Significance
Repeat-associated non-AUG (RAN) translation enables protein translation independent of an AUG start codon. Previous studies identified RAN-translated products in cellular models of CAG repeat expansion diseases, including Huntington’s disease (HD). However, whether RAN-translated products, when expressed at the endogenous level, truly contribute to disease pathogenesis remains unknown. In the present study, we established knock-in mouse models for HD, which enable the expression of mutant Huntingtin (HTT) and RAN-translated products or only RAN-translated products at endogenous levels. We were unable to detect RAN-translated products in these mouse models, and found only the mouse model that expresses expanded polyQ peptides showed HD pathological phenotypes, suggesting that RAN-translated products do not critically contribute to HD pathogenesis.
Keywords: CAG repeat, Huntington’s disease, aggregates, neurodegeneration, gene targeting
Abstract
Identification of repeat-associated non-AUG (RAN) translation in trinucleotide (CAG) repeat diseases has led to the emerging concept that CAG repeat diseases are caused by nonpolyglutamine products. Nonetheless, the in vivo contribution of RAN translation to the pathogenesis of CAG repeat diseases remains elusive. Via CRISPR/Cas9-mediated genome editing, we established knock-in mouse models that harbor expanded CAG repeats in the mouse huntingtin gene to express RAN-translated products with or without polyglutamine peptides. We found that RAN translation is not detected in the knock-in mouse models when expanded CAG repeats are expressed at the endogenous level. Consistently, the expanded CAG repeats that cannot be translated into polyglutamine repeats do not yield the neuropathological and behavioral phenotypes that were found in knock-in mice expressing expanded polyglutamine repeats. Our findings suggest that RAN-translated products do not play a major role in the pathogenesis of CAG repeat diseases and underscore the importance in targeting polyglutamine repeats for therapeutics.
CAG repeat expansion in the exons leads to the translation of mutant proteins with an expanded polyglutamine (polyQ) tract in nine neurodegenerative diseases, including Huntington’s disease (HD) and spinocerebellar ataxias (1). It is believed that polyQ expansion in these proteins could disrupt multiple cellular processes, including gene transcription, protein homeostasis, mitochondria function, and vesicle transport, which eventually lead to neurotoxicity, mainly through gain-of-function mechanisms (2–4). This theory is based on the findings that expanded polyQ causes protein misfolding to yield cytotoxicity. However, this theory is challenged by recent findings that nucleotide-repeat expansions can cause repeat associated non-AUG (RAN) translation via a noncanonical mechanisms of translation initiation.
RAN translation bypasses the need for an AUG start codon and enables protein translation in all three different reading frames (5, 6). Therefore, it is important to clarify the primary pathogenic form for CAG repeat diseases, as either polyQ products or RAN products could be the target for therapeutic interventions.
To date, RAN-translated products have been identified in a number of repeat expansion diseases (7–11), including HD (12). In cells transfected with exon 1 of Huntingtin (HTT) with expanded CAG repeats, RAN translation led to the production of polyalanine (polyAla), polyserine (polySer), polyleucine (polyLeu), and polycystine (polyCys), in addition to mutant HTT with polyQ expansion, and these RAN-translated products reduced cell survival at levels comparable to mutant HTT (12). These results raise the possibility that polyQ diseases are caused by a mixture of repeat peptides. Nonetheless, given that the efficiency of RAN translation is critically dependent on the repeat length (5), which typically ranges from hundreds to thousands of repeats (13, 14), whether RAN translation caused by the moderate CAG repeat expansions is critical to neuropathology remains unknown. In addition, since previous discoveries of RAN translation or frameshift repeat products in polyQ disease models have largely relied on overexpression systems (15–17), a more rigorous test using an in vivo system that expresses the expanded CAG repeats at an endogenous level is needed.
In the present study, we utilized the CRISPR/Cas9-genome editing tool to generate two knock-in (KI) mouse models for HD: One lacks HTT with polyQ expansion but allows for RAN translation, and the other expresses N-terminal HTT with polyQ expansion. We found that 1) RAN-translated products could not be detected in the brains of these KI mice and 2) only the mouse model expressing HTT with polyQ expansion showed typical HD pathological phenotypes. Our results suggest that in mouse models that express mutant HTT at an endogenous level, the contribution of RAN translation to HD pathogenesis is minimal or not critical.
Results
Generation and Characterization of HD KI Mouse Models.
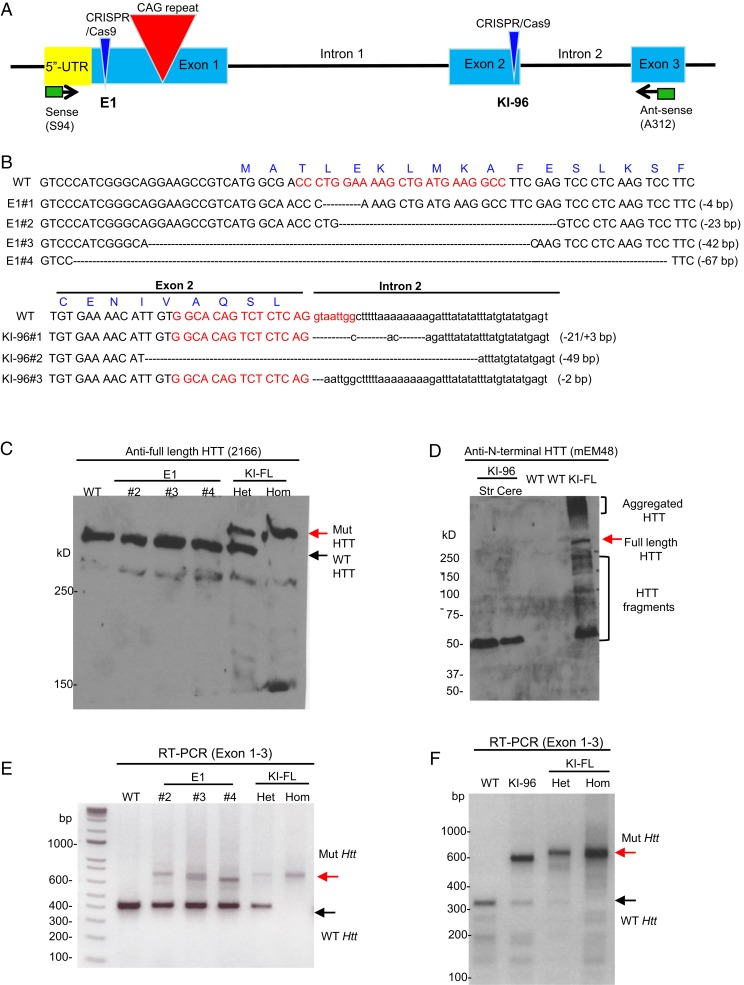
To generate mouse models that harbor expanded CAG repeats and can endogenously express polyQ or RAN (nonpolyQ) products, we performed CRISPR/Cas9-mediated genome editing in the embryos of HD140Q KI mice that express exon 1 of human HTT containing a 140-CAG repeat in the endogenous mouse Htt gene (18). A guide RNA (gRNA) was designed to make mutations upstream of CAG repeats in exon 1 HTT (E1) for expressing RAN products but not full-length HTT. Another gRNA was used to target exon 2 of endogenous Htt (KI-96) for truncating mutant Htt so that only N-terminal HTT containing expanded polyQ repeats is expressed (Fig. 1A). We obtained four lines of E1 mice containing mutations upstream of the intact CAG repeats (Fig. 1B and SI Appendix, Fig. S1A). Among these different lines, E1#1 and E1#2 retained the ATG start codon, but the downstream truncations caused frameshift mutations that blocked the expression of HTT with polyQ expansion; E1#3 and E1#4 lost the ATG start codon, so that proteins could only be produced by RAN translation. As we failed to maintain E1#1 mice, we used E1#2, E1#3, and E1#4 mice for the following biochemical, behavioral, and gene expression studies.
Fig. 1.
Characterization of Htt expression in E1 and KI-96 mouse models. (A) Overview of CRISPR/Cas9 target sites in the Htt gene of HD140Q KI mice and primers (sense, S94, and antisense, A312) used for RT-PCR. (B) Sequencing results showing different deletion mutations caused by CRISPR/Cas9 cutting in four E1 mouse lines and three KI-96 mouse lines. Red letters are used to indicate gRNA sequences; uppercase letters are used to indicate exon sequences, and lowercase letters are used to indicate intron sequences. (C) Western blotting analysis showing full-length HTT in three individual E1 mice. WT, heterozygous (Het), and homozygous (Hom) HD140Q KI (KI-FL) mice were used as references to indicate the location of WT and mutant (Mut) full-length HTT. (D) Western blotting analysis showing N-terminal HTT with polyQ expansion in a KI-96 mouse. HTT fragments could result from mRNA incomplete splicing or the cleavage of HTT. Str, striatum; Cere, cerebellum. (E) RT-PCR analysis showing Htt mRNA expression in three individual E1 mice. (F) RT-PCR analysis showing Htt mRNA expression in a KI-96 mouse and Het and Hom HD140Q KI (KI-FL) mice.
We also obtained three lines of KI-96 mice expressing N-terminal HTT that was terminated in exon 2. These three lines harbor different mutations at the boundary between exon 2 and intron 2. These mutations possibly cause either an early stop codon or disruption of messenger RNA (mRNA) splicing to generate N-terminal HTT that is terminated in exon 2, which contains 96 amino acids in addition to 140 glutamines (Fig. 1B and SI Appendix, Fig. S1B). Alternatively, it is likely that the mutated transcripts got destroyed by nonsense-mediated decay, but an exon 1 transcript that results from incomplete splicing led to the production of exon 1 HTT (19, 20). Since all three KI-96 lines showed similarity in terms of N-terminal HTT expression (SI Appendix, Fig. S2) and animal behaviors, we only maintained the KI-96#3 line for further studies.
We were unable to acquire any homozygous E1 or KI-96 mice through breeding (Table 1), indicating that none of the mutations produced fully functional full-length HTT protein and supporting the previous finding that full-length HTT is essential for embryonic development (21). This assumption was further supported by Western blotting analysis of brain lysates from different mouse lines, using various antibodies against HTT (Fig. 1 C and D and SI Appendix, Fig. S3). Nonetheless, in heterozygous E1 and KI-96 mice, we found that the mRNA expression levels of wild-type (WT) and mutant Htt are comparable to those of HD140Q KI mice, although only KI-96 mice expressed exon1 HTT with an expanded polyQ repeat (Fig. 1 E and F). Thus, heterozygous E1 mice, which express an endogenous level of mutant Htt mRNA but not polyQ-containing mutant HTT protein, provide us with an ideal and rigorous in vivo system to test the role of RAN translation in HD pathogenesis.
Table 1.
Quantification of the number of offspring with different genotypes from heterozygous E1 or KI-96 breeding pairs
| E1 mice | KI-96 mice | |||
| Genotype | Count | Percentage | Count | Percentage |
| WT | 13/39 | 33.33 | 8/21 | 38.1 |
| Heterozygous | 26/39 | 66.67 | 13/21 | 61.9 |
| Homozygous | 0/39 | 0 | 0/21 | 0 |
RAN-Translated Products Were Not Detected in E1 or KI-96 Mice.
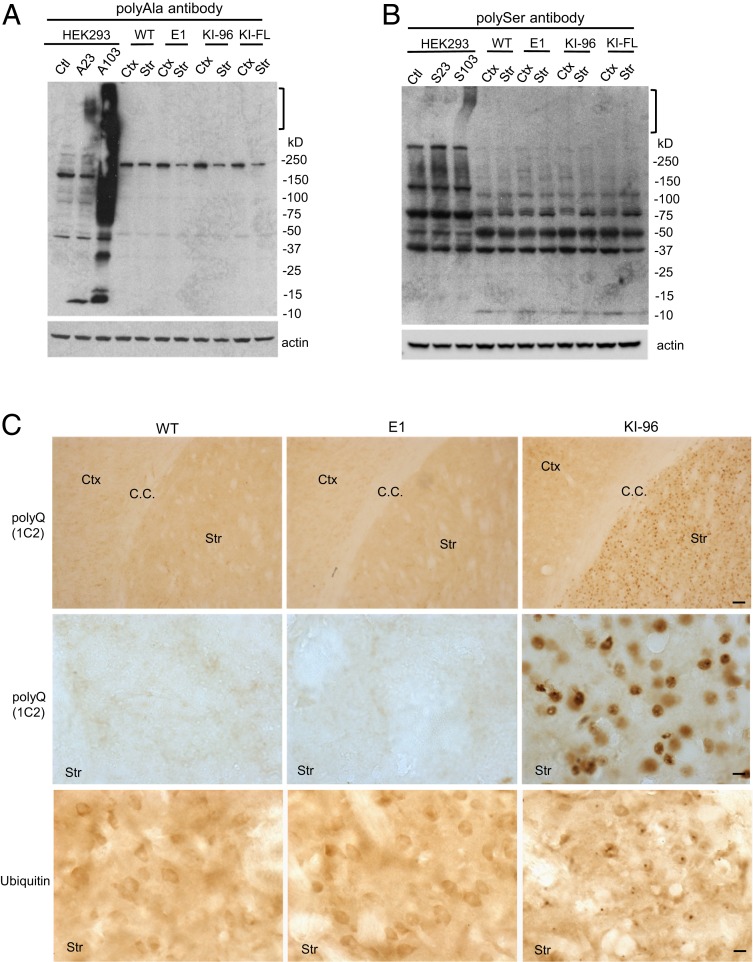
In transfected HEK293 cells, HTT with CAG expansion produces polyAla and polySer peptides through RAN translation (12). Although we successfully detected polyAla and polySer peptides in HEK293 cells using the same antibodies and plasmids described in a previous study (12), we were unable to find such peptides in the brain lysates of E1, KI-96, or HD140Q KI mice (Fig. 2 A and B). Although immunohistochemistry with polyAla and polySer antibodies showed positive staining in postmortem HD brains (12), whether such staining is due to neuronal degeneration or tissue decomposition during postmortem interval remains unknown. Using the same staining protocol, we could not detect any specific labeling in the brains of E1, KI-96, or HD140Q KI mice (SI Appendix, Fig. S4). RAN translation could also enable the expression of polyQ products. Using the polyQ antibody 1C2, however, we failed to detect any positive staining in E1 mice, further suggesting that RAN translation has not occurred. In contrast, KI-96 mice, which express N-terminal HTT containing an expanded polyQ repeat, showed extensive 1C2 labeling, as expected (Fig. 2C). Both mutant HTT and RAN peptides form aggregates that are positive for ubiquitin immunostaining (7, 9, 22). However, we only found ubiquitin positive puncta in the striatum of KI-96 mice, whereas WT and E1 mice showed diffused ubiquitin immunostaining (Fig. 2C).
Fig. 2.
RAN translation products were not detected in the brain of different HD KI mouse models. (A) PolyAla antibody was used to detect RAN-translated polyA peptides in WT, E1#2, KI-96, and HD140Q KI (KI-FL) mice. HEK293 cells transfected with FLAG-104xAla-Ct plasmid were used as a positive control. (B) PolySer antibody was used to detect RAN-translated polyS peptides in WT, E1#2, KI-96, and KI-FL mice. HEK293 cells transfected with FLAG-102xSer-Ct plasmid were used as a positive control. In A and B, aggregated peptides are indicated by brackets. (C) Immunohistochemical staining of WT, E1#2, and KI-96 mouse brains (Ctx, cortex; Str, striatum; C.C., corpus callosum) showing the expression of HTT with polyQ expansion and ubiquitin puncta only in KI-96 mice. (Scale bars: Top, 50 μm; Middle and Bottom, 10 μm.)
KI-96 Mice but Not E1 Mice Showed HD Neuropathological Phenotypes.
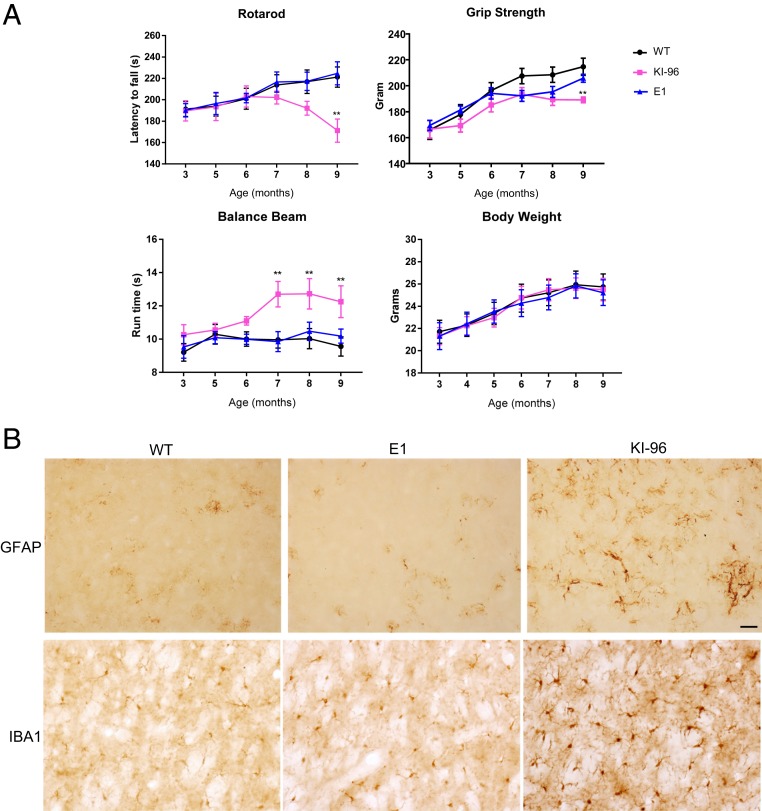
It is possible that endogenous mutant Htt produces a low level of RAN-translated products, which are still neurotoxic, to induce the neurological symptoms and neuropathology seen in HD KI mice. In light of this possibility, we performed a battery of behavioral tests that are commonly used to assess HD pathogenesis (23). Heterozygous KI-96 mice started to show decline in rotarod performance around 7 mo of age, which became statistically significant at 9 mo of age. On the other hand, heterozygous E1 mice had rotarod performances comparable to those of WT mice at all of the time points tested (Fig. 3A). We also examined fine motor skills via balance beam test and muscular functions via grip strength test. In both tests, KI-96 mice displayed significant deficits, whereas the performances of E1 mice resembled those of WT mice. Both E1 mice and KI-96 mice had similar body weight when compared to WT mice (Fig. 3A).
Fig. 3.
KI-96 mice but not E1 mice showed HD-related neuropathological phenotypes. (A) Motor functions of WT, heterozygous E1, and KI-96 mice were evaluated using rotarod, grip strength, and balance beam tests monthly (n = 12 for each group; **P < 0.01 by two-way ANOVA with Bonferroni’s test). (B) Immunohistochemistry result showing increased GFAP and IBA1 expression in the striatum of KI-96 but not WT and E1#2 mice. (Scale bar, 50 μm.)
At the end of the behavioral tests, we killed the mice and examined the neuropathology. Two early hallmarks of HD neuropathology are reactive astrocytes and microglia activation, which can be detected using immunohistochemistry with GFAP and IBA1 antibodies, respectively (24, 25). We found extensive GFAP and IBA1 positive staining in the striatum of KI-96 mice but not in E1 and WT mice (Fig. 3B).
The Transcriptomic Profiles of E1 Mice Closely Resemble Those of WT Mice.
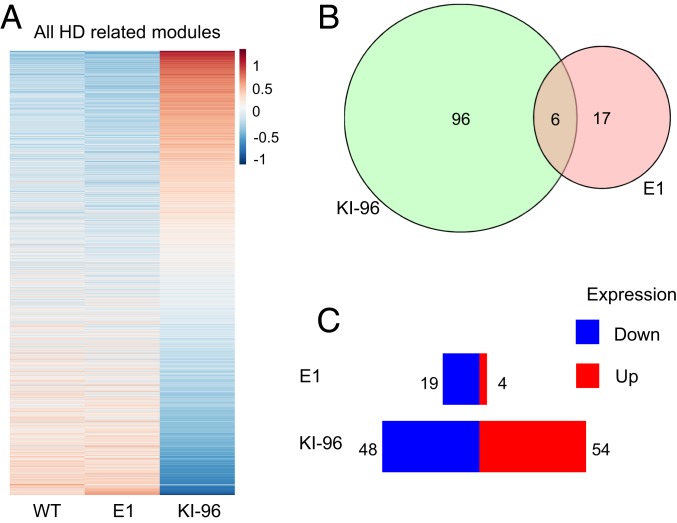
Given that the striatum of HD KI mice showed more pronounced alterations in gene expression than other brain regions (26), we performed RNA sequencing analysis using striatal tissues from 6-mo-old E1 and KI-96 mice. We took advantage of a previously published HD transcriptomic network (26) and focused on the striatal modules that are most relevant to HD pathogenesis (M2, repeat-associated HD signaling; M7, cell death signaling; M9, mitochondria and transport; M20, UBI conjugation; M25, glutamate receptor signaling; M39, DNA repair; M43, fatty acid catabolic process; and M46, glucocorticoid signaling). The transcriptional signatures of E1 mice in all of the modules combined or in each selected module closely resemble those of WT mice, whereas KI-96 mice showed divergent profiling (Fig. 4A and SI Appendix, Fig. S5). In addition, when compared to WT mice in these modules, KI 96 mice showed more genes with significantly altered expression than E1 mice (Fig. 4 B and C and SI Appendix, Fig. S6). Therefore, KI-96 mice but not E1 mice displayed transcriptional features that are found in other HD mouse models.
Fig. 4.
Transcriptional profiling of the striata from E1 and KI-96 mice. (A) Heat map view of gene expression in selected striatum modules consisting of M2 (repeat-associated HD signaling), M7 (cell death signaling), M9 (mitochondria and transport), M20 (UBI conjugation), M25 (glutamate receptor signaling), M39 (DNA repair), M43 (fatty acid catabolic process), and M46 (glucocorticoid signaling) from WT, E1, and KI-96 mice (6 mo old, n = 7 for WT mice, 4 for E1 and KI-96 mice). (B) Overlap of significantly changed genes when comparing E1 or KI-96 mice with WT mice. (C) Overview of significantly changed genes in selected striatum modules when comparing E1 or KI-96 mice with WT mice. More genes in KI-96 mice show altered expression than in E1 mice. Data represent the mean ± SEM.
Discussion
RAN translation represents a novel and provocative mechanism by which protein translation can occur in the setting of nucleotide repeat expansions to produce neurotoxic proteins. Since most of studies of CAG repeat diseases have been focused on the neurotoxicity of expanded polyQ-containing peptides, identification of RAN products in the CAG repeat disease models has opened up a new avenue to investigate the pathogenesis and treatment of these diseases.
However, the in vivo contribution of RAN products to diseases, especially CAG repeat diseases, remains to be rigorously assessed. This is because RAN translation efficiency is greatly influenced by two critical factors: One is the repeat length, and the other is its expression level.
Indeed, the length of CAG repeat is moderately expanded to less than 60 CAGs in most patients, whereas other trinucleotide repeats (such as CGG) are often elongated to several hundreds or thousands to induce cytotoxicity (27, 28). While CGG repeats in the 5′ UTR of FMR1 require an upstream ACG codon to initiate RAN translation (29), CAG repeats in ATXN8 do not (11), suggesting that the mechanism for RAN translation could be repeat or gene specific. The evidence for RAN products in the CAG repeat disease models was largely derived from cultured cells that overexpress CAG repeats (12). However, it has been reported that overexpression of CAG repeats could deplete glutaminyl-transfer RNA, resulting in translational frameshifting and generation of various transframe-encoded species (30). Thus, it is imperative to use a more rigorous system that expresses CAG repeats at the endogenous level to test the theory of RAN translation in CAG repeat diseases.
The KI mouse models we generated allowed us to rigorously examine the role of RAN products in HD. First, KI-96 mice express polyQ products at the endogenous level when exon 2 of Htt is mutated by CRISPR/Cas9. Second, when the upstream sequence of the CAG repeats is disrupted by CRISPR/Cas9, E1 mice express the endogenous level of Htt mRNA with CAG repeat expansion but without producing polyQ products. Using these KI mice that express the same levels of CAG repeat mRNA with or without polyQ products, we can assess whether endogenously expressed RAN products induce any neurological symptoms or neuropathology.
Our findings indicate that when expanded CAG repeats are expressed at the endogenous level, RAN translation was not detected, although it remains questionable whether RAN translation can occur under mildly overexpressed conditions. Consistently, in the HD KI mouse model that does not express expanded polyQ products but allows for RAN translation, we did not find any obvious HD neuropathology. It should be noted that in E1#2 mice, the 23 nucleotides deletion would generate a protein in the alanine frame if the transcript got translated from the start codon. However, we were unable to detect such a protein using the polyAla antibody. It is possible that the protein in the alanine frame is unstable or its conformation prevents the antibody from recognizing the epitope. Nonetheless, our immunohistochemistry result using ubiquitin antibody did not reveal any obvious inclusion bodies, suggesting that the expression of the protein in the alanine frame is at least at a low level. Consistently, we found no obvious neuropathology in E1#2 mice.
Our findings support the idea that expanded polyQ peptides play a critical role in disease pathogenesis. Given that expansion of polyQ repeats in various proteins causes similar protein misfolding and aggregation in different CAG repeat diseases, our findings also strengthen the idea that targeting expanded polyQ repeat is an effective therapeutic strategy (31). Furthermore, our results indicate that generation of RAN translation products is dependent on the repeat length as well as its expression level and highlight the diverse mechanisms underlying nucleotide repeat expansion diseases.
Methods
Antibodies and Plasmids.
Primary antibodies used in this study include HTT (Millipore, MAB1574; Millipore, MAB2166; mEM48, self-produced), actin (Sigma, A5060), GFAP (Cell Signaling, 3670), IBA1 (Wako, 019-19741), and ubiquitin (Dako, Z0458). PolyAla and polySer antibodies and FLAG-23xAla-Ct, FLAG-104xAla-Ct, FLAG-23xSer-Ct, and FLAG-102xSer-Ct plasmids were generously provided by Laura Ranum, University of Florida, Gainesville, FL.
Mouse Lines.
Wild-type C57BL/6J (from Jackson Laboratory) and HD140Q knock-in (a gift from Michael Levine, University of California, Los Angeles, CA) mice were bred and maintained in the Division of Animal Resources at Emory University and were handled according to the policies of the Environmental Enrichment Program for Research Animals. The studies followed the protocol approved by the Animal Care and Use Committee at Emory University.
Generation of E1 and KI-96 mice was performed by Gene Edit Biolab, Morehouse School of Medicine, Atlanta, GA. The mouse zygotes of HD140Q KI mice were injected with mixed mRNA of Cas9 and gRNA (1 µL mixture per zygote). Founders were imported to the Emory animal facility and kept in a quarantine room before releasing to a regular housing room. CRISPR/Cas9-targeted male mice were crossed with wild-type female mice to generate F1 mice. Mice that carried the desired mutations were crossed with wild-type mice to generate heterozygous KI mice.
Mouse Behavioral Tests.
Mouse behavior was assessed using a rotarod apparatus (Rotamex 4/8, Columbus Instruments International). Before the test, the mice were trained on the equipment at 5 rpm for 10 min during 3 consecutive days. In the test, the rotarod was set to gradually accelerate from 0 to 40 rpm over a 5-min period. Each mouse was tested three times. To test the balance beam performances, a 0.8-m-long, 0.6-centimeter-thick wood stick was suspended from a platform on both sides by metal grips. A bright light source at one end and a dark box at the other end were used to motivate the mice to move. Each mouse was trained three times per day for 3 consecutive days and then tested three times. For the grip strength test, the mice were put on the metal grids of a grip meter (Ametek Chatillon) with all their limbs and then gently pulled backward by the tail until they could no longer hold the grids. Each mouse was tested three times. The body weight of the mice was measured monthly using an electronic scale.
Western Blotting and Immunohistochemistry.
Mouse brain tissues used for Western blotting were immersed in ice-cold radioimmunoprecipitation assay buffer containing a Halt protease inhibitor mixture and phosphatase inhibitors (Thermo Fisher Scientific) and lysed by an ultrasonic homogenizer. The lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane. The blot was blocked with 5% milk/phosphate-buffered saline (PBS) for 1 h at room temperature and incubated with selected primary antibodies in 3% bovine serum albumin (BSA)/PBS overnight at 4 °C. On the following day, the blot was washed three times in PBS and incubated with horseradish peroxidase-conjugated secondary antibodies in 5% milk/PBS for 1 h at room temperature. The blot was finally developed using enhanced chemiluminescence prime (GE Healthcare).
For immunohistochemistry, the mice were perfused with 0.9% saline and then 4% paraformaldehyde. The fixed mouse brains were isolated and cryoprotected in 30% sucrose and then sectioned into slices in a cryostat (Leica). The slices were blocked and permeabilized with 3% BSA in PBS supplemented with 0.2% Triton X-100 and then incubated with selected primary antibodies at 4 °C overnight. On the following day, the slices were sequentially treated with biotinylated secondary antibodies and a VECTASTAIN ABC kit (Vector Labs), with PBS washes between each step. The slices were developed in SIGMAFAST 3,3′-Diaminobenzidine tablets (Sigma, D4293). For detecting RAN translation products, harsh antigen retrieval steps were added according to the published protocol (12), including 1 μg/mL proteinase K treatment in 1 mM CaCl2, 50 mM Tris buffer (pH = 7.6) for 40 min at 37 °C, pressure cooking in 10 mM ethylenediaminetetraacetic acid (pH = 6.5) for 15 min, and 95% formic acid treatment for 5 min.
RNAseq Analysis.
RNAs from the striata of WT, KI-96, and E1 mice at 6 mo of age were extract using an RNeasy Lipid Tissue Kit (Qiagen) and then sent to Novogene for sequencing. Each genotype consisted of 4 to 7 mice. Sequencing adapters were trimmed by BBduk (minlen = 100 ktrim = r k = 23 mink = 11 hdist = 1 tpe tbo; https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/). Reads with length under 100 base pairs after trimming were discarded. Clipped reads were aligned to mouse genome GRCm38 (primary assembly) by STAR aligner with default settings. Read counts for genes were obtained by using HTSeq. All mRNA differential expression analyses were performed in R using the Bioconductor package DESeq2, version 1.24.0. We removed one WT sample identified as a potential outlier and used the default settings with sac as a covariate variable. We tested differential expression in two-sample tests of KI (KI-96 and E1) samples versus WT samples.
To draw heat maps, we first applied the variance-stabilizing transformation implemented in DESeq2 to raw counts and removed the effect of sac by using the function removeBatchEffectRaw in the Bioconductor package Limma. Then, sample means corresponding to the three genotypes were calculated based on these transformed values. Finally, a heat map of normalized sample mean values of the three genotypes was generated for genes in selected modules (M2, M7, M9, M20, M25, M39, M43, and M46). For differential expression (DE) tests of different KI genotypes versus wild type (KI-96 vs. WT and E1 vs. WT) in each selected module (M2, M7, M9, M20, M25, M39, M43, and M46), genes were divided into two groups by checking whether the corresponding log fold change was greater than zero or not. The bar plot was generated based on averages of the log10 P value of these grouped genes in different modules for different DE tests.
Data Availability.
RNAseq data used in this study are available in the Gene Expression Omnibus (GEO) database (accession number GSE142603) (32).
Supplementary Material
Acknowledgments
This work was supported by the NIH (Grants R01NS101701, R01NS095279) and the National Natural Science Foundation of China (Grant 81830032). We thank Dr. Laura Ranum at the University of Florida for providing polyAla and polySer antibodies as well as plasmids expressing RAN products.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE142603).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919197117/-/DCSupplemental.
References
- 1.Lieberman A. P., Shakkottai V. G., Albin R. L., Polyglutamine repeats in neurodegenerative diseases. Annu. Rev. Pathol. 14, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riley B. E., Orr H. T., Polyglutamine neurodegenerative diseases and regulation of transcription: Assembling the puzzle. Genes Dev. 20, 2183–2192 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Williams A. J., Paulson H. L., Polyglutamine neurodegeneration: Protein misfolding revisited. Trends Neurosci. 31, 521–528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuccato C., Valenza M., Cattaneo E., Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 90, 905–981 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Cox D. C., Cooper T. A., Non-canonical RAN translation of CGG repeats has canonical requirements. Mol. Cell 62, 155–156 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Wojciechowska M., Olejniczak M., Galka-Marciniak P., Jazurek M., Krzyzosiak W. J., RAN translation and frameshifting as translational challenges at simple repeats of human neurodegenerative disorders. Nucleic Acids Res. 42, 11849–11864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ash P. E., et al. , Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gendron T. F., et al. , Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori K., et al. , The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Todd P. K., et al. , CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78, 440–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zu T., et al. , Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U.S.A. 108, 260–265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bañez-Coronel M., et al. , RAN translation in Huntington disease. Neuron 88, 667–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabczyk E., Usdin K., The GAA*TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 28, 2815–2822 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Mossevelde S., et al. ; Belgian Neurology (BELNEU) Consortium , Clinical evidence of disease anticipation in families segregating a C9orf72 repeat expansion. JAMA Neurol. 74, 445–452 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Gaspar C., et al. , CAG tract of MJD-1 may be prone to frameshifts causing polyalanine accumulation. Hum. Mol. Genet. 9, 1957–1966 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Scoles D. R., et al. , Repeat associated non-AUG translation (RAN Translation) dependent on sequence downstream of the ATXN2 CAG repeat. PLoS One 10, e0128769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stochmanski S. J., et al. , Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models. Hum. Mol. Genet. 21, 2211–2218 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Menalled L. B., Sison J. D., Dragatsis I., Zeitlin S., Chesselet M. F., Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 465, 11–26 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Sathasivam K., et al. , Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl. Acad. Sci. U.S.A. 110, 2366–2370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neueder A., et al. , The pathogenic exon 1 HTT protein is produced by incomplete splicing in Huntington’s disease patients. Sci. Rep. 7, 1307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dragatsis I., Efstratiadis A., Zeitlin S., Mouse mutant embryos lacking huntingtin are rescued from lethality by wild-type extraembryonic tissues. Development 125, 1529–1539 (1998). [DOI] [PubMed] [Google Scholar]
- 22.DiFiglia M., et al. , Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Yang S., et al. , CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 127, 2719–2724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palfi S., et al. , Expression of mutated huntingtin fragment in the putamen is sufficient to produce abnormal movement in non-human primates. Mol. Ther. 15, 1444–1451 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Yang H. M., Yang S., Huang S. S., Tang B. S., Guo J. F., Microglial activation in the pathogenesis of Huntington’s disease. Front. Aging Neurosci. 9, 193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P., et al. , Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 19, 623–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snell R. G., et al. , Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat. Genet. 4, 393–397 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Willemsen R., Levenga J., Oostra B. A., CGG repeat in the FMR1 gene: Size matters. Clin. Genet. 80, 214–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellier C., et al. , Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93, 331–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girstmair H., et al. , Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep. 3, 148–159 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Li Z., et al. , Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature 575, 203–209 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Yang S., Li X.-J., Lack of RAN-mediated toxicity in Huntington’s disease knock-in mice. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142603. Deposited 26 December 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data used in this study are available in the Gene Expression Omnibus (GEO) database (accession number GSE142603) (32).