestional anthrax infections in mice can start in the upper gastrointestinal tract, associated with previous damage to the epithelium, or in the lower gastrointestinal tract, within the lymphatic tissue of the oropharynx or Peyer’s patches, respectively. Stimulation of phagocytic cells, such as dendritic cells and macrophages to engulf spores via the classic complement pathway (CCP), plays an important role in establishing the infection. Interaction between BclA glycoprotein, a major structural component of the B. anthracis exosporium (10), and complement component C1q stimulate both entry into epithelial cells and further activation of CCP, beginning the complement system cascade, marking them for uptake by phagocytic cells providing carriage across the epithelium to adjacent lymphatic tissues (11). After passage past the epithelium, the disease seems to progress very similarly, regardless of the initial route of infection. Spores germinate to vegetative cells, which proliferate and spread through the draining lymphatic system, notoriously involving the spleen, and shortly thereafter become a systemic infection. Hemorrhaging from orifices occurs around the time of death, releasing B. anthracis into the soil and inducing sporulation allowing the pathogen to survive for years in the environment (8).
In Etosha National Park, Namibia, anthrax has been monitored, but not managed, for roughly 40 y, throughout which an effort was made to sample all discovered mortalities. Plains zebra (Equus quagga) are the most common host for B. anthracis in Etosha. Most of these infections likely occur after ingesting spores while grazing at anthrax carcass sites (8), and not from drinking contaminated water (8) nor from inhalation of spores (12). Anthrax mortalities in zebra peak during the rainy season, where enhanced production of forage occurs at nutrient-rich carcass sites (13). Although the majority of the zebra in Etosha have trace levels of antibodies against B. anthracis (indicating a high exposure rate) (14), disease mortality remains quite low, even in outbreak years (<5%), implying few actually succumb to the infection (15).
Our previous study described how increased exposure to high concentrations of the pathogen increases the probability of infection (8). Experimentally, high doses are used to induce gastrointestinal lethal infection in various ungulates, tens to hundreds of millions of spores (8). This is in contrast to the injection route, where LD50s are only tens to hundreds of spores (5), showing that low doses through certain routes can lead to fatal infection.
To investigate these dynamics in nature, we isolated 30 individual colony-forming units (CFUs) from 11 naturally occurring zebra mortalities and genotyped the 330 isolates using multilocus variable number tandem repeat (VNTR) analysis (MLVA) and single-nucleotide repeat (SNR) data, as these markers mutate quickly enough to allow within-host resolution. In conjunction, we conducted a mutation rate experiment to calculate the average number of mutations per gene per generation (µ), treating each VNTR or SNR as a gene. We then designed a joint maximum likelihood (ML) approach for the coalescent process (16) under constant and variable effective population size (17), leveraging the experimental data and the carcass genotyping data to estimate the time to the most recent common ancestor (TMRCA) and effective population sizes (Ne) starting a given infection. The full mathematical and statistical approach detailed in Methods uses recent theory (18, 19), algorithms (17), and ML techniques using Markov chain Monte Carlo (MCMC) for hierarchical models (20–22).
Results
Genotype Data.
SNR and MLVA data yielded 43 unique genotypes from 11 carcasses (30 isolates per carcass) (SI Appendix, Fig. S1 and Table S1). All data are available per request from either corresponding author, N.C.S. or W.C.T.
Laboratory Experiment.
Assuming a constant population size across the laboratory experiment, the ML estimate for the average number of mutations separating a sample size of two genes, (CI: 0.09, 1.42), using the data cloning (DC) methodology described in Methods. Noting that the average number of mutations that separates two genes, , is defined in terms of the mutation rate µ and the effective population size Ne as , we then used MCMC to sample from the conditional distribution of the TMRCA given the ML estimate of , stored the median TMRCA, and computed Ne = 126.5 (CI: 101.3, 181.3) by dividing the duration of the experiment in generations (n = 214) by that median. The mutation rate per gene, per generation, was then computed as (CI: 0.0005, 0.004).
Carcass Sampling.
Assuming constant population size from the zebra carcasses, estimates of θ varied between 0.28 and 1.1, and thus, assuming a mutation rate µ = 0.002, the effective population size of B. anthracis varied between 77.24 and 301.08. TMRCA varied between 24.26 d and 91.46 d (Table 1).
Table 1.
Parameter estimates for both constant size and exponential population growth models
| Constant | Exponential | |||||||
| Zebra no. | θ | Ne | TMRCA | θ0 | β | Ne (1) | TMRCA | CFU |
| 1 | 1.05 | 286.16 | 88.14 | 1.9(1.4,2.63) | 0.69(0.2,1.18) | 215.94(120.18,528.12) | 1.47(0.86,4.35) | 1.71(0.95,4.17) |
| 2 | 1.08 | 294.08 | 89.41 | 1.92(1.39,2.7) | 0.74(0.23,1.23) | 212.13(116.52,514.11) | 1.39(0.83,3.94) | 1.68(0.92,4.06) |
| 3 | 0.79 | 215.5 | 67.18 | 1.88(1.38,2.62) | 0.57(0.1,1.08) | 232.67(118.23,634.15) | 1.74(0.94,7.33) | 1.84(0.93,5.01) |
| 5 | 0.52 | 142.3 | 44.13 | 1.88(1.38,2.61) | 0.36(0.1,0.89) | 295.38(126.63,636.11) | 2.61(1.13,7.22) | 2.34(1.00,5.03) |
| 7 | 1.09 | 297.81 | 90.85 | 1.92(1.38,2.72) | 0.76(0.25,1.26) | 209.15(114.66,508.35) | 1.36(0.81,3.75) | 1.65(0.91,4.02) |
| 8 | 1.03 | 280.55 | 82.05 | 2.02(1.27,3.33) | 0.96(0.29,1.6) | 193.25(88.07,621.59) | 1.11(0.64,3.37) | 1.53(0.70,4.91) |
| 9 | 0.6 | 163.24 | 49.7 | 1.91(1.31,2.88) | 0.58(0.1,1.18) | 235.22(101.56,708.49) | 1.74(0.86,7.23) | 1.86(0.80,5.60) |
| 13 | 1.1 | 301.08 | 91.46 | 1.92(1.38,2.75) | 0.77(0.25,1.28) | 208.01(112.63,509.58) | 1.33(0.8,3.69) | 1.64(0.89,4.03) |
| 14 | 0.52 | 142.3 | 44.13 | 1.88(1.38,2.61) | 0.36(0.1,0.89) | 295.38(126.63,636.11) | 2.61(1.13,7.22) | 2.34(1.00,5.03) |
| 17 | 0.53 | 145.22 | 45.06 | 1.89(1.38,2.66) | 0.45(0.1,1) | 260.27(116.15,650.11) | 2.15(1.01,7.21) | 2.06(0.92,5.14) |
| 19 | 0.28 | 77.24 | 24.26 | 2.42(1.67,3.64) | 1.62(0.63,2.47) | 118.09(52.91,375.73) | 0.73(0.47,1.81) | 0.93(0.42,2.97) |
is the average number of mutations that separates two genes under the coalescent process. It is defined as twice the effective population size Ne times the mutation rate µ. This number remains the same under the constant effective population size model. Under the exponential population growth model, the zebra’s B. anthracis population value of at the moment of death is , and the effective population size changes (from present to past) according to the exponential function , where is the exponential rate parameter and . Accordingly, Ne (1) represents the effective population size of B. anthracis in each zebra at the moment of infection using the experiment’s estimated mutation rate (see full model and statistical analyses description in Methods). Confidence intervals are calculated only for the exponential population growth model, since it was the best fit to the data. TMRCA is expressed in days, assuming a mutation rate of 0.002. The founding size of the population has been converted to CFU from the effective population size from the exponential model.
The exponential model gave radically different results. In that model, it is assumed that the effective population grows exponentially from past to present at a rate β. Under the coalescent process, this exponential growth model for the effective population size is formulated as a change from the present (zebra’s time of death) to the past until the time of infection by a “founder” B. anthracis population using . In this model, changes over time according to (see full description of the model in Methods). Estimates of the B. anthracis population θ at the moment of zebra death are given by the value of at time 0 and are denoted . Its estimates for each zebra varied between 1.88 and 2.42, with β values ranging between 0.36 and 1.62 (Table 1). The effective population size of the founder B. anthracis population (i.e., at the beginning of the infection) is denoted as (see Methods) and was estimated to range between 118.09 and 295.38 (Table 1). Ne values are converted to CFUs using the Ne scaling given by the mutation rate experiment. Since this experiment was started with 1 CFU, we then scaled effective population sizes assuming that Ne of 126.5 = 1 CFU. The CFUs at the moment of infection estimated for all sampled zebras ranged between ∼1 and 3 (Table 1). The estimated TMRCA from the coalescent model was used as an estimate of the elapsed time from the moment of infection with a founder B. anthracis population until death (see Methods). This estimate varied between 0.73 d and 2.61 d for all zebras (Fig. 1). Full results of the estimates of θ, , Ne, and TMRCA and CIs for each parameter are shown in Table 1. Finally, model selection through likelihood ratio tests (LRTs) showed the exponential population growth model was a better fit to the data for all zebras (P value < 0.0001 in every case).
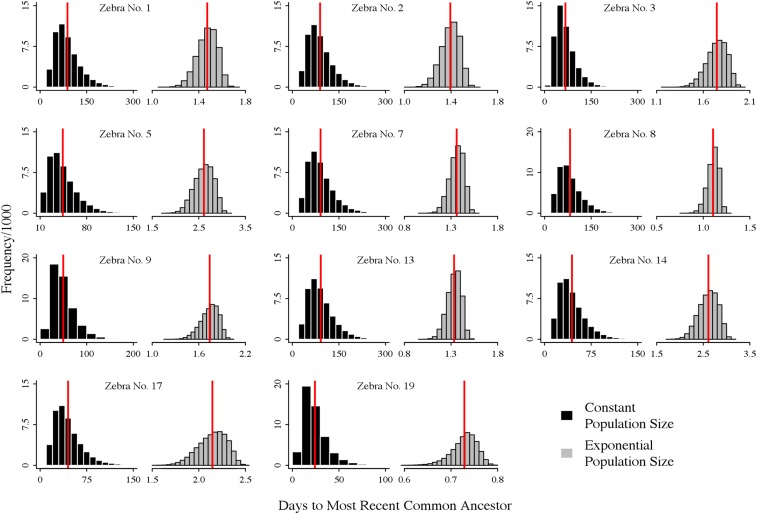
Fig. 1.
Histograms of the TMRCA for 11 zebra carcasses plotted for 50,000 samples of the posterior distribution given the likelihood of the constant population size (black) and exponential population growth (gray) models. According to DC theory, the ML estimate of TMRCA (red vertical line) is given by the mean of these 50,000 samples. The estimates have been rescaled to represent time in days and not coalescent time.
Discussion
Our best results, not surprisingly, were from the exponential model, as this most closely resembles the population growth dynamics of B. anthracis. From these data, we show estimates of parameters of lethal anthrax infections in free-ranging wildlife postmortem. Experimentally, infections have a short duration of infection and, via injection models, low infectious doses (23). Somewhat similar studies have estimated duration of infection for chronic and highly mutable viral pathogens, namely HIV (24). We use this method to estimate both duration of infection and infecting founding population size on a slow-evolving, acute, bacterial pathogen (25). It should be noted that the model used here applies to B. anthracis, as the assumptions we make reflect the biology of this highly clonal pathogen. Stratilo and Bader (26) were the first to describe the use of SNRs to characterize diversity within infections. To date it is likely the only developed typing system using SNRs (27).
Population Dynamics.
B. anthracis populations fluctuate through transmission and infection stages. During an infection, the population increases exponentially and, afterward, goes through three transmission bottlenecks (Fig. 2) to start an infection in a new host. These bottlenecks occur in succession. The first is a slow process of spore decay at carcass sites. This decay may be augmented slightly by some vegetative activity during this telluric process (28); nevertheless, the overall trend is decay (Fig. 2, points C to D), a process taking years (8). The other two bottlenecks occur during the infection process, first, upon ingestion of a subset of spores (ingested dose) from a carcass site, and, finally, a bottleneck as a portion of the ingested population that establishes the infection (founders), which we calculate here in this study (Table 1).
Fig. 2.
Illustration of population dynamics of B. anthracis through infection-transmission cycles for log(N) B. anthracis population (shaded yellow) over time (split into days and years). Point A denotes ingestion: Ungulates grazing at carcass sites ingest a portion of the spores present along with forage and soil, creating a bottleneck. Point B denotes crossing epithelium: After ingestion, only a portion of the ingested cells cross the epithelium, starting the infection. Point C denotes climax population: the population climax, near the time of death. Point D denotes local pathogen extinction: the point where no infectious spores remain at the carcass site.
Grazing and Exposure to B. anthracis (Fig. 2, Point A).
While many vertebrates are suitable hosts for B. anthracis, the foraging behavior and overall ecology of many herbivores leads them to be the major hosts and maintainers of anthrax in natural settings. Here, ingestional anthrax, contracted from grazing at contaminated carcass sites (13), is purportedly the most common pathway of infection in wild and domestic ungulates, although other routes of transmission may occur (7, 8, 29, 30). For E. quagga in Etosha, grazing and ingestion of spores via contaminated plants and soil represents the largest hazard. It is difficult to know how strong of a bottleneck occurs between the ingested dose and the infecting dose, as the dose ingested is likely to be highly variable depending on site age and host behavior. However, simulation models of zebra foraging behavior indicate that there is a high probability of ingesting doses up to 106 spores with even a bite or two of grass at a carcass site within the first 2 y (8). Over 5 y of simulations, there remained a spike in the probability of ingesting doses up to 105 to 106; doses higher than this were highly improbable.
Establishment of the Infection (Fig. 2, Point B).
After ingestion, the process of infection establishment begins. For mouse gastrointestinal animal models, two major locations, the oropharynx (when epithelium is damaged) and/or Peyer’s Patches, are tissues commonly associated with B. anthracis entry from the lumen into the body (9). In wild ungulates, infection establishment has been speculated to be enhanced through damaged tissues caused by rough forage (31, 32) or gut parasites such as helminths due to higher activity of immune cells at these wound sites (32). Entry occurs through phagocytosis of spores by macrophages, carriage across the epithelium, and transport to lymphatic tissue. After phagocytosis, spores germinate and the vegetative cells escape the phagosome, starting the infection (33). High proportions of spores can germinate within hours, but can also be quite staggered, depending on germinates present (34).
Although anthrax establishes via several routes of infection, crossing the epithelium is typically mediated through macrophages, and, from our data and in accordance with Lowe et al. (35), B. anthracis incurs a large population bottleneck starting the infection. Parsimoniously, our data suggest a small population can result in these animals and progress quickly to a lethal infection. The majority of the subsequent population diversity seems to be arising in-host; hence, there are very similar diversity patterns among infections. Likewise, Lowe et al. (35) suggest a similar mechanism creating a bottleneck for an intranasal anthrax model, where a substantial population bottleneck occurs between the inoculum and the founding population in the nasal mucosa-associated lymphoid tissue.
For anthrax, route of infection greatly affects the necessary dose to reach an LD50. This is especially true between oral and injectional routes, where the epithelium acts as an effective barrier to infection. For instance, de Vos (36) reports that kudu (Tragelaphus strepsiceros) ingestional lethal doses were estimated at 1.5 × 107 (range 1 × 106 to 6.5 × 107), while a parenteral (injected) dose of 250 cells proved fatal to impala (Aepyceros melampus). These data also reflect trends for sheep where lethality for ingestional anthrax requires much larger doses and only tens of cells required via injection (23). By our estimates, the founding population reflects the number of spores which crossed the epithelium and successfully germinated to start the infection. Despite our estimated low number of spores, large doses of ingested spores may be required to start gastrointestinal anthrax infections. Where BclA on the outmost coat stimulates the classical complement system (11), a high dose might be needed to produce an adequate innate immune response to stimulate macrophages and dendritic cells to take up spores marked with C3 fragments. Strikingly, infectious doses among zebras in this study were very similar, which reflects pathogen diversity and suggests some common pathology for B. anthracis and/or a shared trait among the individual zebra mortalities, such as genetic, behavioral, or life history, including previous exposure.
The success of using coalescence modeling to estimate Ne and TMRCA depends on having enough genetic resolution within the sampled population. This means having sampled enough individuals from a given population in combination with a high enough diversity, which corresponds to mutation rate. Although pathogens such as B. anthracis, Yersinia pestis, and others are often referred to as “highly clonal” or “slowly evolving,” it is important to make some distinctions. These pathogens are often classified this way due to high sequence similarity in coding regions, yet mutations such as indels (including VNTRs/SNRs) and genomic rearrangements are ignored in this classification. This is especially true with the use of genome sequencing for population studies, where, most often, resequencing and aligning to a reference are used, which often, technically, have hurdles in assembling larger VNTRs and ignore rearrangements in favor of reference synteny. Yet, longer read technology and de novo alignment will make these data available. In conclusion, this method may be quite amendable to other disease systems and even clinical settings, given that these types of markers (VNTRs and SNRs) are used and may yield valuable information for curtailing disease transmission.
Methods
Study Area.
This study was conducted using isolates of B. anthracis collected from anthrax carcasses in central Etosha National Park, Namibia, from 2008 to 2012. Anthrax is endemic in Namibia, and Etosha National Park has regular annual outbreaks of anthrax recorded primarily in grazing herbivores (37, 38). More than 50% of anthrax cases recorded are of plains zebras (E. quagga), and, among the herbivorous species, zebras show the strongest propensity for foraging on grasses at anthrax carcass sites (13).
Isolation of B. anthracis from Blood Swabs.
Culture and isolation of B. anthracis was done at the Etosha Ecological Institute’s pathogen laboratory. Dried, refrigerated carcass swabs from 11 zebra anthrax mortalities with three zebra from 2008, four from 2009, two from 2010, and two from 2012 (SI Appendix, Fig. S2) were used to collect isolates for this study. Swabs were rehydrated in 1.5 mL of sterile distilled water and agitated occasionally for several minutes to suspend spores. Dilutions of 10−2, 10−4, and 10−6 were prepared and plated on PLET (polymyxin-lysozyme-EDTA-thallous acetate) agar using 5 μL of each dilution and the undiluted with an additional 50 μL of sterile, distilled water to spread the sample evenly over the agar. Thirty isolated colonies were selected from among the plates for each carcass. If a particular morphology was in doubt as to whether or not it was B. anthracis, standard confirmation tests (penicillin and Ɣ-phage) on a representative from that morphology were done before picking samples. Entire colonies were transferred from the culture plates to 0.5-mL cryotubes containing 0.25 mL of PLET agar, using sterile toothpicks, and incubated for several days at 37 °C before shipping at ambient temperature to University of Florida in Gainesville.
Mutation Rate Experiment Methods.
An isolate was obtained from a blood swab from a zebra carcass containing the most common genotype in Etosha (genotype 6) according to Beyer et al. (39). This isolate is from A.Br.003 (A.Br.Aust94) using Van Ert et al.’s (40) global classification, and group 5.4 using a new population genomic classification (41). The zebra carcass was found on 22 February 2010 (carcass ID: EB100224-01WT). The colony was placed into 25 mL of Difco nutrient broth in a 50-mL tube and mixed gently in an incubator at 37 °C (range 35 °C to 41 °C) for 24 h. The remaining part of the colony was transferred to a cryotube to preserve as the initial diversity for the experiment. After 24 h, the B. anthracis culture in nutrient broth was diluted to 10−6 in sterile water. We then inoculated 1 µL of 10−6 dilution into 60 50-mL tubes each with 25 mL of nutrient broth. These 60 samples were gently mixed in the incubator at 37 °C for 24 h. From these original 60 tubes, five additional serial transfers were done. Isolates from the 60 lineages and the progenitor were shipped to University of Florida. The starting isolate used for this experiment was sequenced and is available on GenBank (Submission ID: SUB6568587; Sequence accession: SAMN13323522; Bioproject accession: PRJNA590262) (42).
DNA extraction.
At University of Florida, isolates were grown on 5% sheep blood agar for 24 h to 48 h, and DNA was isolated using a modification of the method presented by Van Ert et al. (40).
MLVA-25 genotyping.
MLVA-25 genotyping was performed as described by Lista et al. (43), with minor changes in PCR chemistry and volumes to reduce genotyping costs and adaptations in primer labeling to accommodate analyses on the Applied Biosystems (ABI; Applied Biosystems) instruments. Briefly, cold start, multiplex PCR was performed using 5.0-µL reactions (rxn) containing 0.5 U/rxn Taq DNA Polymerase (Syd Laboratories), 1× Syd Taq Buffer (contains MgCl2), 1× concentration of multiplex primer mix, 0.25 mM each 2′-deoxynucleoside 5′-triphosphates (dNTPs) (Applied Biosystems), and 0.5 µL of template DNA. Thermal cycling conditions were as per Lista et al., with the exception of omitting the initial denaturation step (cold start polymerase). PCR products were diluted 1:40 by the direct addition of 195 µL of molecular-grade water to the PCR plates, and 1.0 µL of diluted product was added to 19.0 µL of a formamide/LIZ 1200 (ABI) size standard mixture (0.285 μL size standard per well) and denatured. Electrophoresis was conducted on an ABI 3730 sequencer and fragment sizes determined using GeneMapper software (Applied Biosystems).
SNR-4 genotyping.
The four SNR loci described in Kenefic et al. (27) were amplified in multiplex. The 10.0-µL PCRs were carried out with final concentrations of the following: 1.0 µL of template DNA per reaction, 1× PCR buffer, 0.5 U per reaction Pyrococcus furiosus (pfu) Polymerase (Agilent Technologies), 3 mM MgCl2*, and 0.25 mM each dNTP. The final primer concentrations in the reaction were 0.1 µM HM-1, 0.15 µM HM-2, 0.1 µM HM-6, and 0.25 µM HM-13. The PCR products were diluted 1:20, and 1.0 μL was mixed with 19.0 μL of a formamide/LIZ 500 (Applied Biosystems) size standard mixture (0.285 μL of standard per rxn) and denatured. Fragment sizing for SNR-4 was performed on an ABI 3730 (Applied Biosystems), and array sizes were determined using GeneMapper software (Applied Biosystems).
Modeling Approach: An Overview.
In what follows, we briefly overview our modeling approach using the coalescent process (16), the rationale of our analyses, and the questions we sought to answer with them. Then, we give a detailed statistical account of our methodologies.
Here we used statistical inference for the coalescent process (16) to leverage the results from the serial passage culturing of B. anthracis, and the MLVA and SNR types sampled from the 11 zebra carcasses. In a landmark paper, Tavaré et al. (44) showed how to use computational sampling methods to estimate the TMRCA from a sample of size n genes and the count of “segregating sites,” or the number of variable loci in these genes. Critical for their inferential approach is the adoption of a mutation model. As these authors mention, a wide variety of models for the mutation process can be incorporated into the coalescent. When the data are DNA sequences, the infinitely-many-sites model (45) may be appropriate. This model is commonly applied to sequence data (e.g., cytochrome b mitochondrial DNA [mtDNA] used in ref. 46 to infer ancestry) and variation at loci among the sampled genes. In this case, we refer to a gene as a sequence from an individual (or sample in our case). Specifically, these datasets consist of the sequence of nucleotides at a specific region of the genome for which individuals are variable at specific loci within the region. The number of these variable loci is the number of segregating sites, which is critical for our calculations. Furthermore, identical sequences within a group of individuals are labeled as haplotypes, and their frequencies in the sample are recorded (see figure 1 in ref. 46).
A careful reading of Watterson (45), Ward et al. (46) and Tavaré et al. (44) suggests the infinitely-many-sites model seems to be equally applicable to MLVA and SNR data structure and nature of polymorphic microsatellites. With respect to the data structure, the analogy is as follows: In our case, the equivalent to one DNA sequence haplotype is a series of the MLVA/SNR alleles at every MLVA/SNR locus found in one sample (e.g., SI Appendix, Table S2A). In what follows, we call each different sequence of MLVA/SNR alleles an MLVA/SNR haplotype. Also, just as with the mtDNA data, we also have the observed frequencies of each one of the MLVA/SNR haplotypes within the samples in each zebra. The annotated table of MLVA/SNR haplotypes and their frequencies is shown in SI Appendix, Table S1. In that table, ni refers to the total number of samples for zebra i (i = 1, 2… 11). For more details about the data structure and notation, see the example in Statistical Analyses.
With respect to the biological justification of the applicability of the infinitely-many-sites model to the MLVA/SNR dataset, the analogy with Watterson’s (45) setting is as follows. Watterson first assumed, as his data unit, a portion of DNA specifying a single polypeptide chain of an enzyme (a functional “gene”). Recombination due to crossing over could be ignored so new alleles only result from mutation. Furthermore, the model does not require accommodating linkage and/or independence among loci. The model name, “infinitely-many-sites,” assumes no two mutations ever occur at the same site (locus), so, at each site, there are only two possible nucleotides: the original wild type and the mutant type. In our case, then, adopting this model assumes the interallelic mutations at each MLVA/SNR locus are symmetrical and identical. Although we recognize this assumption is a simple approximation of reality, it allows a clever MCMC solution by Ewens and Joyce (17) (described in Statistical Analyses) to bypass the integration over all genealogies and target the estimation of the TMRCA, while ignoring the estimation of the topology of the genealogical tree among the MLVA/SNR genes. Having a quick access to the estimation of the TMRCA allowed us to, first, estimate the TMRCA from the serial transfer experiments, calibrate this coalescent time with real time units (in days), and estimate a laboratory effective population size and mutation rate. Second, it allowed us to estimate the time (in days) from initial host infection to host death as the TMRCA between all of the MLVA/SNR variants sampled within a single host, for each host. Third, it allowed us to carry out a test of the hypothesis of within-host exponential growth of the effective population size vs. the usual coalescent assumption of constant effective population size. Infection by B. anthracis undergoes at least two bottlenecks driven by host resistance in specific organs (35), suggesting that a model with exponential growth posterior to initial infection might be a more realistic scenario than the constant population size model. Fourth, adopting the infinitely-many-sites model allowed estimates of the effective population size of the MLVA/SNR genes upon death for each zebra. Finally, our methodology also allowed us to estimate the effective population size for these genes at the onset of host infection. In that sense, the joint estimation of the effective population size and the hypothesis test mentioned above allowed us to distinguish between two hypotheses: 1) Each host was initially invaded with a large B. anthracis load which did not grow significantly; 2) zebra were initially infected with a small B. anthracis load, which grew fast and exponentially during infection. The comparison of the effective population sizes with the laboratory effective population size which underwent various bottlenecks allowed us to discuss the within-host population processes from the time of infection until host death.
In what follows, we delve into the mathematical modeling details, starting with the description of the model parameters and likelihood functions under both models, and detailing the coalescent time scaling transformation to real time units.
Statistical Analyses.
Data structure and general model setting.
Before setting our statistical notation, recall that, here, our functional “gene” unit is the B. anthracis genome, genotyped for 25 MLVA and four SNR sites for any one sample within a zebra. For zebra 2, for example, for which there were 26 samples (our “genes”), four MLVA/SNR sites were variable (see SI Appendix, Table S2 A and B for the table presenting the raw data). These samples have seven distinct MLVA/SNR haplotypes. Heretofore, we will simply say for zebra 2 we have 26 sampled genes and seven MLVA/SNR haplotypes, each one with frequencies shown in SI Appendix, Table S2C.
The key parameter in the coalescent process with neutral mutations is , the average number of mutations separating a sample of size n = 2 genes. Furthermore, , where is the “effective population size” and µ is the mutation rate (per gene, per generation). “N-Coalescent” time is measured retrospectively, with 0 being at present and increasing from present to past. Formally, this stochastic process is a pure death process (16), where the quantity that is “dying” is the number of distinct gene lineages, from present to past. This effective population size is assumed constant over time and is defined as the size of the “population” of genes from which the samples in the present time are taken. This quantity is equal to the census population size in an idealized Wright−Fisher model (19). Although is an abstract parameter, for a real biological population, it is proportional to the rate at which genetic diversity is lost or gained. In the absence of natural selection and if the variation in the number of descendant genes per gene as well as the generation time are known, a census population size can be approximated (47). To date, statisticians working in this field (e.g., ref. 19) adopt a more cautious interpretation of the effective population size and simply see it as a measure of relative genetic diversity (48, 49). In any case, this parameter (Ne) is useful, because, under the coalescent, time is rescaled so one unit of continuous coalescent time is equivalent to generations ( is used in diploid models). With that scaling, we can transform our estimated TMRCA expressed in coalescent time units into real time units.
Several coalescent-based methods for estimation of were derived using stringent and flexible assumptions, such as constant population size, exponentially growing population size, logistic, and piecewise linear. To remove the inflexible conditions imposed by adopting any time-dependent model, Palacios and Minin (19) go so far as to propose a nonparametric, stochastically varying Markov Random Field model for (19). Even this last complex model formulation can be tied to a specific mathematical model of population dynamics: a translated Stochastic Gompertz diffusion model of population size growing under environmental variability (18). Because most implementations of the coalescent under variable population size can be tied to a population dynamics rationale, we opted for testing the applicability of the constant vs. the exponentially growing as way to compromise between biological realism and estimability of parameters in the light of the data. Although most of these methodologies have been implemented and readily available software exists (e.g., “BEAST”) to analyze the data under different models, these programs rely on a set of hard-coded genetic mutation models to carry the likelihood calculation by integrating the genealogy likelihood over all possible genealogies (50). Because we are mainly interested in the estimation of the TMRCA and not in the topology of the within-host genealogies, we used the approach proposed by Ewens and Joyce (17) to deal with this case, to swiftly bypass the topology estimation problem. Although, in their lecture notes, Ewens and Joyce only outline this approach, here we coded it de novo and extended it for the joint estimation of and the TMRCA (scaled to real time units) under a constant effective population size model and an exponentially growing effective population size model. The code was originally written by one of us (J.M.P.) during a mathematical population genetics workshop taught in 2009 by Joyce, Ewens, Krone, and Ponciano at the Center for Research in Mathematics in Guanajuato, Mexico.
The joint distribution of coalescent times.
The coalescent process is a continuous-time, discrete-state Markov death process, which is initiated at the present time by gathering a random sample of n genes from a population of Ne genes. Then, the process models how the number of distinct gene lineages sampled in the present decreases one at a time when we traverse time from the present to the past. When two genes sampled today find a common ancestor j generations back into the past, we say a “coalescence” has occurred. These “coalescent events” happen until all genes in a sample have found a common ancestor. Kingman (16) and multiple authors subsequently described the mathematical properties of the retrospective and random time period elapsed since the moment one finds n genes in a sample until all of these genes have found their most common recent ancestor (TMRCA). Regardless of the assumptions about the size of Ne, TMRCA adopts a probability distribution that can be thought of as the sum of all of the intercoalescent times in a genealogy, which are all of the time periods between two consecutive coalescences in a genealogy. Using stochastic processes terminology, these intercoalescent times are the interevent times of the Markov death process.
One attractive feature of the coalescent model is its mathematical simplicity, which allows an intuitive understanding of the model properties and of the intercoalescent events using simple biological and probabilistic rationales. The number of discrete generations from the present to the past until the first coalescence occurs is modeled using a geometric random variable where the “success” probability p is the probability that, in a sample of n genes, two individuals find a common ancestor one generation in the past. Its complement, 1 − p, is the probability that no coalescence occurs. Thinking of generations as independent trials, the probability of any two genes among these n genes finding a common ancestor j generations back in the past is simply , and the probability of their first common ancestor appearing more than generations ago is . The analytical expression for p is found as follows: The probability any two genes picked at random today have two different ancestors one generation back in the past is , since the first gene has Ne choices for its ancestor and the second has N − 1 choices. The probability that these two genes have a common ancestor one generation back in the past (i.e., that a coalescence occurs) is then simply . This fraction only gives us the value of p for a sample of size 2 genes. Also, note that the expected number of generations until two individuals find their common ancestor is . Iterating the above argument to include three or more genes, it is easy to see that the probability 1 − p that a sample of n genes all find different ancestors one generation back in the past is
and hence the probability that at least one coalescence occurs one generation back in the past is Denoting the intercoalescent, geometrically distributed, random time between k and k − 1 gene ancestors as Uk, it follows that
for constant population size. Now, if Ne is large relative to , coalescent events will occur rarely: Many generations would elapse before a coalescence occurs. It then makes sense to rescale time using a continuous scale instead of discrete generations by measuring it in units of Ne so that coalescent time units (e.g., one coalescent time unit is equivalent to Ne generations). Applying this rescaling is achieved by computing the limit
Thus, measured in continuous time, the intercoalescent time between k and k − 1 gene ancestors can be modeled using an exponential distribution with rate . The TMRCA can be simply modeled as a sum of exponentially distributed intercoalescent times. Using the Markov property, the joint probability distribution of the intercoalescent times is simply written as the product of all of the intercoalescent exponential distributions.
To set notation as well as visualize these intercoalescent times, we plotted a realization of a genealogy under the coalescent process assuming that, at present, a sample of n = 7 genes was gathered (SI Appendix, Fig. S3). In that graph, the ui denote realizations of the (random) intercoalescent times, and ti denote the accumulated time, from the present to the past. Accordingly, under a model of changing effective population size Ne(t), the quantity tk−1 = uk + tk is no longer exponentially distributed. Instead, the pdf of each inter-coalescent time is (50)
and their joint pdf is simply written as the product of these densities, for k = n, n − 1,…, 2. When it is assumed the population grows exponentially from past to present at a rate (or, alternatively, decays exponentially from present to the past), expressed as , then
Mutation in the coalescent.
A mutational model for the coalescent process is derived by thinking once again in discrete generations and then making a continuous time approximation. Let µ denote the probability that the offspring of a gene, from one generation to the next, is a mutant. Let Yr be the total number of mutations accumulated in one gene line of descent after r generations. Under the assumption of independence across lineages, this number of mutations can be modeled with a binomial distribution with probability µ and total number of trials r. Denote S2 the number of mutations separating two individuals. Conditional on the time U2 (in discrete generations) until these two individuals find their most recent common ancestor, , and recalling that , it follows that . Using the same time scale change defined above and replacing r with Net, the binomial probability mass function (pmf) of Yr becomes
as . Thus, mutations in the coalescent are simply modeled with a Poisson process with rate . Critical for this derivation is the conditioning step, and the integration (i.e., calculation of the expected value or average) over all of the possible genealogy lengths separating two individuals. The same integration is needed to compute the overall likelihood functions.
Likelihood function under the coalescent with mutations.
The reader familiar with hierarchical or “state-space models” in biology, will recognize that the coalescent process with mutation is indeed a hierarchical stochastic model. Such models allow researchers to incorporate variability in parameters that otherwise might be unrealistically treated as fixed. In addition, these models allow the incorporation of multiple layers of process and/or observation variability. Until recently, computational difficulties rendered likelihood inference for these models unfeasible, or plainly unreliable. For all but the simplest models, the likelihood function is written as a multidimensional integral. Here we solve this integration problem using DC, which is an efficient and extensively tested computational algorithm to find the ML (20–22, 52–57). The DC theorem allows one to apply a typical Bayesian posterior calculation and MCMC sampling to a number c of copies (clones) of the data (53). When c is large, the sample mean vector of the resulting simulated posterior distribution corresponds to the ML estimates of the parameters. Furthermore, the sample variance−covariance matrix of the posterior, multiplied by c, provides estimates of the variances and covariances of these ML estimates (the inverse of the observed Fisher’s information matrix). Ponciano et al. (22) extended this estimation methodology to a complete inferential approach by proving and demonstrating how DC for hierarchical models can be easily extended to carry model selection, LRTs, and computing profile likelihood intervals with much better coverage than the Wald confidence intervals for small sample sizes. This DC methodology is what we use here. We refer the interested reader to Ponciano et al. (20), who show, step by step, the explicit DC calculations for an analytically tractable example. We favored this methodology because, unlike any available Bayesian software to work with the coalescent process, we can (and did) explicitly and efficiently assess the identifiability and estimability of the model parameters. This assessment is the greatest advantage of using DC for hierarchical models vs. conforming to a Bayesian estimation methodology. Here again, we refer the reader to Ponciano et al. (20) for explicit and extensive accounts of such assessment. In SI Appendix, Table S2, we illustrate the assessment of parameter identifiability using the data coming from one zebra.
With a sample of size n, a total of Sn segregating sites are observed, and the likelihood function is written as the Poisson probability with Sn variants emerging along the genealogy, averaged over all possible genealogies. The joint distribution of the intercoalescent times ui,i = n, n − 1, …, 2 (Fig. 1) is simply the product of their pdfs . For the constant Ne population model, this product is
whereas, for the exponential model where it is assumed the population decays exponentially from the present to the past according to the model ,
Since, along a branch of length u of the genealogy, the number of mutations is distributed Poisson with mean θu/2 for the constant effective population size model, given a particular genealogy (i.e., given an particular set of values of ), the conditional distribution of the total number of mutations along this genealogy is going to be Poisson-distributed with mean θL/2, where
is the total length of a given genealogical tree (SI Appendix, Fig. S3). That is,
For the exponential growth model, the value of changes over time according to , and we arbitrarily assume such changes only occur at the coalescent events, and therefore,
Averaging these Poisson probabilities over all of the possible genealogy lengths gives us the likelihood function as
Both likelihood functions were maximized in the program JAGS (Just Another Gibbs Sampler) (57) using the DC methodology. Our computer code is available at https://github.com/jmponciano/PNAS-Coalescent. After maximizing the likelihood, we used the methodology in Ponciano et al. (22) to compute the ML estimates of the latent variables u2, u2, …, un and of their sum, which is the TMRCA. We also used Ponciano et al.’s (22) DC LRT and model selection tools to test the goodness of fit of the exponential vis-à-vis the constant population size model for the data in all zebra. For the laboratory data, we assumed the constant population size model (59). Joyce et al. (59) demonstrated that the overall dynamics of a serial passage experiment with plasmid-carrying and plasmid-free bacteria mirrored the dynamics during a single day, because bacteria were grown approximately to the same total from one cycle to the next of the experiment. Under these conditions, the bacterial dynamics could be accurately predicted (60) and estimated by assuming a constant bacterial population size at the end of each cycle. The alternative would be to fit a coalescent model with as many bottlenecks as serial passage transfers, which is beyond the scope of this work. The laboratory constant population size assumption allowed us to estimate the laboratory Ne directly from the coalescent time scaling and the known number of elapsed generations throughout the experiment (214, at 6 generations per day). Since our coalescent model fitting gave us the ML estimate of the TMRCA, and one unit of the coalescent time corresponds to Ne discrete generations, we simply obtained our Ne estimate as 214/TMRCA. Since our model fitting also gives us an independent estimate of θ for the laboratory, we could solve for the per generation mutation rate µ = 0.002.
Finally, the value of in the above likelihood can be arbitrarily substituted by θ(t) without affecting the maximum location in parameter space (61–63). After all, both quantities are proportional to each other. After maximization, whenever we fitted the constant population size, we accomplished the transformation from values of θ to values of by dividing by twice the laboratory mutation rate per generation µ. Recalling one unit of coalescent time corresponds to generations for this simple model and knowing the number of generations per day is approximately six, we then transformed the ML estimate of the TMRCA to days and took this value as the estimate of the retrospective number of days from death to infection. For the exponential model, the transformation from coalescent time to generations was accomplished by solving the following question: How many discrete generations j does it take to traverse τ units of exponentially decaying coalescent time, starting from the present to the past?
Suppose the population size j generations back into the past, corresponding to τ coalescent time units is N(j). Because the amount of coalescent time traversed from generation i to i + 1 back in the past is , then, during j generations, the total amount of coalescent time τ is given by
Having an estimate of τ (which, for us, will be the TMRCA), all we did was to solve for j in the above equation, by using the exponential growth model and the integral approximation
Accordingly, .
For both models, we transformed the TMRCA from coalescent time units to real time units assuming two possible values of . First, we estimated Ne using the mutation rate estimated from the laboratory experiment and the ML estimate of θ for each zebra and either the constant population size or exponential population growth models. For the exponential population size model, we then estimated the initial Ne when each zebra was infected using the ML estimates of β in each zebra.
Data Availability.
All data and detailed methods are available upon request to W.C.T. or N.C.S. This includes detailed protocols, data (CFU counts and timetables for the transfer experiment, photos of sampled colonies for the mutation rate experiment genotype data including raw fragment size data, etc.).
Supplementary Material
Acknowledgments
We thank the Ministry of Environment and Tourism in Namibia for permission to conduct research in Etosha National Park, and we are grateful to the scientific staff and managers at the Etosha Ecological Institute for logistical support and assistance. We thank Zoe Barandongo, Claudine Cloete, and Clemens Naomob for laboratory assistance. Funding was provided by NSF Grants OISE-1103054 and DEB-1816161 (to W.C.T.), and the Centre for Ecological and Evolutionary Synthesis (CEES) funded through the Research Council of Norway (RCN) 225031/E31 (to N.C.S.).
Footnotes
The authors declare no competing interest.
Data deposition: The genome data have been deposited on the National Center for Biotechnology Information BioSample (accession no. SAMN13323522) and BioProject (accession no. PRJNA590262). Computer code used in this study is available at GitHub, https://github.com/jmponciano/PNAS-Coalescent.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920790117/-/DCSupplemental.
References
- 1.Koch R., The etiology of anthrax, based on the life history of Bacillus anthracis. Beiträge zur Biologie der Pflanzen 2, 277–310 (1876). [Google Scholar]
- 2.Lee A., et al. , A standardized mouse model of Helicobacter pylori infection: Introducing the Sydney strain. Gastroenterology 112, 1386–1397 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Santos R. L., et al. , Animal models of Salmonella infections: Enteritis versus typhoid fever. Microbes Infect. 3, 1335–1344 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Jirtle R. L., Skinner M. K., Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 8, 253–262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hugh-Jones M., Blackburn J., The ecology of Bacillus anthracis. Mol. Aspects Med. 30, 356–367 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Hugh-Jones M. E., de Vos V., Anthrax and wildlife. Rev. Off. Int. Epizoot. 21, 359–383 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Blackburn J. K., Van Ert M., Mullins J. C., Hadfield T. L., Hugh-Jones M. E., The necrophagous fly anthrax transmission pathway: Empirical and genetic evidence from wildlife epizootics. Vector Borne Zoonotic Dis. 14, 576–583 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Turner W. C., et al. , Lethal exposure: An integrated approach to pathogen transmission via environmental reservoirs. Sci. Rep. 6, 27311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glomski I. J., Piris-Gimenez A., Huerre M., Mock M., Goossens P. L., Primary involvement of pharynx and Peyer’s patch in inhalational and intestinal anthrax. PLoS Pathog. 3, e76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson B. M., Waller L. N., Fox K. F., Fox A., Stewart G. C., The BclB glycoprotein of Bacillus anthracis is involved in exosporium integrity. J. Bacteriol. 189, 6704–6713 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu C., Jenkins S. A., Xue Q., Xu Y., Activation of the classical complement pathway by Bacillus anthracis is the primary mechanism for spore phagocytosis and involves the spore surface protein BclA. J. Immunol. 188, 4421–4431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barandongo Z. R., Mfune J. K. E., Turner W. C., Dust-Bathing behaviors of African herbivores and the potential risk of inhalational anthrax. J. Wildl. Dis. 54, 34–44 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Turner W. C., et al. , Fatal attraction: Vegetation responses to nutrient inputs attract herbivores to infectious anthrax carcass sites. Proc. R. Soc. B Biol. Sci. 281, 20141785 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cizauskas C. A., Bellan S. E., Turner W. C., Vance R. E., Getz W. M., Frequent and seasonally variable sublethal anthrax infections are accompanied by short-lived immunity in an endemic system. J. Anim. Ecol. 83, 1078–1090 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellan S. E., Gimenez O., Choquet R., Getz W. M., A hierarchical distance sampling approach to estimating mortality rates from opportunistic carcass surveillance data. Methods Ecol. Evol. 4, 361–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kingman J. F. C., The coalescent. Stochastic Process. Appl. 13, 235–248 (1982). [Google Scholar]
- 17.Ewens W., Joyce P., “Mathematical population genetics, introduction to the stochastic theory” in Lecture Notes of the Summer School in Probability and Statistics (Center for Research in Mathematics, Guanajuato, Mexico, 2009). https://www.cimat.mx/Eventos/xepe/. Accessed 6 February 2020.
- 18.Ponciano J. M., A parametric interpretation of Bayesian Nonparametric Inference from Gene Genealogies: Linking ecological, population genetics and evolutionary processes. Theor. Popul. Biol. 122, 128–136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palacios J. A., Minin V. N., Gaussian process-based Bayesian nonparametric inference of population size trajectories from gene genealogies. Biometrics 69, 8–18 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Ponciano J. M., Burleigh J. G., Braun E. L., Taper M. L., Assessing parameter identifiability in phylogenetic models using data cloning. Syst. Biol. 61, 955–972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lele S. R., Dennis B., Lutscher F., Data cloning: Easy maximum likelihood estimation for complex ecological models using Bayesian Markov chain Monte Carlo methods. Ecol. Lett. 10, 551–563 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Ponciano J. M., Taper M. L., Dennis B., Lele S. R., Hierarchical models in ecology: Confidence intervals, hypothesis testing, and model selection using data cloning. Ecology 90, 356–362 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Anonymous , Anthrax in Humans and Animals, Turnbull P., Ed. (World Health Organization, ed. 4, 2008). [PubMed] [Google Scholar]
- 24.Dialdestoro K., et al. , Coalescent inference using serially sampled, high-throughput sequencing data from intrahost HIV infection. Genetics 202, 1449–1472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achtman M., Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62, 53–70 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Stratilo C. W., Bader D. E., Genetic diversity among Bacillus anthracis soil isolates at fine geographic scales. Appl. Environ. Microbiol. 78, 6433–6437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenefic L. J., et al. , A high resolution four-locus multiplex single nucleotide repeat (SNR) genotyping system in Bacillus anthracis. J. Microbiol. Methods 73, 269–272 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Braun P., et al. , Microevolution of Anthrax from a young Ancestor (M.A.Y.A.) suggests a soil-borne life cycle of Bacillus anthracis. PLoS One 10, e0135346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turell M. J., Knudson G. B., Mechanical transmission of Bacillus anthracis by stable flies (Stomoxys calcitrans) and mosquitoes (Aedes aegypti and Aedes taeniorhynchus). Infect. Immun. 55, 1859–1861 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basson L., et al. , Blowflies as vectors of Bacillus anthracis in the Kruger National Park. Koedoe 60, a1468 (2018). [Google Scholar]
- 31.Beyer W., Turnbull P. C. B., Anthrax in animals. Mol. Aspects Med. 30, 481–489 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Cizauskas C. A., et al. , Gastrointestinal helminths may affect host susceptibility to anthrax through seasonal immune trade-offs. BMC Ecol. 14, 27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon T. C., Fadl A. A., Koehler T. M., Swanson J. A., Hanna P. C., Early Bacillus anthracis-macrophage interactions: Intracellular survival survival and escape. Cell. Microbiol. 2, 453–463 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Hu H., Emerson J., Aronson A. I., Factors involved in the germination and inactivation of Bacillus anthracis spores in murine primary macrophages. FEMS Microbiol. Lett. 272, 245–250 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Lowe D. E., Ernst S. M. C., Zito C., Ya J., Glomski I. J., Bacillus anthracis has two independent bottlenecks that are dependent on the portal of entry in an intranasal model of inhalational infection. Infect. Immun. 81, 4408–4420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Vos V., The ecology of anthrax in the Kruger National Park, South Africa. Salisbury Med. Bull. 68S, 19–23 (1990). [Google Scholar]
- 37.Turner W. C., et al. , Soil ingestion, nutrition and the seasonality of anthrax in herbivores of Etosha National Park. Ecosphere 4, art13 (2013). [Google Scholar]
- 38.Lindeque P. M., Turnbull P. C., Ecology and epidemiology of anthrax in the Etosha National Park, Namibia. Onderstepoort J. Vet. Res. 61, 71–83 (1994). [PubMed] [Google Scholar]
- 39.Beyer W., et al. , Distribution and molecular evolution of Bacillus anthracis genotypes in Namibia. PLoS Negl. Trop. Dis. 6, e1534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Ert M. N., et al. , Global genetic population structure of Bacillus anthracis. PLoS One 2, e461 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruce S. A., Schiraldi N. J., Kamath P. L., Easterday W. R., Turner W. C., A classification framework for Bacillus anthracis defined by global genomic structure. Evol. Appl., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barandongo Z., Bruce S. and Turner W. C., MIGS Cultured Bacterial/Archaeal sample from Bacillus anthracis. GenBank. https://www.ncbi.nlm.nih.gov/biosample/13323522. Deposited 18 November 2019.
- 43.Lista F., et al. , Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 6, 33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavaré S., Balding D. J., Griffiths R. C., Donnelly P., Inferring coalescence times from DNA sequence data. Genetics 145, 505–518 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watterson G. A., On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975). [DOI] [PubMed] [Google Scholar]
- 46.Ward R. H., Frazier B. L., Dew-Jager K., Pääbo S., Extensive mitochondrial diversity within a single Amerindian tribe. Proc. Natl. Acad. Sci. U.S.A. 88, 8720–8724 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakeley J., Sargsyan O., Extensions of the coalescent effective population size. Genetics 181, 341–345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rambaut A., et al. , The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frost S. D. W., Volz E. M., Viral phylodynamics and the search for an ‘effective number of infections.’ Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1879–1890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Felsenstein J., Inferring Phylogenies (Sinauer Associates, Sunderland, MA, 2004). [Google Scholar]
- 51.Griffiths R. C., Tavare S., Ancestral inference in population-genetics. Stat. Sci. 9, 307–319 (1994). [Google Scholar]
- 52.Bolker B. M., et al. , Generalized linear mixed models: A practical guide for ecology and evolution. Trends Ecol. Evol. (Amst.) 24, 127–135 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Lele S. R., Nadeem K., Schmuland B., Estimability and likelihood inference for generalized linear mixed models using data cloning. J. Am. Stat. Assoc. 105, 1617–1625 (2010). [Google Scholar]
- 54.Solymos P., dclone: Data cloning in R. R J. 2, 29–37 (2010). [Google Scholar]
- 55.Baghishani H., Mohammadzadeh M., A data cloning algorithm for computing maximum likelihood estimates in spatial generalized linear mixed models. Comput. Stat. Data Anal. 55, 1748–1759 (2011). [Google Scholar]
- 56.Campbell D., Lele S., An ANOVA test for parameter estimability using data cloning with application to statistical inference for dynamic systems. Comput. Stat. Data Anal. 70, 257–267 (2014). [Google Scholar]
- 57.Gomez J. P., Robinson S. K., Blackburn J. K., Ponciano J. M., An efficient extension of N-mixture models for multi-species abundance estimation. Methods Ecol. Evol. 9, 340–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plummer M., “JAGS: A program for analysis of Bayesian graphical models using Gibbs sampling” in Proceedings of the Third International Workshop on Distributed Statistical Computing, Leisch F., Hornik K., Zeileis A., Eds. (R Project for Statistical Computing, 2003), pp. 124–125. [Google Scholar]
- 59.Joyce P., et al. , Modeling the impact of periodic bottlenecks, unidirectional mutation, and observational error in experimental evolution. J. Math. Biol. 50, 645–662 (2005). [DOI] [PubMed] [Google Scholar]
- 60.De Gelder L., et al. , Combining mathematical models and statistical methods to understand and predict the dynamics of antibiotic-sensitive mutants in a population of resistant bacteria during experimental evolution. Genetics 168, 1131–1144 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pybus O. G., Rambaut A., Harvey P. H., An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155, 1429–1437 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minin V. N., Bloomquist E. W., Suchard M. A., Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 25, 1459–1471 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paradis E., Paradis M. E., Package ‘coalescentMCMC.’ Biometrika 57, 97–109 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and detailed methods are available upon request to W.C.T. or N.C.S. This includes detailed protocols, data (CFU counts and timetables for the transfer experiment, photos of sampled colonies for the mutation rate experiment genotype data including raw fragment size data, etc.).