Significance
Structural plasticity is a critical property of many protein complexes that has been challenging to study using conventional structural biology tools. Cross-linking mass spectrometry (XL-MS) has become an emergent technology for elucidating architectures of large protein complexes. While effective, current XL-MS methods mostly rely on lysine reactive cross-linking chemistry and have limited capacity in fully defining dynamic structures of protein complexes. Here, we have developed an integrated structural approach based on three MS-cleavable cross-linkers with distinct chemistries. This approach enabled us to obtain highly reliable and comprehensive cross-link data that significantly facilitate integrative structural modeling of dynamic protein complexes. In addition, it has been successfully applied to the COP9 signalosome to determine its structural dynamics associated with its function.
Keywords: COP9 signalosome, cross-linking mass spectrometry, integrative structure modeling, architectures of protein complexes, structural dynamics
Abstract
The COP9 signalosome (CSN) is an evolutionarily conserved eight-subunit (CSN1–8) protein complex that controls protein ubiquitination by deneddylating Cullin-RING E3 ligases (CRLs). The activation and function of CSN hinges on its structural dynamics, which has been challenging to decipher by conventional tools. Here, we have developed a multichemistry cross-linking mass spectrometry approach enabled by three mass spectometry-cleavable cross-linkers to generate highly reliable cross-link data. We applied this approach with integrative structure modeling to determine the interaction and structural dynamics of CSN with the recently discovered ninth subunit, CSN9, in solution. Our results determined the localization of CSN9 binding sites and revealed CSN9-dependent structural changes of CSN. Together with biochemical analysis, we propose a structural model in which CSN9 binding triggers CSN to adopt a configuration that facilitates CSN–CRL interactions, thereby augmenting CSN deneddylase activity. Our integrative structure analysis workflow can be generalized to define in-solution architectures of dynamic protein complexes that remain inaccessible to other approaches.
The COP9 signalosome (CSN) is an evolutionarily conserved and essential multisubunit protein complex involved in diverse cellular and developmental processes in animals and plants (1–3). The CSN functions as a deneddylase, specific for cleaving Nedd8 modification from cullin proteins, the key components of Cullin–RING ubiquitin E3 ligases (CRLs) (4–8). CRLs represent the largest evolutionarily conserved superfamily of multisubunit E3s (5, 6), which embody ∼30% of all human E3 proteins and coordinate degradation of ∼20% of the proteins processed by the proteasome. The dynamic cycle of neddylation and deneddylation of cullins is a critical step in regulating the assembly and activity of CRLs (6, 9, 10). In addition to enzymatic regulation of CRLs, the CSN can inactivate CRLs noncatalytically by direct binding, preventing their association with E2 enzymes and ubiquitination substrates (11–14). While abnormal CRL activity is frequently associated with various human diseases, multiple studies have also identified the CSN as a positive regulator of oncogenes and negative regulator of tumor suppressors (15–19). Moreover, elevated expression of CSN subunits has been found in a number of human tumors, often with poor prognosis (20, 21). Therefore, better understanding of the CSN structure would provide new insights on their function and the regulation of CRLs associated with human pathology.
The canonical CSN complex (hereafter referred to as CSN) typically consists of eight subunits (CSN1–8) (1, 3), including six different PCI (proteasome lid-CSN-initiation factor 3) domain-containing subunits (CSN1 to CSN4, CSN7, and CSN8) and two MPN (MPR1/PAD1 amino-terminal) domain-containing proteins (CSN5 and CSN6). Among them, CSN5 is the catalytic subunit directly responsible for CSN deneddylase activity (4). The CSN complex shares sequence similarities to the 19S proteasome lid subcomplex and the eukaryotic translation initiation complex eIF3, which also contain PCI and MPN domains (1, 3). The X-ray structure of recombinant human CSN has revealed that CSN5 and CSN6 MPN domains form a heterodimer, while the six remaining PCI subunits assemble into a horseshoe-shaped ring from which their arm-like α-helical domains project (22). The PCI subunits provide a scaffold, primarily through CSN2 and CSN4, which facilitates the recruitment of neddylated CRLs. Meanwhile, the two MPN subunits are slightly juxtaposed, exposing the active MPN catalytic core in CSN5 (12, 23–25). All eight subunits are united in a helical bundle formed by their C-terminal carboxyl α-helices, which are stacked between the CSN5–CSN6 dimer and PCI ring. Interestingly, substrate-free CSN exists in an inactive, autoinhibited state (23). Structural and biochemical characterization of CSN–CRL complexes have revealed substrate-induced structural dynamics associated with CSN activation (12, 23–26). Binding of neddylated CRLs to CSN triggers substantial remodeling and extensive conformational changes of the complex, activating the isopeptidase activity of CSN5. Although the structural plasticity of the CSN is important for CSN activation and function in regulating CRL activities in cells, it has not been well characterized due to limitations in existing technologies.
Recently, the ninth CSN subunit, CSN9 (also known as CSNAP [CSN acidic protein]), has been discovered to complex with CSN1–8 stoichiometrically to form a nine-membered noncanonical CSN complex (also known as CSN9-bound CSN, hereafter referred to as CSNn) (27). As canonical CSN subunits (CSN1–8) have a one-to-one correspondence to the subunits of the 19S proteasome lid subcomplex (3, 28), CSN9 is homologous to DSS1, the smallest component of the 19S lid. While CSN9 is not essential for the assembly and catalytic activity of CSN (27), a recent study has suggested that CSN9 reduces the affinity of CSN–CRL interactions, contributing to steric regulation of CRLs (14). The depletion of CSN9 appears to have a global impact on CRL-associated activities, leading to altered reproductive capacity, suppressed DNA damage response, decreased viability, and delayed cell cycle progression (14). It has also been suggested that the C terminus of CSN9 is important in its incorporation within the CSN complex, likely through interactions with CSN3, CSN5, and CSN6 (27). However, due to its small size and highly disordered structure, it remains challenging to accurately determine interaction interfaces between CSN9 and CSN. As a result, no high-resolution structures are available for the CSN9-bound CSN complex. Thus, alternative strategies to dissect the architecture of the noncanonical CSN complex and determine how CSN9 interacts with CSN1–8 are needed to help us uncover structural details underlying the functional importance of CSN9 in cells.
In recent years, cross-linking mass spectrometry (XL-MS) has become a powerful strategy for probing protein–protein interactions (PPIs) (29–31). While effective, XL-MS possesses several inherent challenges, including unambiguous identification of cross-linked peptides due to their complex fragmentation when conventional (i.e., noncleavable) cross-linkers are used. To facilitate MS identification, we have developed a suite of sulfoxide-containing MS-cleavable cross-linkers (e.g., disuccinimidyl sulfoxide [DSSO]) (32–36). These MS-cleavable reagents contain symmetric MS-labile C-S bonds (adjacent to the sulfoxide group) that are selectively and preferentially fragmented prior to peptide backbone cleavage during collision-induced dissociation (CID) (31–36). Such fragmentation has proven robust, thus enabling simplified and accurate identification of cross-linked peptides by MSn analysis. Among them, DSSO is an amine-reactive sulfoxide-containing MS-cleavable cross-linker that has been successfully applied for in vitro studies of purified protein complexes (32, 37, 38) and cell lysates (39, 40). Although lysine-reactive reagents are most popular, they alone cannot provide a full PPI mapping as some interaction interfaces do not contain proximal lysines for cross-linking (31). Therefore, we have developed dihydrazide sulfoxide (DHSO) for acidic residues (35) and bismaleimide sulfoxide (BMSO) for cysteine cross-linking (36), complementing the lysine-reactive DSSO and expanding PPI coverage on residue-specific protein interconnectivity. In addition to PPI mapping, XL-MS data has been successfully used for integrative structure modeling of protein complexes as observed cross-links impose upper distance bounds on pairs of cross-linked residues (41–44). Coupling cross-link data with other biophysical data (43, 44) and utilizing cross-linkers with different reactive chemistries (43) can significantly increase the accuracy of the resulting structures by integrative modeling. In comparison to conventional structural tools, XL-MS approaches can uniquely characterize large, heterogeneous, and dynamic protein assemblies in solution (31).
In this work, we developed and employed a multichemistry XL-MS approach enabled by three MS-cleavable cross-linkers to obtain comprehensive PPI maps of the CSN (CSN9-free) and CSNn (CSN9-bound) complexes to significantly improve precision and accuracy of their models. Based on our cross-link data, X-ray structures, and comparative models of CSN subunits, we computed the complete integrative structures of CSN and CSNn at 16- and 22-Å precisions, respectively. The integrative structures have maintained the core architecture of the known X-ray structure of CSN (PDB ID code 4D10), but importantly revealed additional conformations and configurations of CSN in solution that were absent in the static structure. The integrative structure of CSNn has defined the CSN9 binding site in a cleft formed among CSN1, CSN3, and CSN8, resulting in local subunit reorientations that more likely contribute to CSN9-dependent increase of CSN deneddylase activity in vitro. Collectively, this work not only provides molecular features for us to better determine the structure dynamics of the CSN complex, but also reveals the structural basis underlying the role of CSN9 in CSN-mediated activities. Moreover, the integrated structural approach presented here is effective and can be generalized to define in-solution structures of dynamic protein complexes that remain inaccessible to other approaches.
Results
Multichemistry XL-MS Strategy for CSN Complexes.
To define the architectures of CSN and CSNn complexes, we aimed to perform a comprehensive XL-MS analysis to maximize PPI mapping and to facilitate integrative structure modeling. To this end, we developed a combinatory XL-MS approach based on multiple MS-cleavable cross-linkers that carry specific but complementary cross-linking chemistries. Specifically, we selected three sulfoxide-containing MS-cleavable cross-linkers that target lysines (DSSO) (32), acidic residues (DHSO) (35), and cysteines (BMSO) (36). This combination is based on the critical roles of the selected reactive residues in protein structures, and the complementarity of the resulting cross-links for mapping PPIs. Both lysines and acidic residues are highly prevalent and often enriched at protein interaction interfaces, whereas cysteines are less abundant but can be more selective for targeting specific regions. In addition, no disulfide bonds have been reported for CSN subunits, indicating that cysteine cross-linking would be suited for structural analysis of CSN. Importantly, the usage of these reagents has shown to significantly improve the coverage of PPI mapping even for simple proteins (35, 36). The general workflow of our multichemistry XL-MS strategy is illustrated in SI Appendix, Fig. S1. CSN complexes were purified under reducing condition after coexpression in Escherichia coli (Datasets S1 and S2), which were catalytically active and used for all XL-MS experiments. It is noted that CSN7 exists as two functionally redundant homologs in mammalian cells, CSN7a and CSN7b (45). Here, CSN7b was expressed and incorporated into CSN complexes for structural analysis. Each purified complex was first subjected to DSSO, DHSO, and BMSO cross-linking separately (SI Appendix, Fig. S1). The resulting cross-linked complexes were then enzymatically digested and separated to enrich cross-linked peptides by peptide size-exclusion chromatography (SEC) (46). The cross-links identified by liquid chromatography (LC)-MSn analysis were then used for generating 2D cross-link maps to describe intersubunit interactions and for integrative structure modeling.
Identification of CSN Cross-Linked Peptides.
To illustrate cross-link identification, representative MSn spectra of DSSO, DHSO, and BMSO cross-linked peptides of CSN are displayed in SI Appendix, Fig. S2. As DSSO, DHSO, and BMSO cross-linked peptides all carry two symmetric MS-cleavable bonds adjacent to the central sulfoxide in linker regions, cleavage of either one during MS2 analysis physically separates cross-linked peptide constituents (α and β), resulting in the detection of two characteristic fragment ion pairs modified with complementary cross-linker remnants (αA/βT and αT/βA), regardless of linker chemistries (SI Appendix, Fig. S2 A–C). MS3 analyses of these characteristic MS2 fragment ion pairs enabled accurate identification of their sequences (SI Appendix, Fig. S2 D–I). In combination with MS1 and MS2 data, DSSO, DHSO, and BMSO cross-linked peptides were identified unambiguously. In this work, we have performed at least four biological replicates for each XL-MS experiment. As a result, from all of XL-MS experiments, we identified a total of 682 DSSO, 275 DHSO, and 456 BMSO unique cross-linked peptides of CSN (Datasets S3–S5), and a total of 856 DSSO, 723 DHSO, and 576 BMSO unique cross-linked peptides of CSNn (Datasets S6–S9). Based on the identified cross-linked peptides, residue-to-residue linkages were determined (SI Appendix, Fig. S3). To ensure the validity of subsequent analyses, we decided to only use highly reproducible residue-to-residue linkages that have ≥60% occurrence among all biological replicates of each experiment. Thus, we obtained a total of 452 highly reproducible cross-links for CSN, including 214 K-K, 169 D/E-D/E, and 69 C-C linkages, describing 205 intersubunit (74 DSSO, 91 DHSO, and 40 BMSO) and 247 intrasubunit interactions (140 DSSO, 78 DHSO, 29 BMSO) (Datasets S10–S12). For CSNn, a total of 544 highly reproducible cross-links were acquired with 269 K-K, 167 D/E-D/E, and 108 C-C linkages, representing 244 intersubunit (86 DSSO, 83 DHSO, and 75 BMSO) and 300 intrasubunit interactions (183 DSSO, 84 DHSO, 33 BMSO) (Datasets S13–S15). These high-confidence cross-links were used for subsequent analyses (SI Appendix, Fig. S3).
The CSN Interaction Topology.
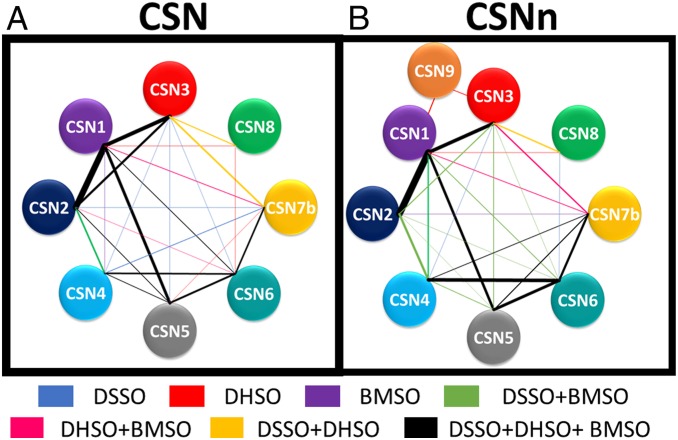
To define intersubunit physical contacts, we generated experimentally derived interaction topology maps of CSN complexes based on the highly reproducible cross-link data (Fig. 1 and Datasets S10–S15). As a result, extensive interaction networks were formulated comprising a total of 26 and 24 unique pairwise interactions for CSN and CSNn, respectively (Datasets S16 and S17). Among the three linkers, DSSO yielded the most connectivity within CSN, indicating lysine reactive reagents best-suited for general assessment of PPIs within CSN. While DHSO and BMSO identified less overall, they did yield additional subunit contacts. Specifically, DSSO alone identified five unique PPIs; in comparison, DHSO and BMSO yielded a total of seven unique PPIs (Dataset S16). To better assess linker-dependent interactions, we constructed DSSO, DHSO, and BMSO PPI maps separately for each CSN complex (SI Appendix, Fig. S4 A–F). Since the majority of CSN subunits possess similar percentages of K, D/E, and C residues in their primary sequences, the number of cross-links representing each intersubunit interaction is more likely dependent on the number of cross-linkable residues at their interaction interfaces, as well as the detectability of resulting cross-linked peptides. This is further illustrated by 2D cross-link maps (SI Appendix, Fig. S4 G–L). For example, for the two smallest subunits of CSN, CSN7b has a relatively high percentage of acidic residues and its interactions were mostly revealed by DHSO, whereas CSN8 interactions were better described by DSSO due to its relatively high percentage of lysines (Fig. 1A and SI Appendix, Fig. S4 A–C).
Fig. 1.
PPI maps of the CSN complexes based on cross-link data from all three linkers (DSSO, DHSO, BMSO). (A) CSN (CSN1–8). (B) CSNn (CSN1–9). Each CSN subunit is represented by colored nodes. The edges between two connected nodes are color-coded to describe PPIs resulted from individual or combinations of cross-linkers: That is, blue, DSSO; red, DHSO; purple, BMSO; lime, DSSO+BMSO; magenta, DHSO+BMSO; gold, DSSO+DHSO; black, DSSO+DHSO+BMSO. Edge thickness was determined by the total number of unique cross-links identified between the interactors.
Similar to the CSN complex, all three linkers have yielded extensive and complementary cross-links to represent subunit interconnectivities of CSNn (Dataset S17). Importantly, 16 CSN9-containing cross-links have been identified (Dataset S14), demonstrating its physical interactions within CSN at the residue level. Specifically, the C-terminal tail of CSN1 and several regions across CSN3 have been found to interact with CSN9. Since CSN9 is highly acidic with few lysine and no cysteine residues, only DHSO was able to capture CSN9 interactions within the CSNn complex. With the addition of CSN9, it appears that CSNn presented unique characteristics in its cross-link maps in comparison to those of CSN (Fig. 1B and SI Appendix, Fig. S4 D–F). This suggests that CSN9 may induce local changes in the CSNn complex that impact cross-link formation. Collectively, our results have demonstrated the effectiveness and complementarity of our combinatory XL-MS strategy in mapping PPIs within CSN complexes. Integration of multichemistry cross-linking not only enabled cross-validation of intersubunit interactions, but also expanded interaction coverage due to the distinct capabilities of uncovering interactions at specific protein regions.
Mapping of CSN Cross-Links to the X-Ray Structure.
To assess whether the cross-links agreed with the X-ray structure, we first mapped the identified K-K, D/E-D/E, and C-C linkages of CSN complexes to the existing CSN X-ray structure (3.8 Å, PDB ID code 4D10) by determining their Cα-Cα spatial distances (Datasets S10–S15). Considering linker spacer arm lengths (i.e., DSSO [10.1 Å], DHSO [12.4 Å], and BMSO [24.2 Å]), side-chain lengths of targeted amino acids (i.e., K [5.4 Å], D/E [2.5/3.7 Å], and C [2.8 Å]), as well as side-chain flexibility and dynamics, we have estimated the maximum Cα-Cα distances spanned by each linker: DSSO at 30 Å, DHSO at 30 Å, and BMSO at 45 Å. Thus, cross-links with distances above these thresholds were considered nonsatisfying or violating. For intersubunit interactions, 60% of DSSO cross-links of CSN were considered violating (SI Appendix, Fig. S5A). This is surprising as usually less than 20% of lysine-reactive cross-links are violated when mapped onto existing high-resolution structures (30, 38, 40). Similar discrepancies with the X-ray structure were observed for DHSO and BMSO data as 55% DHSO and 87% of BMSO intersubunit cross-links were beyond the expected thresholds (SI Appendix, Fig. S5 B and C). In contrast, most intrasubunit cross-links of CSN were satisfied in the X-ray structure, with only 12% of DSSO, 15% of DHSO, and 21% BMSO violating intrasubunit cross-links (SI Appendix, Fig. S5 A–C). Since the high-resolution structure of CSNn has not been resolved, we also mapped CSNn cross-links onto the same CSN structure. Similarly, a significant portion of intersubunit cross-links of CSNn from all three linkers (i.e., 57% of DSSO, 52% of DHSO, and 84% BMSO) were nonsatisfying (SI Appendix, Fig. S5 D–F), whereas for the intrasubunit cross-links, only 10% of DSSO, 10% of DHSO, and 23% BMSO were nonsatisfying (SI Appendix, Fig. S5 D–F). The high proportion of violating intersubunit cross-links is more likely due to the additional conformations that CSN complexes may adopt in solution beyond the one observed in the X-ray structure.
Integrative Structure Modeling of the CSN Complex.
To determine CSN structure in solution, we performed integrative structure modeling using the previously described four-stage workflow (SI Appendix, Supplemental Method and Fig. S6 and Dataset S18) (38, 43, 44, 47–50). The input information included the highly reproducible cross-link datasets (Datasets S10–S15), the X-ray structure of CSN (PDB ID code 4D10), and two comparative models of CSN7b subunit domains based on the structure of the CSN7a subunit in the X-ray structure of CSN. The representation of the system used for modeling of CSN was chosen as follows. First, the helical bundle comprising segments from each of the eight subunits was constrained based on the X-ray structure. Second, the remaining structures of subunits CSN1–8 were represented by 15 rigid bodies, corresponding to different domains of the proteins (SI Appendix, Supplemental Method and Fig. S7H and Dataset S18). Finally, short (4 to 13 residues) segments linking rigid bodies and regions missing in the X-ray structure (2 to 52 residues long) were modeled as flexible strings of 2 to 10 residue beads each. Next, we exhaustively sampled configurations of the 16 rigid bodies (i.e., the helical bundle and the 15 rigid bodies) that satisfy the cross-links as well as sequence connectivity and excluded volume restraints, using a Monte Carlo method that started with a random initial structure. The modeling did not rely on any knowledge of the X-ray structure except for the shapes of the 16 rigid bodies. The sampling yielded 71,350 representative models that sufficiently satisfied the input restraints. The clustering of the ensemble identified a single distinct cluster containing the majority (76%) of the individual models (SI Appendix, Fig. S7 A–D), corresponding to the complete integrative structure of CSN in solution. The precision of the cluster corresponds to the variability among the clustered ensemble and defines the overall precision (uncertainty) of the integrative CSN structure (Fig. 2A and SI Appendix, Fig. S7), which was quantified by the average RMSD with respect to the centroid of 16 Å (SI Appendix, Supplemental Method). The centroid structure is the most similar structure to all of the other structures in the cluster.
Fig. 2.
Integrative structures of CSN. (A) The integrative structure of CSN determined at 16-Å precision when all three cross-link datasets (DSSO+DHSO+BMSO) were used for modeling. For each subunit, the localization probability density of the ensemble of models is shown with a representative structure (the centroid) from the ensemble embedded within it. (B) Integrative modeling of CSN determined using DHSO or DHSO+DSSO datasets yielded models determined at 29- and 24-Å precision, respectively. (C) Graphical representation of determined model precisions with seven combinations of our three cross-link datasets, illustrating that increasing the number of cross-linking chemistries (abscissa axis) for integrative structure modeling leads to increased model precision (ordinate axis). CSN subunit was color-coded as illustrated.
Validation of the Integrative Structure of the CSN Complex.
To validate the integrative structure of CSN, we first assessed how well it satisfied the input cross-links used to compute it. The integrative structure of CSN satisfied 98% of the cross-links. The remaining 2% of the cross-links would be satisfied if the threshold was increased by 10 Å (SI Appendix, Fig. S7F). These violations can be rationalized by experimental uncertainty, coarse-grained representation of the complex, and finite structural sampling.
Next, we evaluated the integrative structure of CSN by cross-validation against different input cross-link datasets. Namely, we independently repeated integrative modeling described above with six different subsets of CSN cross-links (Datasets S10–S12), including: 1) DSSO only, 2) DHSO only, 3) BMSO only, 4) DSSO and DHSO, 5) DSSO and BMSO, and 6) DHSO and BMSO. The results were examined in three ways as follows. First, we gauged how well each of the six CSN model ensembles satisfied different subsets of the cross-links. All six models satisfied more than 95% of all cross-links, whether or not they were used for modeling, thus increasing our confidence in modeling. Second, we showed that increasing the amount of input information improved the precision of the output model when sampling was exhaustive. This result is expected when the choice of model representation (here, the 16 rigid bodies) is appropriate for input information (here, mainly the cross-links) as encoded in the scoring function. In addition to validating the model and the data, the improved precision of the model resulting from increasing the number of cross-linking chemistries demonstrate the complementarity of the three cross-linking datasets (Fig. 2 B and C). Specifically, the model precision increased from 37 Å for BMSO cross-links only to 16 Å for all three types of cross-links (i.e., DSSO+DHSO+BMSO). Third, we calculated the overlaps between the integrative structure ensemble using all cross-links and each of the six model ensembles based on a subset of cross-links. The overlap was quantified by the ratio of the distance between ensemble centroids to three times the sum of the ensemble precisions (SI Appendix, Supplemental Method). The distance between two ensemble centroids is defined by their RMSD. The ensemble precision is defined by the RMSD from the centroid averaged over all models in the ensemble. In particular, two structural aspects were evaluated, including the tertiary structure of each individual subunit (a total of 8 subunits) as described by the intramolecular distances as well as the relative positions and orientations of all pairs of subunits (a total of 28 pairs) in the complex as described by the intermolecular distances. For each of the 8 subunits and each of the 28 pairs of subunits, the integrative structure based on all cross-links overlapped with the integrative structures based on each of the 6 cross-link subsets (SI Appendix, Fig. S8). Therefore, these cross-validations further increased our confidence in the integrative structure of CSN.
Comparison of Integrative and X-Ray Structures of CSN.
To compare the integrative and X-ray structures of CSN, we first examined how well both structures satisfied our cross-link datasets and determined that the integrative structure did much better than the X-ray structure, for both intrasubunit (98% vs. 85%) and intersubunit (99% vs. 39%) cross-links (SI Appendix, Fig. S7F and Datasets S19–S21). These results indicate that the integrative structure ensemble is a better representation of CSN conformations in solution than the X-ray structure.
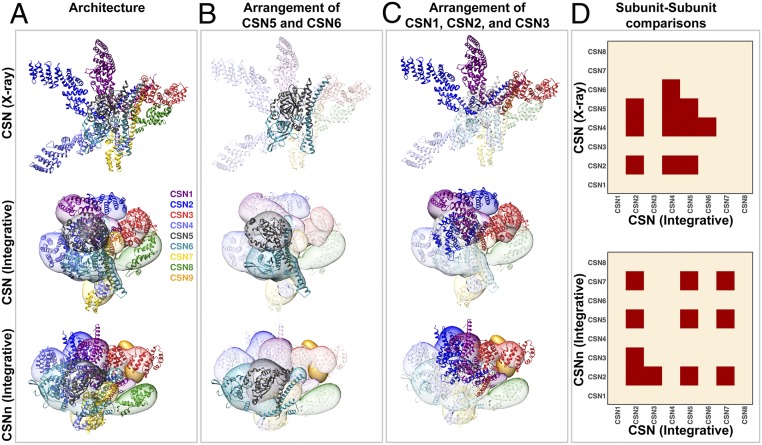
Next, we inspected whether or not the integrative model preserved the core of the previously determined CSN structures, which contains three main features: 1) The PCI ring (in the order of CSN7-CSN4-CSN2-CSN1-CSN3-CSN8), 2) the CSN5–CSN6 dimer, and 3) a helical bundle consisting of a helix from each of the eight subunits (23, 45, 51). During our modeling, while the helical bundle was constrained as a rigid body (Figs. 2A and 3 A and D), the order of the PCI ring and CSN5–CSN6 dimer were not enforced. However, the latter two features emerged from our simulation and resemble those in the X-ray structure (Figs. 2A and 3B and SI Appendix, Fig. S7G). This preservation is important especially for the CSN5–CSN6 dimer, as it is crucial for keeping CSN5 inactive in the absence of a substrate, and releasing CSN5 for activation upon substrate binding (12, 23, 24, 52). The CSN5–CSN6 dimer was well-represented by our cross-link data, resulting in the highest precisions among the 28 pairs of subunits in the integrative structure of CSN (16 Å) (Figs. 2A and 3 B and D and SI Appendix, Fig. S7). Moreover, subunits CSN3 and CSN8 also adopted similar positions and orientations relative to other subunits in both the integrative and X-ray structures (Figs. 2A and 3A and SI Appendix, Fig. S7G), albeit the precision of the CSN3–CSN8 pair in the integrative structure was relatively low (25 Å). In summary, the core of CSN integrative structure in solution is similar to previous X-ray and electron microscopy (EM) structures (23, 45, 51).
Fig. 3.
Comparison of integrative and X-ray structures of the CSN complexes. (A) Overall architectures of CSN: X-ray structure (PBD ID code 4D10) (Top), CSN integrative structure (Middle), and CSNn integrative structure (Bottom). For each subunit in the integrative structures, the localization probability density of the ensemble of models is shown with a representative structure (the centroid) from the ensemble embedded within it. The CSN and CSNn structures show that the models adopt a more condensed state as compared to the X-ray structure, but they generally retain the overall architecture with only the helical bundle being constrained during modeling. (B) The arrangement of the CSN5–CSN6 (MPN domain containing subunits) dimer was an emerging feature in integrative structures; however, a slight shift in the interface was observed in the CSNn model. (C) Models indicate that the arrangement of CSN1, CSN2, and CSN3 was altered in the presence of CSN9; CSN2 moved from a state interacting with CSN3 in CSN to an opened state in the CSNn model, resembling the overall architecture of the CSN X-ray structure. (D) Respective binary subunit–subunit comparison of the CSN integrative structure with the CSN X-ray structure (Upper) and the CSNn integrative structure (Lower), respectively. The structures were compared by calculating their ensemble overlap; the overlap was quantified by the ratio of the distance between ensemble centroids to three times the sum of the ensemble precisions. Differences are shown in red. The CSN subunit was color-coded as illustrated.
Finally, we computed the RMSD between the CSN X-ray and integrative structure centroids to assess whether the RMSD was larger than three times the precision of the integrative structure, as the resolution of the X-ray structure is much higher than that of the integrative structure. The crystallographic structures of three subunits (i.e., CSN2, CSN4, and CSN5) and four pairs of subunits (i.e., CSN2–CSN4, CSN2–CSN5, CSN4–CSN5, and CSN4–CSN6) were found to lie further than three times the integrative structure precision from the ensemble centroid (Fig. 3D), indicating significant differences in these regions between the two compared structures. The observed differences were further supported by the largest RMSDs measured in these regions between the X-ray and integrative structure centroid of CSN (SI Appendix, Fig. S9A). The detected discrepancies are unlikely the result of integrative modeling uncertainty; instead, they likely reflect different functional states in solution or differences between the solution and X-ray structures. Specifically, the C terminus of CSN4 interacts tightly with the C terminus of CSN6 (precision of 20 Å) (Fig. 3D and SI Appendix, Fig. S7G), opposite from CSN5 in the integrative structure (Figs. 2A and 3A). In contrast, CSN4 does not interact with CSN6 in the X-ray structure (Fig. 3D). The relative positions and orientations of CSN2, CSN4, CSN5, and CSN6 in the integrative structure were determined by satisfying all but 1 of the 47 intersubunit cross-links. In contrast, the X-ray structure only satisfied 30 of these cross-links.
Although the arrangement order of CSN1, CSN2, and CSN3 remained unchanged, the N terminus of CSN2 was found to wrap around CSN1 toward CSN3 in the integrative structure (Figs. 2A and 3 A and C), whereas it projected outwards without contacting either CSN1 or CSN3 in the X-ray structure. The relative positions and orientations of CSN1, CSN2, CSN3, and CSN4 in the integrative structure were determined by satisfying all but 1 of the 98 intersubunit cross-links. In contrast, the X-ray structure only satisfied 28 of these cross-links and none of the 16 cross-links between CSN2 and CSN3. Taken together, the results demonstrate that integrative structure modeling of CSN based on our comprehensive cross-link data were able to not only recapitulate the core architecture common to all known CSN structures, but also uncover significant quaternary differences relative to the X-ray structure.
Integrative Structure Modeling of the CSNn Complex.
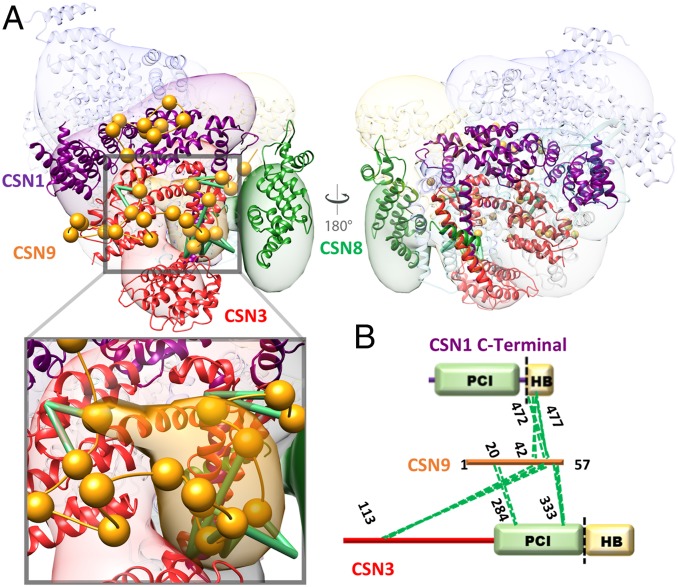
To localize the CSN9 subunit and map its interactions with the CSN complex, we also performed integrative structure modeling of CSNn (CSN9-bound CSN), based primarily on 619 highly reproducible cross-links for CSNn from all three cross-linkers (SI Appendix, Supplementary Method and Fig. S10 and Dataset S22). Integrative structure modeling of CSNn was performed the same way as described above for CSN. The structure of CSN9, a 57 amino acid-long acidic protein, is unknown and cannot be modeled. Therefore, it was represented as a string of flexible beads corresponding to two residues each. The sampling of the CSNn complex yielded 125,750 representative models that sufficiently satisfied the input restraints. The clustering of the ensemble identified a single distinct cluster containing the majority (79%) of the individual models (SI Appendix, Fig. S10), corresponding to the complete integrative structure of CSNn in solution. The precision of the cluster is 22 Å (SI Appendix, Fig. S10 A–D), which is sufficient to map all positions and relative orientations of CSN1–9 subunits (Figs. 3A and 4A and SI Appendix, Fig. S10E). Moreover, the integrative structure of CSNn satisfied 99% of the input cross-links (intersubunit and intrasubunit) (Datasets S23–S25). Importantly, the resulting structure of CSNn has precisely localized CSN9 at a cavity formed by the C terminus of CSN1, all of CSN3, and CSN8 (Fig. 4A). The position of amino acid residues 20 to 57 of CSN9 was specified by satisfying all of the 16 CSN9-containing intersubunit cross-links (Fig. 4 A and B). It is noted that the exact position of the first 19 amino acid residues of CSN9 could not be accurately determined since cross-linked peptides involving this region were not identified. Regardless, we were able to determine the interactions of CSN9 with CSN1–8 in the integrative structure. We consider a contact between CSN9 and any of the CSN1–8 subunit if the two subunits are within 12 Å from each other; a contact is defined as an interaction if the contact frequency across the ensemble is at least 75% (SI Appendix, Fig. S10G). As a result, CSN1 and CSN3 were found in the closest proximity to CSN9 across the ensemble and thus were identified as CSN9 interactors, corroborating well with our cross-link data. Therefore, the CSN9–CSN interactions have been precisely determined by integrative structure modeling (Fig. 4 and SI Appendix, Fig. S10G), providing CSN9’s binding cavity and its interactors.
Fig. 4.
Binding of CSN9 in the CSNn integrative structure. (A) The integrative structure of CSNn determined at 22-Å precision using all three cross-link datasets (DSSO+DHSO+BMSO). For each subunit, the localization probability density of the ensemble of models is shown with a representative structure (the centroid) from the ensemble embedded within it. The higher probable localization of CSN9, corresponding to its C terminal, on the CSNn model is represented by the orange localization probability density, and a representative structure from the ensemble is shown with spheres corresponding to two residues per beads connected by an extrapolated trace of the backbone. CSN9 primarily interacts with the main body of CSN3 (red) while its C-terminal tail also falls into the cavity between CSN1 (purple), CSN3 (red), and CSN8 (green). The Inset displays a closer view of CSN9 interaction. Green lines represent CSN9-containing DHSO cross-links. (B) Two-dimensional DHSO cross-link map linking CSN9 to CSN1 and CSN3 at specific residues.
Comparison of Integrative Structures of the Canonical and Noncanonical CSNs.
To compare the two CSN complexes in light of their precisions, we then examined their structural differences among the conformations of single subunits and configurations of pairs of subunits by assessing whether the differences are larger than the sum of their precisions (Fig. 3D) and by computing the RMSD between their respective centroid (SI Appendix, Fig. S9B). While a large portion of the two compared structures was similar, the conformation of 3 of the 8 subunits (i.e., CSN2, CSN5, and CSN7) and 3 of the 28 pairs of subunits (i.e., CSN2–CSN3, CSN2–CSN5, and CSN2–CSN7) had notable differences in these regions (Fig. 3D and SI Appendix, Fig. S9B). Both the integrative structures of CSN and CSNn maintained similar core structures (i.e., ordering of the PCI ring, the CSN5–CSN6 dimer, and the helical bundle) (Fig. 3B). However, CSN2 changed its conformation and position relative to its neighbors (i.e., CSN3, CSN5, and CSN7) (Fig. 3 A, C, and D and SI Appendix, Fig. S9B). Specifically, in the integrative structure of CSNn, CSN2, and CSN4 localize adjacent to one another, allowing the formation of the CSN9-binding cavity (Figs. 3D and 4). The conformation and relative position of the CSN2 subunit in the integrative structure of CSNn were determined by satisfying all 74 intersubunit cross-links obtained for CSNn. Therefore, our results suggest that CSN2 possesses structural plasticity, enabling its interaction with CSN1 and CSN3 to yield a more open configuration in CSN9-bound CSN than in CSN9-free CSN.
To explore the potential role of CSN9-mediated structural changes, we compared the integrative structures of CSN and CSNn to the cryo-EM structure of the CRL4A-bound CSN complex (at resolution of 6.4 Å) (24). Specifically, we assessed whether the structure of the CSN complexed with neddylated CRL4A overlapped with the two integrative structures. The structure of CRL4A-bound CSN differs from the integrative structure of CSN for one subunit (i.e., CSN2) and two pairs of subunits (i.e., CSN2–CSN4 and CSN2–CSN5) (SI Appendix, Fig. S9C). In contrast, the structure of CRL4A-bound CSN has no significant differences with the integrative structure of CSNn (SI Appendix, Fig. S9D). Similar comparisons were performed with the structure of CRL1-bound CSN (at resolution of 7.2 Å) (25). While the structure of CSN bound to neddylated CRL1 differs from the integrative structure of CSN for two subunits (i.e., CSN2 and CSN5) and three pairs of subunits (i.e., CSN2–CSN4, CSN2–CSN5, and CSN2–CSN6) (SI Appendix, Fig. S9E), it has no significant differences with the integrative structure of CSNn (SI Appendix, Fig. S9D). Collectively, these assessments suggest that CSN9-bound CSN is structurally similar to CRL-bound CSN (24, 25). Upon CSN9 binding, the integrative structure of CSNn displays local structural changes, mainly on the conformation and position of CSN2. Specifically, CSN2 moves closer to CSN4, causing CSN9-bound CSN to adopt a configuration resembling CRL-bound CSN (24, 25).
Biochemical Validation of CSN9 Binding.
In order to validate the interactions of CSN9 with the CSN complex revealed by XL-MS and structural modeling, we performed in vitro binding assays using purified CSN subunits. CSN9 only interacts with CSN1-2-3 and CSN1-2-3-8 subcomplexes, whereas no binding was detected with CSN4-6-7, CSN4-6-7-5, or CSN4-6-7-5-8 subcomplexes (SI Appendix, Fig. S11). These results confirm that CSN1 and CSN3 are present in the subcomplex required for CSN9 binding onto CSN. To understand the importance of CSN9, we have compared in vitro deneddylase activities of CSN and CSNn with neddylated Cullin 1 as the substrate. Similar results were obtained for the same assay performed at different time scales (SI Appendix, Fig. S12), demonstrating that CSNn displayed markedly increased activity over CSN and CSN9 can enhance CSN activity in vitro.
Quantitative Validation of the Structural Dynamics of the CSN Complexes.
To validate the observed structural differences between CSN models with and without CSN9, parallel reaction monitoring (PRM)-based targeted quantitation of CSN cross-links was utilized (53). Since DHSO cross-linking yielded the most intersubunit linkages best describing CSN9-induced structural changes, we individually cross-linked CSN and CSNn with DHSO for PRM experiments. To perform unbiased quantitative analysis, we generated a total of 341 PRM targets based on highly reproducible DHSO cross-linked peptides previously obtained from CSN and CSNn complexes (Datasets S11 and S14). Peptide quantitation was derived from the summation of peak areas of all transitions through Skyline software. As exemplified in Fig. 5A, an intra-CSN4 cross-link (E306–E345) from both CSN and CSNn samples displayed similar abundance, indicating that this interaction is independent of CSN9. In contrast, a CSN2–CSN3 cross-link (CSN2:E63–CSN3:E333) was only observed in CSNn and not in CSN, suggesting a CSN9-induced conformational change. In total, 229 DHSO cross-linked peptides were quantified, which represent 18 intersubunit interactions (Dataset S26). As shown in Fig. 5B, the vast majority of quantified cross-links remained unchanged between CSN and CSNn, confirming that CSN9 does not trigger major organizational changes within the CSN complex during its binding. This corroborates well with the modeling results as both of our CSN models satisfied 99% of DHSO cross-links from both complexes. Apart from unchanged interactions, a total of 22 cross-linked peptides were found with significant changes (>2.5-fold, greater than 3σ) between the two compared complexes (Fig. 5B).
Fig. 5.
PRM-based targeted quantitation of DHSO cross-linked peptides to validate CSN9-induced structural changes in CSN. (A) Skyline outputs for PRM quantitation of a representative DHSO intrasubunit (CSN4:E306–CSN4:E345) (Upper) and an intersubunit (CSN2:E63–CSN3:E333) (Lower) cross-linked peptides to compare their relative abundance in the CSN and CSNn complexes. Based on peak areas, the relative abundance ratio (CSN/CSNn) of the intrasubunit cross-link was determined as 1.11 (Upper), indicating no significant change. In contrast, the relative abundance of the intersubunit cross-link (CSN/CSNn) was determined as 30.15 (Lower), suggesting a significant change. (B) The distribution of cross-link ratios (CSN/CSNn) of 229 DHSO cross-linked peptides (represented as log2 values) determined by PRM quantitation, in which only 22 cross-linked peptides displayed significant changes (>2.5-fold, greater than 3σ), including 4 with decreased ratios (red dots) and 18 with increased ratios (blue dots). The cross-link ratios (CSN/CSNn) describe the relative abundance of cross-linked peptides in the two compared complexes. (C) Abundance of five quantifiable CSN2–CSN3 cross-links (CSN2:D45–CSN3:E333, CSN2:E59–CSN3:E284, CSN2E59–CSN3:E333, CSN2:E63–CSN3:E333, and CSN2:E161–CSN3:E284) detected in the CSN and CSNn complexes. The underlined numbers shown represent relative abundance ratios (CSN/CSNn) of the selected cross-linked peptides between the two complexes, indicating that these interactions are favored in CSN. (D) The five cross-links shown in (C) were mapped on CSN and CSNn integrative structures. The linkages in the CSN model (green) are satisfied within the expected distance (<30 Å), which are not satisfied in the CSNn model (magenta). Details on PRM quantitation of the cross-linked peptides are listed in Dataset S26.
Besides cross-links involving CSN9, two additional cross-linked peptides corresponding to two intersubunit interactions (i.e., CSN4–CSN6 and CSN6–CSN7) have decreased CSN/CSNn ratios, suggesting that these cross-links are favored in CSNn. In contrast, 18 cross-linked peptides describing 7 intersubunit interactions (CSN1–CSN2, CSN1–CSN3, CSN1–CSN5, CSN2–CSN3, CSN2–CSN7, CSN4–CSN5, and CSN6–CSN7) and 1 intra-CSN1 interaction have increased CSN/CSNn ratios, implying that these cross-links are preferably formed in CSN. Apart from CSN9-containing interactions, five quantifiable CSN2–CSN3 cross-links exhibited the most significant changes between the two compared complexes with CSN/CSNn ratios all greater than 10.2 (Fig. 5C), indicating that CSN2–CSN3 interactions were severely disrupted upon CSN9 binding. This is consistent with the structural differences between CSN and CSNn revealed by integrative modeling as these linkages were only satisfied by the CSN models (Fig. 5D). Since CSN1 closely interacts with CSN2, CSN3, and CSN9, the decreased abundance of CSN1–CSN2 and CSN1–CSN3 cross-links in CSNn supports the CSNn model, suggesting that the main body of CSN2 swings away from CSN1 and CSN3 into a more open state. Collectively, PRM-based targeted quantitation of CSN cross-links strongly supports structural similarities and differences between the integrative models of the two CSN complexes.
Discussion
In this work, we have developed a multichemistry XL-MS approach based on three distinct MS-cleavable cross-linkers (i.e., DSSO, DHSO, and BMSO) to comprehensively map PPIs and facilitate integrative structure modeling of CSN complexes. The large number of cross-links identified in this work is highly complementary, allowing expanding PPI coverage and cross-validating results. This approach enables us to obtain the most extensive intrasubunit and intersubunit interaction maps of CSN (CSN9-free) and CSNn (CSN9-bound) complexes. It is noted that CSN9-containing interactions were only identified through DHSO cross-linking, not by DSSO and BMSO, signifying the need of multichemistry XL-MS to fully characterize PPIs of CSN complexes. Importantly, the combinatory XL-MS data enabled structural characterization of the CSN complexes with complete sequences and significantly enhanced the precision of integrative structure modeling, resulting in the precisions of 16 and 22 Å for CSN and CSNn, respectively. These are considerably higher than the precision of models from single and dual cross-linking chemistries (24 to 37 Å). While lysine-to-lysine and acidic-to-acidic residue cross-links have been successfully applied for structural mapping and modeling (31, 35, 38, 46, 54, 55), we demonstrate here that cysteine-to-cysteine cross-links are as effective for structure determination of protein complexes. This is illustrated by the fact that a single integrative structure (i.e., a single cluster of models) satisfies most of the BMSO cross-links, similarly to DSSO and DHSO cross-links (Fig. 2 and SI Appendix, Fig. S7). In addition, we obtained highly overlapping model ensembles based on seven different combinations of the three types of cross-link data (i.e., DSSO, DHSO, and BMSO cross-links) (Fig. 2C and SI Appendix, Fig. S8), confirming the validity and coherence of our cross-link data. Therefore, coupling combinatory XL-MS based on multiple cross-linking chemistries with integrative structure modeling facilitates the determination of the interaction and structure dynamics of CSN complexes. The same strategy can be directly adopted for characterizing architectures of other dynamic protein complexes in solution.
During XL-MS analyses, we have found that although the majority of intrasubunit cross-links of CSN from all three linkers were satisfied by the known X-ray structure (PDB ID code 4D10), most of intersubunit cross-links were classified as violating. This implies that CSN has much more flexible intersubunit than intrasubunit interactions. Since X-ray crystallography only reveals static structures with a single conformation, distance violation of cross-links suggests the presence of multiple conformations and configurations of CSN in solution. Similar results have been obtained for the CSNn complex, further confirming the interaction and structural plasticity of CSN complexes. While CSN is known to carry structural flexibility to allow its interaction with a diverse array of CRLs to regulate their activities (12, 23–25), our XL-MS results provide additional evidence to support CSN structural heterogeneity in solution. Because of this, our cross-link dataset generated here is comprised of a wide range of possible conformations of CSN complexes. Therefore, to minimize complexity, only highly reproducible cross-link data were used to derive structural ensembles that represent major conformations of CSN complexes in solution. The integrative structures of CSN complexes have satisfied 98% of all of the cross-links obtained in this work, considerably better than the X-ray structure. This result further indicates that CSN contains additional accessible states other than the one determined by X-ray crystallography. In contrast to the observed conformational and configurational differences in intersubunit interactions, the core structure of CSN is preserved. Indeed, we have found that the CSN model maintains overall configuration with the presence of the PCI ring and the positioning of CSN5–CSN6 dimer, apart from a rearrangement of CSN2 with respect to CSN1, CSN3, and CSN4 positioning in the complex. The core structure of CSN has also been detected in the CSNn model, which was derived from a completely different set of cross-link data used for CSN modeling. As these core modules are crucial for the CSN assembly, structure, and function (12, 23–25, 45, 51), their determination by integrative modeling based primarily on cross-links further demonstrates the effectiveness of our approach and the validity of the determined integrative structures.
Here, we have determined that CSN9 predominantly interacts with CSN3 and CSN1, and is localized in a cavity formed by CSN1-3-8 in the CSNn structure. Although CSN3–CSN9 interaction has previously been shown biochemically (27), our results have identified interaction contacts between the two interactors. Importantly, we have identified CSN1 as an additional CSN9 interactor and determined CSN9 binding sites within the CSN complex. While it has been suggested that CSN9 may bind to CSN5 and CSN6 (27), no cross-links between CSN9 and CSN5 or CSN6 were identified and the integrative structure of CSNn shows that both subunits are much farther away from CSN9 than CSN1 and CSN3. Interestingly, CSN1, CSN3, and CSN8 form a connected submodule in the integrative and X-ray structures of CSN (23), and the assembled CSN1–3-8 subcomplex can be isolated in mammalian cells (56). It is known that each CSN subunit has a corresponding homolog in the nine-subunit 19S lid complex (27, 57). Recently, the proteasome subunit DSS1/Rpn15, the homolog of CSN9, has been determined to interact with Rpn3 (homolog of CSN3) and Rpn7 (homolog of CSN1), which forms a subcomplex prior to the 19S lid assembly (58), corroborating well with the close interactions of CSN9 with CSN3 and CSN1. These results further indicate interaction similarities between the CSN and the 19S lid complexes.
Apart from similarities in organizational architectures in the CSN integrative and X-ray structures, we have observed structural differences between the integrative structures of CSN and CSNn that may contribute to CSN dynamics. One notable difference is the CSN2–CSN3 interaction and its relative location to CSN1 subunit. Specifically, in the CSN integrative structure, the CSN2 N terminus wraps around CSN1 toward CSN3 and away from CSN4, whereby CSN2 is not readily available to interact with Cullin and Rbx1. This is of importance because CSN2 plays a major role along with CSN4 in stabilizing the CSN–CRL interaction when CSN binds to CRLs (12, 23–25). CSN1 has been shown to bind to the CRL4A adaptor DDB1, which is important in stabilizing Cul4A and required for efficient deneddylation (24). However, CSN1 involvement appears to be specific for CRL4 and not CRL2 and CRL3 complexes (24, 26). While CSN3 has not been shown to directly contact CRL components, overexpression of CSN3 leads to increased amounts of CSN in cells and down-regulation of CSN3 causes the destruction of CSN and cell death (59). Thus, we speculate that the observed changes of interactions among CSN1, CSN2, and CSN3 may represent one of the major conformations of CSN that is needed to interact with specific subsets of CRLs in cells.
While the integrative structures of CSN and CSNn have both maintained the core structure of CSN, CSN9 binding causes a major shift in CSN2 and its interactions with neighboring subunits that have been confirmed by quantitative XL-MS analysis. Given the critical importance of CSN2 in CSN–CRL interactions (12, 24, 25), we suspect that CSN9-induced structural changes may be associated with the augmented CSN in vitro deneddylase activity observed in this work. Comparative analysis has revealed that the major differences between canonical CSN (CSN9-free) and CRL-bound CSN lie in the relative position of CSN2 and its interaction with CSN5 (SI Appendix, Fig. S9 C and E), indicating that CSN2 has to undergo conformational changes to fulfill its role in facilitating CSN binding to CRLs (24, 25). Therefore, the observed structural alterations at CSN2 would be important for the formation of the CSN–CRL complex, the prerequisite for subsequent deneddylation. The structure similarity between CSN9-bound CSN and CRL-bound CSN (SI Appendix, Fig. S9D) strongly supports the biological relevance of CSN9-induced structural changes. Thus, these results prompt us to propose a structural model in which CSN9 causes the canonical CSN to adopt a configuration favorable for interacting with CRLs (Fig. 6). In the absence of CSN9, binding of neddylated CRL to CSN results in a series of conformational changes, among which the initial important steps involve the movement of N-terminal domains of CSN2 and CSN4 toward cullin (12, 24, 25). These rearrangements occur prior to the release and activation of CSN5.
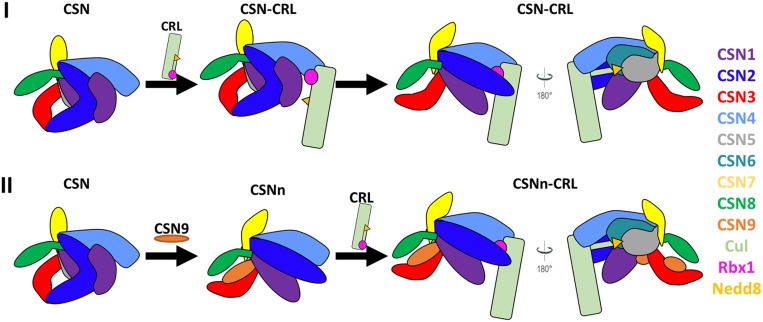
Fig. 6.
The proposed structural model of CSN9 binding to facilitate CSN interaction with neddylated CRLs. CSN and neddylated CRL subunits were color-coded as illustrated. (I) CSN9-free CSN needs to undergo substantial conformational changes upon binding to a neddylated CRL. In comparison, (II) CSN9-bound CSN adopts a configuration better suited for CRL binding.
In contrast, the addition of CSN9 triggers CSN to undergo conformational changes by repositioning the N terminus of CSN2 away from CSN3 but closer to CSN4 (Fig. 6). As the resulting conformation and configuration of CSN9-bound CSN are highly similar to those of CRL-bound CSN, we suspect that CSN9 may enhance the affinity (or recognition) between CSN and its substrate, neddylated CRLs, thus facilitating the assembly of CSN-neddylated CRL complex to enhance CSN activation and deneddylation of CRLs. In addition, the conformation of CSNn may also enable its faster release from deneddylated CRLs as reported (14). In the absence of CSN9, the assembly/disassembly of the CSN–CRL complex would more likely be much slower due to substantial conformational changes required for the activation of CSN upon binding to CRLs, thus leading to slower deneddylation rate. Therefore, the differences in the assembly/disassembly of CSN–CRL complexes more likely contribute to their interaction affinity, and slower disassembly of the CSN–CRL complex could imply tighter interaction. In summary, CSN9-induced conformational changes related to CSN2, are biologically relevant, especially in preparing CSN for associating with neddylated CRLs, thereby contributing to augmenting deneddylation activity of CSN. The integrative structures of CSN complexes determined in this work have established a structural basis for us to further dissect condition-induced structural dynamics of CSN in the future, unraveling molecular insights into its activation, function, and regulation under different physiological and pathological conditions.
Methods
Expression and Purification of CSN Complexes.
Eight of total nine subunits of the human CSN complex, except CSN5, were overexpressed and purified from E. coli. Two three-subunit subcomplexes, CSN1-2-3 and CSN4-6-7, were prepared through coexpression. Briefly, CSN2 was subcloned into a modified pGEX4T1 (Amersham Biosciences) vector containing a GST tag followed by a tobacco etch virus (TEV) protease cleavage site, while both CSN1 and CSN3 were subcloned into a modified pET15b (Novagen) vector containing a chloramphenicol resistance cassette. After coexpression in BL21(DE3) (Novagen), the CSN1-2-3 formed a complex and was purified by glutathione-affinity chromatography. Following TEV cleavage, the CSN1-2-3 subcomplex was further purified by anion exchange and gel-filtration chromatography. CSN4-6-7 was prepared in the same way. CSN8 and CSN9 were subcloned into the pGEX4T1 vector individually and subjected to the same purification procedure. Recombinant full-length CSN5 inserted into a modified GTE vector (Invitrogen). It has a GST tag that was removed during purification and was prepared from insect cells using a baculovirus expression system. Two CSN complexes, with or without CSN9, were reconstituted by incubating the purified subcomplexes and individual subunits in equimolar ratio and polished by SEC. Neddylated Cul1–Rbx1 complex was prepared as described previously (60).
XL-MS Analysis of CSN Complexes.
Affinity-purified human CSN complex with or without CSN9 were cross-linked with DSSO, DHSO, or BMSO, respectively. Each CSN complex was reacted with a selected cross-linker at their optimized molar ratios (protein to linker) respectively: DSSO (1:250), BMSO (1:400), and DHSO (1:30) (32, 35, 36). DMTMM was used to activate acidic residues for DHSO cross-linking (35). All reactions were performed for 1 h at room temperature. The resulting cross-linked proteins were digested by lys-C and trypsin. Cross-linked peptides were enriched by peptide SEC, analyzed by LC MSn, and identified through database searching, as previously described (SI Appendix, Supplemental Method) (35, 36).
PRM Targeted Quantitation of Cross-Linked Peptides.
The 341 PRM targets were obtained based on highly reproducible DHSO cross-linked peptides of CSN and CSNn complexes, as summarized in Datasets S11 and S14. For targeted analysis, the mass spectrometer was operated with the following settings: No survey scan collected, tMS2 resolving power 30,000, AGC target 5e4, maximum injection time 54 ms, isolation window 1 m/z, and CID normalized collision energy of 23%. A total of 341 cross-links were monitored over 3 separate targeted analyses for each sample, along with a set of 16 heavy-labeled AQUA peptides. Targeted analysis of AQUA peptides used the same settings as cross-link ions except were subjected to higher-energy collision dissociation with normalized collision energy of 30%. Transition lists based on expected cross-link fragmentation ions were generated and quantified using Skyline v.4.2.0.19072. Once exported, extracted intensities were normalized within sample sets using relative intensities of AQUA peptides based on quantified b and y ions.
In Vitro Deneddylation Assay.
A mixture containing 5 μM Nedd8-Cul1-Rbx1 and 20 nM CSN was incubated in reaction buffer of 50 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM TCEP. The reactions were carried out at room temperature and stopped by adding SDS/PAGE sample buffer at indicated time points, then analyzed by 9% SDS/PAGE and stained with Coomassie blue.
Biochemical Validation of the CSN9 Interactors.
Purified components of CSN, including CSN5, CSN8, and subcomplex CSN1-2-3 and CSN4-6-7, were used for pull-down assay. His-GB1 fused CSN9 served as the bait protein. The prey samples (different combinations of CSN subunits) were mixed with His-GB1-CSN9 at molar ratio 2:1. After 10-min incubation, His Mag Sepharose Ni beads (GE Healthcare) were added into the samples and suspended by gently tapping the sample tubes for 5 min to immobilize His-GB1-CSN9 and its binding partners. Then the beads were washed with 20 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 20 mM imidazole five times. The beads were further eluted with 200 mM imidazole and the elution was analyzed on a 4 to 15% Mini-PROTEAN TGX Gel (Bio-Rad). To identify the binding partners of CSN9, all of the purified CSN components were loaded on the same gel.
Integrative Structure Modeling.
Integrative structure modeling was carried out to determine the structures of the human canonical and noncanonical CSN complexes (SI Appendix, Supplemental Method and Datasets S18 and S22). Mass spectrometry raw data have been deposited at the PRIDE Archive proteomics data repository site (dataset identifier PXD014673). All of the relevant scripts, data, and results are available at GitHub, https://salilab.org/CSN2019. The integrative structures of CSN and CSNn are deposited at PDB-Dev (https://pdb-dev.wwpdb.org/), with ID codes PDBDEV_00000037 and PDBDEV_00000038, respectively.
Supplementary Material
Acknowledgments
We thank Prof. A. L. Burlingame and Robert Chalkley for their support of the development version of Protein Prospector. This work was supported by National Institutes of Health Grants R01GM074830 and R01GM130144 (to L.H.), and P41GM109824, R01GM083960, and S10OD021596 (to A.S.); National Science Foundation graduate research Fellowship 1650113 (to I.E.C.); and National Science Foundation Grant CHE 1807612 (to S.D.R.). N.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Mass spectrometry raw data have been deposited at the PRIDE Archive proteomics data repository site (dataset identifier PXD014673). All the relevant scripts, data, and results are available at GitHub, https://salilab.org/CSN2019. The integrative structures of CSN and CSNn are deposited at PDB-Dev, https://pdb-dev.wwpdb.org/ (PDB ID codes PDBDEV_00000037 and PDBDEV_00000038).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915542117/-/DCSupplemental.
References
- 1.Wei N., Deng X. W., The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Wolf D. A., Zhou C., Wee S., The COP9 signalosome: An assembly and maintenance platform for cullin ubiquitin ligases? Nat. Cell Biol. 5, 1029–1033 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Wei N., Serino G., Deng X. W., The COP9 signalosome: More than a protease. Trends Biochem. Sci. 33, 592–600 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Cope G. A., et al. , Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Deshaies R. J., Joazeiro C. A., RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Cope G. A., Deshaies R. J., COP9 signalosome: A multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Lee J. E., et al. , The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell. Proteomics 10, M110.006460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skaar J. R., Pagan J. K., Pagano M., Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia L., Sun Y., SCF E3 ubiquitin ligases as anticancer targets. Curr. Cancer Drug Targets 11, 347–356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emberley E. D., Mosadeghi R., Deshaies R. J., Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J. Biol. Chem. 287, 29679–29689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enchev R. I., et al. , Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2, 616–627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer E. S., et al. , The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Füzesi-Levi M. G., et al. , CSNAP, the smallest CSN subunit, modulates proteostasis through cullin-RING ubiquitin ligases. Cell Death Differ., 10.1038/s41418-019-0392-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cope G. A., Deshaies R. J., Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 7, 1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y. H., et al. , Molecular targeting of CSN5 in human hepatocellular carcinoma: A mechanism of therapeutic response. Oncogene 30, 4175–4184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y., Claret F. X., Targeting Jab1/CSN5 in nasopharyngeal carcinoma. Cancer Lett. 326, 155–160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong G., Li H., Shan T., Zhang N., CSN5 silencing inhibits invasion and arrests cell cycle progression in human colorectal cancer SW480 and LS174T cells in vitro. Int. J. Clin. Exp. Pathol. 8, 2809–2815 (2015). [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H., et al. , COPS5 inhibition arrests the proliferation and growth of serous ovarian cancer cells via the elevation of p27 level. Biochem. Biophys. Res. Commun. 493, 85–93 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Lee M. H., Zhao R., Phan L., Yeung S. C., Roles of COP9 signalosome in cancer. Cell Cycle 10, 3057–3066 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson K. S., Zundel W., The emerging role of the COP9 signalosome in cancer. Mol. Cancer Res. 3, 645–653 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer E. S., et al. , Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingaraju G. M., et al. , Crystal structure of the human COP9 signalosome. Nature 512, 161–165 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Cavadini S., et al. , Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531, 598–603 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Mosadeghi R., et al. , Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. eLife 5, e12102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faull S. V., et al. , Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome. Nat. Commun. 10, 3814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozen S., et al. , CSNAP is a stoichiometric subunit of the COP9 signalosome. Cell Rep. 13, 585–598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharon M., et al. , Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17, 31–40 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Sinz A., Arlt C., Chorev D., Sharon M., Chemical cross-linking and native mass spectrometry: A fruitful combination for structural biology. Protein Sci. 24, 1193–1209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitner A., Faini M., Stengel F., Aebersold R., Crosslinking and mass spectrometry: An integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Yu C., Huang L., Cross-linking mass spectrometry: An emerging technology for interactomics and structural biology. Anal. Chem. 90, 144–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao A., et al. , Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10, M110.002212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu C., Kandur W., Kao A., Rychnovsky S., Huang L., Developing new isotope-coded mass spectrometry-cleavable cross-linkers for elucidating protein structures. Anal. Chem. 86, 2099–2106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaake R. M., et al. , A new in vivo cross-linking mass spectrometry platform to define protein-protein interactions in living cells. Mol. Cell. Proteomics 13, 3533–3543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez C. B., et al. , Developing an acidic residue reactive and sulfoxide-containing MS-cleavable homobifunctional cross-linker for probing protein-protein interactions. Anal. Chem. 88, 8315–8322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutierrez C. B., et al. , Development of a novel sulfoxide-containing MS-cleavable homobifunctional cysteine-reactive cross-linker for studying protein-protein interactions. Anal. Chem. 90, 7600–7607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao A., et al. , Mapping the structural topology of the yeast 19S proteasomal regulatory particle using chemical cross-linking and probabilistic modeling. Mol. Cell. Proteomics 11, 1566–1577 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., et al. , Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell. Proteomics 16, 840–854 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F., Rijkers D. T., Post H., Heck A. J., Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat. Methods 12, 1179–1184 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Liu F., Lössl P., Rabbitts B. M., Balaban R. S., Heck A. J. R., The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell. Proteomics 17, 216–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog F., et al. , Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Erzberger J. P., et al. , Molecular architecture of the 40S⋅eIF1⋅eIF3 translation initiation complex. Cell 158, 1123–1135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S. J., et al. , Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rout M. P., Sali A., Principles for integrative structural biology studies. Cell 177, 1384–1403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubiel D., Rockel B., Naumann M., Dubiel W., Diversity of COP9 signalosome structures and functional consequences. FEBS Lett. 589, 2507–2513 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Leitner A., Walzthoeni T., Aebersold R., Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc. 9, 120–137 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Alber F., et al. , Determining the architectures of macromolecular assemblies. Nature 450, 683–694 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Russel D., et al. , Putting the pieces together: Integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sali A., et al. , Outcome of the first wwPDB Hybrid/Integrative Methods Task Force workshop. Structure 23, 1156–1167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneidman-Duhovny D., Pellarin R., Sali A., Uncertainty in integrative structural modeling. Curr. Opin. Struct. Biol. 28, 96–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rockel B., Schmaler T., Huang X., Dubiel W., Electron microscopy and in vitro deneddylation reveal similar architectures and biochemistry of isolated human and Flag-mouse COP9 signalosome complexes. Biochem. Biophys. Res. Commun. 450, 991–997 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Birol M., et al. , Structural and biochemical characterization of the Cop9 signalosome CSN5/CSN6 heterodimer. PLoS One 9, e105688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chavez J. D., et al. , A general method for targeted quantitative cross-linking mass spectrometry. PLoS One 11, e0167547 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X., et al. , Carboxylate-selective chemical cross-linkers for mass spectrometric analysis of protein structures. Anal. Chem. 90, 1195–1201 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Leitner A., et al. , Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9455–9460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heusel M., et al. , Complex-centric proteome profiling by SEC-SWATH-MS. Mol. Syst. Biol. 15, e8438 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharon M., Taverner T., Ambroggio X. I., Deshaies R. J., Robinson C. V., Structural organization of the 19S proteasome lid: Insights from MS of intact complexes. PLoS Biol. 4, e267 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai M., et al. , In-depth analysis of the lid subunits assembly mechanism in mammals. Biomolecules 9, 213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peth A., Berndt C., Henke W., Dubiel W., Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability. BMC Biochem. 8, 27 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu C., et al. , Characterization of dynamic UbR-proteasome subcomplexes by in vivo cross-linking (X) assisted bimolecular tandem affinity purification (XBAP) and label-free quantitation. Mol. Cell. Proteomics 15, 2279–2292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.