Significance
Proteins involved in neurodegenerative disease often aggregate, leading both to amorphous phase separation and to fibrillization. Tau protein involved in Alzheimer’s and Pick’s diseases is in this class. We show that the free-energy landscapes display two channels of tau aggregation: one which leads to more ordered amyloid fibrils and a nonfibrillar channel that leads to amorphous phases. The nonfibrillar oligomers have been suggested to be the pathogenic species. The distinct structural properties of the two channels show they must backtrack to interconvert between the two channels, suggesting a complex interplay between amorphous phase separation and the formation of ordered amyloid fibrils. This interplay is tuned by the way tau is phosphorylated and the specific isoforms at play.
Keywords: neurodegenerative diseases, tau, aggregation, amorphous phase separation, fibrillization
Abstract
Filaments made up of different isoforms of tau protein are associated with a variety of neurodegenerative diseases. Filaments made up of the 4R-tau isoform, which has four repeat regions (R1 to R4), are found in patients suffering from Alzheimer’s disease, while filaments made of the 3R-tau isoform, which contains only three repeat units (R1, R3, and R4), are found in patients with Pick’s disease (frontotemporal dementia). In this work, a predictive coarse-grained protein force field, the associative memory water-mediated structure and energy model (AWSEM), is used to study the energy landscapes of nucleation of the two different fibrils derived from patients with Pick’s and Alzheimer’s diseases. The landscapes for nucleating both fibril types contain amorphous oligomers leading to branched structures as well as prefibrillar oligomers. These two classes of oligomers differ in their structural details: The prefibrillar oligomers have more parallel in-register -strands, which ultimately lead to amyloid fibrils, while the amorphous oligomers are characterized by a near random -strand stacking, leading to a distinct amorphous phase. The landscape topography suggests that there must be significant structural reordering, or “backtracking,” to transit from the amorphous aggregation channel to the fibrillization channel. Statistical mechanical perturbation theory allows us to evaluate the effects of changing concentration on the aggregation free-energy landscapes and to predict the effects of phosphorylation, which is known to facilitate the aggregation of tau repeats.
Many neurodegenerative diseases are characterized by the formation of specific protein deposits within neurons and in the spaces between neurons, often in specific areas of the brain. Neurofibrillary tangles made up largely of tau protein have been found in the brains of patients with Alzheimer’s and Pick’s diseases (frontotemporal dementia) (1). The precise role of these protein aggregates in pathogenesis remains an open question. An attractive idea is that, rather than the characteristic long fibrils, it is small soluble oligomers that impair neuronal function. In this paper, we explore the relation between the small oligomers of tau, an amorphous phase of tau aggregation, and the fully formed fibrils using a realistic computational method that has been shown to capture the key features of the aggregation landscapes and oligomerization routes of A peptides (2, 3), polyglutamine repeats (4), and the Huntingtin proteins (5).
In the human brain, alternative splicing of the tau pre-mRNA results in six molecular isoforms of the protein. These six tau isoforms differ by containing either three (3R taus) or four (4R taus) microtubule binding repeats (R) of 31 to 32 amino acids in the carboxy-terminal half and by having one (1N), two (2N), or zero (0N) amino-terminal inserts of 29 amino acids each; the extra repeat in 4R tau is the second repeat (R2) of 4R taus (6, 7). This alternative splicing of tau pre-mRNA results in the expression of three 3R taus (0N3R, 1N3R, and 2N3R) and three 4R taus (0N4R, 1N4R, and 2N4R) (7). The 2N4R tau is the largest-size human brain tau with a length totaling 441 amino acids (tau441).
In Alzheimer’s disease, hyperphosphorylated tau proteins aggregate into intracellular neurofibrillary tangles, which appear as paired helical filaments (1, 8). Ordinarily, tau is a microtubule-associated protein, which is important in regulating the stability and dynamics of microtubules (MTs) and is involved in neurite outgrowth and axonal transport. Tau can be thought of as having two domains: the projection domain and the microtubule-binding domain. The projection domain is the N-terminal domain of tau, which is both acidic and proline rich. The projection domain interacts with both cellular membranes and the cytoskeleton, where it proves necessary for intracellular cargo transportation. The microtubule association domain binds to microtubules and makes up the fibrillar cores of the neurofibrillary tangles. Neither of the two domains is well structured in the monomeric state.
Different alternative splicing events generate six distinct isoforms of tau, which take part in amyloid formation. The amyloid of the four-repeat (4R) isoform of tau, characteristically found in patients with Alzheimer’s disease, has been structurally characterized by Scheres and coworkers (9). Although the aggregation of A into extracellular plaque also happens in Alzheimer’s disease, the interplay in pathogenesis between the aggregation of A and tau remains largely unclear. The cryo-EM structure of the 4R tau amyloids derived from patient samples with Alzheimer’s disease contains only repeats R3 and R4 in the well-resolved fibrillar core and displays the usual parallel in-register form seen in amyloids from other peptides. Outside the fibrillar core, at both N- and C-terminal ends, the structures of the tangles appear to remain disordered. These terminal regions of the peptide form a fuzzy coat outside the core. Cryo-EM structures of the R1/3/4 (3R) isoform amyloids, which are found in patients with Pick’s disease, have also been determined and also display a parallel in-register topology (10). The fibrillar core of the R1/3/4 isoform deposit is structurally quite different from that of the R3/4 deposits. In this paper, we explore the consequences of these structural differences between the isoforms for the oligomerization and nucleation of large aggregates.
The regulation of tau aggregation is quite sophisticated and involves a range of posttranslational modifications including phosphorylation, truncation, nitration, glycation, acetylation, and methylation (1). These posttranslational modifications regulate the ability of tau to bind and assemble microtubules. The modifications also affect the aggregation of the isoforms into neurofibrillary tangles. There are 80 serine/threonine and 5 tyrosine residues in the tau isoforms that can potentially be phosphorylated (11). Phosphorylation in the repeat domains leads to the disassociation of tau from the microtubules. Phosphorylation also facilitates aggregation. The level of phosphorylation found in the brains of those manifesting the disease is higher than the level of phosphorylation found in healthy brains (1). In vitro tau proteins can aggregate to form fluid droplets. Phosphorylation influences liquid–liquid phase separation (LLPS) in tau (12). Liquid–liquid phase separation has been used to explain how membraneless structures that display a liquid-like behavior form in eukaryotic cells (13). Such amorphous structures include P granules, nucleoli, and stress granules. These membraneless assemblies are often enriched in intrinsically disordered proteins and enriched with proteins that contain multiple modular domains that allow them to assemble in an amorphous fashion through multiple valency (13, 14). The tau protein thus provides an interesting example of the dichotomy between amorphous assembly and assembly into more ordered fibrils.
The simulation of protein aggregation is challenging due to the large size of the systems that need to be put together (15). Atomistic simulations require typically large computational resources. This difficulty has led us to use a coarse-grained but realistic force field to carry out a survey of the energy landscapes of oligomers and fibrils of many proteins involved in neurodegenerative diseases. We have previously used the coarse-grained model, the associative memory water-mediated structure and energy model (AWSEM) (16), to study the initial stages of misfolding and aggregation of titin (17, 18), 40 (2), 42 (3), polyglutamine repeats (4), and Huntingtin Exon1 fragments (5). The same force field is employed in the AWSEM-Amylometer algorithm (19), a simple scanning calculation which has proved able to predict with considerable accuracy both the aggregation propensity and the topology of fibrils of a variety of proteins and peptides. These results encourage confidence that the ingredients of the AWSEM force field are adequate for understanding many aspects of amyloid formation and the kinetics of protein aggregation.
In this work, we construct the energy landscapes for forming ordered fibrils and oligomers of the two different isoforms of tau protein that have been derived from patients with Pick’s and Alzheimer’s diseases. The aggregation landscape shows that there exist two channels of assembly, one made up of amorphous oligomers and the other involving prefibrillar oligomers. These two classes of oligomers possess distinct structural details: The prefibrillar oligomers have more parallel in-register -strands, which ultimately lead to forming macroscopic amyloid fibrils, while the amorphous oligomers are filled with random -strand stacking, which eventually leads to a macroscopically amorphous state typical of the liquid–liquid phase separation seen in membraneless organelles. The distinct structural differences suggest that significant structural reordering, or backtracking, must take place to change from the amorphous channel to the prefibrillar channel of assembly.
Aggregation Landscapes of R3/4 in Tau and the Existence of Kinetic Backtracking
We first study the aggregation landscape of the R3/4 repeat peptide, which is found in the fibrils of the 4R isoform derived from patients with Alzheimer’s disease. The solved structure by Scheres and coworkers (9) possesses a fibrillar core of 73 residues (residues 306 to 378). This core is made out of the third and fourth repeats. The core apparently is surrounded by a disordered fuzzy coat made up of the additional residues. There are actually two different polymorphic fibrils whose structures have been determined by Scheres and coworkers (9), one for the paired helical filaments (PHFs) and the other for the straight filaments (SFs). The two polymorphs, however, possess a common C-shaped architecture for the monomeric ribbons of which they are made (9).
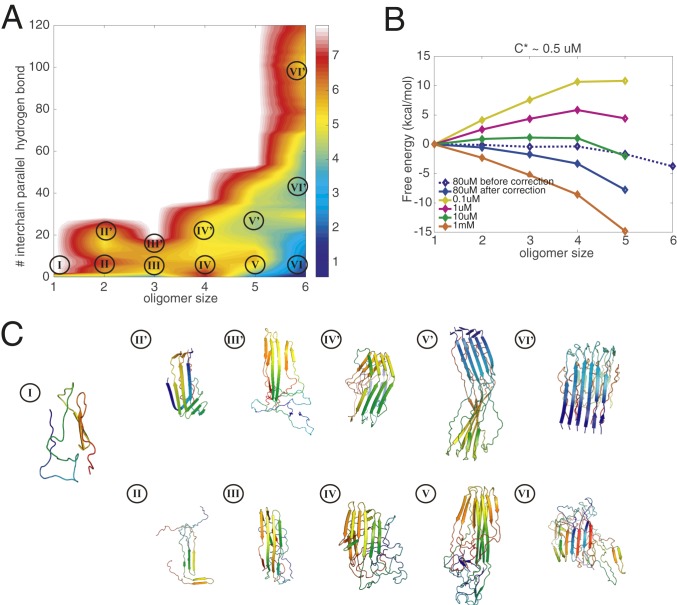
We, therefore, first construct the aggregation free-energy profile for six R3/4 monomers in a simulation box at the nominal laboratory concentration of around 80 M. Umbrella-sampling simulations using the fraction of native contacts based on the determined structure (PDB ID: 5O3T) as the biasing coordinate are then carried out to construct the free-energy profiles. As shown in Fig. 1A, the energy landscape can be projected onto a 2D free-energy surface using as simultaneous progress variables the oligomer size and the number of interchain parallel hydrogen bonds among the six monomers (signifying the formation of fibrillar-like -strands). Various oligomeric species are sampled in the simulations and labeled on the free-energy surface in Fig. 1A. Apart from the monomeric species, which is made up of predominantly disordered segments (structure I in Fig. 1C), oligomers of each size can be found either as an amorphous species (which has fewer parallel hydrogen bonds) or as a prefibrillar species (more parallel bonds, most of which are in-register), as shown in Fig. 1C. The amorphous species are hydrogen bonded; however, the hydrogen bond patterns in the amorphous channel are less ordered than in the fibrillar channel. Ultimately, the amorphous species will lead to the formation of a macroscopic amorphous phase. As shown on the free-energy surface, at the nominal simulation concentration, the amorphous hexamer (basin VI) is the most favored species thermodynamically. Formation of the amorphous hexamer is downhill at the simulated concentration of 80 M. Further growth of the prefibrillar oligomers is not as thermodynamically favored as the growth along the amorphous channel. Also, we see there are distinct free-energy barriers disfavoring the transition between the two structural classes of oligomers. At the simulation concentration, the barriers for the growth of both amorphous oligomers and the prefibrillar oligomers are smaller than the barriers for interconversions between the channels. The difficulty in making transitions between the amorphous species and the fairly ordered prefibrillar oligomers inferred from the free-energy surface suggests that there are two kinetically independent channels of oligomerization: one involving growth of the prefibrillar oligomers finally reaching the amyloid fiber and the other coexisting channel of growth involving amorphous oligomers leading eventually to an amorphous, macroscopic, liquid–liquid phase.
Fig. 1.
Grand canonical free-energy profiles for R3/4 at a temperature of 300 K. (A) The 2D free-energy surface at the concentration of 80 M is plotted using the number of intermolecular parallel hydrogen bonds and the oligomer size as the two dimensions. The number of intermolecular parallel hydrogen bonds monitors the association of oligomers into fibrillar -strands. Size is used to label the local basins for different oligomer states. (B) The grand canonical free energy as corrected for concentration changes indicates the saturation concentration of free monomers. (C) Representative structures are shown in each basin: Each monomer is colored with a rainbow spectrum from red to blue, N terminus to C terminus. The monomeric state of tau is largely extended and disordered.
In simulations using a finite number of monomers within a finite box, the cache of monomers becomes depleted as clusters grow. This effect is negligible in laboratory experiments that study the early phase of aggregation but we must correct for this simulation artifact. As in our previous papers, we adopt the Reiss physical cluster theory (20) as detailed in Chen and Wolynes (5) to correct for the finite-size effect of the simulation box. The grand canonical free-energy profile, , is downhill at the nominal simulation concentration (80 M) (blue line in Fig. 1B), showing that the growth of oligomers is favored. By carrying out the correction, we can find the concentration dependence of the free-energy profile and then estimate solubility limits. The solubility is estimated by scanning a range of concentrations and finding the lowest concentration at which the free-energy profile becomes downhill at the largest oligomer sizes that were reliably sampled. The solubility of R3/4 in the core of K18 aggregates as estimated from our calculation is around 0.5 M. This value agrees well with experimental determinations of the solubility of K18 [0.37 M (21)]. We can also find the critical nucleus size for any given concentration by finding the size of oligomer that corresponds to the peak of the free-energy profile. The observed critical nucleus size is around 2 to 3 in the low-concentration regime, also agreeing with experimental observation (Fig. 1B). To compare the relative favorability of the amorphous oligomerization channel with that of the fibrillization channel, we also computed separately grand canonical free-energy profiles for the growth of prefibrillar oligomers (SI Appendix, Fig. S1A) and for the growth of amorphous oligomers (SI Appendix, Fig. S1B), each considered as the only possibility. For the R3/4 construct, we find that the amorphous oligomers are more insoluble than are the specifically ordered fibrils. The growth through the amorphous channel is thus typically more favored in terms of free energy under the same laboratory concentrations (SI Appendix, Fig. S1 A and B), suggesting that the R3/4 region of K18 is more likely to use the channel leading toward phase separation rather than to fibrillize (12). Admittedly, the inferred free-energy surfaces of the amorphous oligomers are somewhat noisy because the umbrella-sampling simulations are performed using only the similarity to the fibrillar form as the biasing coordinate. Nevertheless, the collected data are sufficiently adequate for us to infer that the amorphous channel should appear as a significant branch in the aggregation of tau.
Direct transitions from amorphous oligomers to prefibrillar oligomers involve transient disassociation of antiparallel hydrogen bonds and the reformation of parallel hydrogen bonds (SI Appendix, Fig. S6B). These kinetic constraints will create a barrier that needs to be to crossed (SI Appendix, Fig. S6A). During the simulations, no direct transitions between amorphous oligomers and their prefibrillar counterparts were observed, but the association and disassociation of different oligomers, thus changing their size, is frequently observed. The idea of kinetic backtracking explains the separation into the two pools of oligomers following distinct channels in the free-energy landscape: The amorphous oligomers generally need to disassociate into monomers before they can reassociate to become prefibrillar forms and contribute to form amyloid fibers (2). This proposed backtracking mechanism is in harmony with the experimental evidence showing that two types of oligomers in tau form during aggregation, one of which can directly associate into fibrils and the other of which transforms into basic monomers to form fibrils (22).
Aggregation Landscapes of the R1/3/4 Isoform of Tau and the Existence of Kinetic Backtracking
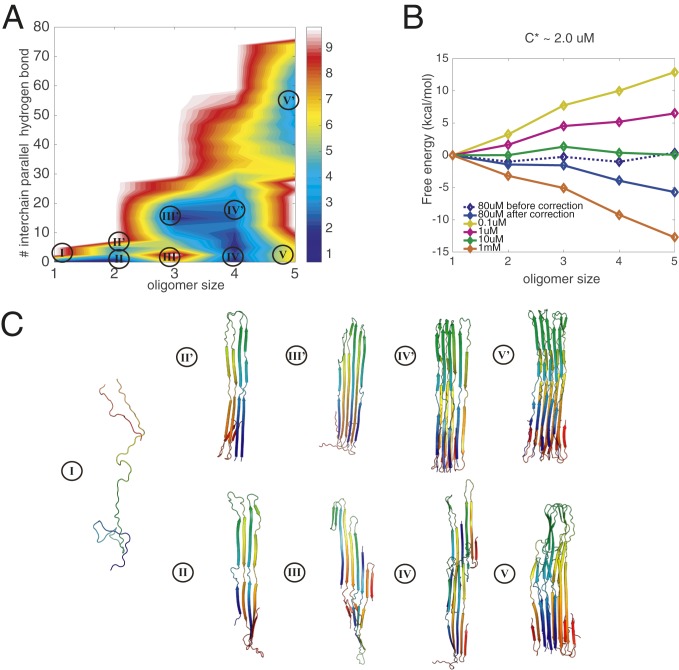
The cryo-EM structure of tau amyloids derived from patients with Pick’s disease has also been determined. The fibrillar core is composed of the R1/3/4 repeats, also called K19 (10). The tau filaments formed in Pick’s disease adopt a fold that is distinct from the Alzheimer’s fold, which has used the R3/4 repeats as the fibrillar core. To elucidate the differences between the landscapes for forming these two distinct disease-related folds, we simulated the aggregation free-energy landscape for six R1/3/4 monomers in a simulation box at the nominal laboratory concentration of around 80 M. Again, we find two types of oligomers for each size (Fig. 2A). While the amorphous oligomers now show a trace of parallel hydrogen bonds, the prefibrillar forms are also now much richer in parallel-in-register -strands (Fig. 2C). Similar to the landscapes found for the Alzheimer’s tau fold, the landscape for the Pick’s disease tau fold displays two routes of oligomerization. Again, there are modest barriers discouraging interconversion between the two structural classes on the 2D free-energy surface. Different from what was found for the Alzheimer’s tau fold, the prefibrillar pentamer is now thermodynamically more favored than the amorphous pentamer basin. The free-energy profile along the growth pathway of the amorphous oligomers is not as strongly favored as was found for R3/4, indicating a relatively weaker amorphous oligomerization propensity. This observation is consistent with the experimental evidence that the 3R construct (K19) possesses a lower tendency to undergo amorphous phase separation than does K18 (12), whose fibrillar core is R3/4.
Fig. 2.
Grand canonical free-energy profiles for R1/3/4 at a temperature of 300 K. (A) The 2D free-energy surface at the concentration of 80 M is plotted using the number of intermolecular parallel hydrogen bonds and the oligomer size as the two dimensions. The number of intermolecular parallel hydrogen bonds monitors the association of oligomers into fibrillar -strands. Size is used to label the local basins for different oligomer states. (B) The grand canonical free energy as corrected for concentration changes indicates the saturation concentration of free monomers. Few hexamers were formed in the simulations, so we do not extend the landscapes to the hexamer in the diagram owing to the resulting noise in the free-energy estimation. (C) Representative structures are shown in each basin: Each monomer is colored with a rainbow spectrum from red to blue, N terminus to C terminus. The monomeric state of tau is largely extended and disordered.
The free-energy profile obtained after making the correction for the changing concentration of free monomers in the simulation exhibits a downhill behavior at the nominal concentration (Fig. 2B). The predicted critical concentration for aggregation is around 2 M, and this value compares well with experiments that indicate the solubility limit for 3R tau is around 1.3 M at 303 K. We also computed the separate grand canonical free-energy profiles for the growth of prefibrillar oligomers (SI Appendix, Fig. S2A) and amorphous oligomers (SI Appendix, Fig. S2B) for R1/3/4. Different from what was seen for R3/4, the growth of the amorphous oligomers is not as favored in terms of free energy under the same laboratory concentrations for K19 (SI Appendix, Fig. S2 A and B). Apparently it is harder for K19 to undergo amorphous oligomerization into an amorphous phase than for the R3/4 construct to do so. Again, this result from simulation agrees with experimental observations (12) that also indicate that the growth of amorphous oligomers is harder for K19. Between the concentrations of 1 M and 10 M, prefibrillar oligomers readily associate into larger fibrils, while the amorphous oligomers will still remain largely trapped in the early stages of oligomerization. When the initial concentration is more than 10 M, amorphous oligomers can now again readily associate into larger sizes along the amorphous channel and phase separate as fluid droplets. This simulation result is in harmony with the observations of distinct separated phases of K19 at 10 M in Zweckstetter and coworkers (12).
We also compared the structural differences between the two pools of oligomers using the number of parallel hydrogen bonds and the number of antiparallel hydrogen bonds as two discriminators. As shown in SI Appendix, Fig. S7A, the landscape shows two oligomerization channels, corresponding to the amorphous oligomers (with fewer parallel hydrogen bonds) and prefibrillar oligomers. To convert between the two structural channels, in general, oligomers need to cross a free-energy barrier of at least 4 kcal/mol. Estimates of free-energy barriers using approximate reaction coordinates serve as lower bounds, and the true barrier may thus be significantly higher. The distinct structural features of the amorphous oligomers and the prefibrillar oligomers suggest again the necessity of backtracking to undergo structural interconversions (SI Appendix, Fig. S7B). Experiments using fluorescence energy transfer of labeled tau peptide have shown distinct kinetics for oligomers with different fluorescence resonance energy transfer (FRET) efficiencies (22). In SI Appendix, we show the distance distribution for the corresponding labeled pairs (pairs of cysteine 260, in the first repeat, on different tau monomers) in the simulated ensemble chosen for the two distinct channels in R1/3/4. The fibrillization channel is characterized by a decidedly narrower distribution of FRET pair distances in the largest oligomers (SI Appendix, Fig. S5). A more detailed comparison of the simulated ensembles with the FRET results is difficult to make because, of the two constructs that we simulated, only one (R1/3/4) contains C260 and neither one contains the region around K280, which was deleted in the K18K280 construct that was studied by FRET.
The Effect of Phosphorylation on Tau Aggregation
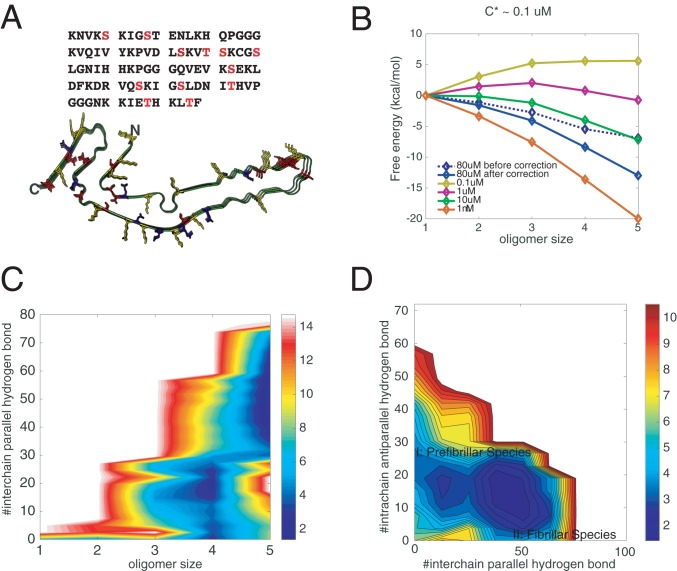
To perform its functions, tau undergoes phosphorylation. The most common sites of phosphorylation in tau are serine, threonine, and tyrosine (23), comprising as many as 85 possible sites of phosphorylation in full-length tau, about 20% of the amino acids in the protein (SI Appendix, Fig. S8A and Fig. 3A) (24). Only a few of the sites actually become phosphorylated, however, at any given time. Apparently, the actual sites that are phosphorylated are random. This heterogeneity of phosphorylation poses a problem for studying the molecular mechanisms of phosphorylation in the aggregation of tau. Phosphorylation of tau occurs before the formation of insoluble tau deposits. In vivo, it is believed that multiple kinases, such as MAPK, GSK-3, MARK, cdk2, and cdk5, can carry out the phosphorylations (25). In vivo, multiple phosphorylated versions of the tau isoforms exist, depending on the order of phosphorylation events from the various kinases.
Fig. 3.
Effects of phosphorylations on the aggregation of R1/3/4. (A) The amino acid sequence of R1/3/4 in tau. All of the potential phosphorylated serines/threonines are shown in red. The solved fibrillar structure of R1/3/4 is also illustrated, with positively charged residues colored in yellow, negatively charged residues in blue, and potentially phosphorylated sites in red. Three of the 12 residues are randomly phosphorylated during recalculating the free-energy surfaces using thermodynamic perturbation theory (SI Appendix, Fig. S2). (B) The perturbation-based grand canonical free-energy profiles for different oligomer states corrected for the monomer concentration changes in the fixed number simulation. (C) Perturbation-based grand canonical free-energy surface at the concentration of 80 M at 300 K is plotted using the number of intermolecular parallel hydrogen bonds and the oligomer size as the two dimensions. (D) The recalculated grand canonical free-energy surface is also plotted using the number of intermolecular parallel hydrogen bonds and the number of intramolecular antiparallel hydrogen bonds as the two dimensions to be compared with the landscape for the unphosphorylated assembly.
We have computed the free-energy landscapes of both R3/4 and R1/3/4 for several of their specific phosphorylated forms, using thermodynamic perturbation theory (see Methods for details). In comparison with the nonphosphorylated versions, phosphorylation thermodynamically facilitates oligomerization (SI Appendix, Fig. S8 B and C and Fig. 3 B and C). In the laboratory, Zweckstetter and coworkers (12) found that phosphorylation of K18 favors phase separation into the amorphous form. We note that the random pattern of phosphorylation also encourages forming amorphous aggregates rather than a fiber. Disorder in the phosphorylation pattern has primarily a first-order effect on the free energy of the ordered fibril with a fixed structure on average, but would allow the amorphous forms to adjust their distribution of structures to further lower the free energy. Indeed, it is possible that a separate phase transition can occur, involving separating tau proteins that have different phosphorylation patterns. This effect would show up more strongly in second-order thermodynamic perturbation theory. It would be interesting to see whether such demixing of aggregates with different phosphorylation patterns occurs in the natural deposits of tau protein.
After phosphorylation in our simulations, we find that the change in electrostatics favors the fibrillar-like states by neutralizing the charge environments of the phosphorylated sites (SI Appendix, Fig. S8A and Fig. 3A), but on the other hand the nonelectrostatic tertiary contact interactions after phosphorylation (which changes serines into glutamates effectively) slightly disfavor the fibrillar states (SI Appendix, Fig. S3 B and C).
To separate the effects of phosphorylation on the two distinct oligomerization channels, we separately computed the perturbed 1D free-energy profiles along both the amorphous oligomer channel and the prefibrillar oligomer growth channel. For R3/4, phosphorylation decreases the free-energy barrier for forming larger oligomers for both of the routes. The solubility, therefore, also is decreased (SI Appendix, Fig. S1 C and D). As a result, aggregates of both phosphorylated forms are more readily formed thermodynamically than when the proteins are unphosphorylated. This is in harmony with experimental observations on K18. Again for R1/3/4, the larger oligomers of both structural forms are favored by phosphorylation (SI Appendix, Fig. S2 C and D). Phosphorylation favors oligomerization thermodynamically. Phosphorylation, however, does not lower the barrier for forming the larger prefibrillar oligomers in R1/3/4 (SI Appendix, Fig. S2C). This suggests that fibrillar growth can be kinetically manipulated by phosphorylation.
Discussion
A Universal Role for Backtracking in Protein Aggregation?
The partial unfolding of a protein along its folding pathway to overcome topological frustration has been called backtracking. The backtracking mechanism, while uncommon in the context of globular protein folding, does sometimes occur for the nearly perfectly funneled landscapes of protein monomers: Ubiquitin’s assembly mechanism is an example where folding is not purely progressive but involves such backtracking. Backtracking has also turned out to be a key feature in the aggregation of A, and critical mutations in the backtracking region of A have been shown to affect aggregation kinetics and thermodynamics (2). For tau, the distinct structural properties of the amorphous oligomers and the prefibrillar oligomers, which use the backtracking mechanism to interconvert between the species, suggest a complex interplay between amorphous phase separation and the formation of more ordered amyloid fibrils. The backtracking mechanism, giving rise to separate channels of aggregation, may well be a universal feature in protein aggregation landscapes. Its prevalence is consistent with the fact that liquid–liquid phase separation has been observed for several protein systems that still form macroscopic fibrils that are ordered in the end.
Liquid–Liquid Phase Separation and the Role of Phosphorylation.
Protein inclusions such as the neurofibrillary tangles in Alzheimer’s and Pick’s diseases are solid-like. Within the tangles, full-length tau and its more aggregation-prone fragments are locked into cross–-structures, resulting in their long-term maintenance within the cell. In contrast, a characteristic property of liquid-like cellular compartments, which are not bounded by closed membranes, is the rather rapid exchange of molecules between the interior of these compartments and the surrounding cytoplasm. Liquid–liquid phase separation has been described in various other systems that are also amyloid prone; however, little has been said concerning the connections between the phase separation phenomenon and amyloid formation. The results of the simulation for both R3/4 (the fibrillar core of K18) and R1/3/4 (K19) overlap well with the experimental evidence that R3/4 readily forms a separated amorphous phase through amorphous oligomerization, while the R1/3/4 isoform apparently does not form such a well-defined liquid phase even at a higher concentration. We see here that there is a bridge between the existence of two distinct channels for oligomerization and what is seen macroscopically. For both systems, phosphorylation encourages forming larger oligomers thermodynamically through the electrostatic interactions, which generally also favors the formation of amorphous phases. We note that the role of phosphorylation in aggregation has surfaced in studies of multiple systems (26, 27). Phosphorylation of fibrillar core residues disrupts aggregation of FUS also through electrostatic interactions, implying that the specific role of phosphorylation strongly depends on the specific protein being studied (26, 28).
Two Channels of Tau Aggregation on the Aggregation Landscape.
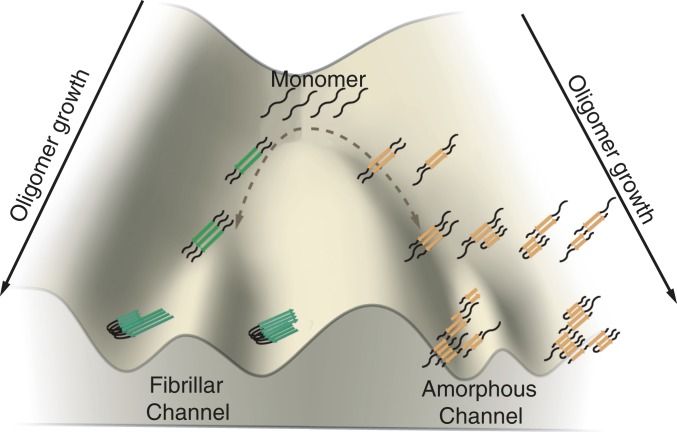
Developing kinetic models of protein aggregation and fibril growth is difficult due to the complexity arising from the diversity of species involved. Several different mechanisms of rearrangement are possible. Kinetic models of tau aggregation have been constructed by Klenerman, Knowles, and coworkers (29) that employ a minimal scheme where monomers can form type A oligomers that can lead to fibrils, while a distinct set of type B oligomers cannot form complete fibrils. Their work focused on a different form of the protein from the constructs studied here. Nevertheless, our present theoretical work harmonizes with the idea of the existence of two oligomerization channels: a prefibrillar channel which leads to amyloid fibrils and a nonfibrillar channel that leads to amorphous phases and nonfibrillar oligomers (Fig. 4). In the experimental work, fluorescence resonance energy transfer of labeled protein was used to infer the existence of two channels. As we have seen in the simulations, FRET efficiency distributions are predicted to be different in the two channels.
Fig. 4.
Schematic diagram of the growth of oligomers along the prefibrillar channel and the amorphous channel. The oligomers from the two channels are colored differently, and different polymorphs are also shown to illustrate the diverse species that could be formed in the two channels.
In our landscape analyses of the simulations, we not only observe an ordered growth of prefibrillar species along the fibril axis adopting a parallel-in-register form; we also notice the complicated patterns of oligomerization along the nonfibrillar channel. Since many segments in tau repeats are predicted to be amyloidogenic according to AWSEM-Amylometer calculations, these segments, when mixed together, have a propensity to associate together by stacking those segments in a larger variety of ways, which ultimately leads to disordered larger oligomers (17, 18). By such a branching process, oligomer growth will lead to a loosely packed but concatenated aggregation of molecules in a fluid phase. The structural heterogeneity within the amorphous oligomer channel might also arise from slow rearrangement of the specific monomeric contacts within the oligomers, which leads to still more complexity in modeling formation kinetics. Even in the prefibrillar channel which leads to ordered amyloids finally, however, amyloids may take on a range of polymorphic forms. We have previously discussed this end-stage polymorphism for A through a “ribbon folding” algorithm that approximates two-dimensional protein folding (30). Accurately predicting the kinetic details of the aggregation of tau isoforms is a challenge for the future.
Methods
A detailed description of the materials and methods is given in SI Appendix, SI Methods. Briefly, the simulations were carried out using the AWSEM force field for proteins in the LAMMPS open-source software package.
Data Availability.
The simulation and analysis codes underlying Figs. 1–4 are available online at https://github.com/chemlover/Tau_PNAS- (31).
Supplementary Material
Acknowledgments
This work was initially supported by Grant R01 GM44557 from the National Institute of General Medical Sciences. Additional support was also provided by the D. R. Bullard-Welch Chair at Rice University, Grant C-0016. We thank the Data Analysis and Visualization Cyberinfrastructure funded by NSF Grant OCI-0959097, as well the Center for Theoretical Biological Physics sponsored by NSF Grant PHY-1427654.
Footnotes
Competing interest statement: W.E. and P.G.W. coauthored a letter to PNAS in 2017.
Data deposition: The simulation and analysis codes underlying Figs. 1–4 are available online at https://github.com/chemlover/Tau_PNAS-.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921702117/-/DCSupplemental.
References
- 1.Wang Y., Mandelkow E., Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 22–35 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zheng W., Tsai M.-Y., Chen M., Wolynes P. G., Exploring the aggregation free energy landscape of the amyloid- protein (1–40). Proc. Natl. Acad. Sci. U.S.A. 113, 11835–11840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng W., Tsai M.-Y., Wolynes P. G., Comparing the aggregation free energy landscapes of amyloid beta (1–42) and amyloid beta (1–40). J. Am. Chem. Soc. 139, 16666–16676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M., Tsai M. Y., Zheng W., Wolynes P. G., The aggregation free energy landscapes of polyglutamine repeats. J. Am. Chem. Soc. 138, 15197–15203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen M., Wolynes P. G., Aggregation landscapes of Huntingtin exon 1 protein fragments and the critical repeat length for the onset of Huntington’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, 4406–4411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Espinoza M., De Silva R., Dickson D. W., Davies P., Differential incorporation of tau isoforms in Alzheimer’s disease. J. Alzheimer’s Dis. 14, 1–16 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iqbal K., Liu F., Gong C.-X., Grundke-Iqbal I., Tau in Alzheimer disease and related tauopathies. Curr. Alzheimer Res. 7, 656–664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buée L., Bussière T., Buée-Scherrer V., Delacourte A., Hof P. R., Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev. 33, 95–130 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Fitzpatrick A. W. P., et al. , Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcon B., et al. , Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prokopovich D. V., Whittaker J. W., Muthee M. M., Ahmed A., Larini L., Impact of phosphorylation and pseudophosphorylation on the early stages of aggregation of the microtubule-associated protein tau. J. Phys. Chem. B 121, 2095–2103 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Ambadipudi S., Biernat J., Riedel D., Mandelkow E., Zweckstetter M., Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein tau. Nat. Commun. 8, 275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morriss-Andrews A., Shea J. E., Computational studies of protein aggregation: Methods and applications. Annu. Rev. Phys. Chem. 66, 643–666 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Papoian G. A., Ulander J., Eastwood M. P., Luthey-Schulten Z., Wolynes P. G., Water in protein structure prediction. Proc. Natl. Acad. Sci. U.S.A. 101, 3352–3357 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng W., Schafer N. P., Wolynes P. G., Frustration in the energy landscapes of multidomain protein misfolding. Proc. Natl. Acad. Sci. U.S.A. 110, 1680–1685 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W., Schafer N. P., Wolynes P. G., Free energy landscapes for initiation and branching of protein aggregation. Proc. Natl. Acad. Sci. U.S.A. 110, 20515–20520 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M., Schafer N. P., Zheng W., Wolynes P. G., The associative memory, water mediated, structure and energy model (AWSEM)-Amylometer: Predicting amyloid propensity and fibril topology using an optimized folding landscape model. ACS Chem. Neurosci. 9, 1027–1039 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Reiss H., Bowles R. K., Some fundamental statistical mechanical relations concerning physical clusters of interest to nucleation theory. J. Chem. Phys. 111, 7501–7504 (1999). [Google Scholar]
- 21.Zhong Q., Congdon E. E., Nagaraja H. N., Kuret J., Tau isoform composition influences rate and extent of filament formation. J. Biol. Chem. 287, 20711–20719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjaergaard M., et al. , Oligomer diversity during the aggregation of the repeat region of tau. ACS Chem. Neurosci. 9, 3060–3071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alzheimer’s Association, Alzheimer’s disease facts and figures. Alzheimers Dementia 11, 332–384 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Lindwall G., Cole R. D., Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 259, 5301–5305 (1984). [PubMed] [Google Scholar]
- 25.Hanger D. P., Anderton B. H., Noble W., Tau phosphorylation: The therapeutic challenge for neurodegenerative disease. Trends Mol. Med. 15, 112–119 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Murray D. T., et al. , Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang A., et al. , A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37, e97452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monahan Z., et al. , Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36, 2951–2967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shammas S. L., et al. , A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat. Commun. 6, 7025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M., Schafer N. P., Wolynes P. G., Surveying the energy landscapes of A fibril polymorphism. J. Phys. Chem. B 122, 11414–11430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Chen M., Schafer N. P., Wolynes P. G.. The data of Tau paper on PNAS. Github. https://github.com/chemlover/Tau_PNAS-. Deposited 23 January 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The simulation and analysis codes underlying Figs. 1–4 are available online at https://github.com/chemlover/Tau_PNAS- (31).