Significance
The QKI-5 is a major isoform of QKIs and exerts tumor-suppressive functions in non-small cell lung cancer (NSCLC). MicroRNA dysregulation occurs frequently in cancers, and miR-196b-5p has been reported to have dual functions in NSCLC. We found that miR-196b-5p was up-regulated in NSCLC and was negatively regulated by QKI-5. We demonstrated that miR-196b-5p promoted cellular migration, proliferative ability, and tumor growth both in vitro and in vivo by directly targeting the tumor suppressors, GATA6 and TSPAN12. QKI-5 bound to miR-196b-5p and reduced its stability, thus inhibiting miR-196b-5p expression. In addition, up-regulated miR-196b-5p expression in NSCLC was partially controlled by hypomethylation of its promoter region. Our study suggests that the QKI5∼miR-196b-5p∼GATA6/TSPAN12 pathway might be a promising therapeutic strategy in treating NSCLC.
Keywords: QKI, miR-196b, NSCLC, GATA6, TSPAN12
Abstract
Lung cancer is the leading cause of cancer-related deaths worldwide and non-small cell lung cancer (NSCLC) accounts for over 80% of lung cancer cases. The RNA binding protein, QKI, belongs to the STAR family and plays tumor-suppressive functions in NSCLC. QKI-5 is a major isoform of QKIs and is predominantly expressed in NSCLC. However, the underlying mechanisms of QKI-5 in NSCLC progression remain unclear. We found that QKI-5 regulated microRNA (miRNA), miR-196b-5p, and its expression was significantly up-regulated in NSCLC tissues. Up-regulated miR-196b-5p promotes lung cancer cell migration, proliferation, and cell cycle through directly targeting the tumor suppressors, GATA6 and TSPAN12. Both GATA6 and TSPAN12 expressions were down-regulated in NSCLC patient tissue samples and were negatively correlated with miR-196b-5p expression. Mouse xenograft models demonstrated that miR-196b-5p functions as a potent onco-miRNA, whereas TSPAN12 functions as a tumor suppressor in NSCLC in vivo. QKI-5 bound to miR-196b-5p and influenced its stability, resulting in up-regulated miR-196b-5p expression in NSCLC. Further analysis showed that hypomethylation in the promoter region enhanced miR-196b-5p expression in NSCLC. Our findings indicate that QKI-5 may exhibit novel anticancer mechanisms by regulating miRNA in NSCLC, and targeting the QKI5∼miR-196b-5p∼GATA6/TSPAN12 pathway may enable effectively treating some NSCLCs.
Lung cancer is the most common cancer and the leading cause of cancer-related deaths worldwide (1). Non-small cell lung cancer (NSCLC) accounts for over 80% of lung cancer cases, and many NSCLC patients are diagnosed at the advanced stage due to late onset of clinical symptoms and inadequate screening methods. Despite advancements and improvements in surgical and medical treatments, the 5-y survival rate of lung cancer patients remains frustratingly poor (2). Thus, novel and sensitive biomarkers for early NSCLC detection and target molecules for developing new therapies are urgently needed.
The RNA binding protein Quaking (QKI) is a member of the signal transduction and activation of RNA (STAR) family. The QKI gene encodes four major alternatively spliced messenger RNAs (mRNAs) (QKI-5, QKI-6, QKI-7, and QKI-7b) with different C terminals. QKI-5 has a nuclear localization signal and thus shuttles between the nucleus and cytoplasm. In contrast, QKI-6 and QKI-7 are mostly localized in the cytoplasm (3, 4). QKI plays multifunctional roles for target RNAs, including location, stability, translational efficiency, and microRNA (miRNA) processing, by binding to the QKI responsive element [QRE, 5′-A(C/A)UAA-3] during physiological and pathological processes (5–8). QKI-5 is reported to be a major isoform of QKIs and is predominantly expressed in lung cancer. Down-regulated QKI-5 expression is involved in lung cancer cell proliferation and metastasis by influencing alternative mRNA splicing of NUMB and inhibiting β-catenin (8, 9). Although accumulating evidence indicates the importance of QKI-5 in NSCLC progression, the underlying mechanisms of QKI-5 in lung cancer pathogenesis remain unclear. Considering the critical roles of miRNAs in lung cancer progression and metastases, we speculated that QKI-5 might impact the functions of miRNAs and therefore also affect NSCLC progression and metastasis.
miRNAs are small endogenous noncoding RNAs that negatively regulate mRNA stability and/or repress mRNA translation by binding the complementary sequences in the target gene’s 3′-untransrated region (UTR) (10). Most miRNAs function as tumor suppressor or oncogenes in a tissue-specific manner, and dysregulated miRNA expression is closely related to human cancer development and progression (11). Chemically modified antisense oligonucleotides efficiently and specifically inhibit miRNA function. In contrast, miRNA mimics could functionally replenish lost miRNA expression in specific diseases, thereby highlighting the potential for developing novel therapeutic methods (12–14). A mimic of the tumor suppressor, miR-34, has reached phase 1 clinical trials for treating multiple solid tumors, and antimiR-122 has reached phase 2 clinical trials for treating hepatitis C (15).
GATA6 is a key transcription factor involved in differentiation of the distal epithelium during lung morphogenesis (16). The expression of GATA6 was generally down-regulated in lung cancer and associated with lung cancer metastasis and prognosis (17). Several miRNAs have been reported to target GATA6 involving the pathogenesis of various cancers (18–20), including lung cancer. TSPAN12, a member of the tetraspanin family, was reported to be involved in retinal vascular development in a mouse model (21). In cancer, TSPAN12 functions as an oncogene or tumor suppressor depending on the cancer type. Abolishment of TSPAN12 in breast cancer cells inhibits primary tumor growth but increases metastasis in vivo via the canonical Wnt signaling pathway (22). Knockdown of TSPAN12 in cancer-associated fibroblasts inhibits lung cancer cell proliferation and invasion (23). In addition, overexpression of TSPAN12 in colorectal cancer and NSCLC promotes tumor growth (24, 25). However, the underlying mechanisms of TSPAN12 in lung cancer progression are largely unknown.
In the present study, we showed that QKI-5 negatively regulated miR-196b-5p and increased miR-196b-5p expression in NSCLC promoted cell migration and proliferation by directly targeting GATA6 and TSPAN12. QKI-5 binding to miR-196b-5p influences miR-196b-5p stability, resulting in enhanced miR-196b-5p expression. Up-regulated miR-196b-5p expression in NSCLC is closely related to hypomethylation of its promoter region. Our in vitro and in vivo analyses demonstrated that the QKI-5∼miR-196b-5p∼GATA6/TSPAN12 axis is a potential therapeutic target in NSCLC.
Results
QKI-5 Is Down-Regulated in NSCLC and Associated with Lung Cancer Cell Growth.
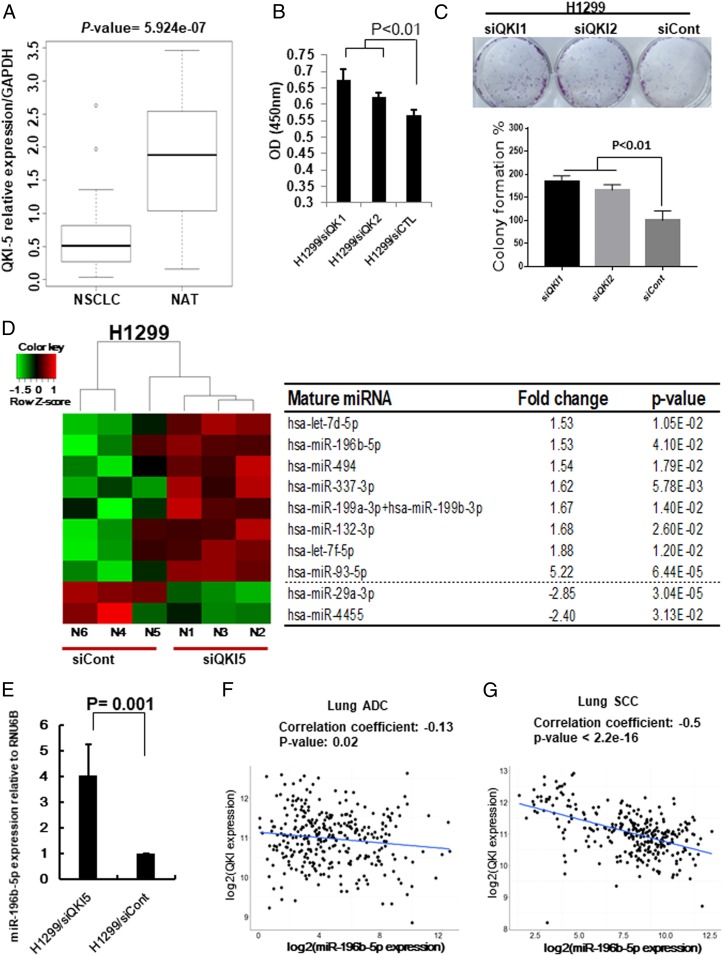
First, we checked QKI expression in 334 lung adenocarcinoma (ADC) patients with 57 matched normal adjacent tissues (NATs) (SI Appendix, Fig. S1A) and 349 lung squamous cell carcinoma (SCC) patients with 51 matched NATs (SI Appendix, Fig. S1B) from The Cancer Genome Atlas (TCGA) RNA-sequencing (RNA-seq) data. Significantly down-regulated QKI expression was observed in both lung ADC (P < 2.2e-16) and lung SCC (P < 2.2e-16). We further analyzed QKI-5 expression in 30 NSCLC tissues and 30 corresponding NATs from Zhoushan Hospital of Wenzhou Medical University and found that QKI-5 expression was significantly down-regulated in NSCLC tissues compared to the NATs (Fig. 1A). Subsequent Kaplan–Meier survival analysis using 1,882 available NSCLC patients from the Kaplan–Meier plotter showed that high expression of QKI is significantly associated with favorable prognosis of NSCLC patients (SI Appendix, Fig. S1C). In addition, our qRT-PCR results demonstrated that most lung cancer cell lines showed reduced expression of QKI-5 compared to the normal lung cell lines (SI Appendix, Fig. S1D). Since QKI-5 is markedly down-regulated in NSCLC and associated with the patient’s survival, we further investigated the role of QKI-5 in lung cancer cell growth and colony formation. We selected H1299 and U2020 cells, which have relatively higher expression of QKI-5, to knock down QKI-5 expression. The expression levels of QKI-5 after knockdown were confirmed by Western blot analysis (SI Appendix, Fig. S2 A and B). In response to knockdown of QKI-5, H1299 cell’s proliferative and colony-formation ability were significantly increased (Fig. 1 B and C). Similar results were also observed in QKI-5 knockdown U2020 cells (SI Appendix, Fig. S2 C and D). All together, these results indicate that QKI-5 is bona fide tumor suppressor in NSCLC.
Fig. 1.
QKI-5 down-regulation promotes miR-196b-5p expression. (A) Expression level of QKI-5 in 60 paired NSCLC tissues and their matched NATs. The RNA samples were extracted from 30 NSCLC tissues and 30 corresponding NATs. The RNAs were subject to qRT-PCR with a QKI-5 probe and the expression was normalized by GAPDH. (B and C) Cell proliferation assay (B) and colony formation assay (C) for QKI-5 knockdown H1299 cells. The cell growth rates were measured by cell counting kit 8. The values present mean ± SD as determined by quintuplet assays. Colony-forming areas were measured by ImageJ software. The average values were derived from three random areas. (D) Results of NanoString miRNA assay by comparing QKI-5 knockdown H1299 cells (H1299/siQKI) and control (H1299/siCont) cells. (E) qRT-PCR measure miR-196b-5p expressions in H1299/siQKI and H1299/siCont cells. (F and G) QKI expression from TCGA RNA-seq data and miR-196b-5p expression from miR-seq data were used to examine correlation between miR-196b-5p and QKI expressions in lung ADC dataset (n = 306) (F) and lung SCC dataset (n = 289) (G).
QKI-5 Downregulation Promotes miR-196b-5p Expression.
To identify QKI-5–regulated miRNAs, we conducted a NanoString nCounter miRNA expression assay using QKI-5 knockdown H1299 cells (H1299/siQKI) and control cells (H1299/siCont). We selected 10 differentially expressed miRNAs with P values less than 0.05 and the fold changes larger than 1.5 (Fig. 1D). Among 10 differentially expressed miRNAs, 8 were up-regulated in H1299/siQKI and 2 were down-regulated. Since the role of miR-196b-5p in lung cancer remained controversial, we selected miR-196b-5p for further analyses. Subsequent qRT-PCR analysis validated that miR-196b-5p was up-regulated in H1299/siQKI cells (Fig. 1E). In addition, Pearson correlation analyses using TCGA lung ADC (n = 306) or lung SCC (n = 289) samples that had both QKI-5 and miR-196b-5p expressions showed that miR-196b-5p was significantly negatively correlated with QKI-5 in lung ADC (r = −0.13, P = 0.02) and lung SCC (r = −0.5, P < 2.2e-16) (Fig. 1 F and G). These results suggest that QKI-5 might negatively regulate the miR-196b-5p expression in NSCLC.
miR-196b-5p Promotes Lung Cancer Cell Growth In Vitro and In Vivo.
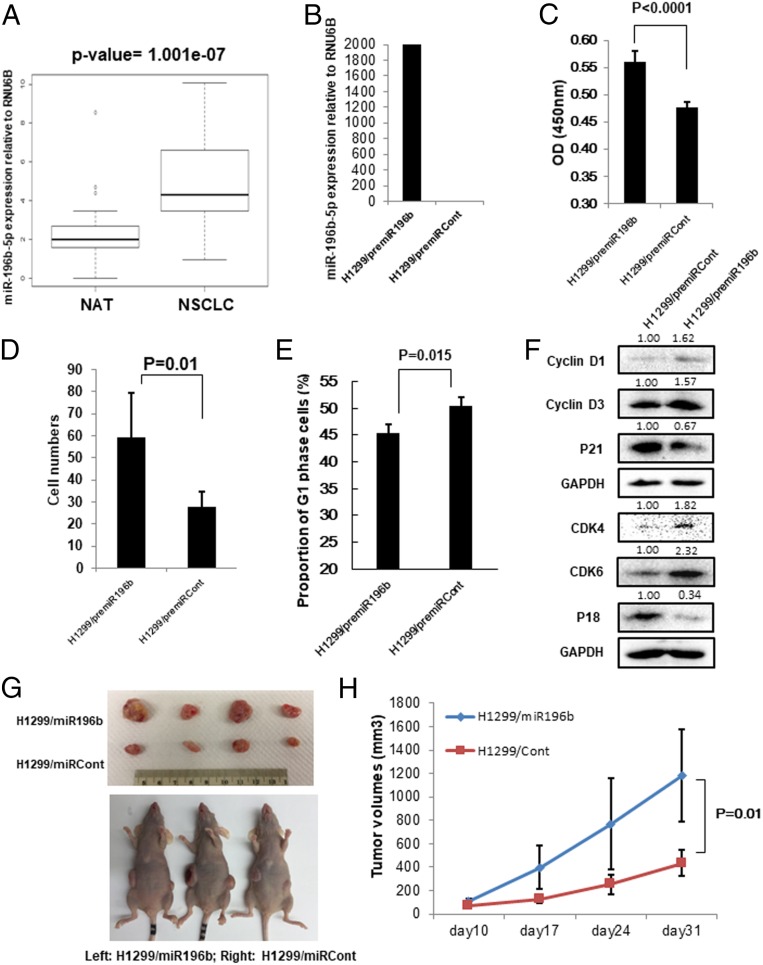
Our results showed that QKI-5 expression was significantly down-regulated in NSCLC tissues and negatively correlated with miR-196b-5p expression. We next examined miR-196b-5p expression using the TCGA miRNA-seq dataset including 468 lung ADC, 198 lung SCC, and 86 NATs and found that miR-196b-5p was markedly up-regulated in both lung ADC (SI Appendix, Fig. S3A) and SCC (SI Appendix, Fig. S3B) compared to the NATs. Reanalysis of miR-196b-5p expression in 70 paired NSCLC tissues and NATs from The Ohio State University Comprehensive Cancer Center Tissue Procurement Shared Resource confirmed that miR-196b-5p expression was significantly up-regulated in NSCLC tissues compared to the corresponding NATs (Fig. 2A and SI Appendix, Fig. S3C). Given the high expression of miR-196b-5p in NSCLC tissues, we speculate that miR-196b-5p might play a role in the pathogenesis of NSCLC. To investigate the function of miR-196b-5p in lung cancer cells, we transiently overexpressed PremiR-196b precursor miRNA in two NSCLC cell lines, H1299 and A549 cells, since these two cell lines have relatively low expression of miR-196b-5p compared to other lung cancer cell lines (SI Appendix, Fig. S3D). The expression levels of miR-196b-5p after overexpression were confirmed by qRT-PCR (Fig. 2B and SI Appendix, Fig. S4A). In response to overexpression of miR-196b-5p, the H1299 (Fig. 2 C and D and SI Appendix, Fig. S4D) and A549 (SI Appendix, Fig. S4 B and C) cells’ proliferative and migratory ability were significantly increased. In addition, miR-196b-5p overexpression in H1299 cells promoted cell cycle G1-to-S transition (Fig. 2E), indicating that increased lung cancer cell proliferative ability by miR-196b-5p might be due to promoted cell cycle progression. As expected, we found that overexpression of miR-196b-5p in H1299 cells induced G1 phase-related proteins expression including cyclin D1, cyclin D3, CDK4, and CDK6 and suppressed CDK inhibitor 1A (P21) and 2C (P18) (Fig. 2F). Similar results were also observed in the miR-196b-5p–overexpressing A549 cells (SI Appendix, Fig. S4E). To investigate the functions of miR-196b-5p in tumor growth in vivo, the stable miR-196b-5p–overexpressing H1299/miR-196b cells and corresponding control cells were subcutaneously (s.c.) injected into the flanks of nude mice. Overexpression of miR-196b-5p in H1299 cells significantly increased tumor growth in vivo compared to the control cells, indicating that miR-196b-5p functions as an onco-miRNA in lung cancer in vivo (Fig. 2 G and H).
Fig. 2.
miR-196b-5p plays oncogenic functions in NSCLC. (A) Expression level of miR-196b-5p in 70 paired NSCLC tissues and their matched NATs. The RNA samples were extracted from 35 NSCLC tissues and 35 corresponding NATs. The RNAs were subject to qRT-PCR with an miR-196b-5p probe and the expression was normalized by U6B. Significantly up-regulated miR-196b-5p was observed in NSCLC tissues compared with their matched NATs. (B) qRT-PCR measure miR-196b-5p expression in premiR-196b–overexpressing H1299 cells and control cells. (C) Cell proliferation assay for premiR-196b–overexpressing H1299 cells and control cells. The cell growth rates were measured by cell counting kit 8. The values present mean ± SD as determined by quintuplet assays. (D) Cell migration assay for premiR-196b–overexpressing H1299 cells and control cells using transwell membranes. The average counts were derived from six random microscopic fields. (E) Proportion of cells in cell cycle G1 phase in premiR-196b–overexpressing H1299 cells and control cells were determined by flow cytometry analysis. The values present mean ± SD as determined by triple assays. (F) Western blot analysis of cell cycle G1 phase-related proteins (Cyclin D1, Cyclin D3, CDK4, CDK6, P18, and P21) in premiR-196b–overexpressing H1299 cells and control cells. The bands were quantified using ImageJ software and relative values were obtained by normalizing to the value of each corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (G and H) Effects of miR-196b-5p on tumor growth in mouse model. Stable miR-196b-5p–overexpressing H1299 cells (H1299/miR196b) and control cells (H1299/miRCont) were s.c. injected into the flanks of nude mice. (G) Representative photographs of the tumors at day 31 after inoculation with either the H1299/miR196b or H1299/miRCont cells. (H) Tumor growth in nude mice s.c. injected into flanks with H1299/miR196b or H1299/miRCont. Data are presented as mean ± SD (n = 5 per group).
GATA6 and TSPAN12 Are Direct Targets of miR-196b-5p.
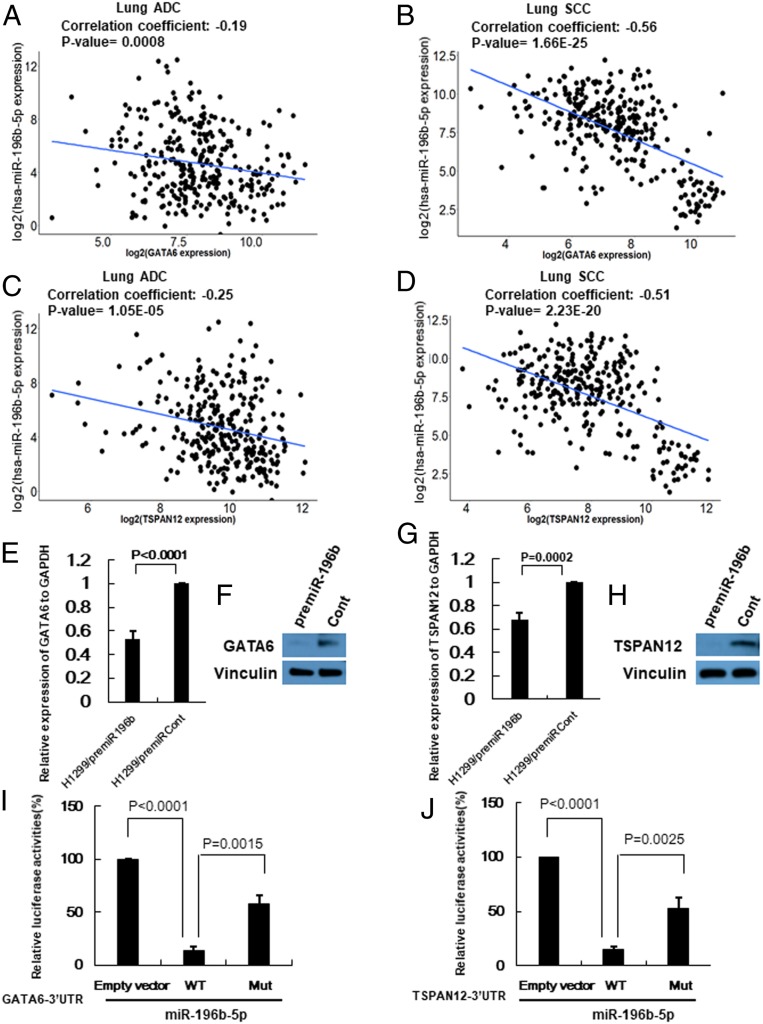
To better understand the underlying mechanisms of miR-196b-5p in NSCLC, we performed bioinformatics analyses to identify possible targets of miR-196b-5p. First, we used seven available miRNA target prediction websites to predict miR-196b-5p targets and selected possible targets with scores ≥5 (SI Appendix, Table S1). Next, we conducted Pearson correlation analysis using 289 lung SCC (n = 289) samples from TCGA with expression data for both miR-196b-5p and its target genes. Among 43 possible targets of miR-196b-5p, 12 genes were significantly negatively correlated with miR-196b-5p, including known targets, FAS and AQP4 (SI Appendix, Table S2) (26, 27). Since both GATA6 and TSPAN12 were most significantly negatively correlated with miR-196b-5p and the 3′UTRs of both transcripts contained conserved sequences complementary to the miR-196b-5p seed sequence (SI Appendix, Fig. S5A), we selected these two genes for further analyses. Pearson correlation analysis of 306 lung ADC samples and 289 lung SCC samples from the TCGA that had both miR-196b-5p and GATA6 or TSPAN12 expression data showed that miR-196b-5p was significantly negatively correlated with both GATA6 (r = −0.19, P = 0.0008 in lung ADC; r = −0.56, P = 1.66e-25 in lung SCC) and TSPAN12 (r = −0.25, P = 1.05e-05 in lung ADC; r = −0.51, P = 2.23e-20 in lung SCC) (Fig. 3 A–D). To investigate the effects of miR-196b-5p on the expression of target mRNAs and proteins, we conducted overexpression analysis. Overexpressing miR-196b-5p significantly reduced both mRNA and protein expressions of GATA6 and TSPAN12 in H1299 cells (Fig. 3 E–H). Next, to determine whether GATA6 and TSPAN12 are direct targets of miR-196b-5p, we performed luciferase reporter assays. miR-196b-5p consistently reduced the luciferase activity for the 3′UTRs of GATA6 and TSPAN12 after cotransfecting each of the 3′UTRs and miR-196b-5p mimics in 293T cells. To further confirm target specificity, we used the QuikChange Mutagenesis kit to generate a mutated form of the 3′UTRs in the miR-196b-5p binding sites where binding sites were destroyed. Each 3′UTR of GATA6 and TSPAN12 had one well-conserved miR-196b-5p binding site. The primer sequences for 3′UTRs mutagenesis are shown in SI Appendix, Table S3. Cotransfecting miR-196b-5p with mutated forms of the 3′UTRs (GATA6 mut 3′UTR and TSPAN12 mut 3′UTR) markedly attenuated the reduction of luciferase activities on the wild-type 3′UTRs (Fig. 3 I and J), suggesting that miR-196b-5p bound specifically to its target 3′UTRs. These results suggest that miR-196b-5p directly targets GATA6 and TSPAN12, and elevated miR-196b-5p expression may reduce GATA6 and TSPAN12 expressions in NSCLC.
Fig. 3.
GATA6 and TSPAN12 are direct targets of miR-196b-5p. (A and B) GATA6 expression from TCGA RNA-seq data and miR-196b-5p expression from miR-seq data were used to examine correlation between miR-196b-5p and GATA6 expressions in lung ADC dataset (n = 306) (A) and lung SCC dataset (n = 289) (B). (C and D) TSPAN12 expression from TCGA RNA-seq data and miR-196b-5p expression from miR-seq data were used to examine correlation between miR-196b-5p and TSPAN12 expressions in lung ADC dataset (n = 306) (C) and lung SCC dataset (n = 289) (D). (E and F) qRT-PCR and Western blot to measure GATA6 mRNA and protein levels in lung cancer cells transfected with premiR-196b or control. (G and H) qRT-PCR and Western blot to measure TSPAN12 mRNA and protein levels in lung cancer cells transfected with premiR-196b or control. (I and J) Luciferase reporter constructs containing wild-type or mutated form of GATA6 (I) and TSPAN12 (J) 3′UTRs were cotransfected with miR-196b-5p mimic into the 293T cells. Data are presented as mean ± SD as determined by triple assays.
GATA6 and TSPAN12 Play Crucial Roles in miR-196b-5p–Mediated Lung Cancer Progression.
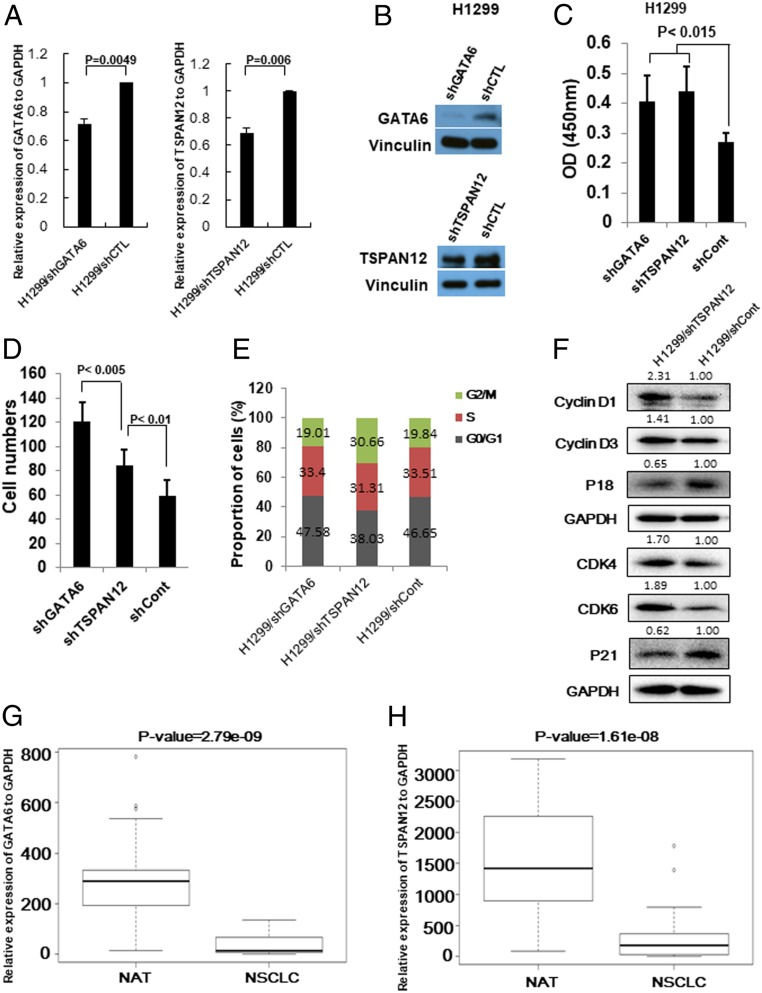
First, we investigated the functions of GATA6 and TSPAN12 in lung cancer cell proliferation and migration. We knocked down GATA6 or TSPAN12 in H1299 lung cancer cells by short hairpin RNAs. The mRNA (Fig. 4A) and protein (Fig. 4B) expression levels of GATA6 and TSPAN12 after knockdown were confirmed by qRT-PCR analysis and Western blot analysis. Knocking down GATA6 or TSPAN12 significantly increased H1299 cells’ proliferative (Fig. 4C) and migratory (Fig. 4D and SI Appendix, Fig. S5B) ability. Interestingly, knocking down GATA6 more efficiently promoted lung cancer cell migration than knocking down TSPAN12 (SI Appendix, Fig. S5B). In addition, cell cycle analysis showed that knocking down GATA6 did not affect each cell cycle proportion of cells; however, knocking down TSPAN12 drastically promoted transition of G1 to S cell cycle (Fig. 4E). Accordingly, cell cycle G1 phase-related proteins were consistently changed after knocking down TSPAN12 (Fig. 4F), and these changes were completely the same as after overexpression of miR-196b-5p. These results indicate that both GATA6 and TSPAN12 were involved in miR-196b-5p–induced cell proliferation and migration. GATA6 tends to affect miR-196b-5p–mediated cell migration while TSPAN12 tends to affect cell proliferation via regulating transition of G1 to S cell cycle. Next, we evaluated GATA6 and TSPAN12 expressions in 60 paired NSCLC tissues and NATs from Zhoushan Hospital of Wenzhou Medical University and confirmed that both GATA6 and TSPAN12 expressions were significantly down-regulated in NSCLC tissues compared to the corresponding NATs (Fig. 4 G and H and SI Appendix, Fig. S5 C and D). We further examined both GATA6 and TSPAN12 expressions in lung ADC and lung SCC using TCGA dataset. A total of 334 lung ADC tissues with 57 matched NATs and 349 lung SCC tissues with 51 matched NATs with available gene expression data were assayed and found that both GATA6 and TSPAN12 were significantly down-regulated in both lung ADC and lung SCC when compared with the NATs. In lung ADC, the GATA6 (P < 2.2e-16) and TSPAN12 (P < 2.2e-16) expressions were dramatically reduced in primary tumor tissues (SI Appendix, Fig. S5 E and F). Similar reductions of GATA6 (P < 2.2e-16) and TSPAN12 (P < 2.2e-16) expression levels were also observed in lung SCC (SI Appendix, Fig. S5 G and H). The Kaplan–Meier survival analysis using 3021 available NSCLC patients from the Kaplan–Meier plotter showed that high expressions of GATA6 or TSPAN12 were significantly associated with favorable prognosis of NSCLC patients (SI Appendix, Fig. S5 I and J), indicating that both GATA6 and TSPAN12 play critical role in the progression of NSCLC.
Fig. 4.
GATA6 and TSPAN12 play a crucial role in NSCLC progression. (A) qRT-PCR measures GATA6 and TSPAN12 mRNA levels in lung cancer cells transfected with shGATA6 or shTSPAN12 plasmids. (B) Western blot analysis measures GATA6 and TSPAN12 protein levels in lung cancer cells transfected with shGATA6 or shTSPAN12 plasmids. (C) Cell proliferation assay for GATA6 or TSPAN12 knockdown H1299 lung cancer cells. The cell growth rates were measured by cell counting kit 8. The values present mean ± SD as determined by quintuplet assays. (D) Cell migration assay for GATA6 or TSPAN12 knockdown H1299 cells using transwell membranes. The average counts were derived from six random microscopic fields. (E) Proportion of cells in each cell cycle phase in H1299/shGATA6, H1299/shTSPAN12, and H1299/shCont cells were determined by flow cytometry analysis. (F) Western blot analysis of cell cycle G1 phase-related proteins (Cyclin D1, Cyclin D3, CDK4, CDK6, P18, and P21) in H1299/shTSPAN12 cells and control cells. The bands were quantified using ImageJ software and relative values were obtained by normalizing to the value of each corresponding GAPDH. (G) Expression level of GATA6 in 60 paired NSCLC tissues and their matched NATs. The RNA samples were extracted from 30 NSCLC tissues and 30 corresponding NATs. The RNAs were subject to qRT-PCR with a GATA6 probe and the expression was normalized by GAPDH. (H) Expression level of TSPAN12 in 60 paired NSCLC tissues and their matched NATs. The RNA samples were extracted from 30 NSCLC tissues and 30 corresponding NATs. The RNAs were subject to qRT-PCR with a TSPAN12 probe and the expression was normalized by GAPDH.
CpG172 Methylation Regulates miR-196b-5p Expression.
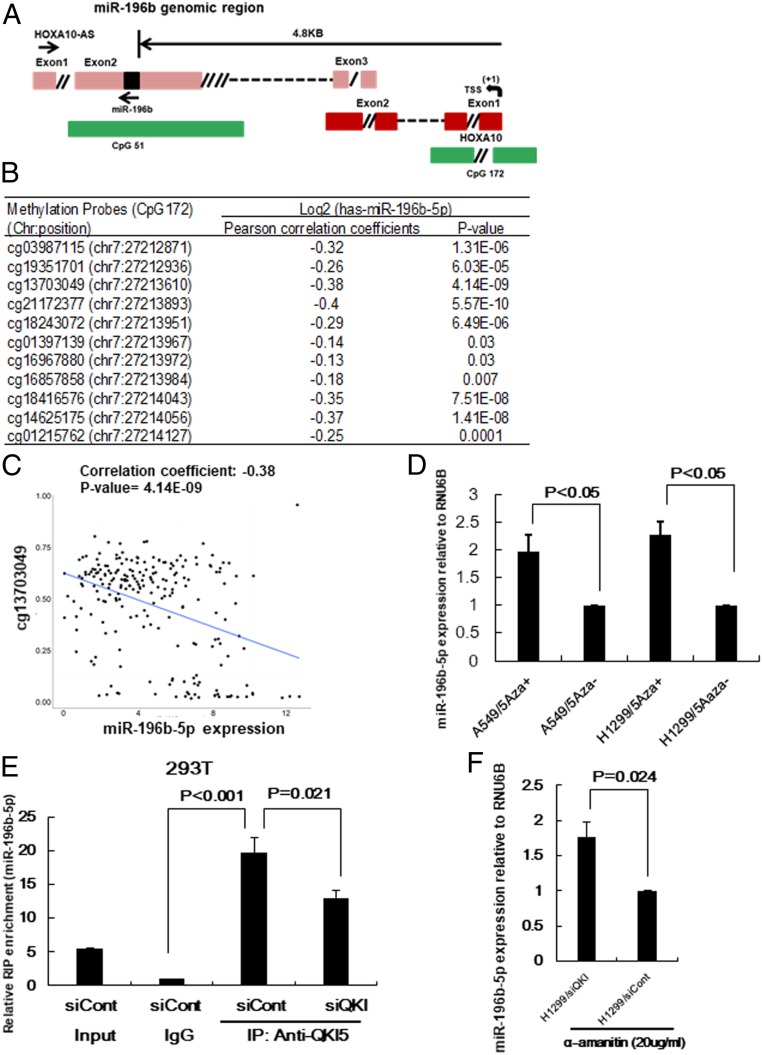
Epigenetic changes and/or transcriptional activation in the promoter region are reported to be closely related to miRNA expression (28). miR-196b-5p is located on exon 2 of the long noncoding RNA, HOXA10-AS, and directionally antisense to the HOXA10-AS. HOXA10 is located near the miR-196b-5p genomic region and has the same transcriptional direction as that of miR-196b-5p. miR-196b-5p is on the CpG island, CpG51, and the HOXA10 promoter region has another CpG island, CpG172 (Fig. 5A). We first determined whether up-regulated miR-196b-5p expression in NSCLC is associated with hypomethylation of CpG51. Eleven methylation probes located on CpG51 with available methylation data were selected from TCGA Illumina Infinium Human DNA Methylation 450k beadchip data to evaluate the correlation between miR-196b-5p expression and CpG51 methylation status. TCGA NSCLC samples (n = 221) having both methylation and miR-196b-5p expression data were extracted for Pearson correlation analyses. Among 11 probes on CpG51, only one probe showed significant anticorrelation with miR-196b-5p expression (SI Appendix, Fig. S6A), indicating that methylation status of CpG51 might not be associated with miR-196b-5p expression in NSCLC. To investigate whether miR-196b-5p and HOXA10 transcriptionally coregulated under the same promoter, we evaluated the expression pattern of these two genes using TCGA dataset and found significant positive correlation between miR-196b-5p and HOXA10 (r = 0.69, P = 2.2e-16), suggesting that these two genes might share the same promoter (SI Appendix, Fig. S6B). Next, we examined whether up-regulated miR-196b-5p expression in NSCLC is associated with hypomethylation of its promoter region’s CpG island, CpG172. Pearson correlation analysis using TCGA NSCLC samples having both methylation and miR-196b-5p expression data showed that all 11 methylation probes in CpG172 were significantly anticorrelated with the expression of miR-196b-5p in NSCLC (Fig. 5B), further indicating that promoter’s methylation could be involved in the repression of miR-196b-5p transcription. Anticorrelation between the methylation probe cg13703049 on the CpG172 and miR-196b-5p expression in NSCLC (r = −0.38, P = 4.14e-09) are shown in Fig. 5C. In addition, treatment of lung cancer cells by the demethylation reagent 5-aza-CdR for 3 d significantly increased miR-196b-5p (Fig. 5D) as well as pri-miR-196b (SI Appendix, Fig. S6C) expression. Taken together, these results suggest that hypomethylation at miR-196b-5p promoter-associated CpG island at least partially contributes to the higher miR-196b-5p expression in NSCLC.
Fig. 5.
Underlying mechanisms of up-regulated miR-196b-5p expression in NSCLC. (A) Schematic structure of miR196b-5p∼HOXA10-AS ∼HOXA10 genomic locus (not to scale). miR-196b-5p is located in exon 2 of HOXA10-AS. TSS indicates putative transcription start site and green boxes show the CpG islands. CpG51 expands exon 2 of HOXA10-AS and CpG172 expands exon 1 of HOXA10. (B) Correlation between miR-196b-5p expression from TCGA dataset and methylation probes in the promoter region of miR-196b-5p (CpG172) from TCGA Illumina Infinium Human DNA Methylation 450k beadchip in NSCLC. (C) Representative figure of correlation between miR-196b-5p expression and methylation probe cg13703049 (chr 7: 27213610) in NSCLC. (D) qRT-PCR to measure miR-196b-5p expression in lung cancer cell lines A549 and H1299 cells after treatment with 7.5µM 5-aza-CdR for 3 d. The values present mean ± SD as determined by triple assays. (E) Lysates from QKI-5 knockdown 293T cells or control cells were subjected to RIP analysis. The cell extracts were subjected to immunoprecipitation with IgG or anti-QKI5 antibody. Pull-down RNA was analyzed by qRT-PCR using specific probe for miR-196b-5p. Data are presented as mean ± SD as determined by triple assays. (F) RNAs were extracted from QKI-5 knockdown 293T cells or control cells treated with 20 μg/mL α-amanitin for 9 h and then subjected to qRT-PCR with a miR-196b-5p probe. U6B probe was used for normalization. Data are presented as mean ± SD as determined by triple assays.
QKI-5 Interacts with miR-196b-5p and Negatively Regulates Its Expression.
Our NanoString miRNA profiling data showed that inhibiting QKI-5 promoted miR-196b-5p expression in lung cancer cells. To further investigate the underlying mechanisms by which QKI-5 regulates miR-196b-5p, we first determined the interaction between QKI-5 and miR-196b-5p by RNA-binding protein immunoprecipitation (RIP) assay using an antibody against QKI-5. Both miR-196b-5p (Fig. 5E) and primiR-196b (SI Appendix, Fig. S6D) were enriched in the RIP samples using the QKI-5 antibody compared with the RIP samples using immunoglobulin G (IgG). Interestingly, knocking down QKI-5 significantly reduced miR-196b-5p enrichment in the RIP samples compared with the control samples using an antibody against QKI-5. Of note, QKI-5 exhibited stronger binding ability to miR-196b-5p than to primiR-196b as the miR-196b-5p enrichment was ∼20 times higher in the RIP samples with QKI-5 antibody than in those with IgG. However, primiR-196b was only approximately three times higher in the RIP samples with QKI-5 antibody than in those with IgG. As expected, enrichment of the internal control, U6B, did not differ between RIP samples that used IgG and those that used QKI-5 antibody (SI Appendix, Fig. S6E), suggesting that QKI-5 bound specifically to miR-196b-5p. To investigate the function of QKI-5 on miR-196b-5p and primiR-196b stability, QKI-5 knockdown H1299 cells and control cells were treated with the RNA polymerase II inhibitor α-amanitin. After 9 h of treatment with α-amanitin, miR-196b-5p (Fig. 5F) was more abundant in the QKI-5 knockdown H1299 cells than in the control cells, while primiR-196b was not (SI Appendix, Fig. S6F). These results indicate that QKI-5 regulates miR-196b-5p but not primiR-196b expression at least partially by interacting with and decreasing the stability of miR-196b.
Effects of GATA6 and TSPAN12 on Tumor Growth In Vivo.
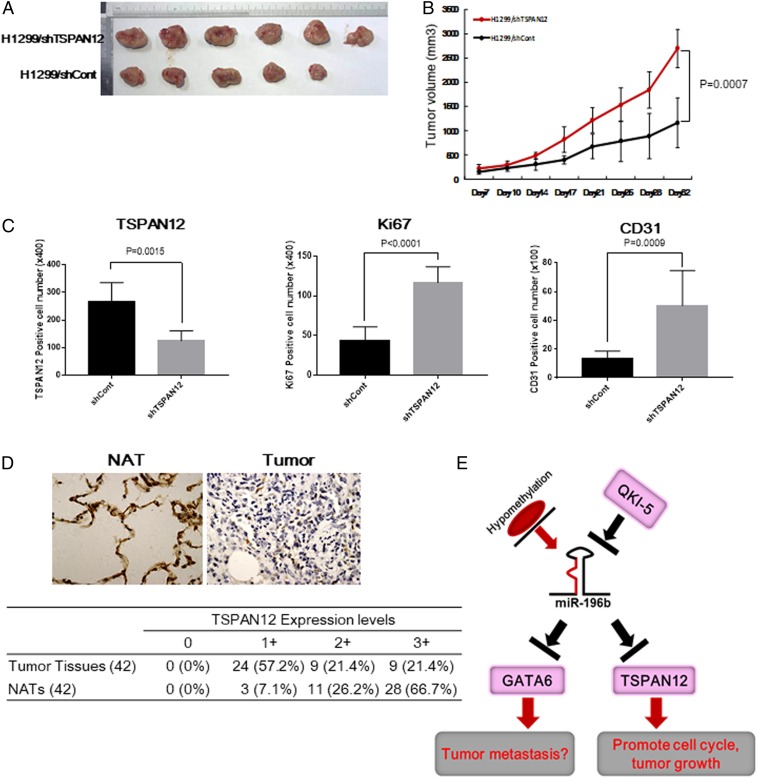
Our in vitro analyses showed that both GATA6 and TSPAN12 were involved in NSCLC cell proliferation and migration. To understand the role of GATA6 and TSPAN12 down-regulation in tumor growth in vivo, the stable GATA6 or TSPAN12 knockdown H1299/shGATA6 or H1299/shTSPAN12 and the corresponding control cells were s.c. injected into the flanks of nude mice. Knockdown of TSPAN12 in H1299 cells significantly increased tumor growth in vivo compared with that in the control cells (Fig. 6 A and B); however, knockdown of GATA6 in H1299 cells did not affect tumor growth in vivo (SI Appendix, Fig. S7A), indicating that enhanced tumor growth via miR-196b-5p overexpression was mainly due to down-regulated expression of TSPAN12 in vivo. Immunohistochemical analyses for the expressions of TSPAN12, Ki67, and murine CD31 protein showed that both Ki67- and CD31-positive cells were significantly increased, while TSPAN12-positive cells were markedly down-regulated in the tumor tissues derived from H1299/shTSPAN12-injected mice (Fig. 6C and SI Appendix, Fig. S7B), indicating that down-regulation of TSPAN12 might promote cell proliferation and angiogenesis in vivo. In addition, we further analyzed TSPAN12 protein expression levels in tissue samples from 42 NSCLC patients by immunohistochemical staining. The NSCLC tissues displayed significantly down-regulated TSPAN12 immunoreactivity compared with that of the NATs from the same patients. Of note, 92.9% of the NAT specimens showed moderate to strong TSPAN12 expression; however, only 42.8% of NSCLC tissue specimens presented moderate to strong TSPAN12 expression (Fig. 6D). These results indicate that both mRNA and protein levels of TSPAN12 were down-regulated in NSCLC tissues and TSPAN12 might play a critical role in NSCLC carcinogenesis.
Fig. 6.
(A and B) Effects of TSPAN12 on tumor growth in a mouse model. (A) Representative photographs of the tumors at day 32 after inoculation with either of the H1299/shTSPAN12 or H1299/shCont cells. (B) Tumor growth in nude mice s.c. injected into flanks with H1299/shTSPAN12 or H1299/shCont cells. Data are presented as mean ± SD (n = 6 per group). (C) TSPAN12-, CD31-, and Ki67-positive cells in tumors tissues derived from H1299/shTSPAN12 or H1299/shCont cells. Paraffin sections of tumors developing in nude mice were stained with anti-TSPAN12, anti-CD31, or anti-ki67 antibodies. The number of TSPAN12-positive cells, CD31-positive microvessels, or ki67-positive tumor cells in eight fields of tumors that demonstrated the highest reactivities with antibodies were counted at ×100 or ×400, respectively, and presented as mean ± SD. (D, Upper) Representative immunohistochemical staining for TSPAN12 in NSCLC tissues and NATs from the same patient. (D, Lower) Summary of tissue immunohistochemical staining data for TSPAN12 in 42 pairs of clinical NSCLC tissues and NATs. (E) Proposed dysregulated miR-196b-5p mediated down-regulation of GATA6 and TSPAN12 is involved in lung cancer pathogenesis in our research.
Discussion
miRNA has tissue-specific expression patterns, and its expression is often dysregulated in most cancers. Numerous miRNAs function as oncogenes or tumor suppressors in most cancers, depending on the cancer tissue type (29). miR-196b-5p functions in cancer pathogenesis in a context-dependent manner (30–33) but remains controversial in lung cancer. Recent reports indicate that miR-196b-5p inhibits lung cancer progression and metastasis by targeting LIN28 and Runx2 (33, 34). It has also been reported that miR-196b-5p was epigenetically silenced in the premalignant stage of lung cancer (35), indicating tumor-suppressive functions of miR-196b-5p. On the other hand, miR-196b-5p has been reported to increase aggressiveness of NSCLC by direct targeting of HOXA9 (36). miR-196b-5p also promotes lung cancer cell migration and invasion by targeting GATA6 (18); however, detailed mechanisms of miR-196b-5p and GATA6 in clinical patient samples and mouse model are unclear. These facts clearly indicate that the functions of miR-196b-5p in the progression and metastasis of lung cancer remain to be further determined.
Regarding the dual functions of miR-196b-5p, our current study claims four findings to support the oncogenic functions of miR-196b-5p in NSCLC. First, the tumor suppressor, QKI-5, was significantly down-regulated in NSCLC, and knocking down QKI-5 in lung cancer cells increased miR-196b-5p expression. The miR-196b-5p expression was significantly negatively correlated with QKI-5 expression and was up-regulated in NSCLC tissues. Second, a series of in vitro and in vivo experiments consistently showed that miR-196b-5p functions as an onco-miRNA in NSCLC. Third, miR-196b-5p directly targeted GATA6 and TSPAN12. The expressions of GATA6 and TSPAN12 were markedly down-regulated in NSCLC tissues and negatively correlated with the expression of miR-196b-5p. Knockdown analyses showed that GATA6 more strongly inhibited lung cancer cell migration, while TSPAN12 tends more to restrain lung cancer cell proliferation through cell cycle arrest. Accordingly, mouse xenograft models demonstrated that knockdown of TSPAN12 promoted in vivo tumor growth of lung cancer cells, while knockdown of GATA6 did not. Fourth, we analyzed the association between promoter methylation and miR-196b-5p expression in two CpG islands on the two possible miR-196b-5p promoters. CpG172 methylation levels on the promoter region of miR-196b-5p were negatively correlated with the expression of miR-196b-5p in NSCLC. In addition, QKI-5 bound to miR-196b-5p and negatively regulated its stability, resulting in up-regulated miR-196b-5p expression in NSCLC (Fig. 6E).
Our in vitro experiments showed that GATA6 could inhibit lung cancer cell proliferative and migratory abilities but did not affect the cell cycle. In contrast with the in vitro analyses, our mouse xenograft model indicated that knockdown of GATA6 did not affect tumor growth in vivo. Importantly, knockdown of GATA6 exhibited stronger and superior migratory abilities than did the TSPAN12 knockdown in vitro. These results suggest that GATA6 might primarily function in lung cancer cell metastasis rather than in primary tumor growth. Further studies are needed to validate this.
The functions of TSPAN12 in cancer are largely unknown. A few studies have reported that TSPAN12 plays oncogenic roles in lung cancer (24, 37). However, no plausible mechanisms have been investigated for the TSPAN12-mediated regulation of lung cancer progression. Inconsistent with previous reports regarding the oncogenic functions of TSPAN12, we claim that TSPAN12 plays tumor-suppressive roles in NSCLC. Using a TCGA dataset and our NSCLC samples, we confirmed that the expression of TSPAN12 was significantly down-regulated in NSCLC tissues compared with those of the NATs. Increased expression of TSPAN12 was associated with favorable prognosis in patients with NSCLC. In addition, knockdown of TSPAN12 increased proliferative and migratory abilities of lung cancer cells and promoted cell cycle transition. Consistent with the in vitro study, mouse xenograft models also showed that knockdown of TSPAN12 accelerated tumor growth in vivo accompanied with increased angiogenesis, indicating that TSPAN12 has novel, unrecognized functions in NSCLC.
We propose that QKI-5 mediated up-regulation of miR-196b-5p enhanced NSCLC progression by directly targeting GATA6 and TSPAN12. The current study revealed that the anticancer mechanisms of QKI-5 may involve regulating miRNAs. The study provided important insight into understanding the dual functions of miR-196b-5p in NSCLC and suggested that targeting the QKI-5∼miR-196b-5p∼GATA6/TSPAN12 pathway may be a potential therapeutic strategy for treating NSCLC.
Materials and Methods
Patients and Tissue Samples.
This study was approved by the Institutional Research Human Ethical Committee of the Wenzhou Medical University for the use of clinical biopsy specimens and informed consent was obtained from the patients before starting. Sixty paired frozen tissue specimens from patients with NSCLC were obtained from Zhoushan Hospital of Wenzhou Medical University. Tissue samples were flash-frozen using liquid nitrogen within 2 h of surgical resection and stored at −80 °C until analyses. For the miRNA expression, 35 cases with stage 1 of human NSCLC samples were histologically confirmed according to hematoxylin/eosin staining and formalin-fixed, paraffin-embedded tissues were sent to The Ohio State University Pathology Core Facility to microdissect out cancerous tissues and adjacent normal tissues for total RNA isolation by RecoverALL Total Nucleic Acid Isolation kit (Ambion) as described previously (38). Tissues were obtained under an Ohio State University-approved Institutional Review Board protocol and written informed consent was obtained from patients before sample analyses.
Animal Study.
All animal experimental procedures complied with the Wenzhou Medical University’s Policy on the Care and Use of Laboratory Animals. The detailed procedures are described in SI Appendix, SI Materials and Methods.
RIP.
The RIP assay was performed using Magna RIP kit (Millipore) according to the manufacturer's instructions. Briefly, the QKI-5 knockdown 293T cells and control cells were scraped into phosphate-buffered saline containing protease inhibitors and then resuspended in RIP lysis buffer (Millipore) containing protease and RNase inhibitors. Protein A/G Magnetic Beads were incubated overnight with 5 μg of rabbit monoclonal anti-QKI5 antibody (ab232502; Abcam) or normal Rabbit IgG (Millipore) as negative control. After incubation, the samples were added to antibody–bead complexes and incubated overnight. After washings, immune complexes and input were eluted and treated with proteinase K and heated at 55 °C for 30 min to digest the protein. RNA was purified with phenol/chloroform extraction followed by ethanol precipitation. RNAs obtained from RIP samples were subjected to qRT-PCR using specific Taqman probes.
A detailed description of the materials and methods used in this study is given in SI Appendix, SI Materials and Methods. For additional details on the NanoString nCounter assay, virus infection and transfection, qRT-PCR, cell migration and proliferation assays, flow cytometry analysis, Western blot analysis, TCGA dataset analysis, luciferase reporter assay, demethylation analysis, target analysis, animal study, immunohistochemical staining, and statistical analysis see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China grant 81672305 to R.C. and National Cancer Institute grant 1R35CA197706-01 to C.M.C.
Footnotes
The authors declare no competing interest.
Data deposition: The miRNA profiling data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE135348).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917531117/-/DCSupplemental.
References
- 1.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Gridelli C., et al. , Non-small-cell lung cancer. Nat. Rev. Dis. Primers 1, 15009 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Ebersole T. A., Chen Q., Justice M. J., Artzt K., The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 12, 260–265 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Wu J., Zhou L., Tonissen K., Tee R., Artzt K., The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 274, 29202–29210 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Chénard C. A., Richard S., New implications for the QUAKING RNA binding protein in human disease. J. Neurosci. Res. 86, 233–242 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Wang F., et al. , The RNA-binding protein QKI5 regulates primary miR-124-1 processing via a distal RNA motif during erythropoiesis. Cell Res. 27, 416–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Vogel G., Yu Z., Richard S., The QKI-5 and QKI-6 RNA binding proteins regulate the expression of microRNA 7 in glial cells. Mol. Cell. Biol. 33, 1233–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong F. Y., et al. , The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 10, e1004289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X., et al. , Quaking-5 suppresses aggressiveness of lung cancer cells through inhibiting β-catenin signaling pathway. Oncotarget 8, 82174–82184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel D. P., MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Calin G. A., Croce C. M., MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Li Z., Rana T. M., Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 13, 622–638 (2014). [DOI] [PubMed] [Google Scholar]
- 13.van Rooij E., Kauppinen S., Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 6, 851–864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Rooij E., Olson E. N., MicroRNA therapeutics for cardiovascular disease: Opportunities and obstacles. Nat. Rev. Drug Discov. 11, 860–872 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rupaimoole R., Slack F. J., MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Yang H., Lu M. M., Zhang L., Whitsett J. A., Morrisey E. E., GATA6 regulates differentiation of distal lung epithelium. Development 129, 2233–2246 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Cheung W. K., et al. , Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell 23, 725–738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Feng C., Shi S., miR-196b promotes lung cancer cell migration and invasion through the targeting of GATA6. Oncol. Lett. 16, 247–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yunqi H., et al. , miR-455 functions as a tumor suppressor through targeting GATA6 in colorectal cancer. Oncol. Res. 27, 311–316 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X., Zhang W., Ji W., miR-181a targets GATA6 to inhibit the progression of human laryngeal squamous cell carcinoma. Future Oncol. 14, 1741–1753 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Junge H. J., et al. , TSPAN12 regulates retinal vascular development by promoting Norrin- but not Wnt-induced FZD4/beta-catenin signaling. Cell 139, 299–311 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Knoblich K., et al. , Tetraspanin TSPAN12 regulates tumor growth and metastasis and inhibits β-catenin degradation. Cell. Mol. Life Sci. 71, 1305–1314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otomo R., et al. , TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc. Natl. Acad. Sci. U.S.A. 111, 18691–18696 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z., Hou D., Wang X., You Z., Cao X., TSPAN12 is overexpressed in NSCLC via p53 inhibition and promotes NSCLC cell growth in vitro and in vivo. OncoTargets Ther. 11, 1095–1103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J., Chen C., Li G., Chen D., Zhou Q., Upregulation of TSPAN12 is associated with the colorectal cancer growth and metastasis. Am. J. Transl. Res. 9, 812–822 (2017). [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Wang L., He F., Li B., Han R., Long noncoding RNA LINC00629 restrains the progression of gastric cancer by upregulating AQP4 through competitively binding to miR-196b-5p. J. Cell. Physiol. 235, 2973–2985 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Li Z., et al. , miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 3, 688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Leva G., Garofalo M., Croce C. M., MicroRNAs in cancer. Annu. Rev. Pathol. 9, 287–314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croce C. M., Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer S. E., et al. , miR-196b target screen reveals mechanisms maintaining leukemia stemness with therapeutic potential. J. Exp. Med. 215, 2115–2136 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porretti J., et al. , CLCA2 epigenetic regulation by CTBP1, HDACs, ZEB1, EP300 and miR-196b-5p impacts prostate cancer cell adhesion and EMT in metabolic syndrome disease. Int. J. Cancer 143, 897–906 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Shao L., et al. , Methylation of the HOXA10 promoter directs miR-196b-5p-dependent cell proliferation and invasion of gastric cancer cells. Mol. Cancer Res. 16, 696–706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stiegelbauer V., et al. , miR-196b-5p regulates colorectal cancer cell migration and metastases through interaction with HOXB7 and GALNT5. Clin. Cancer Res. 23, 5255–5266 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Bai X., et al. , MicroRNA-196b inhibits cell growth and metastasis of lung cancer cells by targeting Runx2. Cell. Physiol. Biochem. 43, 757–767 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Tellez C. S., et al. , miR-196b is epigenetically silenced during the premalignant stage of lung carcinogenesis. Cancer Res. 76, 4741–4751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S. L., et al. , Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear Factor-kappa B activity in non-small cell lung cancer cells. Mol. Carcinog. 55, 1915–1926 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Ye M., et al. , TSPAN12 promotes chemoresistance and proliferation of SCLC under the regulation of miR-495. Biochem. Biophys. Res. Commun. 486, 349–356 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Peng Y., et al. , Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 15043–15048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.