Significance
Single-cell transcriptomics has emerged as a powerful means to define the molecular heterogeneity of brain neurons. However, which of the neurons with known transcriptomes interact with each other in specific neural circuits is largely unknown. Here, we devised a strategy, termed “Connect-seq,” which combines retrograde viral tracing and single-cell transcriptomics to determine the molecular identities of individual upstream neurons in a defined circuit. Using Connect-seq, we uncovered a large variety of signaling molecules expressed in neurons upstream of hypothalamic neurons that control physiological responses to stress. Information obtained by Connect-seq can be used to overlay molecular maps on anatomical neural circuit maps and generate molecular tools for probing the functions of individual circuit components.
Keywords: single-cell RNA-seq, neural circuits, stress
Abstract
The mouse brain contains about 75 million neurons interconnected in a vast array of neural circuits. The identities and functions of individual neuronal components of most circuits are undefined. Here we describe a method, termed “Connect-seq,” which combines retrograde viral tracing and single-cell transcriptomics to uncover the molecular identities of upstream neurons in a specific circuit and the signaling molecules they use to communicate. Connect-seq can generate a molecular map that can be superimposed on a neuroanatomical map to permit molecular and genetic interrogation of how the neuronal components of a circuit control its function. Application of this method to hypothalamic neurons controlling physiological responses to fear and stress reveals subsets of upstream neurons that express diverse constellations of signaling molecules and can be distinguished by their anatomical locations.
The brain contains a multitude of neural circuits that control diverse functions, but the specific neuronal components of most circuits and their respective roles are largely a mystery (1–3). The application of single-cell RNA sequencing techniques has allowed rapid advances in defining the transcriptomes of brain neurons (1–6). However, much less is known about which of the vast numbers of individual neurons with known transcriptomes are interconnected to control specific functions. Here we devised a strategy, “Connect-seq,” to obtain additional information about the neuronal components of specified neural circuits. The strategy couples the use of conditional neurotropic viruses that cross synapses with transcriptome analysis of single upstream neurons infected with virus to obtain the molecular identities of individual neurons connected in a circuit.
To test the strategy, we focused on neural circuits that transmit signals to corticotropin-releasing hormone neurons (CRHNs) in the paraventricular nucleus of the hypothalamus (PVN) (7). In response to danger, CRHNs induce increases in blood stress hormones that act on multiple tissue systems to coordinate appropriate physiological responses. Surges in blood stress hormones occur in response to a variety of external and internal dangers or “stressors,” including predator odors, tissue injury, inflammation, and emotional and psychological stress (7, 8). Studies using classical neuroanatomical and neurophysiological approaches have provided a large body of information on PVN inputs that may affect CRHNs and the effects of classical neurotransmitters on CRHN activity and function (7, 9–12). However, the exact identities of upstream neurons that control CRHN functions and the mechanisms by which they do so have not been defined.
Viral tracing studies indicate that CRHNs receive direct synaptic input from neurons in 31 different brain areas (8). These include 19 areas of the hypothalamus, a brain area that integrates information from the body and external environment and organizes fitting behavioral and physiological responses. Neurons two or more synapses upstream of CRHNs are seen in additional areas, including one small area of the olfactory cortex that has proved key to stress hormone responses to predator odors (8). The viral tracing studies provide an anatomical map of neurons directly presynaptic to CRHNs, but the molecular identities of those neurons are unknown.
If one could identify genes whose expression defines subsets of neurons upstream of CRHNs in different brain areas, one would have molecular tools to explore which subsets mediate responses to different stressors and to manipulate those subsets to gain insight into how they function. Information regarding neurotransmitters and neuromodulators used by the upstream neurons to communicate with CRHNs would, in addition, provide information relevant not only to an understanding of neural circuit functions, but also potential insights into pharmacological interventions to modify the functions of specific circuit components.
To test the Connect-seq strategy, we infected Cre recombinase-expressing CRHNs with a Cre-dependent Pseudorabies virus (PRV) that travels retrogradely across synapses. Using flow cytometry, we isolated single virus-infected neurons and then used single-cell RNA sequencing (RNA-seq) to define the transcriptomes of individual upstream neurons. These experiments revealed a large diversity of neurotransmitter and neuromodulator signaling molecules in neurons directly upstream of CRHNs, including more than 40 different neuropeptides. Many individual neurons coexpressed multiple different signaling molecules. These included neurons coexpressing neuropeptides with neurotransmitters or biogenic amines, as well as neurons coexpressing different neuropeptides. Upstream neurons expressing specific signaling molecules mapped to selected brain areas, demonstrating that Connect-seq can provide molecular tools with which to dissect the functions of individual neuronal components of neural circuits.
Results
Transcriptome Analysis of Neurons Upstream of CRHNs.
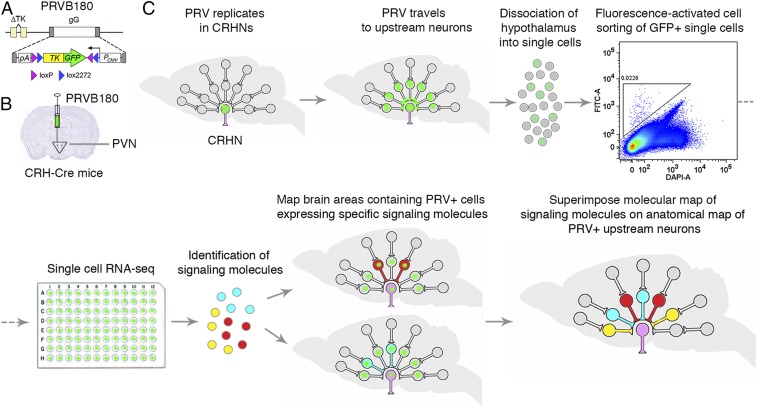
We first developed a Pseudorabies virus, PRVB180, which has Cre recombinase-dependent expression of thymidine kinase (TK) fused to green fluorescent protein (GFP) (Fig. 1A). After infecting neurons expressing Cre, this virus will travel retrogradely across synapses to infect upstream neurons. We injected PRVB180 into the PVN of CRH-IRES-Cre (CRH-Cre) mice (Fig. 1B), which express Cre in CRHNs (13). Following infection with PRVB180, immunostaining of brain sections for GFP indicated that the virus travels retrogradely at the same rate as PRVB177, which expresses TK-HA (hemagglutinin-tagged TK) instead of TK-GFP and travels to directly presynaptic neurons on day 3 postinfection (d3pi) and then to neurons two synapses upstream on d4pi (8).
Fig. 1.
Connect-seq method. (A) PRVB180 has Cre recombinase-dependent expression of a thymidine kinase–green fluorescent protein fusion protein (TK-GFP). (B) CRHNs were infected with PRVB180 by injecting the virus into the PVN of CRH-Cre mice. (C) Following PRVB180 replication in CRHNs and travel of virus to upstream neurons, the hypothalamus was isolated and dissociated to obtain single cells. Flow cytometry was used to deposit cells with high GFP (FITC-A) and low DAPI-A staining, one per well, in multiwell plates. RNA-seq was used to analyze their transcriptomes and the signaling molecules (neurotransmitters/neuromodulators) they expressed. The locations of PRV+ upstream neurons expressing selected signaling molecules were then determined, allowing a molecular map of upstream neurons to be superimposed on the existing anatomical circuit map.
We next conducted transcriptome analysis of neurons upstream of CRHNs. We infected CRHNs with PRVB180 and, on d3pi, isolated the hypothalamus and dissociated the tissue into a single-cell suspension (Fig. 1C). Using flow cytometry, we isolated GFP+ (PRV-infected) cells, one per well, in 96-well plates (14) (Materials and Methods and SI Appendix, Fig. S1). Since they were isolated on d3pi, the neurons isolated should be presynaptic to CRHNs, but the possibility that a minor fraction are two synapses upstream cannot be absolutely excluded. Microscopic examination of cells sorted into microtiter plates confirmed that the methods used deposited only a single cell per well (Materials and Methods and SI Appendix, Fig. S2). cDNAs were prepared from each sorted cell and tested for GFP expression by PCR. We then conducted RNA-seq (15, 16) on individual GFP-expressing cells by Illumina-sequencing cDNAs prepared from those cells (SI Appendix, Fig. S3A). Gene expression in individual cells (SI Appendix, Fig. S3B) was determined by alignment to the mouse genome using standard methods (17, 18). Infection of cells with PRV was confirmed by aligning sequence data to PRV genome data (Materials and Methods and SI Appendix, Fig. S3 C and D). We then examined the transcriptome of each cell for expression of markers of neurons or glia (Materials and Methods) and proceeded to analyze 117 cells that expressed neuronal markers.
Neurons were sequenced at an average of ∼6.5 million reads per cell. Comparison of sequenced reads and numbers of genes expressed in neurons did not indicate a correlation (Pearson’s coefficient = −0.06; SI Appendix, Fig. S3E). To evaluate the effect of PRV infection on the number of genes detected, we used Pearson’s correlation. Comparison of the number of genes expressed with the percentage of PRV transcripts in individual neurons indicated a weak correlation (−0.22), suggesting that PRV infection can reduce the number of genes detected to a small extent (SI Appendix, Fig. S3F).
Neurons Upstream of CRHNs Express Diverse Neurotransmitters and Neuromodulators.
To characterize the neurons upstream of CRHNs, we focused on signaling molecules that neurons use to convey information to their downstream partners (19). These include classical “fast neurotransmitters,” which act via ligand-gated ion channels on downstream neurons to rapidly activate or inhibit those neurons, and numerous neuromodulators that bind to G protein-coupled receptors (GPCRs) on downstream neurons to modulate their excitability. We reasoned that the large number of different signaling molecules in the brain and their differential expression among neurons could serve to 1) optimize the discovery of molecular identifiers that would distinguish different upstream neurons and 2) provide potential insight into the molecular mechanisms used by upstream neurons to communicate with CRHNs.
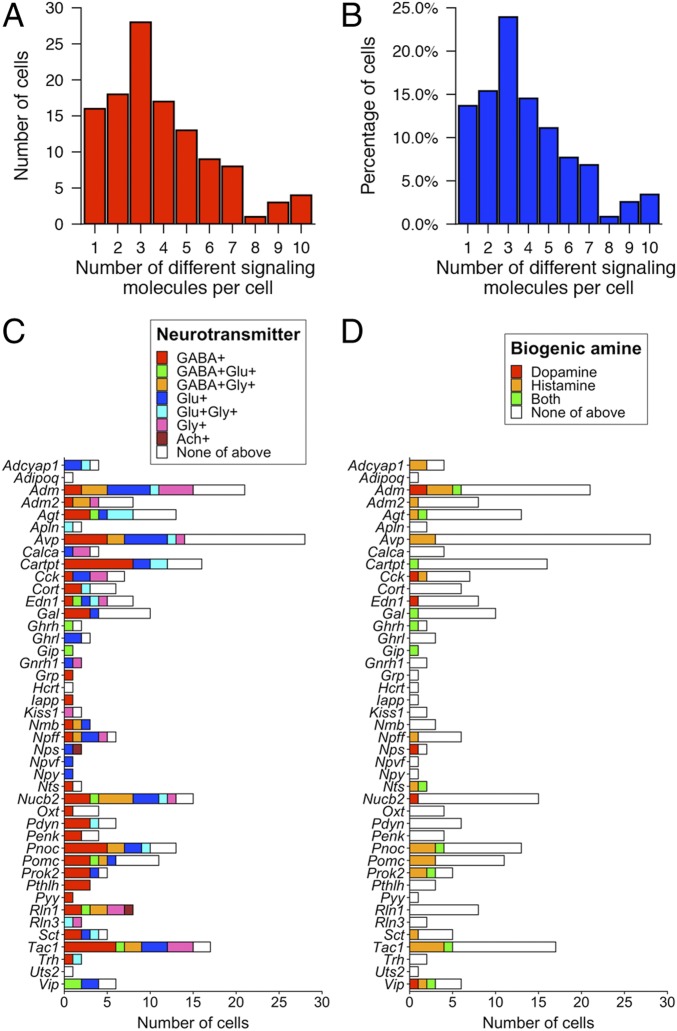
We identified a large number and variety of neurotransmitters and neuromodulators expressed by PRV-infected neurons upstream of CRHNs (Fig. 2). We used a conservative threshold of 1 FPKM (fragments per kilobase of transcript per million mapped reads) to define expressed genes in individual cells. All of the infected neurons that were analyzed expressed at least one signaling molecule. The signaling molecules included glutamate and GABA, which act via ligand-gated ion channels and are the major excitatory and inhibitory neurotransmitters in the brain, respectively. We also identified two other fast neurotransmitters that signal through ligand-gated ion channels, acetylcholine and glycine. All of these neurotransmitters can also act as neuromodulators by binding to specific GPCRs on downstream neurons.
Fig. 2.
Neurons upstream of CRHNs express a large array of signaling molecules. (A) Pie charts show percentages of upstream neurons expressing different neurotransmitters, biogenic amines, neuropeptides, gaseous neuromodulators, and other neuromodulators. (B) Heat map illustrating expression levels of different signaling molecules expressed in 117 individual upstream neurons. Different neurons are indicated on the x-axis. The y-axis shows different signaling molecules, neuronal markers, and housekeeping genes and their encoding or indicator genes.
The neurotransmitters were expressed in 65% of upstream neurons analyzed (Fig. 2A), but the number of neurons expressing different neurotransmitters varied (Fig. 2B and SI Appendix, Fig. S4A). Glutamate and GABA were expressed in 24.8% and 36.8% of the neurons, respectively, while the other neurotransmitters were expressed in far fewer neurons (1.7% for acetylcholine and 17.1% for glycine). CRHNs have ligand-gated channels for both glutamate (20) and GABA (21, 22) and receive direct synaptic input from both neurotransmitters (11, 23, 24). Our results suggest that CRHNs can be activated by numerous glutamatergic presynaptic neurons, but that many other upstream neurons may suppress CRHN activity via GABA transmission. Previous studies indicate that upstream GABAergic neurons can provide tonic inhibition to CRHNs, the release of which can lead to CRHN activation by disinhibition (11, 25). In addition, some upstream GABAergic neurons might be involved in stimulus-induced suppression of CRHN responses, as previously reported for certain odors (26, 27).
Small numbers of upstream neurons expressed biogenic amines (dopamine, histamine; Fig. 2 and SI Appendix, Fig. S4B). These molecules, which signal through GPCRs, have widespread effects in the brain, can also act on peripheral tissues, and are the targets of numerous pharmacological agents (28). By identifying the functions of different biogenic amine-expressing neurons upstream of CRHNs and determining how the amines affect CRHNs, it is possible that small molecule agonists or antagonists for specific biogenic amine receptors or their transporters could ultimately be employed to block or enhance the effects of particular stimuli on CRHNs.
Less common types of neurotransmitters/neuromodulators were also identified in small numbers of upstream neurons. These included the gaseous neuromodulators carbon monoxide (CO) and nitric oxide (NO), as well as d-serine, and the purines ATP and adenosine (Fig. 2 and SI Appendix, Fig. S4C).
Remarkably, neuropeptide expression was seen in 88.9% of upstream neurons (Fig. 2A). A total of 43 different neuropeptides were identified in neurons upstream of CRHNs (Fig. 2 and SI Appendix, Fig. S4D). None of the neurons expressed Crh, suggesting that infected CRHN (“starter cells”) had died by d3pi. This is consistent with observations that neurons can exhibit normal physiological properties for 48, but not 72, hours after PRV infection (29, 30).
Neuropeptides can act as neuromodulators to enhance or dampen neuronal excitability by binding to GPCRs on downstream neurons (31, 32). The number of neurons expressing different neuropeptides ranged from a single neuron (e.g., Grp [gastrin releasing peptide] and Npy [neuropeptide Y]) to 28 neurons (Avp [arginine vasopressin]; Fig. 2B and SI Appendix, Fig. S4D). The large number of neuropeptides identified in upstream neurons and their differential expression among the upstream neurons provides a rich set of tools for superimposing a molecular map on the anatomical map of CRHN upstream circuits and for dissecting the functions of different subsets of upstream neurons.
Upstream Neurons Can Express Multiple Signaling Molecules.
Many of the upstream neurons expressed more than one neurotransmitter and/or neuromodulator (Figs. 2B and 3 and SI Appendix, Fig. S5). All analyzed neurons expressed at least one of the signaling molecules examined. Strikingly, 86.3% of the neurons expressed two or more signaling molecules and 47% expressed four or more (Fig. 3 A and B).
Fig. 3.
Combinatorial expression of signaling molecules in neurons upstream of CRHNs. (A and B) Graphs show the number (A) and percentage (B) of upstream neurons expressing different numbers of neurotransmitters and/or neuromodulators. (C and D) Graphs show the number of cells that coexpressed different neuropeptides with the excitatory neurotransmitter glutamate or the inhibitory neurotransmitter GABA or glycine or acetylcholine (C) or with different neuromodulatory biogenic amines (D).
Many upstream neurons coexpressed a given neuropeptide together with glutamate or GABA, or with glutamate and GABA in different neurons (Figs. 2B and 3C). In some cases, a neuropeptide was coexpressed with a biogenic amine (Figs. 2B and 3D) or other neuromodulator (Fig. 2B and SI Appendix, Fig. S5). This is consistent with previous reports that a neuropeptide (Pomc [proopiomelanocortin] or Ghrh [growth hormone-releasing hormone]) can be coexpressed with glutamate in some neurons, but GABA in others (33–35). In our experiments, of seven neurons expressing Cck (cholecystokinin), two coexpressed glutamate and one coexpressed GABA, and, of 17 neurons expressing Tac1 (tachykinin1), three coexpressed glutamate, eight coexpressed GABA, and one coexpressed both. We also found Cck and Tac1 coexpressed with either glutamate or GABA in different neurons in RNA-seq data from 898 hypothalamic neurons, which were available in the Gene Expression Omnibus database (accession no. GSE74672) (5).
It is known that neurons expressing a neuropeptide or biogenic amine can also express a classical neurotransmitter (4, 5, 32, 36, 37). These results further indicate that many neuropeptides can be coexpressed with glutamate in some upstream neurons but with GABA in others. Of 39 neuropeptides coexpressed with glutamate or GABA in our studies, 17 were coexpressed with glutamate in some neurons and GABA in others (Fig. 3C), and, interestingly, among these, nine were coexpressed with glutamate and GABA in the same neurons.
These results raise the possibility that individual neuropeptides could exert different modulatory effects on CRHNs depending on whether they are released onto CRHNs together with glutamate versus GABA. For example, if a neuropeptide acts to enhance CRHN excitability, it may further enhance glutamate stimulation of CRHNs, but dampen GABAergic suppression of CRHNs. Our observations are consistent with a complex patterning of stimulatory, inhibitory, and neuromodulatory effects that act at the level of synaptic input to CRHNs from individual neurons to fine tune the effects of upstream inputs on the CRHNs.
Upstream Neurons Can Coexpress Multiple Neuropeptides.
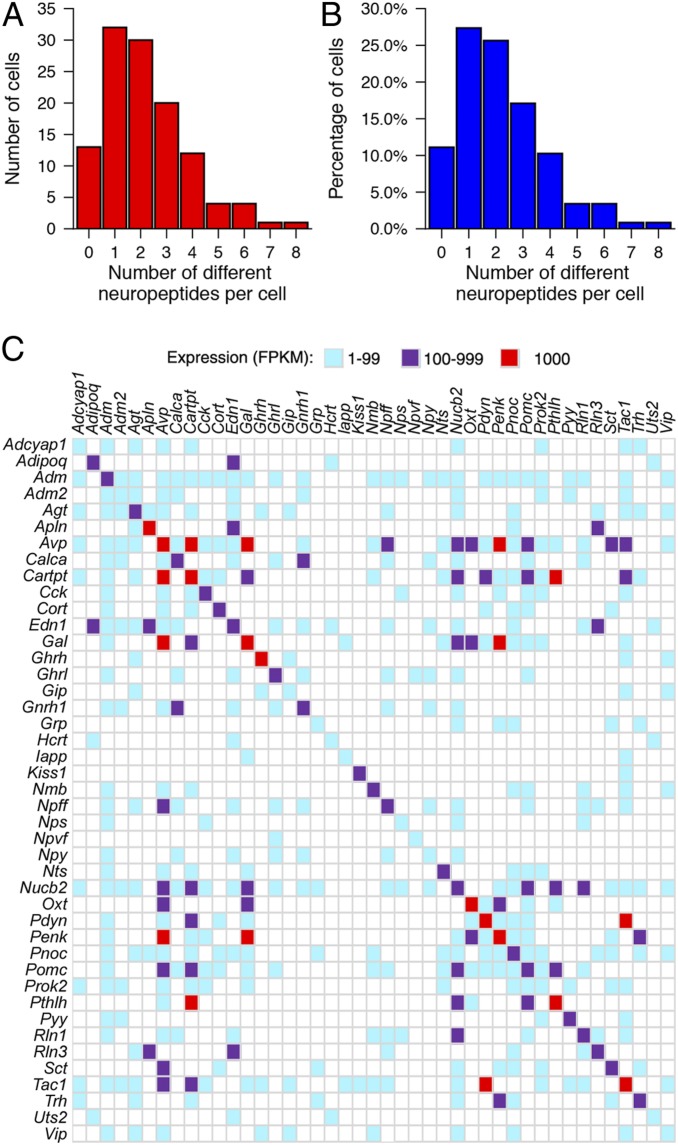
These studies also revealed many upstream neurons expressing more than one neuropeptide. As noted earlier, neuropeptides were expressed in 88.9% of the neurons upstream of CRHNs. While 27.4% of upstream neurons expressed a single neuropeptide, 61.5% coexpressed two or more, 6.8% coexpressed five or six neuropeptides, and a few neurons expressed seven or eight neuropeptides each (Fig. 4 A and B). Since single-cell RNA-seq is subject to the loss (“drop-out”) of low copy number mRNAs that can vary among cells, coexpression of different neuropeptides in additional neurons cannot be excluded.
Fig. 4.
Upstream neurons can express more than one neuropeptide. (A and B) Graphs show the number (A) and percentage (B) of upstream neurons that expressed different numbers of neuropeptides. (C) Coexpression of specific neuropeptides in single neurons. Neuropeptides detected in upstream neurons are indicated on the x- and y-axes. Colored boxes indicate pairs of neuropeptides coexpressed in at least one neuron at 1 to 99 FPKM (blue), 100 to 999 FPKM (purple), or ≥1,000 FPKM (red).
These results are consistent with previous reports that neurons expressing a given neuropeptide can also express other neuropeptides (35, 38, 39), although the large number of neuropeptides that could be coexpressed in single neurons was unexpected. Nonetheless, we also found individual neurons expressing multiple neuropeptides in available transcriptome data for 898 single hypothalamic neurons in a previous study (as detailed earlier) (5). In addition, we compared RNA-seq data on PRV+ Pomc+ neurons isolated by Connect-seq versus uninfected Pomc+ neurons manually isolated from Pomc-eGFP mice (SI Appendix, Fig. S6). In both cases, individual Pomc+ neurons coexpressed one or more other neuropeptides with Pomc.
Comparisons of different neuropeptides coexpressed in individual neurons indicate that a given neuropeptide can be coexpressed with a variety of other neuropeptides (Fig. 4C). In some cases, this was only one other neuropeptide, but, in others, a neuropeptide was coexpressed with numerous other neuropeptides, although often in different neurons. The level of expression of individual neuropeptides and other signaling molecules varied among neurons. However, high-level expression of two neuropeptides in the same neuron was relatively rare. For example, Penk (proenkephalin) was coexpressed with 12 other neuropeptides, but high levels (≥1,000 FPKM) of only two of those neuropeptides were detected in a neuron(s) expressing a high level of Penk (Fig. 4C).
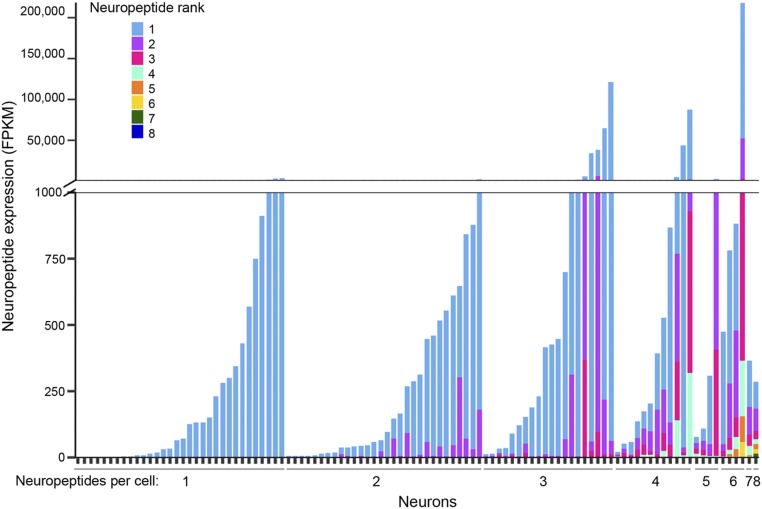
To further investigate the coexpression of neuropeptides in single neurons, we compared the levels of expression of different neuropeptides in individual neurons (Fig. 5). At the level of single neurons, a few neurons expressed two or three neuropeptides at similar levels. However, many other neurons expressed one neuropeptide at a much higher level than one or more others expressed in the same cell. These results suggest that, while a neuron might express multiple neuropeptides, the downstream effects of one might predominate over the effects of others. Some neuropeptides, as well as other signaling molecules, were expressed at very different levels in different cells (Fig. 5 and SI Appendix, Fig. S7), raising the possibility that they might play a more important role in signaling to CRHNs by some upstream neurons than others. One intriguing question is whether the relative expression levels of different signaling molecules in a neuron can be altered according to physiological state, as previously reported for one neuropeptide (40).
Fig. 5.
Coexpression of neuropeptides in individual neurons. Expression levels of different neuropeptides in single neurons varied. Cells are arranged on the x-axis by the number of neuropeptides they expressed (1–8). Vertical bars indicate expression of neuropeptides in each cell, with different neuropeptides indicated in different colors and their expression in FPKM shown on the y-axis.
Upstream Neurons Expressing Specific Neuromodulators Map to Selected Brain Areas.
The major goal of these studies was to find a way to uncover molecular identifiers of upstream neurons that would allow for a molecular map to be superimposed on the anatomical map of neurons upstream of CRHNs. To investigate this possibility, we examined neurons upstream of CRHNs for the expression of neuromodulators we had identified by transcriptome analysis of upstream neurons.
In our studies, we identified numerous neuropeptides in upstream neurons isolated from the hypothalamus. Viral tracing studies previously showed neurons directly upstream of CRHNs in 19 hypothalamic areas. Using in situ hybridization data available in the Allen Brain Atlas (http://mouse.brain-map.org/) (41), we confirmed that a number of the neuropeptides we had identified in upstream neurons are indeed expressed in one or more of those 19 hypothalamic areas (SI Appendix, Fig. S8). Of course, it was unclear whether neurons seen in the atlas are upstream of CRHNs.
To investigate this question, we analyzed the locations of upstream neurons expressing specific neuromodulators. We first infected Cre-expressing CRHNs with PRVB177, which has Cre-dependent expression of hemagglutinin (HA)-tagged TK (8). After 3 days, when the virus had crossed one synapse to infect immediately upstream neurons, we costained brain sections with riboprobes for specific neuromodulators and anti-HA antibodies to detect PRV-infected neurons expressing detectable levels of neuromodulators (Fig. 6).
Fig. 6.
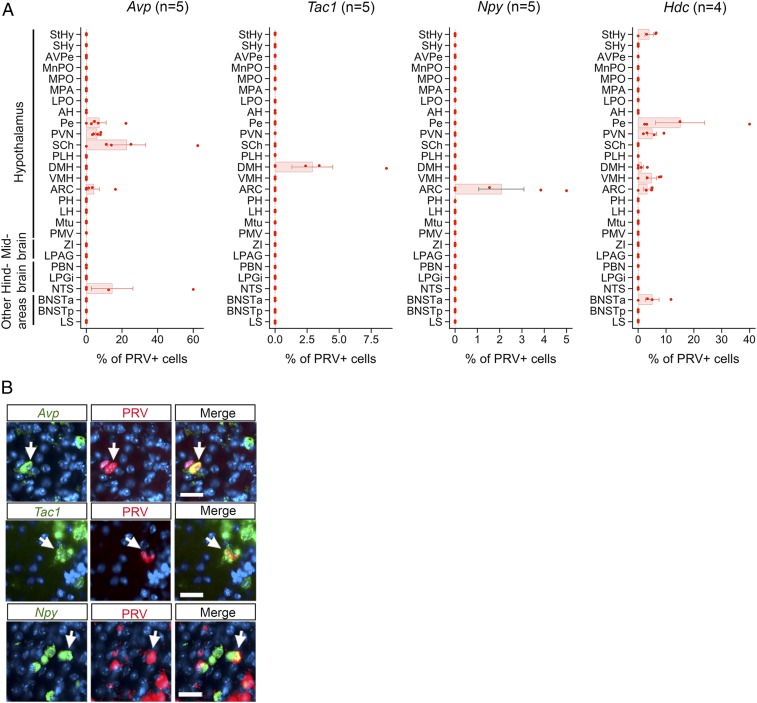
Upstream neurons expressing individual signaling molecules map to specific brain area(s). (A) Graphs show the percentage of PRV+ neurons labeled for different neuromodulators (Avp, Tac1, Npy, or Hdc) in different brain areas following CRHN infection with PRVB177. Error bars indicate SEM. Parenthesized numbers indicate the number of animals (“n”) per condition. (B) CRHNs were infected with PRVB177 and brain sections costained on d3pi with neuropeptide riboprobes (green) and anti-HA antibodies (red; PRV+ cells). Arrows indicate colabeled neurons. (Scale bars, 25 μM.)
Avp was detected in PRV+ neurons in five brain areas, including four areas of the hypothalamus (Fig. 6 A and B). In contrast, Tac1 and Npy were each seen in PRV+ neurons in only one hypothalamic area, the DMH for Tac1 and the ARC for Npy. Hdc (histidine decarboxylase), a marker for the expression of the biogenic amine, histamine, was seen in upstream neurons in six hypothalamic areas and one other brain area.
The selected expression of neuromodulators we identified in upstream neurons in only certain brain areas confirms that data obtained using Connect-seq can be used to superimpose a molecular map on the anatomical map of neural circuits upstream of CRHNs (SI Appendix, Fig. S9). This information can ultimately be used to map functional responses to particular stressors to subsets of upstream neurons in specific brain areas, thereby providing insight into the functional meaning of the anatomical/molecular mapping and suggesting tools to dissect the roles of specific neuronal subsets in particular functions.
Discussion
The brain contains a vast number of neural circuits that govern diverse functions, but the neuronal components of those circuits and their individual contributions to circuit function are largely undefined (1–3, 19). Although recent studies using single-cell RNA-seq have defined the transcriptomes of a multitude of mouse brain neurons (3–6, 14, 35, 42), how those neurons are interconnected in neural circuits that control distinct functions is still poorly understood. Recent technical advances have led to methods for genetically tagging and defining the transcriptomes of neuronal subsets responsive to specific stimuli as well as subsets that innervate a given region or subset of neurons (43–49). However, the diversity of neurons that innervate a particular genetically defined neuronal subset has not previously been investigated at the level of single upstream neurons.
In the present studies, we devised a strategy, termed Connect-seq, which utilizes a combination of viral tracing and single-cell transcriptomics to uncover the molecular identities of individual neurons upstream of a specific set of neurons and reveal the neurotransmitters and neuromodulators they use to communicate with their downstream synaptic targets. As a proof of concept, we used Connect-seq to gain insight into circuits that transmit information to CRHNs, which control blood levels of stress hormones.
We examined a relatively small number of neurons upstream of CRHNs compared to the numbers used in large-scale transcriptome census studies of the brain. Even so, the upstream neurons expressed a large number and variety of neurotransmitters and neuromodulators, including dozens of different neuropeptides. While some neurons expressed only one of these signaling molecules, others expressed different combinations. Since the upstream neurons could synapse with many other downstream neurons in addition to CRHNs, it is possible that only some of these signaling molecules are used to communicate with CRHNs. Nonetheless, these findings suggest the potential for a complex array of inputs to CRHNs that could excite or inhibit CRHNs or modulate their excitability.
Localization experiments revealed subsets of upstream neurons expressing individual neuromodulators we had identified by Connect-seq in distinct brain areas. In conclusion, Connect-seq enables the construction of a molecular map that can be superimposed on an anatomical map of neural circuits, thereby allowing the investigation of roles played by individual neuronal components of those circuits under normal conditions and in disease.
Materials and Methods
Mice.
CRH-IRES-Cre (CRH-Cre) mice were generated as described previously (13). All procedures involving mice were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee. Pomc-eGFP (stock no. 009593) and C57BL/6J mice were purchased from The Jackson Laboratory.
Pseudorabies Viral Vectors.
Construction of PRVB177 was described previously (8). PRVB180 was made according to the methods described previously (8). Briefly, PRVB180 was constructed using homologous recombination between a targeting vector (PRVTK-GFP) and genomic DNA of PRV TK-BaBlu, a thymidine kinase (TK)-deleted PRV Bartha strain derivative with a LacZ insertion into the gG locus (50). For generating PRVTK-GFP, a flexstop-flanked sequence (51) encoding a PRV TK fused at its C terminus to enhanced green fluorescent protein (eGFP), obtained from the pEGFP-N1 vector (Clontech), was first cloned with an inverse orientation into an eGFP-deleted pEGFP-N1 vector (Clontech). Next, following restriction digestion, NsiI fragments containing a CMV promoter, the flexstop-flanked coding sequence, and an SV40 polyadenylation signal were cloned between gG locus sequences matching those 5′ and 3′ to the lacZ sequence in PRV TK-BaBlu to give the final targeting vector (PRVTK-GFP). The vector was then linearized and cotransfected with PRV TK-BaBlu genomic DNA into HEK 293T cells (ATCC). Recombinant virus clones were selected and confirmed following methods described previously (52).
PRVs were propagated by infecting PK15 cells (ATCC) with the viruses using a multiplicity of infection (m.o.i.) of 0.1 to 0.01. After ∼2 days of infection, cells showed a prominent cytopathic effect. Cells were scraped from the dishes, pelleted by centrifugation, and frozen using liquid nitrogen and then quickly thawed in a 37 °C water bath. After three freeze–thaw cycles, cell debris was removed by centrifugation twice at 1,000 × g for 5 min, and the supernatant was aliquoted and stored at −80 °C until use. The titer of viral stocks was determined using standard plaque assays on PK15 cells (53), with titers expressed in plaque-forming units (p.f.u.).
Stereotaxic Injections.
Viruses were injected into the PVN of CRH-Cre mice as described previously (8). All injections were done under inhalation anesthesia of 2% isoflurane. Briefly, 1 µL of PRVs (PRVB180, PRVB177; 1 to 1.5 × 106 p.f.u.) were loaded into a 1-µL syringe and injected bilaterally into the brain at a rate of 100 nL/min using a Stereotaxic Alignment System (David Kopf Instruments). The needle was inserted into the PVN based on a stereotaxic atlas (54). After recovery, animals were singly housed with regular 12-h dark/light cycles, and food and water were provided ad libitum.
Isolation of Single Cells.
For Connect-seq experiments, we used a total of 34 adult (6 to 14 wk old) virgin male and female mice for 34 independent experiments. Eleven adults (five males and six females) yielded 698 GFP+ cells, and 384 cells were sequenced. For female mice, the day of estrous cycle was not determined. Adult CRH-Cre mice were injected with PRVB180. After 3 days, mice were euthanized by cervical dislocation, and the brain quickly removed and submerged in ice-cold Hibernate-A medium (A1247501; Thermo Fisher Scientific). The hypothalamus was carefully microdissected under a microscope by obtaining a single coronal section extending from the optic chiasm to the posterior end of the hypothalamus (rostrocaudal axis) and then the hypothalamus between the anterior commissures (laterally and dorsally), which can include ventral portions of adjacent BNST. The isolated tissue was dissected into tiny pieces in dissociation buffer [Hibernate-A medium with papain (10 U mL−1, PAP2; PDS kit; Worthington Biochemical) and DNase (200 U mL−1, DNase vial, D2; PDS kit; Worthington Biochemical)]. Dissected tissue fragments were transferred into 5 mL of dissociation buffer and incubated for 30 min at room temperature. After incubation, tissue pieces were gently triturated 2 to 3 times through a series of fire-polished glass Pasteur pipettes with decreasing diameters of ∼600 µm, ∼300 µm, and ∼150 µm to dissociate tissue into a cloudy suspension. Cells were sieved using a 70-µm cell strainer into 50-mL tubes containing 10 mL of ice-cold Hibernate-A medium with 1% BSA. Cells were then centrifuged at 800 rpm for 5 min at 4 °C, and the cell pellet obtained was resuspended in ∼500 to 1,000 µL of Hibernate-A medium containing 2% B27 (Thermo Fisher) and DAPI (0.5 ng mL−1) to stain for dead cells. Cells were sieved using a 40-µM cell strainer to obtain single-cell suspension and then subjected to flow cytometry.
PRV-infected single cells were isolated based on the fluorescence emitted by TK-GFP using flow cytometry (FACSAria II; BD Biosciences) in a “single-cell sorting mode” according to methods described previously (14) with minor modifications. Cells were sorted using a 100-μM nozzle at a sheath pressure of 20 psi. Cells were first sorted based on their size and granularity using forward (FSC-A) and side scatter (SSC-A) pulse area parameters to exclude debris, followed by exclusion of aggregates or doublets using pulse width and height parameters (SSC-W vs. SSC-H and FSC-W vs. FSC-H), and then, single cells with high GFP and low DAPI (to obtain live cells) were individually placed in wells of 96-well plates containing 3 μL of RLT Plus lysis buffer (no. 1053393; Qiagen). Single-cell suspensions obtained from control samples, such as cells dissociated from the hypothalamus of a wild type C57BL6/J mouse or cortex of CRH-Cre mice injected with PRVB180, were analyzed first in the initial experiments to determine the threshold for fluorescence, and then single-cell suspensions from CRH-Cre mice were analyzed to sort GFP+ fluorescent cells.
Single cells were obtained from Pomc-eGFP mice as previously described (55). In brief, adult mice were euthanized by cervical dislocation and the arcuate nucleus of the hypothalamus was quickly microdissected and dissociated using papain (10 U mL−1, PAP2, PDS kit; Worthington Biochemical) and DNase (200 U mL−1, DNase vial, D2, PDS kit; Worthington Biochemical). After incubation, tissue pieces were gently triturated 2 or 3 times through a series of fire-polished glass Pasteur pipettes with decreasing diameters of ∼600 µm, ∼300 µm, and ∼150 µm to dissociate tissue into a cloudy suspension. Dissociated cells were plated on coverslips, and fluorescent single cells were visualized by fluorescence microscopy and transferred to individual tubes using a microcapillary pipet and then processed to generate cDNA libraries as described previously (55).
Single-Cell RNA Sequencing.
For Connect-seq experiments, FACS-sorted single cells were processed immediately to generate cDNA libraries using SMART-seq2 (15, 16) with minor modifications. After sorting cells one per well into 96-well plates containing RLT Plus lysis buffer, polyadenylated mRNAs in each well were isolated using magnetic beads with Streptavidin (11205D, Dynabeads; Thermo Fisher) coupled to a modified biotinylated oligo-dT primer (Eurofins Genomics). Beads were added to each well and incubated for 20 min at room temperature. After incubation, the plates were placed on a magnet, retaining the beads with mRNAs at the bottom, and washed twice with wash buffer containing 1× SuperScript II first-strand buffer, 2 U μL−1 RNase inhibitor (30281, Lucigen), and 0.5% (vol/vol) Tween 20 (P9416; Sigma-Aldrich) in nuclease-free water (AM997; Thermo Fisher) at room temperature.
The beads were resuspended in 11 μL of reverse transcription mix containing 1× SuperScript II first-strand buffer, 1 mM dNTP (R1092; Thermo Fisher), 6 mM MgCl2, 1 M Betaine (B0300; Sigma-Aldrich), 5 mM DTT, 1 μM template-switching oligo (TSO; AAGCAGTGGTATCAACGCAGAGTGAATrGrG+G, RNase-free HPLC purified; Exiqon), 0.5 U μL−1 RNase inhibitor, and 10 U μL−1 SuperScript II reverse transcriptase (no. 18064–014; Thermo Fisher) in nuclease-free water. mRNAs were then reverse-transcribed by incubation at 42 °C for 90 min followed by 10 cycles of 50 °C for 2 min and 42 °C for 2 min, and then incubated at 70 °C for 15 min. The resulting cDNAs were then amplified using 1× KAPA HiFi HotStart ReadyMix (KK2602; KAPA Biosystems) and IS PCR primer (AAGCAGTGGTATCAACGCAGAGT, HPLC purified; Eurofins Genomics) for 30 PCR cycles.
Amplified cDNAs were then purified using an equal volume of AMPure XP beads (A63880; Beckman Coulter). Aliquots of cDNA libraries were verified for the presence of PRV transcripts by PCR using primers for GFP (5′, GAGCAAGGCGAGGAGCTGTT; 3′, GGTCAGCTTGCCGTAGGTG). Purified cDNA libraries were next quantified using the Qubit dsDNA HS assay kit (Q32854; Thermo Fisher Scientific). Sequencing libraries were then generated using 1/4 reaction volume of the Nextera XT DNA library prep kit (Illumina) according to the manufacturer’s instructions. Briefly, 1.25 μL of single-cell cDNAs (∼0.25 ng μL−1) were used for tagmentation reaction, and then PCR amplified to insert sequencing adaptors and cell-specific barcodes.
Single-cell cDNA libraries for Pomc+ (GFP+) neurons isolated from Pomc-eGFP mice were prepared as described previously (55). Oligo dT-primed cDNAs were generated from each cell using reverse transcriptase, and a poly(A) extension was added to the 3′ end of each cDNA using deoxynucleotidyl transferase. One third of each cDNA library was then used for amplification with a universal primer. Sequencing libraries were prepared using an Illumina TruSeq DNA Sample Prep Kit as described previously (55).
The resulting libraries were then subjected to Illumina deep sequencing. Sequencing was performed using an Illumina HiSeq 2500 instrument in rapid mode employing a paired-end, 50-base read length (PE50) sequencing strategy. Image analysis and base calling were performed using Illumina Real Time Analysis v1.18 software, followed by “demultiplexing” of indexed reads and generation of FASTQ files using Illumina’s bcl2fastq conversion software (v1.8.4, ref. 56). Reads of low quality were discarded prior to adapter trimming using Trim Galore (v.0.4.4, ref. 57) with the options “–adapter AAGCAGTGGTATCAACGCAGAGTAC–stringency 8–quality 0 -e 0.15–length 20–paired–retain_unpaired”.
The default options in TopHat (17) (v2.1.0) were used to align reads to the mouse genome (UCSC mm10 assembly, using GENCODE M15 release gene models) as well as to the Pseudorabies virus genome (NC_006151.1). Cufflinks (18) (v.2.2.1) was used to estimate gene expression profiles in units of fragments per kilobase of transcript per million mapped reads (FPKM) by first running the “cuffquant” tool on the aligned reads for each cell with the “-u” option, which performs additional algorithmic steps designed to better assign ambiguously mapped reads to the correct gene of origin. Per-cell gene-expression profiles were subsequently normalized with the “cuffnorm” utility, using the “classic-fpkm” normalization method, for use in downstream analysis.
A total of 384 PRV-infected cells were sequenced at a depth average of 6,674,000 reads (median, ∼6,700,000; range, ∼17,500 to ∼13,300,000). A total of 347 cells that expressed at least 500 genes per cell were considered for downstream analysis. These included 117 neurons (as detailed later), which were sequenced at an average depth of 6,530,693 reads (median, 6,696,084; range, 34,414 to 12,211,210) and expressed an average of 3,272 genes (median, 3,215; range, 548 to 10,391).
Expression of genes encoding signaling molecules in single-cell transcriptome data were analyzed using a binary operator “%in%” in R (58, 59). Briefly, lists of gene names of different signaling molecules and the transcriptome data on upstream neurons were uploaded into R and converted to a vector and a data frame, respectively. The data frames with expression data contained gene names in rows and different cells arranged in columns. Using the binary operator function, first, the column in the expression data that contained gene names and the vector with gene names encoding signaling molecules were analyzed to find a match, and, as a result, a new data frame was generated that contained the expression data of signaling molecules in individual upstream neurons.
Similarly, to analyze the gene expression of signaling molecules in previous sequencing data, we used the binary operator function in R. Previous sequencing data were obtained from Gene Expression Omnibus (accession no. GSE74672) (5), which contained expression data of 898 neurons, as defined by their cluster algorithm. Marker genes that expressed at least one FPKM or at least one molecule (for UMI data) were considered for further analysis. Plots for illustration were generated in RStudio (59) using ggplot2 package (60).
Analysis of Marker Genes.
Previous studies established that PRVs spread faithfully between functionally connected neurons by direct cell-to-cell contact, but not by diffusion via extracellular space (61, 62). Neuronal infection with PRVs can activate neighboring astrocytes and microglia, which isolate severely infected and dying neurons. However, glial cells lack the machinery required for the production and release of infectious (enveloped) virions (63, 64).
We first manually inspected transcriptome data for transcripts of different cell type markers in order to include neurons, but exclude nonneuronal cells. We used a stringent threshold of ≥10 FPKM for cell type markers as in our previous analysis of olfactory sensory neurons (55). Cells were classified using a criterion that a cell must express two or more of six different marker genes of a cell type but not more than one of any other cell type. We used the following genes to classify cells: Snap25, Map2, Syp, Kif5c, Dlg2, and Dlg4 for neurons; Aldh1l1, Aldoc, Aqp4, Gfap, S100b, and Sparcl1 for astrocytes; Adgre1, Aif1, Tmem119, Cxcr1, Hexb, and Mafb for microglia; and Cldn11, Mbp, Mobp, Mog, Pdgfra, and Gpr17 for oligodendrocytes. If a cell remained unclassified but expressed one marker of neurons but not another cell type, we additionally examined markers for the excitatory neurotransmitter glutamate (Slc17a/6/7/8) and the inhibitory neurotransmitter GABA (Gad1/2) and included cells with one or more of those markers in our analyses of neurons. Using these criteria, we classified 117 cells as neurons, 53 cells as astrocytes, 31 cells as microglia, and 6 cells as oligodendrocytes. We excluded from further analyses an additional 29 (“mixed”) cells that expressed markers for more than one cell type and 111 (“unclassified”) cells that expressed one marker of one of the cell types or none of the markers.
For neurotransmitters and neuromodulators, we used marker genes that encode rate-limiting enzymes required for their biosynthesis or vesicular transporters that package synaptic vesicles with specific neurotransmitters. For neuropeptides, we used genes encoding known ligands of GPCRs obtained from the International Union of Basic and Clinical Pharmacology (IUPHAR)/British Pharmacological Society (BPS) website (https://www.guidetopharmacology.org/) and those with evidence for having an electrophysiological effect on neurons.
In Situ Hybridization.
Dual labeling studies to examine the expression of markers in PRV-infected cells were performed essentially as described previously (8, 55). Coding region fragments of Avp, Tac1, Npy, and Hdc were PCR-amplified from mouse brain cDNA or genomic DNA and cloned into the pCR4 Topo vector (Thermo Fisher). Digoxigenin (DIG)-labeled riboprobes were prepared using the DIG RNA Labeling Mix (Roche). The PVN of CRH-Cre mice aged 2 to 4 months was injected with PRVB177, as described earlier. After 3 days, mice were euthanized by cervical dislocation, and brains were frozen in OCT (Sakura) on dry ice and stored at −80 °C. Coronal cryostat sections of 20 μm were hybridized to DIG-labeled riboprobes at 56 °C for 13 to 16 h.
After hybridization with riboprobes, sections were washed twice for 5 min at 63 °C in 5× SSC followed by twice for 30 min at 63 °C in 0.2× SSC. Sections were then incubated with horseradish peroxidase (POD)-conjugated sheep anti-DIG antibodies (1:2,000; 1207733910; Roche) and biotinylated anti-HA antibodies (1:300, no. 901505; BioLegend) diluted in blocking buffer (1% Blocking reagent, FP1012; Perkin-Elmer) for 2 h at 37 °C. Sections were then washed three times for 5 min at RT in TNT (0.1 M Tris⋅HCl, pH 7.5, 0.5M NaCl, 0.05% Tween) buffer and then treated using the TSA-plus FLU kit (Perkin-Elmer). Sections were then incubated with 0.5 μg mL−1 DAPI and Alexa555-Streptavidin (1:1,000, no. 32355; Thermo Fisher) at room temperature for 1 h and then coverslipped with Fluoromount-G (no. 0100–01; Southern Biotech).
Cell Counting.
Sections were analyzed essentially as described previously (8). Briefly, images were collected using an AxioCam camera and AxioImager.Z2 microscope with an apotome device (Zeiss). Images were acquired using the auto-exposure setting because of the variability in background signals between sections in different animals. No additional postprocessing was performed on collected images for counting. Counting was performed blindly. A mouse brain atlas was used to identify brain structures microscopically and in digital photos. Every fifth section was analyzed for all experiments. Brain areas containing colabeled cells for a given signaling molecule (at least 10 colabeled cells in a given area) in at least two animals were included.
Abbreviations for Brain Areas.
Abbreviations used for brain areas are according to a mouse brain atlas (54) and our previous report (8): AH, anterior hypothalamic area; ARC, arcuate hypothalamic nucleus; AVPe, anteroventral periventricular nucleus; BNSTa, bed nucleus of the stria terminalis, anterior part; BNSTp, bed nucleus of the stria terminalis, posterior part; DMH, dorsomedial hypothalamic nucleus; LH, lateral hypothalamic area; LPAG, lateral periaqueductal gray; LPGi, lateral paragigantocellular nucleus; LPO, lateral preoptic area; LS, lateral septal nucleus; MnPO, median preoptic nucleus; MPA, medial preoptic area; MPO, medial preoptic nucleus; MTu, medial tuberal nucleus; NTS, nucleus of the solitary tract; PBN, parabrachial nucleus; Pe, periventricular nucleus of the hypothalamus; PH, posterior hypothalamic nucleus; PLH, peduncular part of lateral hypothalamus; PMV, premammillary nucleus, ventral part; SCh, suprachiasmatic nucleus; SHy, septohippocampal nucleus; StHy, striohypothalamic nucleus; VMH, ventromedial hypothalamic nucleus; and ZI, zona incerta.
Supplementary Material
Acknowledgments
We thank J. Delrow, A. Marty, A. Dawson, and R. Meredith at the Fred Hutchinson Cancer Research Center (FHCRC) Genomics Facility for their assistance with RNA-seq and A. Berger, S. Dozono, and B. Raden, at the FHCRC Flow Cytometry Facility for their assistance with flow sorting. We thank X. Ye, A. Spray, and E. Albrecht for technical assistance and members of the Buck Laboratory for helpful discussions. This work was supported by the Millen Literary Trust (E.J.L.), the Howard Hughes Medical Institute (L.B.B.), NIH Grants R01 DC015032 (L.B.B.), R01 DC016442 (L.B.B.), and DP2 HD088158 (C.T.), the Paul G. Allen Frontiers Foundation (through the Allen Discovery Center for Cell Lineage Tracing), and an Alfred P. Sloan Fellowship (C.T.). L.B.B. is on the Board of Directors of International Flavors & Fragrances.
Footnotes
The authors declare no competing interest.
Data deposition: Raw sequencing data related to this study have been archived in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database https://www.ncbi.nlm.nih.gov/geo (accession no. GSE139923).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912176117/-/DCSupplemental.
References
- 1.Luo L., Callaway E. M., Svoboda K., Genetic dissection of neural circuits: A decade of progress. Neuron 98, 865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng H., Mesoscale connectomics. Curr. Opin. Neurobiol. 50, 154–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng H., Sanes J. R., Neuronal cell-type classification: Challenges, opportunities and the path forward. Nat. Rev. Neurosci. 18, 530–546 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Zeisel A., et al. , Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romanov R. A., et al. , Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulin J.-F., Tasic B., Hjerling-Leffler J., Trimarchi J. M., Awatramani R., Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Ulrich-Lai Y. M., Herman J. P., Neural regulation of endocrine and autonomic stress responses. Nat. Rev. Neurosci. 10, 397–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondoh K., et al. , A specific area of olfactory cortex involved in stress hormone responses to predator odours. Nature 532, 103–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senst L., Bains J., Neuromodulators, stress and plasticity: A role for endocannabinoid signalling. J. Exp. Biol. 217, 102–108 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Myers B., Scheimann J. R., Franco-Villanueva A., Herman J. P., Ascending mechanisms of stress integration: Implications for brainstem regulation of neuroendocrine and behavioral stress responses. Neurosci. Biobehav. Rev. 74, 366–375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson C. S., Bains J. S., Watts A. G., Neurotransmitter diversity in pre-synaptic terminals located in the parvicellular neuroendocrine paraventricular nucleus of the rat and mouse hypothalamus. J. Comp. Neurol. 526, 1287–1306 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilera G., Liu Y., The molecular physiology of CRH neurons. Front. Neuroendocrinol. 33, 67–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krashes M. J., et al. , An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature 507, 238–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasic B., et al. , Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picelli S., et al. , Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Macaulay I. C., et al. , G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C., et al. , Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buck L. B., Bargmann C. I., “Smell and taste: The chemical senses” in Principles of Neuroscience, Kandel E., Schwartz J., Jessell T., Siegelbaum S., Hudspeth A. J., Eds. (McGraw-Hill, New York, 2012), pp. 712–742. [Google Scholar]
- 20.Aubry J. M., Bartanusz V., Pagliusi S., Schulz P., Kiss J. Z., Expression of ionotropic glutamate receptor subunit mRNAs by paraventricular corticotropin-releasing factor (CRF) neurons. Neurosci. Lett. 205, 95–98 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Cullinan W. E., GABA(A) receptor subunit expression within hypophysiotropic CRH neurons: A dual hybridization histochemical study. J. Comp. Neurol. 419, 344–351 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Mody I., Maguire J., The reciprocal regulation of stress hormones and GABA(A) receptors. Front. Cell. Neurosci. 6, 4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wamsteeker Cusulin J. I., Füzesi T., Watts A. G., Bains J. S., Characterization of corticotropin-releasing hormone neurons in the paraventricular nucleus of the hypothalamus of Crh-IRES-Cre mutant mice. PLoS One 8, e64943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bains J. S., Wamsteeker Cusulin J. I., Inoue W., Stress-related synaptic plasticity in the hypothalamus. Nat. Rev. Neurosci. 16, 377–388 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Cole R. L., Sawchenko P. E., Neurotransmitter regulation of cellular activation and neuropeptide gene expression in the paraventricular nucleus of the hypothalamus. J. Neurosci. 22, 959–969 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami T., et al. , Stress-related activities induced by predator odor may become indistinguishable by hinokitiol odor. Neuroreport 23, 1071–1076 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Matsukawa M., Imada M., Murakami T., Aizawa S., Sato T., Rose odor can innately counteract predator odor. Brain Res. 1381, 117–123 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Wacker D., Stevens R. C., Roth B. L., How ligands illuminate GPCR molecular pharmacology. Cell 170, 414–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Granstedt A. E., Szpara M. L., Kuhn B., Wang S. S., Enquist L. W., Fluorescence-based monitoring of in vivo neural activity using a circuit-tracing pseudorabies virus. PLoS One 4, e6923 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarthy K. M., Tank D. W., Enquist L. W., Pseudorabies virus infection alters neuronal activity and connectivity in vitro. PLoS Pathog. 5, e1000640 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hökfelt T., et al. , Neuropeptides–An overview. Neuropharmacology 39, 1337–1356 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Nusbaum M. P., Blitz D. M., Marder E., Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci. 18, 389–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dicken M. S., Tooker R. E., Hentges S. T., Regulation of GABA and glutamate release from proopiomelanocortin neuron terminals in intact hypothalamic networks. J. Neurosci. 32, 4042–4048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hentges S. T., Otero-Corchon V., Pennock R. L., King C. M., Low M. J., Proopiomelanocortin expression in both GABA and glutamate neurons. 29, 13684–13690 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R., Wu X., Jiang L., Zhang Y., Single-cell RNA-Seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Mestikawy S., Wallén-Mackenzie A., Fortin G. M., Descarries L., Trudeau L. E., From glutamate co-release to vesicular synergy: Vesicular glutamate transporters. Nat. Rev. Neurosci. 12, 204–216 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Tritsch N. X., Granger A. J., Sabatini B. L., Mechanisms and functions of GABA co-release. Nat. Rev. Neurosci. 17, 139–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hahn T. M., Breininger J. F., Baskin D. G., Schwartz M. W., Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat. Neurosci. 1, 271–272 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Elias C. F., et al. , Leptin activates hypothalamic CART neurons projecting to the spinal cord. Neuron 21, 1375–1385 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Zelikowsky M., et al. , The neuropeptide Tac2 controls a distributed brain state induced by chronic social isolation stress. Cell 173, 1265–1279.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lein E. S., et al. , Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Campbell J. N., et al. , A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pomeranz L. E., et al. , Gene expression profiling with cre-conditional pseudorabies virus reveals a subset of midbrain neurons that participate in reward circuitry. J. Neurosci. 37, 4128–4144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen W. E., et al. , Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakurai K., et al. , Capturing and manipulating activated neuronal ensembles with CANE delineates a hypothalamic social-fear circuit. Neuron 92, 739–753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ekstrand M. I., et al. , Molecular profiling of neurons based on connectivity. Cell 157, 1230–1242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenthner C. J., Miyamichi K., Yang H. H., Heller H. C., Luo L., Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 78, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knight Z. A., et al. , Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tasic B., et al. , Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeFalco J., et al. , Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291, 2608–2613 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Schnütgen F., et al. , A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Banfield B. W., Bird G. A., Construction and analysis of alphaherpesviruses expressing green fluorescent protein. Methods Mol. Biol. 515, 227–238 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Card J. P., Enquist L. W., Transneuronal circuit analysis with pseudorabies viruses. Curr. Protoc. Neurosci. 9, 1.5.1–1.5.28 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franklin K., Paxinos G., The Mouse Brain in Stereotaxic Coordinates (Elsevier Inc., ed. 3, 2008). [Google Scholar]
- 55.Hanchate N. K., et al. , Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science 350, 1251–1255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.bcl2fastq conversion software (version 1.8.4) (Illumina, Inc., San Diego, CA).
- 57.Krueger F., Galore Trim, Version 0.4.4. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed 14 March 2017.
- 58.R Core Team , R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). https://www.R-project.org/. Accessed 22 March 2017.
- 59.RStudio Team (2015). RStudio: Integrated development for R. Version 1.2.1206. http://www.rstudio.com/. Accessed 12 December 2018.
- 60.Wickham H., ggplot2: Elegant graphics for data analysis (Springer-Verlag, New York, NY, 2016). [Google Scholar]
- 61.Ekstrand M. I., Enquist L. W., Pomeranz L. E., The alpha-herpesviruses: Molecular pathfinders in nervous system circuits. Trends Mol. Med. 14, 134–140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ugolini G., Advances in viral transneuronal tracing. J. Neurosci. Methods 194, 2–20 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Rinaman L., Card J. P., Enquist L. W., Spatiotemporal responses of astrocytes, ramified microglia, and brain macrophages to central neuronal infection with pseudorabies virus. J. Neurosci. 13, 685–702 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Card J. P., et al. , Pseudorabies virus infection of the rat central nervous system: Ultrastructural characterization of viral replication, transport, and pathogenesis. J. Neurosci. 13, 2515–2539 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.