Significance
Here we show that the histone demethylase JMJD2B is induced in endothelial cells by EndMT provoking stimuli and thereby contributes to the acquirement of a mesenchymal/smooth muscle phenotype. Silencing of JMJD2B inhibited EndMT in vitro and reduced the induction of EndMT after myocardial infarction in vivo. Inhibition of JMJD2B prevents the demethylation of repressive trimethylated histone H3 at lysine 9 (H3K9me3) at promoters of mesenchymal and EndMT-controlling genes, thereby reducing EndMT. Together, our study reports a crucial role for JMJD2B in controlling histone modifications during the transition of endothelial cells toward a mesenchymal phenotype.
Keywords: EndMT, JMJD2B, epigenetics, H3K9me3, SULF1
Abstract
Endothelial cells play an important role in maintenance of the vascular system and the repair after injury. Under proinflammatory conditions, endothelial cells can acquire a mesenchymal phenotype by a process named endothelial-to-mesenchymal transition (EndMT), which affects the functional properties of endothelial cells. Here, we investigated the epigenetic control of EndMT. We show that the histone demethylase JMJD2B is induced by EndMT-promoting, proinflammatory, and hypoxic conditions. Silencing of JMJD2B reduced TGF-β2-induced expression of mesenchymal genes, prevented the alterations in endothelial morphology and impaired endothelial barrier function. Endothelial-specific deletion of JMJD2B in vivo confirmed a reduction of EndMT after myocardial infarction. EndMT did not affect global H3K9me3 levels but induced a site-specific reduction of repressive H3K9me3 marks at promoters of mesenchymal genes, such as Calponin (CNN1), and genes involved in TGF-β signaling, such as AKT Serine/Threonine Kinase 3 (AKT3) and Sulfatase 1 (SULF1). Silencing of JMJD2B prevented the EndMT-induced reduction of H3K9me3 marks at these promotors and further repressed these EndMT-related genes. Our study reveals that endothelial identity and function is critically controlled by the histone demethylase JMJD2B, which is induced by EndMT-promoting, proinflammatory, and hypoxic conditions, and supports the acquirement of a mesenchymal phenotype.
Endothelial cells (ECs) covering the inner layer of vessels control the exchange of oxygen and nutrients with underlying tissues and play an essential role in maintaining homeostasis of the vessel wall. In response to environmental and mechanical stimuli, fully differentiated ECs can profoundly change their phenotype and function. For example, transforming growth factor beta (TGF-β) and to an extent hypoxia or disturbed blood flow, can promote the transition of ECs to a mesenchymal phenotype (1–3). This process is termed endothelial-to-mesenchymal transition (EndMT). The transition is associated with a reduction of prototypical endothelial genes and with a concomitant de novo expression of mesenchymal marker genes leading to impaired endothelial cell function (4, 5). Cellular transitions are essential physiological processes in the development of differentiating tissues, as well as in the maintenance and repair of adult tissues. During cardiovascular development, EndMT was shown to contribute to cardiac valve formation, developmental intima thickening of the pulmonary artery, and stabilization of newly formed vasculature (6, 7). In adults, EndMT plays an important role in wound healing after tissue injury, where cells that have undergone EndMT can have stabilizing functions and act in a fibroblast-like manner (8). However, EndMT can also contribute to pathophysiological conditions, e.g. by enhancing renal or cardiac fibrosis after tissue injury or ischemia, which is accompanied by a reduction in capillary density (9–11). Additionally, ECs undergoing EndMT are commonly found in atherosclerotic lesions, particularly in complex and unstable lesions in humans (12, 13). Given this importance, many studies addressed the regulatory control of EndMT. Here, activating and inhibitory pathways, such as the TGF-β/Smad2/3 axis (3) and fibroblast growth factor 2 (FGF2) signaling cascade (12, 14, 15) controlling EndMT were identified. Although several studies suggested that the related process of epithelial-to-mesenchymal transition (EMT) is regulated by epigenetic mechanisms such as DNA methylation and posttranslational histone modifications (16, 17), little is known regarding the epigenetic control of EndMT (3).
Jumonji C (JmjC) domain-containing proteins are a class of histone demethylases that epigenetically regulate gene transcription (18). JmjC domain-containing proteins function as important regulators of pluripotency and control differentiation of embryonic stem cells (19–22). Several Jumonji proteins have been either implicated in the regulation of cancer progression or were suggested as a biomarker for several cancer types, like JMJD1A for gastric or JMJD3 for colorectal cancer (23, 24). Moreover, recent studies have highlighted the importance of JmjC domain-containing proteins in the regulation of EMT, which is an underlying mechanism for tumor invasion and metastasis (25–31). One member of this protein family, Jumonji domain-containing protein 2B (JMJD2B, also known as KDM4B) transcriptionally activates gene expression by demethylation of the repressive histone mark H3K9me3 and further coordinates the methylation of the activating histone mark H3K4me3 (32–34). JMJD2B enhances human gastric cancer metastasis by physically interacting with β-catenin, thereby promoting EMT progression (28). However, the role of JMJD2B in endothelial cell plasticity has not been investigated so far. Since JMJD2B expression was significantly induced during endothelial and cardiac differentiation of embryonic stem cells (35), we hypothesize that JMJD2B might play a role in epigenetically controlling the endothelial phenotype identity and investigated its role in EndMT.
Results
JMJD2B Is Induced by EndMT-Promoting Stimuli.
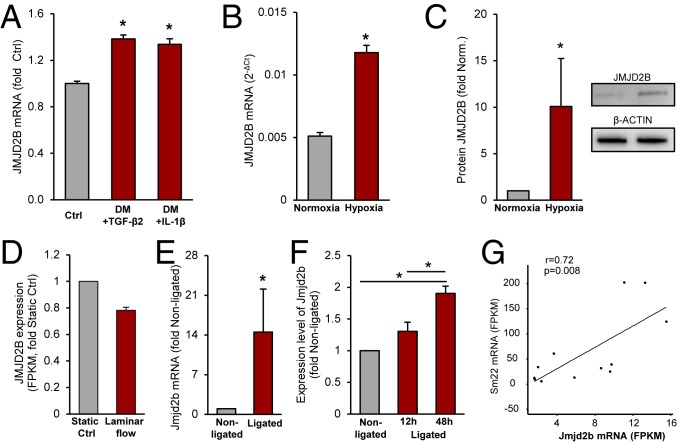
Regulation of JMJD2B was first assessed in vitro in cultured human umbilical vein endothelial cells (HUVECs). The EndMT-promoting stimuli TGF-β2 and interleukin-1β (IL-1β) both induced JMJD2B expression (Fig. 1A). JMJD2B mRNA and protein level were also up-regulated by hypoxia (Fig. 1 B and C), which is known to induce EndMT (2, 36, 37). Furthermore, we assessed the impact of mechanical activation of ECs. We show that JMJD2B is reduced by laminar flow, which was shown to prevent EndMT (1, 38) (Fig. 1D). Furthermore, in comparison to the ECs exposed to stable blood flow (nonligated controls), exposure of ECs to disturbed flow by partial carotid ligation induced the expression of JMJD2B in vivo in a time-dependent manner (Fig. 1 E and F). Thereby, JMJD2B expression correlated with the induction of mesenchymal markers (Fig. 1G and SI Appendix, Fig. S1A). These data collectively demonstrate that EndMT-promoting stimuli induce JMJD2B expression, whereas EndMT-preventing stimuli reduced JMJD2B expression.
Fig. 1.
Regulation of JMJD2B by EndMT-promoting stimuli in vitro and in vivo. (A) JMJD2B mRNA was measured in untreated (Ctrl) or with differentiation medium (DM) + TGF-β2- or IL-1β-treated HUVECS by RT-qPCR and depicted as fold to Ctrl (n = 3). (B) HUVECs were cultivated for 24 h under 0.2% O2 (hypoxia) or 21% O2 (normoxia), and mRNA level were determined by RT-qPCR and normalized to RPLP0 (2-ΔCt) (n = 3). (C) Representative immunoblot (IB) and densiometric quantification of JMJD2B protein level in hypoxia (1% O2) or normoxia, β-ACTIN served as a loading control (n = 5). (D) RNA expression levels of JMJD2B in HUVECs incubated under static and laminar flow conditions determined by RNA sequencing (n = 2). (E–G) Partial carotid artery mice model (nonligated, right carotid artery; ligated, left carotid artery) after 12 h and 48 h of ligation. (E) Jmjd2b mRNA levels were determined by RT-qPCR; values were normalized to 18s rRNA level (n = 4). (F–G) Jmjd2b mRNA levels were determined by RNA sequencing; values are depicted as fold to nonligated (F) or Fragments Per Kilobase Million (FPKM) values as Pearson’s correlation (G). Data are depicted as mean ± SEM. Statistical significance was determined using Student’s t test or ANOVA Bonferroni post hox test (A–F), and Pearson’s correlation (G); *P < 0.05.
Silencing of JMJD2B Reduces EndMT.
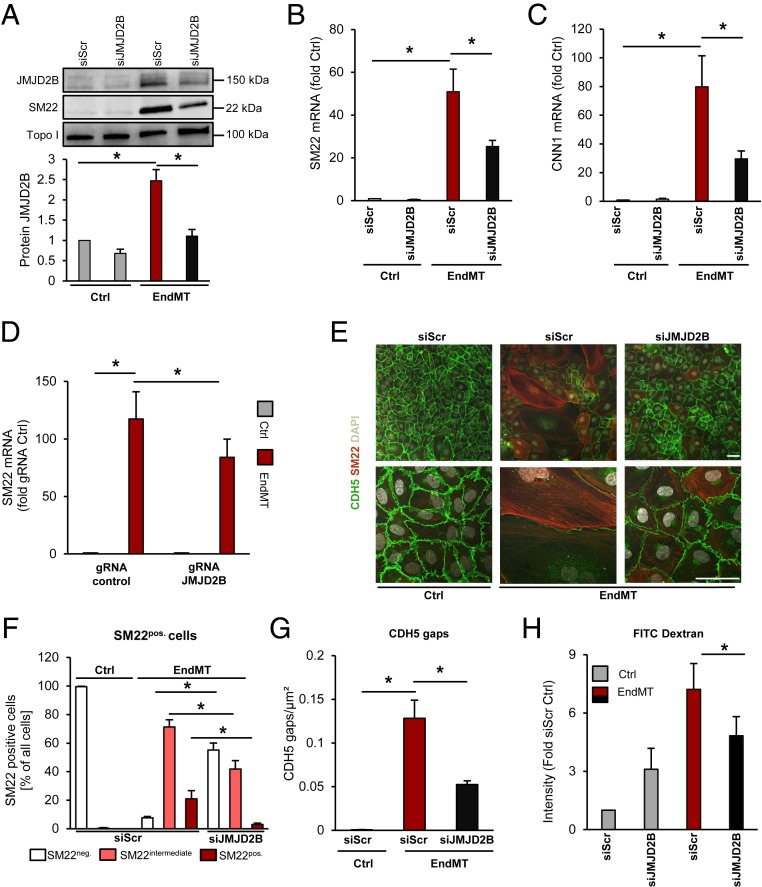
To determine the role of JMJD2B in EndMT, we silenced JMJD2B by pooled siRNAs, which significantly reduced JMJD2B protein levels (Fig. 2A). Knockdown of JMJD2B resulted in a significant decrease of smooth muscle and mesenchymal genes such as smooth muscle protein 22-alpha (SM22) and calponin (CNN1) on mRNA and protein level during TGF-β2-induced EndMT in human ECs (Fig. 2 A–E and SI Appendix, Fig. S2A). Additionally, protein expression of the mesenchymal marker Vimentin was up-regulated upon EndMT induction, and knockdown of JMJD2B prevented these changes (SI Appendix, Fig. S2 B and C). Similar findings were observed when using three alternative siRNA sequences directed against JMJD2B (SI Appendix, Fig. S2D) and CRISPR-Cas9–mediated gene knockdown (Fig. 2D and SI Appendix, Fig. S2E). Morphological and immunohistochemical (IHC) analysis showed that the percentage of SM22pos. cells was reduced after knockdown of JMJD2B compared to control transfected cells (Fig. 2 E and F). Furthermore, siJMJD2B-treated ECs showed preserved CDH5 protein expression and the typical CDH5 organization at adherens junctions after EndMT induction (SI Appendix, Fig. S2F and Fig. 2 E and G). Silencing of JMJD2B further reduced the EndMT-associated increase of endothelial permeability (Fig. 2H), as well as hypoxia-enhanced EndMT (SI Appendix, Fig. S2G). To address whether inhibition of the enzymatic activity of JMJD2B exhibits similar effects, we used the previously described JMJD2B inhibitor CCT365599 (39). Treatment with CCT365599 reduced TGF-β2-stimulated SM22 expression in a dose-dependent manner (SI Appendix, Fig. S2H). Single-cell RNA sequencing (scRNA-seq) confirmed that silencing of JMJD2B reduced the number of mesenchymal marker gene-expressing ECs (SI Appendix, Fig. S3 A–F). The role of JMJD2B as a positive regulator of EndMT was further emphasized in an additional model of EndMT induction by a combination of IL-1β and TGF-β1 (SI Appendix, Fig. S4 A–E) (40).
Fig. 2.
Function of JMJD2B in EndMT. (A) SM22 and JMJD2B protein after siRNA-mediated knockdown of JMJD2B or scrambled siRNA followed by EndMT induction. Topoisomerase I served as loading control. Quantification is shown below, n = 4. (B) SM22 and (C) CNN1 mRNA levels after siJMJD2B or siScr in HUVECs, followed by EndMT induction, determined by RT-qPCR, normalized to RPLP0 mRNA (2-ΔCt) (n = 4). (D) Knockdown of JMJD2B in HUVECS using the CRISPR-Cas9 technology. mRNA of SM22 was quantified via qRT-PCR (n = 7). (E) SM22 immunofluorescence (IF) staining after siJMJD2B or siScr followed by EndMT. Endothelial marker VE-cadherin (CDH5) appears in green, nuclei in white (DAPI), and SM22 in red. (Scale bar, 50 μm.) (F) Percentage of SM22pos. cells per population after siJMJD2B or siScr followed by EndMT, depicted as SM22 negative (white), intermediate (pink), and positive (red) cells (n = 3). (G) CDH5 cell gaps per square micrometer measured using IF staining after siJMJD2B and EndMT (n = 4). (H) Permeability of HUVECs treated with siRNAs and EndMT (n = 4). Data are depicted as mean ± SEM. Statistical significance was determined using Student’s t test or ANOVA Bonferroni post hoc test; *P < 0.05.
In Vivo Regulation of EndMT in Jmjd2bfl/fl Cdh5-iCre Mice.
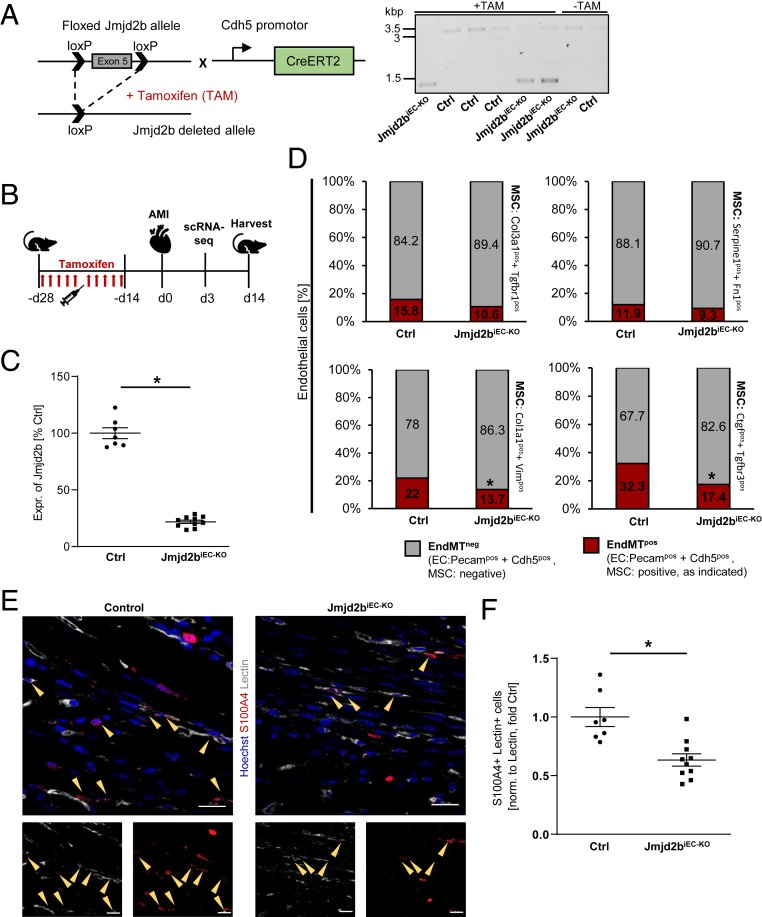
To confirm the causal involvement of Jmjd2b in regulation of EndMT in vivo, we generated an inducible EC-specific knockout mouse model of Jmjd2b (C57BL/6J Cdh5-iCre; Jmjd2bfl/fl mice) (Fig. 3 A and B). Tamoxifen (TAM) treatment efficiently reduced Jmjd2b expression in ECs isolated from Jmjd2bfl/flCdh5Cre+ (Jmjd2biEC-KO) mice compared to Jmjd2bfl/flCdh5Cre− (Ctrl) mice (Fig. 3C). To assess the effect of Jmjd2b on EndMT, we induced myocardial infarction (MI), which is known to augment EndMT (11). First, we performed scRNA-seq of Jmjd2biEC-KO and control mice 3 d after MI and assessed the percentage of EndMT-positive ECs. Single-cell RNA sequencing revealed that Jmjd2b knockout resulted in a relative reduction of ECs with additional mesenchymal marker gene expression (Fig. 3D). The reduction of mesenchymal transition in Jmjd2biEC-KO was consistent for various EndMT marker combinations, such as Col3a1 and Tgfbr1, Serpine1 and Fn1, Col1a1 and Vim, Ctgf and Tgfbr3 (Fig. 3D and SI Appendix, Fig. S5A). These results were further supported by histological assessment of EndMT postinfarction, demonstrating that Jmjd2biEC-KO mice showed a significant reduction of ECs in the myocardium, which were positive for the fibroblast marker S100A4 (Fig. 3 E and F). Of note, despite a significant inhibition of EndMT in Jmjd2biEC-KO mice, we only observed modest effects on cardiac function 2 wk after infarction (SI Appendix, Fig. S5 B and C).
Fig. 3.
Jmjd2b is a positive regulator of EndMT in vivo. (A) Knockout strategy and DNA gel of the floxed Jmjd2b and the Cdh5-CreERT2 allele. Recombination after TAM treatment led to the deletion of exon 5 in the Jmjd2b ORF. Jmjd2b DNA in total heart of Ctrl (3.1 kbp) or Jmjd2biEC-KO (1.4 kbp) mice, with or w/o TAM. (B) Schema of TAM-induced EC deletion of Jmjd2b with a subsequent AMI induction. (C) Jmjd2b mRNA of isolated EC of control (Ctrl) or Jmjd2biEC-KO mice, determined by RT-qPCR, depicted as percentage of Ctrl, n = 7–10. (D) Single-cell RNA sequencing showing percentage of cells positive [≥1 unique molecular identifiers (UMI) counts] for EC marker (Pecam1 and Cdh5) only (gray) and EC plus mesenchymal cell marker (MSC) gene expression (red) 3 d post-AMI. (E) Representative images of IHC stainings of Ctrl and Jmjd2biEC-KO mice hearts 14 d post-AMI. Hoechst appears in blue, lectin in white, and S100A4 in red, scale bar = 20 μM. (F) S100A4+ and lectin+ cells were counted in the sections of Ctrl or Jmjd2biEC-KO mice, normalized to area of lectin, depicted as fold Ctrl, n = 7–10. Data are depicted ±SEM. Student’s t test (C and F), χ2 test with Yates correction (D); *P < 0.05.
Epigenetic Regulation of EndMT.
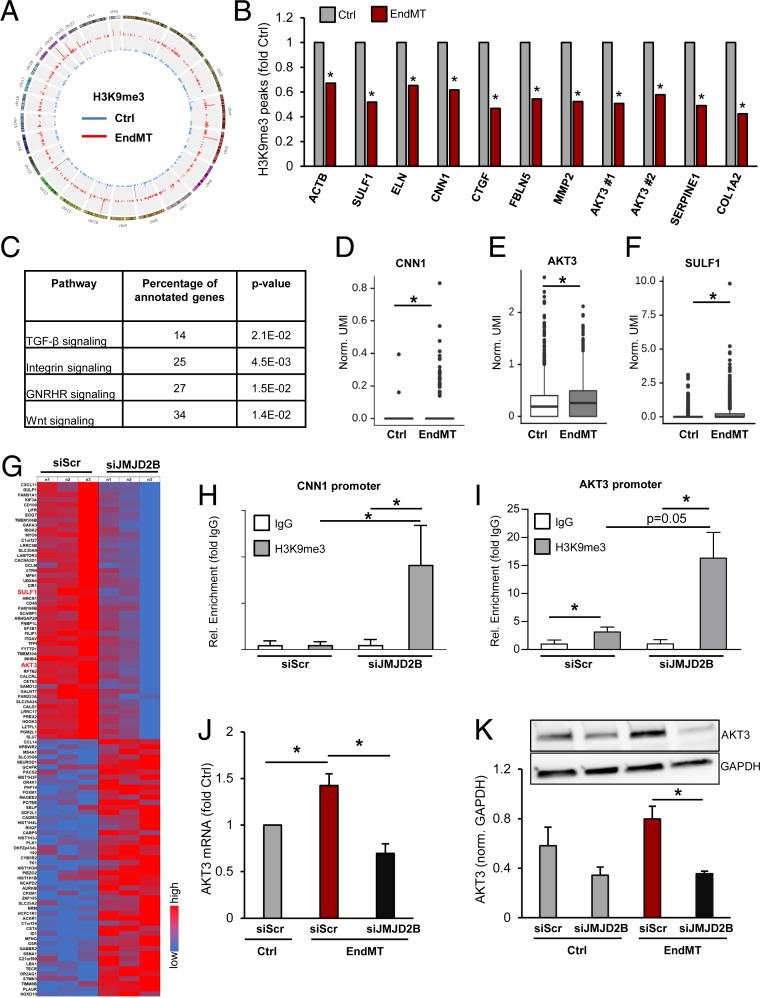
To identify yet undescribed global and gene-specific changes in histone methylation in EndMT, we performed immunoblotting and chromatin immunoprecipitation sequencing (ChIP-Seq) analysis after TGF-β2-mediated induction of EndMT in ECs. Whereas global levels of the repressive histone mark H3K9me3 and the activating mark H3K4me3 were unchanged after EndMT (SI Appendix, Fig. S6 A and B), ChIP sequencing revealed gene-specific reduction of the repressing histone mark H3K9me3 following EndMT (Fig. 4 A and B and SI Appendix, Fig. S7A). Subsequent gene ontology analysis showed that the TGF-β, Integrin, Wnt, and the Gonadotropin signaling pathways are most profoundly associated with the gene-specific changes in H3K9me3 level (Fig. 4C). A selection of EndMT- or EMT-related genes showing significantly reduced H3K9me3 peak levels after EndMT induction are depicted in Fig. 4B and SI Appendix, Fig. S7 B–F. The mesenchymal marker gene CNN1 was among the genes with high H3K9me3 marks at baseline, which were reduced following EndMT (Fig. 4B and SI Appendix, Fig. S7C). Consistently, CNN1 mRNA expression was augmented by EndMT-promoting conditions as shown by bulk RNA and single-cell RNA sequencing (SI Appendix, Fig. S8 A and B and Fig. 4D). Silencing of JMJD2B augmented H3K9me3 marks at the CNN1 promoter (Fig. 4H) and reduced CNN1 mRNA expression levels (Fig. 2C), demonstrating an epigenetic regulation of this mesenchymal gene by JMJD2B.
Fig. 4.
JMJD2B is a positive regulator of EndMT-related genes in endothelial cells. (A) Genomewide CircosPlot showing H3K9me3 ChIP-Seq peaks in control or EndMT-treated cells. Blue, control; red, EndMT. (B) H3K9me3 level in EndMT fold control of selected EndMT-associated genes determined by ChIP-Seq (P < 0.05, n = 3). (C) Gene ontology (http://pantherdb.org) analysis of H3K9me3 ChIP-Seq gene-specific reduced peaks in EndMT. (D–F) Differential gene expression of (D) CNN1, (E) AKT3, (F) SULF1 between Ctrl (n = 2227 cells), and EndMT (n = 1038 cells) single-cell RNA sequencing libraries. Data display individual gene UMI counts normalized to the total UMI count of all genes per cell. (G) Microarray RNA sequencing data after silencing of JMJD2B in TGF-β2-stimulated ECs (n = 3). Heatmap, depicting high expressed genes. High expression is given in red and low expression in blue. (H and I) ChIP-RT-qPCR showing level of H3K9me3 at (H) CNN1 and (I) AKT3 promoter, normalized to IgG (n = 5). (J) AKT3 mRNA expression after silencing of JMJD2B in TGF-β2-stimulated ECs using microarray RNA sequencing (n = 3). (K) Representative IB and densitometric quantification of protein levels of AKT3 after siRNA-mediated knockdown of JMJD2B. GAPDH served as loading control (n = 4). Data are depicted as mean ± SEM. Statistical significance was determined using Student’s t test, or likelihood-ratio test (B and D–F); *P < 0.05.
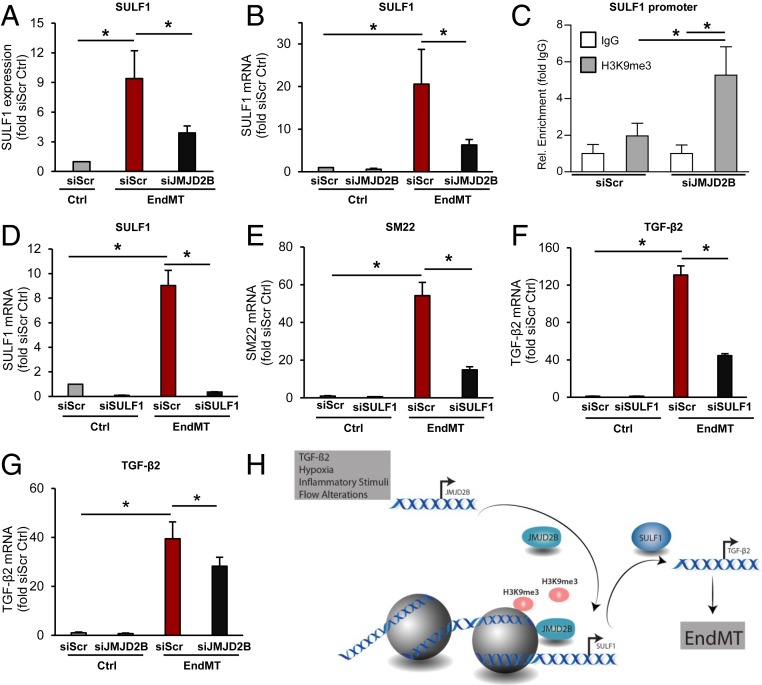
To identify additional putative direct JMJD2B-regulated genes, we analyzed the microarray-based gene expression after silencing of JMJD2B, which revealed significant changes in EndMT-related genes and pathways (Fig. 4G and SI Appendix, Fig. S8C). Direct candidates were then narrowed down by selecting those genes which additionally exhibited reduced H3K9me3 marks at the promoter after silencing of JMJD2B (SI Appendix, Fig. S8D). We identified 148 overlapping candidates, which were associated with cardiovascular and metabolic diseases (SI Appendix, Fig. S8E). Among these candidate genes, we selected the extracellular matrix protein sulfatase 1 (SULF1), which has been implicated in TGF-β signaling, fibrosis, and wound repair (41–43), and the serine/threonine kinase 3 (AKT3), which is part of the noncanonical TGF-β signaling pathway, for further validation. Reduced H3K9me3 peak levels at the SULF1 and AKT3 promoters are shown in Fig. 4B and SI Appendix, Fig. S7 D and E). Consistently, expression of AKT3 and SULF1 mRNA was significantly augmented by EndMT induction as shown by scRNA-seq (Fig. 4 E and F), microarray analysis (Figs. 4G and 5A), RT-qPCR (Figs. 4J and 5B), and on the protein level (Fig. 4K). After silencing of JMJD2B, both genes showed augmented H3K9me3 marks at their promoter (Figs. 4I and 5C). This was coinciding with the repression of AKT3 (Fig. 4 J and K) and SULF1 (Fig. 5 A and B) expression in JMJD2B-silenced ECs.
Fig. 5.
Analysis of JMJD2B target genes in EndMT. (A) SULF1 mRNA expression after silencing of JMJD2B in TGF-β2-stimulated ECs using microarray RNA sequencing (n = 3). (B) mRNA levels of SULF1 after siRNA-mediated knockdown of JMJD2B using RT-qPCR, normalized to RPLP0 (n = 4). (C) ChIP-Seq RT-qPCR showing H3K9me3 levels at promoter of SULF1, normalized to IgG (n = 5). (D–F) mRNA levels after siRNA-mediated knockdown of SULF1 or siScr siRNA in HUVECs followed by EndMT. Total RNA was isolated and mRNA levels were determined by RT-qPCR, normalized to GAPDH (2-ΔCt). (D) SULF1, (E) SM22, (F) TGF-β2 expression levels are depicted as fold siScr Ctrl (n = 3). (G) TGF-β2 mRNA level after siRNA-mediated knockdown of JMJD2B in HUVECs. mRNA levels were determined by RT-qPCR, normalized to RPLP0 (2-ΔCt), depicted as fold to siScr Ctrl (n = 9). (H) Cartoon summarizing our findings. Data are depicted as mean ± SEM. Statistical significance was determined using Student’s t test; *P < 0.05.
To assess whether these genes causally contribute to JMJD2B-mediated augmentation of EndMT, we silenced SULF1 and AKT3 expression by siRNAs. Only siRNAs directed against SULF1, but not against AKT3, reduced TGF-β2-induced expression of mesenchymal marker genes (Fig. 5 D and E and SI Appendix, Fig. S9), confirming the importance of SULF1 in EndMT. Interestingly, silencing of SULF1 also reduced the induction of TGF-β2 upon EndMT treatment (Fig. 5F). TGF-β2 was down-regulated upon JMJD2B silencing (Fig. 5G), but did not show H3K9me3 marks (SI Appendix, Fig. S7F). Together, these data suggest that SULF1 expression is epigenetically controlled by JMJD2B and subsequently activates TGF-β2 expression.
Discussion
The present study demonstrates that the histone demethylase JMJD2B regulates TGF-β1-, TGF-β2-, IL-1β-, and hypoxia-induced EndMT in vitro and in vivo. EndMT is associated with a change in H3K9me3 marks at distinct sets of promoters, whereas global histone trimethylation at H3K9 was not significantly changed. This study provides evidence of an epigenetic pathway that controls EndMT on the level of histone H3K9 demethylation (Fig. 5H).
Cellular differentiation and plasticity is critically regulated by epigenetic control mechanisms, including DNA methylation as well as histone modifications. While multiple studies investigated the involvement of epigenetic regulators in EMT, little is known regarding the role of epigenetic mechanisms in EndMT. DNA hypermethylation of the promoter of Ras-Gap-like protein 1 was shown to mediate hypoxia-induced EndMT via DNMT3A (37). At the level of histones, the H3K27me3-specific histone methyltransferase EZH2 interferes with TGF-β- and IL-1β-induced EndMT (44), whereas the histone deacetylase HDAC3 reprograms aortic ECs into mesenchymal cells (45). Using ChIP sequencing, we observed a change in H3K9me3 marks at various promoters upon induction of EndMT. Affected genes preferentially belonged to the TGF-β, Integrin, and Wnt signaling pathways, all of which are known regulators of EndMT (46). Particularly, TGF-β and its down-stream canonical SMAD proteins and noncanonical proteins, e.g. the phosphatidylinositol 3-kinase (PI3K) or c-Jun N-terminal kinase, are major mediators of EndMT induction. Likewise, Wnt/β-catenin act synergistically with TGF-β to induce EndMT during heart cushion development (47).
Among the specific gene loci, which showed changes in H3K9me3 marks upon EndMT and JMJD2B silencing, we selected CNN1, AKT3, and especially SULF1 for further analysis. CNN1 is a well-established mesenchymal/smooth muscle marker gene and our data suggest that CNN1 is repressed in ECs by H3K9me3 marks. This repression can be reversed by JMJD2B-mediated demethylation during EndMT-promoting conditions, followed by the subsequent activation of gene expression. AKT3 belongs to the PI3K pathway, which has been shown to be relevant for both EMT (48, 49) and EndMT (50). During EndMT, PI3K is induced by TGF-β2 and its activation results in a SMAD-independent induction of EndMT (51). Whether or to which extent AKT3 contributes to this process is unknown. Our results now suggest that AKT3 is not essential for TGF-β2-induced EndMT in cultured primary ECs. However, it has been shown that the AKT pathway and specifically AKT3 regulates valve formation during embryonic development (52). Therefore, a contribution to EndMT in this context might be possible.
The third gene of interest, SULF1, is a heparan sulfate 6-O-endosulfatase (53), which is important for migration of epithelial cells during wound repair and is activated by liver damage (42, 54–56). Heparan sulfate proteoglycans localize to the cell surface and the basement membrane and are a component of the extracellular matrix. Modification by 6-O-sulfation is important for protein interaction and cytokine signaling (57, 58). In this regard, the 6-O-sulfation of heparan sulfate was shown to regulate TGF-β, FGF, and VEGF signaling (43, 59). A dose- and time-dependent induction of SULF1 expression by TGF-β was previously demonstrated in human lung fibroblasts and in murine lungs in vivo (43). SULF1 was further reported to inhibit FGF signaling and FGF-induced angiogenesis in vivo (60) and to increase the availability of TGF-β1 in carcinoma cells (42). Additionally, our data demonstrate that SULF1 controls the expression of TGF-β2. Thus, SULF1 affects at least two crucial pathways in EndMT: it promotes TGF-β2 expression and signaling (42) and posttranscriptionally inhibits the EndMT-impeding action of FGF (60). JMJD2B epigenetically controls this crucial modulator and augments SULF1 expression by reducing H3K9me3 marks.
Endothelial plasticity and EndMT are critically regulated by various environmental factors, including growth factors, cytokines, oxygen tension, as well as mechanical forces (46). The present study shows that many of these factors regulate JMJD2B expression. Thus, hypoxia and disturbed flow induced JMJD2B expression, indicating that JMJD2B may be one common regulator of the endothelial phenotype and identity. However, how JMJD2B is regulated on a molecular level is not well known. JMJD2B transcription was shown to be induced in a HIF1-dependent manner in HeLa cells (61). Conserved hypoxia response elements binding HIF1 were identified in the promoter of Jmjd2b in humans and mice and were functionally validated by luciferase reporter and ChIP assays (61). Thus, JMJD2B may be regulated by several pathways to synergize with TGF-β2 signaling to induce EndMT in vitro and in vivo.
In summary, we identified JMJD2B as an important epigenetic regulator of EndMT in vitro and in vivo, which is elevated by disturbed flow and hypoxia in the endothelium. Although our study uses histological immunostainings and single-cell RNA sequencing to document the effects of endothelial-specific Jmjd2b deletion on EndMT in vivo, lineage tracing of endothelial cells certainly would have further substantiated these findings. Moreover, although endothelial-specific deletion of Jmjd2b reduced EndMT induction after myocardial infarction, we did only observe modest effects on heart function. This may relate to the complex role of EndMT after infarction. While several studies suggested that EndMT contributes to cardiac fibrosis (11, 50, 62, 63), more recent lineage tracing studies suggest that repopulating fibroblasts mainly derive from preexisting fibroblasts and not from other sources (64). However, a transient induction of EndMT may still impair endothelial cell functions contributing to a worsening of heart function after injury. Changes in endothelial phenotypes further play important roles in various pathologies, which might be controlled by epigenetic mechanisms such as histone demethylation.
Materials and Methods
A brief description of the materials and methods can be found below, whereas an expanded methods section can be found in SI Appendix.
Endothelial-to-Mesenchymal Transition Assay.
For the EndMT assay, two distinct media were used, referred to as either full medium (“Ctrl”) or differentiation media (“DM”). HUVECs were incubated in Ctrl media or DM with either TGF-β2 (referred to as “EndMT”), or TGF-β1 and/or IL-1β. After 48 h of incubation, the treatment was repeated.
JMJD2B Inhibitor Treatment.
Final concentrations of 1, 5, or 10 µM inhibitor (CCT365599) or DMSO as a control was added to the medium. CCT365599 was provided by Julian Blagg, Cancer Research UK Cancer Therapeutics Unit at the Institute of Cancer Research, London, UK.
CRISPR-Cas9–Based Knockdown of JMJD2B.
Lentivirus stocks were produced in HEK 293T cells using pCMVΔR8.91 as packaging plasmid and pMD2.G (Addgene No. 12259). Empty backbone vectors were used as control.
Small Interfering RNA Knockdown.
For siRNA-mediated gene silencing, HUVECs were transfected with GeneTrans II.
RNA Sequencing.
Total HUVEC RNA was used as input for whole transcriptome RNA-Seq library preparation (TruSeq Stranded Total RNA, Illumina) following low throughput protocol. Sequencing was performed on NextsEq. 500 (Illumina) using V2 chemistry and a 75-bp single-end setup. RNA expression levels in partial carotid ligation operated age-matched male C57BL/6 mice was performed by the laboratory of H.J. The cDNA libraries were sequenced on an Illumina 2000 HiSeq using v3 chemistry following low throughput protocol; 50 million 100-bp paired-end reads were generated per library. All differentially expressed genes were identified using Cuffdiff2. RNA expression levels under static and laminar flow conditions determined by RNA sequencing were analyzed using published dataset GSE54384.
Microarray Analysis.
Gene-expression analysis was performed using the GeneChip Human Exon 1.0 ST array. Data were analyzed by using the Exon Array Analyzer (EAA) Web interface (http://eaa.mpi-bn.mpg.de/).
ChIP-Seq.
The 4 × 106 cells per IP were cross-linked with a final concentration of 1% formaldehyde for 10 min. Shearing was done using a Covaris S220. DNA was amplified and sequenced on Illumina NextSeq. Trimmed sequences were mapped to GRCH38 with STAR 2.4.2a 4. Peaks were called using homer findPeaks version 4.0311.
Animals.
All animal experiments were performed in accordance with the animal welfare guidelines and German national laws and were authorized by the competent authority (Regierungspräsidium Darmstadt, Hessen, Germany). C57BL/6J Jmjd2bfl/fl mice were obtained from Hitoshi Okada (Kindai University, Osaka-Sayama, Japan) (65) and crossed with C57BL/6J Cdh5-CreERT2 mice. Mice were treated with tamoxifen for 2 wk. After an additional 2 wk, myocardial infarction was performed.
MI.
MI was performed in 12- to 14-wk-old male and female Cdh5-iCre; Jmjd2bfl/fl mice. MI was induced by permanent ligation of the left anterior descending coronary artery (LAD). After 3 d post MI, the heart and the liver was harvested for single-cell RNA sequencing (heart) or for knockout check (liver), 14 d post-AMI for immunohistochemistry. Echocardiography was performed on days 0, 7, and 14.
scRNA-Seq.
Cellular suspensions were loaded on a 10X Chromium Controller (10X Genomics). Libraries were prepared using the Chromium Single Cell 3′ v2 Reagent Kit and Chromium Single Cell 3′ v3 Reagent Kit, respectively. Indexed libraries were equimolarly pooled and sequenced using paired-end 26 × 98 bp as sequencing mode by GenomeScan. Single-cell RNA-Seq outputs were processed using the Cell Ranger suite versions 2.1.1 (in vitro control vs. EndMT samples, mapped to GRCh38) or 3.0.1 (in vivo samples; mapped to mm10, in vitro JMJD2B knockdown samples, mapped to GRCh38). We used Seurat 2.3.4 or 3.0.2 for secondary analysis, following the distributer’s tutorial (satijalab.org).
Statistics.
Mann–Whitney U test or Student’s t test was used to test for statistical differences between two groups as appropriate. For more groups ANOVA with Dunnett’s multiple comparison test was used. A value of P <0.05 was considered statistically significant. To test potential associations, Pearsons’s correlation was used. Significance for differential gene expression in scRNA-Seq analysis was calculated with bimodal maximum likelihood ratio test and χ2 statistic. Microsoft Excel and GraphPad Prism 6 were used to calculate statistical differences.
Data Availability.
The RNA sequencing, ChIP sequencing, and single-cell RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE143148). The microarray has been deposited in the GEO database (accession no. GSE143150).
Supplementary Material
Acknowledgments
We thank Marius Klangwart and Felix Vetter for excellent technical assistance, and Prof. Julian Blagg (Cancer Research UK Cancer Therapeutics Unit at The Institute of Cancer Research, London, UK) for providing CCT365599. The study was supported by the Deutsche Forschungsgemeinschaft (SFB834, Project B5 and SFB1366, Project B4) to S.D. and Project B9 to R.A.B.; Dr. Rolf Schwiete Stiftung to S.D.; and the DZHK, Säule B to S.D. and D.H.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA sequencing, ChIP sequencing, and single-cell RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE143148). The microarray has been deposited in the GEO database (accession no. GSE143150).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913481117/-/DCSupplemental.
References
- 1.Mahler G. J., Frendl C. M., Cao Q., Butcher J. T., Effects of shear stress pattern and magnitude on mesenchymal transformation and invasion of aortic valve endothelial cells. Biotechnol. Bioeng. 111, 2326–2337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X., et al. , Snail is a direct target of hypoxia-inducible factor 1α (HIF1α) in hypoxia-induced endothelial to mesenchymal transition of human coronary endothelial cells. J. Biol. Chem. 290, 16653–16664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovacic J. C., et al. , Endothelial to mesenchymal transition in cardiovascular disease: JACC state-of-the-Art review. J. Am. Coll. Cardiol. 73, 190–209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arciniegas E., Sutton A. B., Allen T. D., Schor A. M., Transforming growth factor beta 1 promotes the differentiation of endothelial cells into smooth muscle-like cells in vitro. J. Cell Sci. 103, 521–529 (1992). [DOI] [PubMed] [Google Scholar]
- 5.DeRuiter M. C., et al. , Embryonic endothelial cells transdifferentiate into mesenchymal cells expressing smooth muscle actins in vivo and in vitro. Circ. Res. 80, 444–451 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Kovacic J. C., Mercader N., Torres M., Boehm M., Fuster V., Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition: From cardiovascular development to disease. Circulation 125, 1795–1808 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arciniegas E., Neves C. Y., Carrillo L. M., Zambrano E. A., Ramírez R., Endothelial-mesenchymal transition occurs during embryonic pulmonary artery development. Endothelium 12, 193–200 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Agarwal S., et al. , Local and circulating endothelial cells undergo endothelial to mesenchymal transition (EndMT) in response to musculoskeletal injury. Sci. Rep. 6, 32514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potenta S., Zeisberg E., Kalluri R., The role of endothelial-to-mesenchymal transition in cancer progression. Br. J. Cancer 99, 1375–1379 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J., Xu Y., Koya D., Kanasaki K., Role of the endothelial-to-mesenchymal transition in renal fibrosis of chronic kidney disease. Clin. Exp. Nephrol. 17, 488–497 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Zeisberg E. M., et al. , Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 13, 952–961 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Chen P.-Y., et al. , Endothelial-to-mesenchymal transition drives atherosclerosis progression. J. Clin. Invest. 125, 4514–4528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evrard S. M., et al. , Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat. Commun. 7, 11853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P.-Y., Qin L., Tellides G., Simons M., Fibroblast growth factor receptor 1 is a key inhibitor of TGFβ signaling in the endothelium. Sci. Signal. 7, ra90 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Chen P.-Y., et al. , FGF regulates TGF-β signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2, 1684–1696 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serrano-Gomez S. J., Maziveyi M., Alahari S. K., Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 15, 18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Y., Dong C., Zhou B. P., Epigenetic regulation of EMT: The snail story. Curr. Pharm. Des. 20, 1698–1705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukada Y., et al. , Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Zhu H., Hu S., Baker J., JMJD5 regulates cell cycle and pluripotency in human embryonic stem cells. Stem Cells 32, 2098–2110 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Ishimura A., et al. , Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression. Development 139, 749–759 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Peng J. C., et al. , Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139, 1290–1302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., et al. , The histone demethylase JMJD2C is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol. Reprod. 82, 105–111 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Yang H., et al. , Elevated JMJD1A is a novel predictor for prognosis and a potential therapeutic target for gastric cancer. Int. J. Clin. Exp. Pathol. 8, 11092–11099 (2015). [PMC free article] [PubMed] [Google Scholar]
- 24.Tokunaga R., et al. , The prognostic significance of histone lysine demethylase JMJD3/KDM6B in colorectal cancer. Ann. Surg. Oncol. 23, 678–685 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Aprelikova O., et al. , The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis. Clin. Epigenetics 8, 38 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., et al. , KDM6B induces epithelial-mesenchymal transition and enhances clear cell renal cell carcinoma metastasis through the activation of SLUG. Int. J. Clin. Exp. Pathol. 8, 6334–6344 (2015). [PMC free article] [PubMed] [Google Scholar]
- 27.Tang B., et al. , JARID1B promotes metastasis and epithelial-mesenchymal transition via PTEN/AKT signaling in hepatocellular carcinoma cells. Oncotarget 6, 12723–12739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L., et al. , JMJD2B promotes epithelial-mesenchymal transition by cooperating with β-catenin and enhances gastric cancer metastasis. Clin. Cancer Res. 19, 6419–6429 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Enkhbaatar Z., et al. , KDM5B histone demethylase controls epithelial-mesenchymal transition of cancer cells by regulating the expression of the microRNA-200 family. Cell Cycle 12, 2100–2112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramadoss S., Chen X., Wang C.-Y., Histone demethylase KDM6B promotes epithelial-mesenchymal transition. J. Biol. Chem. 287, 44508–44517 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang B., et al. , Aberrant JMJD3 expression upregulates slug to promote migration, invasion, and stem cell-like behaviors in hepatocellular carcinoma. Cancer Res. 76, 6520–6532 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Fodor B. D., et al. , Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L., et al. , Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 7541–7546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedersen M. T., et al. , The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Mol. Cell. Biol. 34, 1031–1045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boeckel J.-N., et al. , JMJD8 regulates angiogenic sprouting and cellular metabolism by interacting with pyruvate kinase M2 in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 36, 1425–1433 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Choi S.-H., et al. , A hypoxia-induced vascular endothelial-to-mesenchymal transition in development of radiation-induced pulmonary fibrosis. Clin. Cancer Res. 21, 3716–3726 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Xu X., et al. , Hypoxia-induced endothelial-mesenchymal transition is associated with RASAL1 promoter hypermethylation in human coronary endothelial cells. FEBS Lett. 590, 1222–1233 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Moonen J.-R. A. J., et al. , Endothelial-to-mesenchymal transition contributes to fibro-proliferative vascular disease and is modulated by fluid shear stress. Cardiovasc. Res. 108, 377–386 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Hatch S. B., et al. , Assessing histone demethylase inhibitors in cells: Lessons learned. Epigenetics Chromatin 10, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong J., et al. , A metabolic basis for endothelial-to-mesenchymal transition. Mol. Cell 69, 689–698.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamouille S., Xu J., Derynck R., Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dhanasekaran R., et al. , Activation of the transforming growth factor-β/SMAD transcriptional pathway underlies a novel tumor-promoting role of sulfatase 1 in hepatocellular carcinoma. Hepatology 61, 1269–1283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue X., et al. , Transforming growth factor-beta1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J. Biol. Chem. 283, 20397–20407 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maleszewska M., Gjaltema R. A. F., Krenning G., Harmsen M. C., Enhancer of zeste homolog-2 (EZH2) methyltransferase regulates transgelin/smooth muscle-22α expression in endothelial cells in response to interleukin-1β and transforming growth factor-β2. Cell. Signal. 27, 1589–1596 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Zeng L., et al. , Histone deacetylase 3 unconventional splicing mediates endothelial-to-mesenchymal transition through transforming growth factor β2. J. Biol. Chem. 288, 31853–31866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dejana E., Hirschi K. K., Simons M., The molecular basis of endothelial cell plasticity. Nat. Commun. 8, 14361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebner S., et al. , Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J. Cell Biol. 166, 359–367 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bakin A. V., Tomlinson A. K., Bhowmick N. A., Moses H. L., Arteaga C. L., Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 275, 36803–36810 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Grille S. J., et al. , The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 63, 2172–2178 (2003). [PubMed] [Google Scholar]
- 50.Widyantoro B., et al. , Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation 121, 2407–2418 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Medici D., Potenta S., Kalluri R., Transforming growth factor-β2 promotes Snail-mediated endothelial-mesenchymal transition through convergence of Smad-dependent and Smad-independent signalling. Biochem. J. 437, 515–520 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang Z.-Z., et al. , Dosage-dependent effects of Akt1/protein kinase Balpha (PKBalpha) and Akt3/PKBgamma on thymus, skin, and cardiovascular and nervous system development in mice. Mol. Cell. Biol. 25, 10407–10418 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frese M.-A., Milz F., Dick M., Lamanna W. C., Dierks T., Characterization of the human sulfatase Sulf1 and its high affinity heparin/heparan sulfate interaction domain. J. Biol. Chem. 284, 28033–28044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaman G., et al. , Expression of Sulf1 and Sulf2 in cartilage, bone and endochondral fracture healing. Histochem. Cell Biol. 145, 67–79 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Maltseva I., Chan M., Kalus I., Dierks T., Rosen S. D., The SULFs, extracellular sulfatases for heparan sulfate, promote the migration of corneal epithelial cells during wound repair. PLoS One 8, e69642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graham K., Murphy J. I., Dhoot G. K., SULF1/SULF2 reactivation during liver damage and tumour growth. Histochem. Cell Biol. 146, 85–97 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Esko J. D., Lindahl U., Molecular diversity of heparan sulfate. J. Clin. Invest. 108, 169–173 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakato H., Kimata K., Heparan sulfate fine structure and specificity of proteoglycan functions. Biochim. Biophys. Acta 1573, 312–318 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Uchimura K., et al. , HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: Effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 7, 2 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S., et al. , QSulf1, a heparan sulfate 6-O-endosulfatase, inhibits fibroblast growth factor signaling in mesoderm induction and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 4833–4838 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beyer S., Kristensen M. M., Jensen K. S., Johansen J. V., Staller P., The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283, 36542–36552 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okayama K., et al. , Hepatocyte growth factor reduces cardiac fibrosis by inhibiting endothelial-mesenchymal transition. Hypertension 59, 958–965 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Ghosh A. K., et al. , Genetic deficiency of plasminogen activator inhibitor-1 promotes cardiac fibrosis in aged mice: Involvement of constitutive transforming growth factor-beta signaling and endothelial-to-mesenchymal transition. Circulation 122, 1200–1209 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Fu X., et al. , Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J. Clin. Invest. 128, 2127–2143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawazu M., et al. , Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE 6, e17830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing, ChIP sequencing, and single-cell RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE143148). The microarray has been deposited in the GEO database (accession no. GSE143150).