Significance
Dietary fat circulates in the blood in triglyceride-rich lipoproteins (chylomicrons and very-low-density lipoproteins). These lipoproteins dock along capillary endothelial cells by binding to a functional unit composed of lipoprotein lipase (LPL) and GPIHBP1. This unit hydrolyzes triglycerides, releasing free fatty acids for use by parenchymal cells (e.g., myocytes or adipocytes). ANGPTL proteins 3 and 4 regulate triglyceride hydrolysis by catalyzing the unfolding of LPL. For more than a decade, these ANGPTLs were assumed to inhibit LPL by dissociating catalytically active LPL homodimers into inactive monomers. Our study shows that trapped LPL monomers are stable and that ANGPTL4 inhibits LPL by unfolding of the LPL monomers. Our findings change the paradigm for the regulation of triglyceride metabolism by the ANGPTL proteins.
Keywords: HDX-MS, intravascular lipolysis, lipoprotein lipase, GPIHBP1, surface plasmon resonance
Abstract
The binding of lipoprotein lipase (LPL) to GPIHBP1 focuses the intravascular hydrolysis of triglyceride-rich lipoproteins on the surface of capillary endothelial cells. This process provides essential lipid nutrients for vital tissues (e.g., heart, skeletal muscle, and adipose tissue). Deficiencies in either LPL or GPIHBP1 impair triglyceride hydrolysis, resulting in severe hypertriglyceridemia. The activity of LPL in tissues is regulated by angiopoietin-like proteins 3, 4, and 8 (ANGPTL). Dogma has held that these ANGPTLs inactivate LPL by converting LPL homodimers into monomers, rendering them highly susceptible to spontaneous unfolding and loss of enzymatic activity. Here, we show that binding of an LPL-specific monoclonal antibody (5D2) to the tryptophan-rich lipid-binding loop in the carboxyl terminus of LPL prevents homodimer formation and forces LPL into a monomeric state. Of note, 5D2-bound LPL monomers are as stable as LPL homodimers (i.e., they are not more prone to unfolding), but they remain highly susceptible to ANGPTL4-catalyzed unfolding and inactivation. Binding of GPIHBP1 to LPL alone or to 5D2-bound LPL counteracts ANGPTL4-mediated unfolding of LPL. In conclusion, ANGPTL4-mediated inactivation of LPL, accomplished by catalyzing the unfolding of LPL, does not require the conversion of LPL homodimers into monomers. Thus, our findings necessitate changes to long-standing dogma on mechanisms for LPL inactivation by ANGPTL proteins. At the same time, our findings align well with insights into LPL function from the recent crystal structure of the LPL•GPIHBP1 complex.
The lipolytic processing of triglyceride-rich lipoproteins (TRLs) by lipoprotein lipase (LPL) is the central event in plasma lipid metabolism, releasing fatty acids for uptake by vital tissues, including the heart, skeletal muscle, and adipose tissue (1). A complex between LPL and an endothelial membrane protein, glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1), is responsible for margination of TRLs in the capillary lumen and for the subsequent unloading of triglycerides by ester hydrolysis, releasing free fatty acids (2, 3). In contrast to free LPL, the hydrolase activity of the LPL•GPIHBP1 complex persists in the presence of physiological LPL inhibitors—angiopoietin-like proteins 3, 4, and 8 (ANGPTLs). Such selective inhibition serves to focus triglyceride hydrolysis at the surface of endothelial cells where it is needed for TRL processing (4–7). The physiological relevance of this pathway is set in stone by the severe hypertriglyceridemia that develops when the LPL•GPIHBP1 complex is absent or dysfunctional due to deleterious missense mutations, gene deletions, or inhibitory autoantibodies (8–14). Moreover, genome-wide association studies revealed a strong correlation between ANGPTL variants displaying attenuated LPL inhibition, lower plasma triglyceride levels, and reduced coronary artery disease (15–19). It is therefore evident that the interplay between LPL, GPIHBP1, and ANGPTL proteins 3, 4, and 8 plays central roles in plasma triglyceride metabolism.
We previously showed that the enzymatic activity of purified LPL preparations spontaneously loses activity due to an inherent instability of LPL’s catalytic α/β-hydrolase domain (20). Of note, we also found that ANGPTL4 and ANGPTL3 catalyze the irreversible unfolding of LPL’s hydrolase domain, resulting in a parallel loss of both triglyceride hydrolase activity and esterase activity (4). For decades, the functional unit of LPL was accepted to be a head-to-tail homodimer (21–28). In line with this paradigm, ANGPTL4 was thought to inactivate LPL by converting LPL homodimers into monomers, rendering them highly susceptible to unfolding and inactivation (26, 27, 29, 30). At first glance, the crystal structure of LPL seemed compatible with that view, inasmuch as LPL was arranged in a head-to-tail homodimer topology (31, 32). However, the interaction between the partner protomers involved only small reciprocal contacts, and these contacts occurred between the hydrophobic entrance to the catalytic pocket of one protomer and the tryptophan-rich lipid-binding motif in the other protomer, as illustrated in Fig. 1B (31, 32). Because the interacting regions are critical for LPL function, we suspected that the LPL dimers in the crystal structure were inactive and formed to minimize the entropic penalty of having hydrophobic loops exposed to an aqueous solvent. Commonly used conditions to preserve LPL enzymatic activity [i.e., high protein concentrations and high salt (33, 34)] would favor hydrophobic interactions and likely would promote the formation of LPL homodimers.
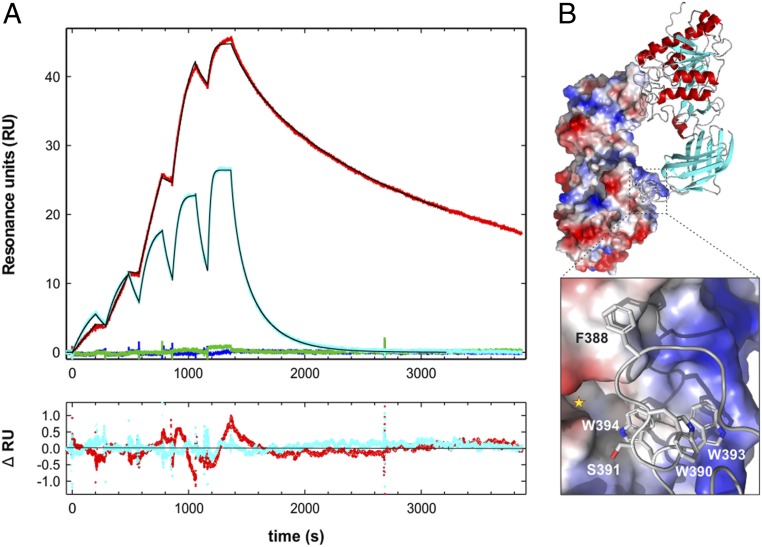
Fig. 1.
Epitope for mAb 5D2 on LPL defined by SPR. (A) Single-cycle SPR sensorgrams for the interaction between immobilized mAb 5D2 and various synthetic peptides corresponding to amino acid residues 383 to 396 of human LPL (KSDSYFSWSDWWSS). Red curve, the wild-type (wt) peptide sequence (1 to 16 nM); blue curve, Trp390→Ala (8 to 128 nM); cyan curve, Trp393→Ala (2 to 32 nM); green curve, Trp394→Ala (8 to 128 nM). Thin black lines represent fits to the data recorded for the wt and Trp393→Ala peptides. Residuals of these fits are shown at the bottom. (B) The structure of an LPL homodimer depicting the reciprocal interactions of two partner monomers, as revealed by X-ray crystallography (31). (Top) One LPL protomer in an electrostatic potential surface representation and the other in a cartoon representation. The hatched square highlights the location of one of the reciprocal LPL homodimer contacts. (Bottom) The corresponding close-up, depicting the involvement of residues 383 to 396 of human LPL in one of two low-energy models. The yellow asterisk marks the entrance to the catalytic site of the LPL protomer.
Recent studies implying that freshly secreted LPL is active as a monomer (35), combined with doubts about the physiologic relevance of the homodimer conformation in the crystal structure (31, 32, 36), prompted us to reconsider mechanisms by which ANGPTL4 inactivates LPL. We found one scenario attractive—that ANGPTLs directly catalyze the unfolding of LPL’s hydrolase domain irrespective of any dimer−monomer transition. Difficulties in isolating LPL monomers represented a roadblock to such studies. Based on the crystal structure of the LPL dimer, we hypothesized that shielding LPL’s lipid-binding loop from solvent exposure would disrupt homodimer formation—even in the setting of high LPL protein concentrations. In an attempt to drive LPL into the monomeric state, we took advantage of the LPL-specific monoclonal antibody (mAb) 5D2, which binds to the hydrophobic lipid-binding region of LPL with high affinity and thereby abolishes the binding of lipoproteins as well as hydrolysis of emulsified triglyceride substrates (37, 38).
With various biophysical measurements, we demonstrate that the Fab fragment of 5D2 does indeed disrupt LPL homodimers, resulting in homogenous preparations of LPL monomers. Further biophysical studies on these preparations reveal 1) that ANGPTL-catalyzed unfolding of LPL occurs independently of LPL homodimer–monomer transitions, 2) that ANGPTL4 acts directly to unfold LPL monomers, and 3) that these monomers do not exhibit increased conformational instability in the absence of ANGPTL4. Our studies showing that ANGPTL4 functions to unfold LPL monomers are entirely consistent with the recent proposal that LPL is active as a monomer (35, 36).
Results
Mapping Hotspot Residues for mAb 5D2 Binding to LPL.
The mouse mAb 5D2, which binds both bovine and human LPL, is widely used in immunoassays to quantify LPL protein mass (39). The 5D2 inhibits LPL-mediated triglyceride hydrolase activity against long-chain triacylglycerol emulsions but has little effect on LPL’s esterase activity (hydrolysis of soluble short-chain substrates) (21). Previous studies suggested that the epitope for 5D2 involved two linear segments within LPL’s C-terminal domain (37), including a tryptophan-rich motif required for lipid- and lipoprotein binding (38). In the crystal structure of LPL, the tryptophan-rich loop of one LPL molecule interacts in a reciprocal fashion with the catalytic pocket of a second LPL molecule, generating a head-to-tail LPL homodimer (31, 32). We suspected that the binding of 5D2 to the tryptophan-rich loop would disrupt the reciprocal LPL•LPL interactions and promote the formation of LPL monomers. To explore this possibility, we took advantage of surface plasmon resonance (SPR) studies to revisit the mAb 5D2 epitope, with the goal of mapping residues that are crucial for mAb 5D2 binding. We immobilized 5D2 on a CM5 sensor chip and measured the real-time binding kinetics of the synthetic tetradecamer peptide KSDSYFSWSDWWSS (human LPL residues 383 to 396; amino acid numbering in our studies begins with the first residue in the mature protein). As shown in Fig. 1A, the synthetic peptide bound 5D2 tightly with a dissociation constant (Kd) of 0.28 nM and a dissociation rate constant (koff) of 1.49 × 10−3 s−1 with a half-life (t1/2) of ∼7.8 min (Table 1).
Table 1.
Epitope Mapping of mAb 5D2 by SPR
| Residues 383 to 396 | kon, 106 M−1⋅s−1 | koff, 10−3 s−1 | Kd, nM | Stoichiometry |
| KSDSYFSWSDWWSS | 5.41 ± 0.001 | 1.49 ± 0.002 | 0.28 | 1.5 |
| KSDSAFSWSDWWSS | 2.34 ± 0.004 | 1.50 ± 0.003 | 0.67 | 1.5 |
| KSDSYASWSDWWSS | NA | NA | 0.76 × 103 | 1.3 |
| KSDSYFAWSDWWSS | 5.73 ± 0.013 | 7.56 ± 0.18 | 1.32 | 1.4 |
| KSDSYFSASDWWSS | NA | NA | 0.36 ×103 | 1.7 |
| KSDSYFSWADWWSS | 0.88 ± 0.004 | 12.86 ± 0.18 | 14.6 | 1.4 |
| KSDSYFSWSDAWSS | 3.38 ± 0.024 | 18.78 ± 0.14 | 14.6 | 1.4 |
| KSDSYFSWSDWASS | NA | NA | 2.9 × 103 | 1.7 |
| GFAIQWIRRVKAGE | NA | NA | 3.9 × 103 | 0.3 |
| CTD | 2.56 ± 0.007 | 0.015 ± 0.00002 | 6.0 × 10−3 | 1.2 |
Shown are the kinetic rate constants derived from single-cycle SPR with immobilized mAb 5D2 (2,300 resonance units ∼ 15.3 fmol/mm2) and LPL synthetic peptides (corresponding to residues 383 to 396 in human LPL) in solution (Fig. 1). The stoichiometry of mAb 5D2 binding represents the ratio between immobilized 5D2 and the calculated Rmax for analyte binding (femtomoles per square millimeter) by fitting the data to a 1:1 binding model. NA signifies that the determination of binding kinetics is not applicable because of very weak binding affinity; only the equilibrium dissociation constant (Kd) could be derived. To quantify the tight interaction between 5D2 and the CTD of human LPL (residues 313 to 448), we used long dissociation times (30,000 s or 500 min) to allow for sufficient curvature in the dissociation phase to calculate koff (1.5 × 10−5 s−1; t1/2 ∼ 770 min). Positions replaced by alanine are highlighted by a bold underlined A.
Next, using SPR and synthetic peptides containing single-site alanine replacements, we defined the hotspot residues for 5D2 binding. This screen identified three essential residues for 5D2 binding (Phe388, Trp390, and Trp394) and two additional residues (Ser391 and Trp393) with significant effects on the affinity of 5D2 binding (Table 1). Replacing Phe388, Trp390, or Trp394 with Ala resulted in a >1,000-fold increase in Kd. Each of those amino acids is buried in the LPL•LPL interface forming the reciprocal contacts that create the LPL homodimer in the crystal structure (31). These observations further support our suspicion that LPL would be unlikely to maintain a homodimeric structure in the presence of 5D2.
We were unable to confirm the importance of a second linear stretch of residues (LPL amino acids 398 to 410; GFAIQWIRRVKAGE) that was proposed to participate in 5D2 binding, albeit with low affinity (37, 40). However, we found that mAb 5D2 binds with even higher affinity to intact LPL than to the synthetic peptide KSDSYFSWSDWWSS, implying that the synthetic peptide does not fully recapitulate the entire epitope present in properly folded LPL (Table 1).
mAb 5D2 Binding Drives LPL into a Monomeric Conformation.
Having shown that the “hotspot residues” in the 5D2 epitope participate directly in the binding interface of LPL homodimers, we next investigated the impact of 5D2 on homodimer formation. For these studies, we generated Fab fragments of both mAb 5D2 (37, 40) and the human GPIHBP1-specific mAbs RF4 and RE3 (41, 42). The epitope for RF4 is centered on Arg53−Leu54 downstream from GPIHBP1’s disordered acidic domain (43), whereas RE3 binds GPIHBP1’s Ly6/uPAR domain and prevents GPIHBP1 from binding LPL (41). We demonstrated, using native polyacrylamide gel electrophoresis (PAGE), that it is possible to generate trimolecular complexes between LPL, GPIHBP1, and either Fab-5D2 or Fab-RF4 (Fig. 2, lanes 1–4). The hydrodynamic volumes of the complexes and the very low isoelectric point of GPIHBP1 determine the migration of these complexes in native gels. Since the electrophoretic mobilities of LPL•GPIHBP1, Fab-RF4•GPIHBP1, and Fab-RE3•GPIHBP1 are similar (Fig. 2C), it is likely that LPL•GPIHBP1 represents a bimolecular complex with a 1:1 stoichiometry rather than a 2:2 stoichiometry. Likewise, the GPIHBP1•LPL•Fab-5D2 complex appeared to represent a 1:1:1 stoichiometry as its migration resembles that of the GPIHBP1•Fab-RF4•Fab-RE3 complex (compare lanes 5 and 6 in Fig. 2C). The low ionic strength that follows the application of the electric field would likely disfavor hydrophobic interactions and promote dissociation of LPL homodimers. For that reason, we also performed orthogonal biophysical methods at normal ionic strength to confirm the binding stoichiometry of these LPL complexes (discussed in GPIHBP1•LPL•Fab Complexes Analyzed by SEC-MALS and Size and Shape of the GPIHBP1•LPL•Fab-5D2 Complex Assessed by SEC-SAXS).
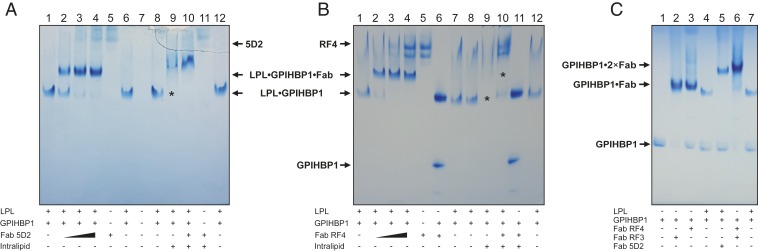
Fig. 2.
Native PAGE of human LPL complexed with GPIHBP1 and various Fab fragments. (A) Analysis of human LPL (1 µM) in the presence of equimolar amounts of GPIHBP1 (lanes 1−4, 6, 8, 9, 10, and 12) and increasing amounts of Fab-5D2 (0.7 µM in lane 2; 1.4 µM in lane 3; 2.1 µM in lanes 4 and 10). The experimental control is 2.1 µM Fab-5D2 alone (lanes 5 and 11). A lipid emulsion of mixed long-chain triacylglycerols (0.2% Intralipid) was added to lanes 9 to 11 immediately before electrophoresis. The lipid emulsion interferes with the migration of GPIHBP1•LPL (lane 9, asterisk) but not GPIHBP1•LPL•Fab-5D2 (lane 10). (B) Analysis of human LPL (1.5 µM) in the presence of 1 µM GPIHBP1 (lanes 1−4, 7−10, and 12) with increasing concentrations of the Fab fragment from the GPIHBP1-specific mAb RF4 (0.8 µM in lane 2; 1.6 µM in lane 3; 2.4 µM in lanes 4 and 10). The molar excess of LPL minimizes formation of Fab-RF4•GPIHBP1 complexes, simplifying migration patterns. The controls without added LPL were 2.4 µM Fab-RF4 (lane 5) and 2.4 µM Fab-RF4 and 5 µM GPIHBP1 (lanes 6 and 11); 0.2% Intralipid was added to lanes 9–11 immediately before electrophoresis. The electrophoretic mobility of the GPIHBP1•LPL complex and the GPIHBP1•LPL•Fab-RF4 complex were impaired by the lipid emulsion (the absence of a complex is marked by asterisks). (C) Migration of GPIHBP1 alone (lane 1) and complexed to Fab-RE3 (lane 2), Fab-RF4 (lane 3), or both (lane 6). For comparison, the electrophoretic migrations of GPIHBP1•LPL (lanes 4 and 7) and GPIHBP1•LPL•Fab-5D2 (lane 5) are shown.
Next, we tested the ability of LPL to enter the native gel in the presence of triglyceride emulsion particles (0.2% [vol/vol] Intralipid), which are known to bind to the tryptophan-rich loop in the carboxyl-terminal domain (CTD) of LPL. Because they bind to LPL, the triglyceride emulsion particles prevent LPL from migrating into the native gel (compare lanes 8 and 9 in Fig. 2 A and B). The binding of Fab-5D2 to LPL prevented the interactions between LPL’s tryptophan-rich CTD loop and the triglyceride emulsion particles, permitting Fab-5D2•LPL complexes to enter the gel (Fig. 2A, lane 10). In contrast, Fab-RF4 did not prevent the sequestration of LPL by triglyceride emulsion particles (Fig. 2B, lane 10). These studies demonstrate that other hydrophobic regions within LPL, for example LPL’s catalytic pocket and lid, do not mediate strong interactions with the triglyceride emulsion particles. Furthermore, our studies show that the tryptophan-rich loop in LPL is accessible for binding by triglyceride emulsion particles. The reciprocal interactions between two partner LPL protomers are almost certainly transient (26), allowing for the tryptophan-rich loop to bind to the triglyceride emulsion particles.
GPIHBP1•LPL•Fab Complexes Analyzed by SEC-MALS.
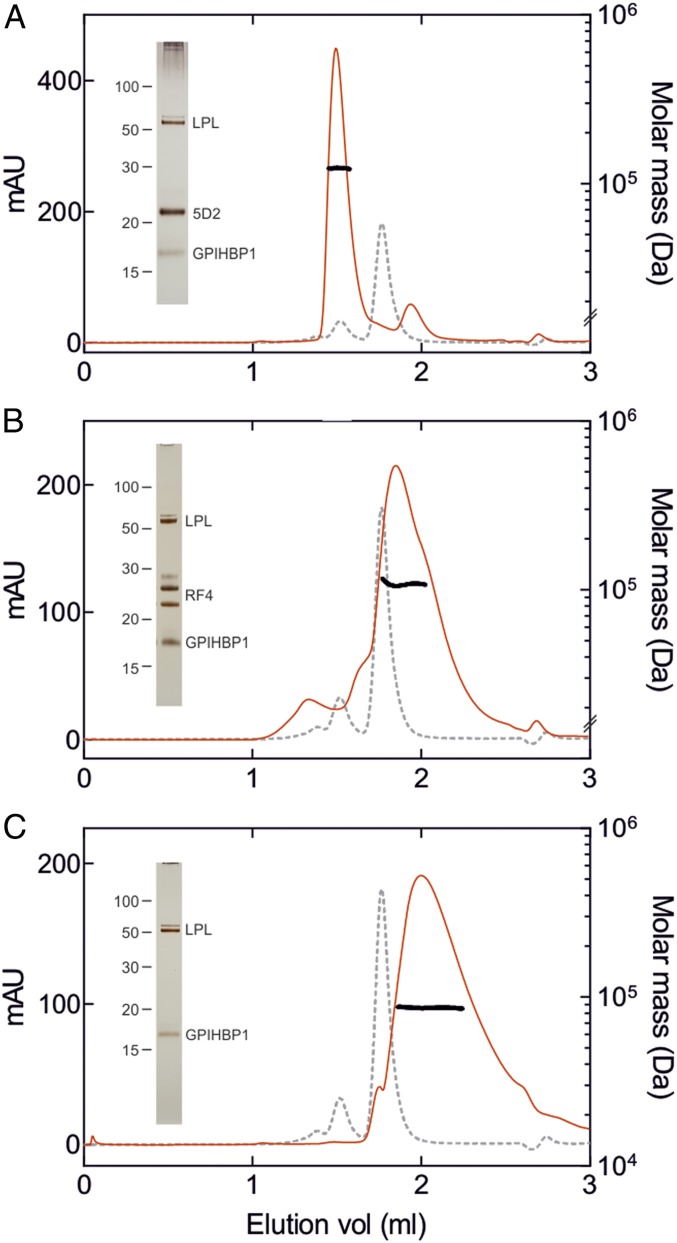
To assess the stoichiometry of GPIHBP1•LPL•Fab-5D2 with orthogonal methods, we examined complexes by size-exclusion chromatography with a Superdex200 Increase column, followed by in-line mass detection by multiangle light scattering (SEC-MALS) or small-angle X-ray scattering (SEC-SAXS). Analyses by sodium dodecyl sulfate (SDS)/PAGE of the peak fractions for bovine LPL (bLPL) (Fig. 3) and human LPL (hLPL) (SI Appendix, Fig. S1) reveal that they contain LPL, GPIHBP1, and Fab fragments, validating that they represent trimolecular complexes. The molecular mass, as judged by MALS, was between 100 and 140 kDa for all samples (Table 2). These data provide strong support for a 1:1:1 stoichiometry of trimolecular complexes with Fab-5D2 or Fab-RF4, despite their distinct elution volumes from the 2.9-mL Superdex200 Increase column (1.5 mL for the Fab-5D2 complex; 1.8 mL for Fab-RF4 complex). It is not clear why the LPL dimer interface appears compromised in the GPIHBP1•LPL•Fab-RF4 complex, but the late elution and peak broadening of that particular complex from the SEC column may provide some insights. Transient solvent exposures of LPL’s lipid-binding loop could allow nonspecific column interactions, delaying the elution of the complex, weakening the LPL•LPL dimer interface, and promoting the dissociation of LPL homodimers into protomers. In hindsight, these sorts of LPL•column interactions could help to explain the 1:1 stoichiometry that we and others observed for the LPL•GPIHBP1 complex by SEC-MALS (Fig. 3 and SI Appendix, Fig. S1) but not by batch SAXS analyses (31, 32).
Fig. 3.
SEC of bLPL complexes with MALS detection. (A) Elution of GPIHBP1•LPL•Fab-5D2 after SEC on a Superdex200 Increase column in 10 mM Tris, 150 mM NaCl, 0.05% (wt/vol) CHAPS, 10% (vol/vol) glycerol, and 0.05% (wt/vol) NaN3, pH 7.2. (B) Elution of GPIHBP1•LPL•Fab-RF4. (C) Elution of GPIHBP1•LPL. The red lines show the absorbance profiles at 280 nm for the samples under investigation; the light gray hatched lines show the BSA control (monomer, dimer, and trimer). The molecular masses determined using refractive indices as well as scattering profiles are shown as solid black lines. The presence of all of the various proteins (LPL, GPIHBP1, and Fabs) in the peak fractions was verified by SDS/PAGE of reduced and alkylated samples, followed by silver staining (insets). The presence of two bands in reduced and alkylated Fab-RF4 is confirmed by ESI-MS of reduced Fab-RF4 (24,189.4 Da and 25,732.0 Da, respectively) and unreduced Fab-RF4 having a mass of 49,915.8 Da.
Table 2.
Mass determinations from SEC-MALS and SEC-SAXS
| Protein samples | ElutionSEC, mL | MassMALS, kDa | MassSAXS/DAM, kDa | MassSAXS/Guinier, kDa | MassMS, Da |
| bLPL•GPIHBP1• Fab-5D2 | 1.49 | 123.0 ± 0.4 | — | — | 117,341 |
| bLPL•GPIHBP1• Fab-RF4 | 1.85 | 108.0 ± 0.1 | — | — | 119,245 |
| bLPL•GPIHBP1 | 2.00 | 86.4 ± 0.8 | — | — | 69,184 |
| hLPL•GPIHBP1• Fab-5D2 | 1.52 | 148.5 ± 34 | 117 ± 10 | 100 ± 10 | 118,445 |
| hLPL•GPIHBP1• Fab-RF4 | 1.67 | 141.3 ± 1.1 | — | — | 120,349 |
| hLPL•GPIHBP1 | 1.97 | 81.9 ± 1.5 | — | — | 70,288 |
Molecular masses of the individual components forming the complex were determined by ESI-MS and are shown for a 1:1 GPIHBP1•LPL complex and 1:1:1 GPIHBP1•LPL•Fab complex. Protein concentrations for I0 normalization of SAXS data were determined from UV peak integration, using the ε0.1% (280 nm) calculated from the sequence with the ProtParam tool (https://web.expasy.org/protparam/). SI Appendix, Table S1 contains detailed information on size and mass determinations based on SAXS data.
Size and Shape of the GPIHBP1•LPL•Fab-5D2 Complex Assessed by SEC-SAXS.
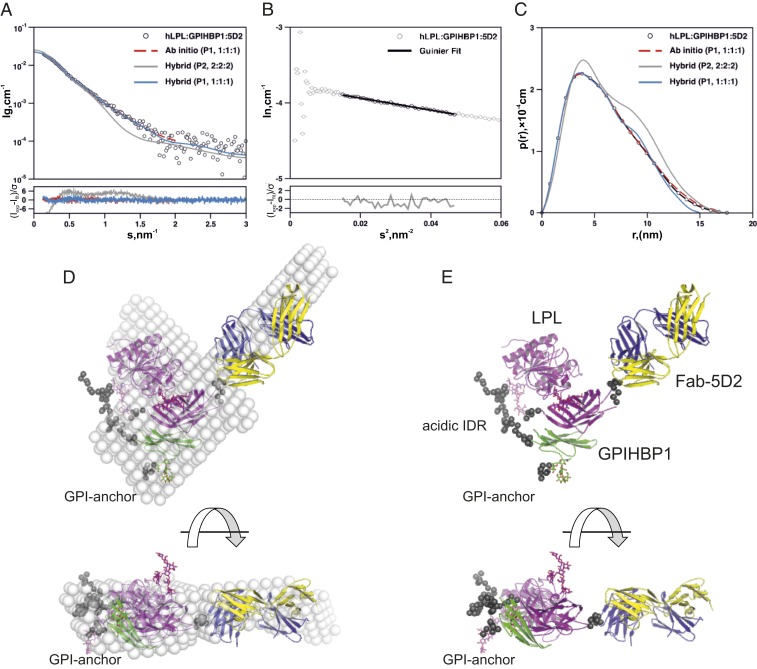
By SEC-SAXS, the scattering properties of the GPIHBP1•LPL•Fab-5D2 complex were also consistent with a trimolecular assembly with 1:1:1 stoichiometry (Table 2 and SI Appendix, Table S1). The size and shape parameters derived from the SEC-SAXS data were incompatible with a 2:2:2 complex. Ab initio shape models were reconstructed from the SEC-SAXS data, revealing an elongated particle volume (Fig. 4) that accommodated a 1:1:1 trimolecular complex but not larger assemblies. Hybrid rigid body modeling based on the available crystal structures (and distance constraints between the tryptophan-rich motif of LPL and Fab-5D2) also yielded 1:1:1 trimolecular models that fit the experimental data (χ2 < 1.1) and closely resembled the ab initio shape. We attempted to generate models with 2:2:2 stoichiometry, but none fit the experimental data for GPIHBP1•LPL•Fab-5D2 (χ2 > 9.5; gray lines in Fig. 4).
Fig. 4.
Ab initio and hybrid molecular modeling of the GPIHBP1•LPL•Fab-5D2 complex. (A) SAXS data recorded in-line for a GPIHBP1•LPL•Fab-5D2 complex eluting from a Superdex200 Increase column are shown as open circles. Also shown are the fit of an ab initio model reconstructed using DAMMIF with P1 symmetry (broken red line), the fit to a P1 hybrid rigid body model reconstructed using CORAL (solid blue line; χ2 < 1.1), and the fit to a P2 model (light gray line; χ2 > 9.5). Residuals plot corresponding to the model fits is shown at the bottom of the panel. (B) A Guinier plot of the SAXS data [ln I(s) vs. s2], where the linearity signifies that the sample is monodisperse without aggregation (correlation = −0.99 with residuals for the fit shown at the bottom of the panel ). (C) Real-space distance distribution of the SAXS data for the GPIHBP1•LPL•Fab-5D2 complex and that of the models. (D) Ab initio surface model of the scattering data and the superimposed P1 rigid body model of the 1:1:1 GPIHBP1•LPL•Fab-5D2 complex. (E) P1 hybrid rigid body model in a cartoon representation; LPL in purple, GPIHBP1 in green, and Fab-5D2 in yellow and blue. The position of the GPI anchor is indicated. The intrinsically disordered acidic region (IDR) of GPIHBP1 is represented by gray beads. The disordered C-terminal peptide linking GPIHBP1 to the GPI-anchor is also shown as beads, as is the tryptophan-rich lipid-binding region of LPL.
Dynamics of LPL in Different Complexes.
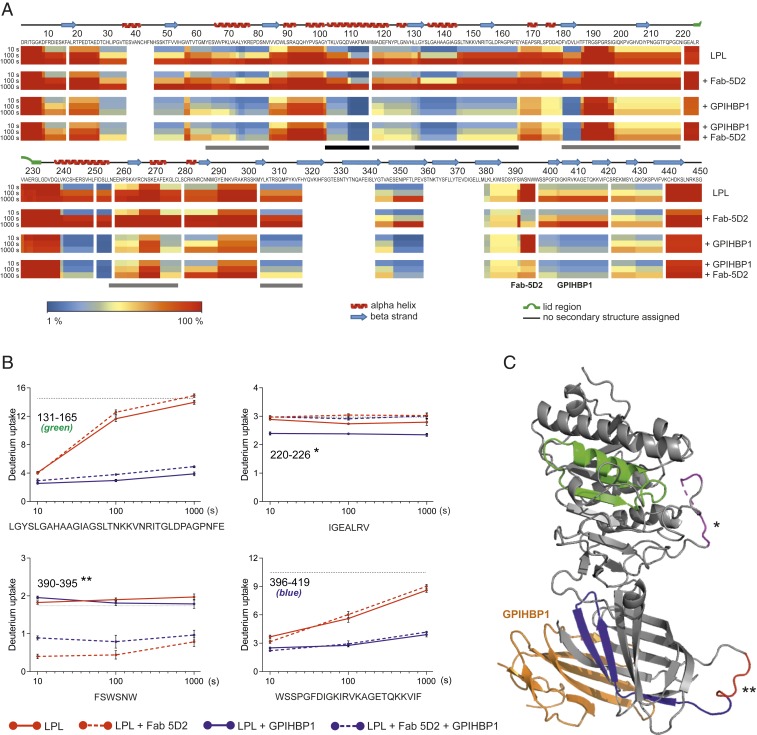
The instability of LPL’s catalytic activity is often ascribed to homodimer dissociation, followed by rapid and irreversible unfolding/aggregation of the protomers (26, 27). According to that view, the LPL monomers formed in the presence of 5D2 should be unstable. To explore this possibility, we used a continuous hydrogen–deuterium labeling protocol to assess the protein dynamics of LPL—both alone and when complexed to Fab-5D2 and/or GPIHBP1. In these studies, we used mass spectrometry to assess the time-dependent exchange of protein backbone hydrogens with deuterium in the presence of deuterium oxide. We examined LPL complexes formed by incubating high concentrations of LPL (10 µM) with equimolar amounts of Fab-5D2 and/or GPIHBP1. Under these conditions, both LPL alone and LPL•GPIHBP1 complexes form LPL homodimers, whereas GPIHBP1•LPL•Fab-5D2 is a 1:1:1 trimolecular complex. After in-line pepsin cleavage of the quenched samples, we recovered 93 unique LPL peptic peptides corresponding to a sequence coverage of 88.9% (SI Appendix, Fig. S2). The progressive uptake of deuterium into 34 individual peptic peptides is shown in SI Appendix, Fig. S3, relative to a 100% deuterium control. From these data, we constructed heat maps depicting LPL protein dynamics when bound to Fab-5D2 and/or GPIHBP1 (Fig. 5A). The binding of Fab-5D2 to LPL markedly reduced deuterium incorporation in residues 390 to 395 (Fig. 5 and SI Appendix, Figs. S3 and S4). Those residues correspond to the 5D2 epitope that we had identified by SPR (Fig. 1). However, the important observation is that there was no significant alteration in protein dynamics in LPL’s α/β-hydrolase domain. Had the 5D2-induced dissociation of homodimers into monomers triggered unfolding of LPL, we would have observed a substantial increase in deuterium uptake in peptides spanning LPL’s α/β-hydrolase domain. We did observe slightly increased amounts of deuterium uptake in LPL bound to Fab-5D2, for example in residues 196 to 219 (SI Appendix, Fig. S4), but the absence of major changes in deuterium uptake indicated that the stability of LPL’s α/β-hydrolase domain is largely independent of transitions from LPL homodimers to Fab-5D2−trapped monomers. Interestingly, disrupting LPL’s dimer interface by the binding of Fab-5D2 had little effect on deuterium uptake in the lid region of LPL (residues 220 to 226 and 227 to 238) (Fig. 5 and SI Appendix, Figs. S3 and S4). This finding indicates that the lid region remains solvent-exposed when Fab-5D2 disrupts the dimer interface or when GPIHBP1 stabilizes LPL (i.e., the lid does not fold back and form a tight complex with the entrance to the catalytic site).
Fig. 5.
Changes in LPL dynamics on binding Fab-5D2 and/or GPIHBP1. (A) Uptake of deuterium into 10 µM bovine LPL alone or when complexed to Fab-5D2 and/or GPIHBP1 was measured 10, 100, and 1,000 s after incubation in deuterium oxide at 25 °C. Lowering pH and temperature quenched deuterium exchange. The deuterium content in 93 individual peptic peptides, representing 88.9% sequence coverage of LPL (SI Appendix, Fig. S2), was determined by MS and is shown as heat maps relative to a 100% deuterium control. Regions in blue have low deuterium uptake, whereas regions in red have high uptake. Secondary structure assignments (shown above the primary sequence) are based on the crystal structure of human LPL (31). Binding sites deduced for Fab-5D2 and GPIHBP1 are indicated below the heat map. Certain areas in LPL exhibit peak broadening or bimodal isotope envelopes (44), demonstrating coexisting conformations of folded and unfolded LPL, and these are highlighted by gray and black bars, respectively. (B) Time-dependent deuterium uptake in four different peptic peptides representing the catalytic triad (residues 131 to 165, green), the lid region (residues 220 to 226, *), the 5D2 epitope (residues 390 to 395, **), and GPIHBP1 binding site (residues 396 to 419, blue). Deuterium uptake plots are shown for LPL (solid red lines), LPL•GPIHBP1 (solid blue lines), LPL•Fab-5D2 (broken red lines), and LPL•Fab-5D2•GPIHBP1 (broken blue lines). The deuterium content represents the average mass of the isotope envelopes and is the mean of three replicates. (C) Location of the catalytic triad, the lid region, the 5D2 epitope, and the GPIHBP1 binding site in the crystal structure of the human LPL•GPIHBP1 complex (GPIHBP1 in orange).
As noted earlier (20), the appearance of bimodal isotope envelopes in peptic peptides from LPLs α/β-hydrolase domain (e.g., peptides 131 to 165) provides an unambiguous signature of protein unfolding (Fig. 5 and SI Appendix, Fig. S5) (4, 20, 44). Binding of GPIHBP1, but not Fab-5D2, abrogates the unfolding of LPL’s α/β-hydrolase domain, manifested by disappearance of the bimodal isotope envelopes (SI Appendix, Fig. S5).
ANGPTL4-Catalyzed Inactivation Is via Direct Unfolding of LPL Monomers.
Unfolding of LPL’s α/β-hydrolase domain is central to regulation of LPL catalytic activity. Earlier hydrogen–deuterium exchange mass spectrometry (HDX-MS) studies revealed that two physiologic inhibitors of LPL, ANGPTL3 and ANGPTL4, function by catalyzing the unfolding of large portions of LPL’s hydrolase domain (4). In the presence of these inhibitors, the N-terminal hydrolase domain unfolds, resulting in the appearance of bimodal isotope envelopes in peptic peptides spanning LPL’s hydrolase domain (Fig. 5A). The unfolding of LPL’s hydrolase domain is mirrored by the loss of triglyceride hydrolase activity (4).
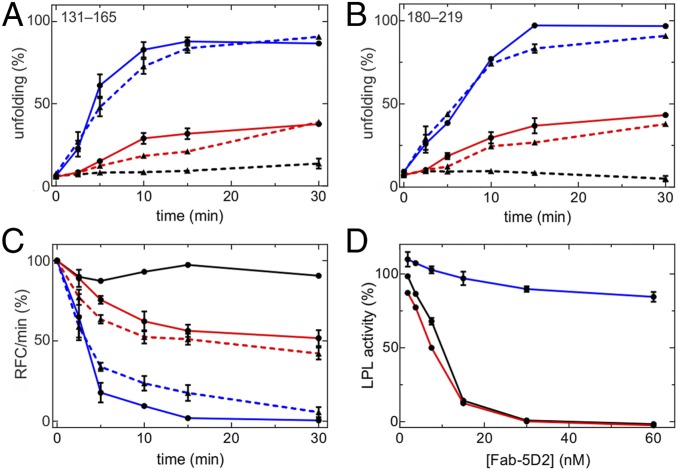
In the current studies, we asked whether LPL monomers (trapped in the monomer conformation by Fab-5D2) are susceptible to ANGPTL4-catalyzed unfolding. To address this issue, we used pulse-labeling HDX-MS studies to assess bimodal isotope envelopes in peptic peptides spanning LPL residues 131 to 165 (containing a large portion of LPL’s catalytic pocket) and residues 181 to 219. In this experiment, we incubated 10 µM LPL alone or in the presence of 1 µM ANGPTL4, 12 µM Fab-5D2, or 20 µM GPIHBP1 in the following combinations: LPL + ANGPTL4, LPL + Fab-5D2, LPL + Fab-5D2 + ANGPTL4, and LPL + Fab-5D2 + GPIHBP1 + ANGPTL4 in protiated solvents at 25 °C. For each combination, we collected aliquots at different time points and recorded snapshots of the average LPL unfolding by measuring deuterium uptake with 10-s pulse labeling in deuterium oxide (SI Appendix, Fig. S6). These studies revealed that the binding of Fab-5D2 to LPL did not alter the stability of the α/β-hydrolase domain, as judged by similar bimodal isotope envelopes for peptides derived from LPL alone and LPL•Fab-5D2 (Fig. 6 A and B). Moreover, we found that the kinetics of LPL unfolding by substoichiometric amounts of ANGPTL4 were comparable for LPL alone and LPL•Fab-5D2 complexes. In contrast to the situation with LPL alone and LPL•Fab-5D2, the LPL•Fab-5D2•GPIHBP1 complex was largely insensitive to ANGPTL4-catalyzed unfolding, underscoring the ability of GPIHBP1 to preserve LPL structure—even in the presence of ANGPTL4 (Fig. 6 A and B).
Fig. 6.
Time-dependent inactivation of LPL by unfolding in the presence of Fab-5D2. (A and B) Unfolding of LPL’s α/β-hydrolase domain as monitored by the emergence of bimodality in the isotope envelopes for peptides 131 to 165 and 181 to 219. In brief, 10 µM LPL was incubated alone (solid red line) or in the presence of 12 µM Fab-5D2 (broken red line), 1 µM ANGPTL4 (solid blue line), 12 µM Fab-5D2 and 1 µM ANGPTL4 (broken blue line), or with 12 µM Fab-5D2, 1 µM ANGPTL4, and 20 µM GPIHBP1 (broken black line) at 25 °C for 2.5 to 30 min. The conformational state of LPL was probed by dilution in deuterium oxide-containing solvents for 10 s followed by quenching of hydrogen–deuterium exchange by lowering the pH and temperature. Isotope envelopes corresponding to residues 131 to 165 are shown in SI Appendix, Fig. S6. (C) Time-dependent inactivation of LPL’s esterase activity with a soluble short-chain substrate (DGGR). The preincubation conditions and labeling are identical to those outlined in A and B, except that the solid black line represents LPL with GPIHBP1. (D) Fab-5D2–mediated inhibition of triglyceride hydrolase activity of 15 nM LPL (red line) or 15 nM LPL with 150 nM GPIHBP1 (black line) with Intralipid emulsion or 15 nM LPL (blue line) with a DGGR substrate. In all panels, the mean and SEM are shown for three to five replicates. RFC, relative fluorescence counts.
Because 5D2 binding shields LPL’s tryptophan-rich lipid-binding motif, we were unable to measure the catalytic activity of LPL•Fab-5D2 complexes in assays using long-chain triacylglycerol emulsions (Fig. 6D). Therefore, in order to examine the impact of ANGPTL4 on the catalytic activity of Fab-5D2–bound LPL, we measured lipase activity with a short-chain esterase substrate, 1,2-di-O-lauryl-rac-glycero-3-glutaric acid 6′-methylresofurin ester (DGGR). Fab-5D2−bound LPL remained active in the DGGR-based assay (Fig. 6D). The time-dependent loss of LPL’s enzymatic activity in this assay mirrored the unfolding of LPL’s hydrolase domain (as judged by pulse-labeled HDX-MS) whether occurring spontaneously or catalyzed by ANGPTL4 (Fig. 6C).
Discussion
The biological importance of ANGPTLs in regulating plasma triglyceride homeostasis by inhibiting LPL activity has been firmly established by human and mouse genetics (15–19) and by pharmacological studies (7, 45–47), but the molecular mechanism for LPL inactivation has remained controversial. One view holds that ANGTPL4 is a reversible and noncompetitive LPL inhibitor with an inhibition constant (Ki) of 0.9 to 1.7 µM (48, 49), but that view relies on studies performed in the presence of deoxycholate (a detergent that stabilizes LPL) (4, 50). Moreover, the low inhibitory efficacy of ANGPTL4 in those studies is difficult to reconcile with the fact that low nanomolar concentrations of ANGPTL4 readily inhibit triglyceride hydrolysis by low nanomolar concentrations of LPL (4–6, 29, 30, 43, 51). An alternative model proposed by Olivecrona and coworkers (29) was that ANGPTL4 catalyzes an irreversible inhibition of LPL by a mechanism they coined the “molecular unfolding chaperone” effect. That view aligns well with the fact that substoichiometric amounts of ANGPTL4 are sufficient to unfold LPL and permanently abolish LPL activity (4, 29). LPL complexed to GPIHBP1 is refractory to ANGPTL4-mediated inhibition (5), but once inhibited by ANGPTL4 subsequent addition of GPIHBP1 could not resurrect LPL activity, as would be expected if the inhibition was reversible (4). The observation by Mysling et al. (20) that spontaneous inactivation of LPL is due to unfolding of LPLs α/β-hydrolase domain naturally raised the possibility that the irreversible inhibition of LPL by ANGPTL4 might also be due to LPL unfolding (20). Indeed, subsequent studies established that ANGTPL4 catalyzes the unfolding of LPLs α/β-hydrolase domain (4). These observations align well with the “molecular unfolding chaperone” mechanism proposed by Olivecrona and coworkers (29).
While it is clear that ANGPTL4 unfolds LPL, the underlying molecular mechanism remains poorly defined. It is commonly assumed that ANGPTLs catalyze the dissociation of LPL homodimers and that the resulting protomers are intrinsically unstable and highly susceptible to inactivation (29, 52). However, the recently solved crystal structures of LPL (31, 32) raised doubts about the physiologic relevance of LPL homodimers (31, 35), necessitating greater scrutiny about how ANGPTL4 inactivates LPL. While the crystal structure of LPL revealed the predicted head-to-tail homodimer conformation, the contact areas between the protomers were small, raising doubts about the stability of homodimers under physiologic conditions (31, 32). Indeed, purified LPL homodimers have been shown to undergo rapid protomer exchange in vitro (26). Prompted by the small interface in the LPL homodimer, we designed experiments to trap protomers by disrupting the dimer interface via binding to Fab-5D2. We verified 5D2-mediated disruption of LPL homodimers with both SEC-MALS and SEC-SAXS at high protein concentrations and normal ion strengths (Figs. 3 and 4 and Table 2).
The ability of Fab-5D2 to disrupt LPL homodimer formation provided us with a unique opportunity to define the intrinsic stability of LPL protomers and assess their susceptibility to ANGTPL-catalyzed unfolding. Clarification of these issues is central to the initial delineation of the modus operandi for ANGPTL4-catalyzed inhibition of LPL. Taking advantage of continuous deuterium labeling and HDX-MS, we showed that free LPL and LPL•Fab-5D2 complexes exhibit similar deuterium uptake, reflecting comparable conformational flexibilities (Fig. 5). Adding GPIHBP1 stabilized LPLs α/β-hydrolase domain, exactly as described previously (20), and occurred irrespective of Fab-5D2 binding (Fig. 5 and SI Appendix, Fig. S4). Pulse-labeled HDX-MS revealed that LPL and LPL•Fab-5D2 unfold spontaneously with similar kinetics, arguing that trapping LPL in a monomeric state does not make it more unstable (Fig. 6). We estimate the t1/2 for unfolding of the trapped LPL monomer to be 45 to 60 min (Fig. 6), which aligns well with the t1/2 of 53 min estimated by compartmental modeling of the decay in LPL activity (26). Our observations are therefore not consistent with the notion that the dissociation of LPL dimers results in protomers that are intrinsically unstable, which is one of the key assumptions underlying the prevailing model for how ANGTPL4 inactivates LPL. We found that LPL monomers are stable and that ANGPTL4 unfolds free LPL and LPL•Fab-5D2 complexes with comparable efficiency (Fig. 6). Combined, our results favor a model in which ANGPTL4 acts by directly targeting the LPL monomers, thus catalyzing their irreversible inactivation by unfolding.
The finding that ANGPTL4 catalyzes the unfolding of LPL monomers sets the stage for elucidating the molecular mechanism by which ANGPTL4 binds LPL and initiates the chain of events leading to the irreversible unfolding of the α/β-hydrolase domain. Solving this conundrum is far from trivial as the mere binding of ANGPTL4 leads inexorably to an allosteric unfolding in LPL. Should HDX-MS experiments be used to map the binding site for ANGPTL4 on LPL it will be essential to differentiate between correlated and uncorrelated exchange mechanisms (4, 20, 44, 53). A recent HDX-MS–based study proposed that ANGPTL4 interacts with the lid (226 to 238) and a nearby α-helix (89 to 102) in LPL (49), but that finding is subject to important caveats. Primarily, the study provided no information on the presence of bimodal isotopic envelopes appearing as a consequence of ANGPTL-induced unfolding of LPL (i.e., coexisting LPL conformations). Second, the concentration of ANGPTL4 used in those studies would have, at least in our hands, resulted in irreversible inhibition of LPL before the first time point. Third, there was no time dependence in deuterium uptake within the putative ANGPTL4 interaction sites, which is surprising given the high Ki that was reported (49). Similar caveats also confound the findings in a second HDX-MS–based study (54), which concludes that ANGPTL4 binds to another region in LPL that harbors the catalytic site (i.e., residues 130 to 162). Of major concern, this particular region exhibits a pronounced bimodality in all our HDX-MS studies signifying that it undergoes unfolding upon ANGPTL-catalyzed LPL inactivation in the absence of GPIHBP1. In our opinion, more studies are needed to map the initial ANGPTL4 encounter site on LPL.
Recently, Beigneux and coworkers (35, 36) challenged the dogma holding that the catalytic activity of LPL was confined to homodimers. This shift in paradigm has a number of important functional implications. First, the role of LMF-1, an endoplasmic reticulum protein required for LPL secretion, may need revision. LMF-1 has been presumed to act by promoting the formation of catalytically active secretion-competent LPL homodimers (55–57), but the insights suggesting that LPL is largely in the form of monomers raises the possibility that LMF-1 simply serves as a chaperone to ensure proper folding of LPL monomers. Recently, the tryptophan-rich lipid binding region in LPL was proposed to play an essential role in trans-Golgi sorting and secretion of LPL by interacting with sphingomyelin-rich microdomains of the Golgi membrane (58). Presumably, such interactions would not be possible with the LPL homodimers documented in the crystal structure. Second, our studies indicate that ANGPTL4, a crucial physiologic regulator of LPL activity, serves to catalyze the unfolding LPL monomers rather than serving to dissociate catalytically active LPL homodimers into intrinsically unstable LPL monomers, as has been assumed (29, 52).
Materials and Methods
Chemicals and Purified Proteins.
bLPL was purified from fresh bovine milk (34). Recombinant hLPL was purified from the media of Chinese hamster ovary cells stably transfected with LPL and LMF1 (31). The CTD313−448 of human LPL and a soluble version of human GPIHBP11–131/R38G were expressed in Drosophila S2 cells and purified as described (20). A truncated version of ANGPTL41–159, representing the coiled-coil domain, was expressed in Escherichia coli (59). SpeB from Streptococcus pyogenes, expressed recombinantly in E. coli, was from Genovis (Sweden).
Generation of Fab Fragments.
To generate monovalent Fab fragments of mAbs 5D2, RF4, and RE3 we used the SpeB enzyme from S. pyogenes. Purified monoclonal antibodies (3 to 4 mg) were incubated with 2,000 units SpeB enzyme for 90 min at 37 °C in 10 mM phosphate buffer containing 150 mM NaCl and 40 mM l-cysteine, pH 7.2. Fab fragments were initially purified on the CaptureSelect resin LC-kappa (Thermo Scientific). Subsequent size-exclusion chromatography on a Superdex S200 yielded pure Fab fragments (removing trace amounts of intact IgG).
Native Gel PAGE.
Native polyacrylamide gels resolve bi- and trimolecular complexes of hLPL, GPIHBP1, and Fab fragments of 5D2, RF4, and RE3 (due to changes in their electrophoretic mobility at pH 8.4) (43). In brief, sample mixtures were loaded on 4 to 16% native polyacrylamide gels (Novex, Thermo Fisher) at 1 to 2 µM (well above the Kd of the binding partners) and then subjected to a field gradient of 100 V for 10 min, 200 V for 30 min, and 300 V for 20 min at 4 °C in a Tris-glycine buffer (pH 8.4). To visualize the protein migration pattern, the gel was stained with Coomassie G-250.
HDX with Continuous Labeling.
HDX provides information on the inherent dynamics/flexibility of LPL in the presence of different binding partners. HDX was performed at 25 °C using 10 mM Hepes and 150 mM NaCl in either H2O or D2O, adjusted to pH 7.4 and pD 7.4 (i.e., pHread is 7.0). Purified bLPL was diluted in protiated Hepes buffer to a final concentration of 10 µM. When LPL complexes were analyzed, 10 µM bLPL was preincubated for 30 min on ice followed by 2 min at 25 °C in the presence of 12 µM Fab-5D2 and/or 10 µM GPIHBP1. To initiate isotopic labeling the protein solutions were diluted in deuterated Hepes buffer, resulting in a final concentration of 70% D2O. After 10, 100, or 1,000 s, aliquots were removed and deuterium exchange quenched by acidification and adding one volume ice-cold quenching buffer [100 mM Na2HPO4, 0.8 M Tris-(2-carboxyethyl)phosphine, and 2 M urea in H2O, pH 2.5]. The quenched samples were placed in an ice bath for 2 min to allow reduction of disulfide bonds and subsequently snap-frozen in liquid nitrogen and stored at −80 °C until the mass spectrometry analyses were performed.
Nondeuterated controls were prepared as described earlier, except that protiated solvents were used in all steps. Fully deuterated controls were prepared by diluting the samples with deuterated Hepes buffer containing 2 M deuterated urea (10 mM Hepes, 150 mM NaCl, and 2 M urea-d4 in D2O, pHread 7.0). To accomplish a fully exchanged state, samples were incubating at 37 °C for 48 h before quenching with the buffer described earlier, except no urea was added in order to achieve an identical solvent composition in all samples. All labeling experiments were performed in triplicate.
HDX with Pulse Labeling.
This labeling protocol provides information on time-dependent changes in bLPL conformation in the presence of binding partners. Buffers and labeling conditions were identical to those described above. Five different bLPL samples were prepared in protiated Hepes buffer: 1) 10 µM bLPL, 2) 10 µM bLPL with 12 µM Fab-5D2, 3) 10 µM bLPL with 1 µM ANGPTL4, 4) 10 µM bLPL with 12 µM Fab-5D2 and 1 µM ANGPTL4, and 5) 10 µM bLPL with 12 µM Fab-5D2, 1 µM ANGPTL4, and 20 µM GPIHBP1. After 5 min on ice, the incubation continued at 25 °C for 0, 2.5, 5, 10, 15, or 30 min. At these time points, the conformation of LPL was probed by a 10-fold dilution in protiated solvent for 10 s at 25 °C (pulse labeling). Adding one volume of ice-cold quench buffer prevented further deuterium exchange (as outlined for the continuous labeling protocol). Labeling was performed in triplicate (or more) for each sample combination.
Deuterium uptake in LPL was analyzed by in-line pepsin digestion and subsequent ultraperformance liquid chromatography–electrospray ionization (ESI) MS with a quadrupole time-of-flight mass spectrometer (Synapt G2; Waters) as described (60). We used HX-Express2 to analyze deuterium uptake in peptides displaying EX1-like kinetics by binomial distribution fitting to obtain the deuterium content of both envelopes of the bimodal isotopic distribution (61).
Catalytic Activity of LPL.
The hydrolase activity of bLPL alone or in complex with Fab-5D2 and/or GPIHBP1 was measured with the fluorescent substrate DGGR, since the shielding of the lipid-binding motif in LPL by 5D2 precludes using emulsified lipid substrates. In brief, different mixtures of 10 µM bLPL, 12 µM Fab-5D2, 20 µM GPIHBP1, and 1 µM ANGPTL4 were preincubated at 25 °C for various time points (identical to those used for pulse-labeled HDX-MS). The residual LPL activity was measured at 1-min intervals for 10 min on aliquots diluted to 30 nM bLPL in 20 µM DGGR in 50 mM Tris, 120 mM NaCl, 10 mg/mL BSA, 0.5% (vol/vol) Triton X-100, pH 7.2, including 200 nM GPIHBP1 at 25 °C. Adding GPIHBP1 to the buffer served to prevent further unfolding of LPL during the enzymatic activity determination. Finally, the specific LPL activity was calculated as the slopes of the linear progression in fluorescence (emission 615 nm/excitation 535 nm) using an Envision (PerkinElmer).
SEC-MALS.
GPIHBP1•LPL•Fab and GPIHBP1•LPL complexes were subjected to SEC with in-line measurements of refractive index and multiangle light scattering with Optilab T-rEX and miniDAWN Treos II detectors from Wyatt Technology. In brief, 20 to 50 µL of a 1 to 3 mg/mL protein solution was applied to a Superdex 200 Increase column (5 mm × 150 mm) from GE Healthcare at a flow rate of 0.1 mL/min in 10 mM Tris, 150 mM NaCl, 0.05% CHAPS (0.8 mM), 10% (vol/vol) glycerol, and 0.05% NaN3, pH 7.2. The Astra software from Wyatt Technology calculated the molecular mass of the protein complex eluting from the SEC using the refractive index and light scattering with dn/dc of 0.161 mL/g (estimated from preceding buffer adjustment).
Data Availability.
SAXS data and rigid modeling for the GPIHBP1•LPL•Fab5D2 complex have been deposited at the Small Angle Scattering Biological Data Bank, https://www.sasbdb.org/ (accession no. SASDHF4).
Supplementary Material
Acknowledgments
We thank Gry Ellis Rasmussen for excellent technical assistance. SAXS data were collected at the EMBL P12 beamline of the storage ring PETRA III and the EMBL X33 beamline of the storage ring DORIS (DESY, Hamburg, Germany). This work was supported by grants HL090553, HL087228, and HL125335 from the National Heart, Lung, and Blood Institute; Transatlantic Network grant 12CVD04 from the Leducq Fondation; Lundbeck Foundation grant R230–2016–2930; and Novo Nordisk Foundation grants NNF17OC0026868 and NNF18OC0033864.
Footnotes
Competing interest statement: M.M. is an employee of Takeda and a Takeda stockowner. G.O. is a shareholder and board member in Lipigon Pharmaceuticals. Neither Takeda nor Lipigon was involved in the research described in this paper. R.Z. and S.G.Y. participated in the same Leducq Foundation Transatlantic Network Grant from 2014–2019, but did not engage in scientific collaborations as part of that program.
Data deposition: Small-angle X-ray scattering data and rigid modeling for the GPIHBP1•LPL•Fab5D2 complex have been deposited at the Small Angle Scattering Biological Data Bank, https://www.sasbdb.org/ (accession no. SASDHF4).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920202117/-/DCSupplemental.
References
- 1.Young S. G., Zechner R., Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 27, 459–484 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goulbourne C. N., et al. , The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 19, 849–860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies B. S., et al. , GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12, 42–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mysling S., et al. , The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife 5, e20958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenburg W. K., et al. , GPIHBP1 stabilizes lipoprotein lipase and prevents its inhibition by angiopoietin-like 3 and angiopoietin-like 4. J. Lipid Res. 50, 2421–2429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovrov O., Kristensen K. K., Larsson E., Ploug M., Olivecrona G., On the mechanism of angiopoietin-like protein 8 for control of lipoprotein lipase activity. J. Lipid Res. 60, 783–793 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kersten S., Angiopoietin-like 3 in lipoprotein metabolism. Nat. Rev. Endocrinol. 13, 731–739 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Beigneux A. P., et al. , Autoantibodies against GPIHBP1 as a cause of hypertriglyceridemia. N. Engl. J. Med. 376, 1647–1658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plengpanich W., et al. , Multimerization of glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) and familial chylomicronemia from a serine-to-cysteine substitution in GPIHBP1 Ly6 domain. J. Biol. Chem. 289, 19491–19499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beigneux A. P., et al. , GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ. Res. 116, 624–632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu X., et al. , GPIHBP1 autoantibodies in a patient with unexplained chylomicronemia. J. Clin. Lipidol. 11, 964–971 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ariza M. J., et al. , Novel mutations in the GPIHBP1 gene identified in 2 patients with recurrent acute pancreatitis. J. Clin. Lipidol. 10, 92–100.e1 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Fong L. G., et al. , GPIHBP1 and plasma triglyceride metabolism. Trends Endocrinol. Metab. 27, 455–469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brahm A. J., Hegele R. A., Chylomicronaemia–Current diagnosis and future therapies. Nat. Rev. Endocrinol. 11, 352–362 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Dewey F. E., et al. , Inactivating variants in ANGPTL4 and risk of coronary artery disease. N. Engl. J. Med. 374, 1123–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgadottir A., et al. , Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat. Genet. 48, 634–639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stitziel N. O., et al. ; Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators , Coding variation in ANGPTL4, LPL, and SVEP1 and the risk of coronary disease. N. Engl. J. Med. 374, 1134–1144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romeo S., et al. , Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Invest. 119, 70–79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stitziel N. O., et al. ; PROMIS and Myocardial Infarction Genetics Consortium Investigators , ANGPTL3 deficiency and protection against coronary artery disease. J. Am. Coll. Cardiol. 69, 2054–2063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mysling S., et al. , The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife 5, e12095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong H., et al. , Lipoprotein lipase domain function. J. Biol. Chem. 269, 10319–10323 (1994). [PubMed] [Google Scholar]
- 22.Wong H., et al. , A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 94, 5594–5598 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Tilbeurgh H., Roussel A., Lalouel J. M., Cambillau C., Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: Consequences for heparin binding and catalysis. J. Biol. Chem. 269, 4626–4633 (1994). [PubMed] [Google Scholar]
- 24.Kobayashi Y., Nakajima T., Inoue I., Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur. J. Biochem. 269, 4701–4710 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Hayne C. K., et al. , We FRET so you don’t have to: New models of the lipoprotein lipase dimer. Biochemistry 57, 241–254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lookene A., Zhang L., Hultin M., Olivecrona G., Rapid subunit exchange in dimeric lipoprotein lipase and properties of the inactive monomer. J. Biol. Chem. 279, 49964–49972 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Osborne J. C. Jr, Bengtsson-Olivecrona G., Lee N. S., Olivecrona T., Studies on inactivation of lipoprotein lipase: Role of the dimer to monomer dissociation. Biochemistry 24, 5606–5611 (1985). [DOI] [PubMed] [Google Scholar]
- 28.Peterson J., Fujimoto W. Y., Brunzell J. D., Human lipoprotein lipase: Relationship of activity, heparin affinity, and conformation as studied with monoclonal antibodies. J. Lipid Res. 33, 1165–1170 (1992). [PubMed] [Google Scholar]
- 29.Sukonina V., Lookene A., Olivecrona T., Olivecrona G., Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc. Natl. Acad. Sci. U.S.A. 103, 17450–17455 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shan L., et al. , The angiopoietin-like proteins ANGPTL3 and ANGPTL4 inhibit lipoprotein lipase activity through distinct mechanisms. J. Biol. Chem. 284, 1419–1424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birrane G., et al. , Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 116, 1723–1732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora R., et al. , Structure of lipoprotein lipase in complex with GPIHBP1. Proc. Natl. Acad. Sci. U.S.A. 116, 10360–10365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng C. F., Bensadoun A., Bersot T., Hsu J. S., Melford K. H., Purification and characterization of human lipoprotein lipase and hepatic triglyceride lipase. Reactivity with monoclonal antibodies to hepatic triglyceride lipase. J. Biol. Chem. 260, 10720–10727 (1985). [PubMed] [Google Scholar]
- 34.Bengtsson-Olivecrona G., Olivecrona T., Phospholipase activity of milk lipoprotein lipase. Methods Enzymol. 197, 345–356 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Beigneux A. P., et al. , Lipoprotein lipase is active as a monomer. Proc. Natl. Acad. Sci. U.S.A. 116, 6319–6328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young S. G., et al. , GPIHBP1 and lipoprotein lipase, partners in plasma triglyceride metabolism. Cell Metab. 30, 51–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang S. F., Reich B., Brunzell J. D., Will H., Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J. Lipid Res. 39, 2350–2359 (1998). [PubMed] [Google Scholar]
- 38.Lookene A., Groot N. B., Kastelein J. J., Olivecrona G., Bruin T., Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J. Biol. Chem. 272, 766–772 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Babirak S. P., Iverius P. H., Fujimoto W. Y., Brunzell J. D., Detection and characterization of the heterozygote state for lipoprotein lipase deficiency. Arteriosclerosis 9, 326–334 (1989). [DOI] [PubMed] [Google Scholar]
- 40.Liu M. S., Ma Y., Hayden M. R., Brunzell J. D., Mapping of the epitope on lipoprotein lipase recognized by a monoclonal antibody (5D2) which inhibits lipase activity. Biochim. Biophys. Acta 1128, 113–115 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Hu X., et al. , Monoclonal antibodies that bind to the Ly6 domain of GPIHBP1 abolish the binding of LPL. J. Lipid Res. 58, 208–215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyashita K., et al. , An enzyme-linked immunosorbent assay for measuring GPIHBP1 levels in human plasma or serum. J. Clin. Lipidol. 12, 203–210.e1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kristensen K. K., et al. , A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 115, E6020–E6029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trelle M. B., et al. , An asymmetric runaway domain swap antithrombin dimer as a key intermediate for polymerization revealed by hydrogen/deuterium-exchange mass spectrometry. Anal. Chem. 89, 616–624 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Ahmad Z., et al. , Inhibition of angiopoietin-like protein 3 with a monoclonal antibody reduces triglycerides in hypertriglyceridemia. Circulation 140, 470–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geldenhuys W. J., Lin L., Darvesh A. S., Sadana P., Emerging strategies of targeting lipoprotein lipase for metabolic and cardiovascular diseases. Drug Discov. Today 22, 352–365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kersten S., New insights into angiopoietin-like proteins in lipid metabolism and cardiovascular disease risk. Curr. Opin. Lipidol. 30, 205–211 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Lafferty M. J., Bradford K. C., Erie D. A., Neher S. B., Angiopoietin-like protein 4 inhibition of lipoprotein lipase: Evidence for reversible complex formation. J. Biol. Chem. 288, 28524–28534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutgsell A. R., Ghodge S. V., Bowers A. A., Neher S. B., Mapping the sites of the lipoprotein lipase (LPL)-angiopoietin-like protein 4 (ANGPTL4) interaction provides mechanistic insight into LPL inhibition. J. Biol. Chem. 294, 2678–2689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bengtsson G., Olivecrona T., Apolipoprotein CII enhances hydrolysis of monoglycerides by lipoprotein lipase, but the effect is abolished by fatty acids. FEBS Lett. 106, 345–348 (1979). [DOI] [PubMed] [Google Scholar]
- 51.Köster A., et al. , Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: Regulation of triglyceride metabolism. Endocrinology 146, 4943–4950 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Yau M. H., et al. , A highly conserved motif within the NH2-terminal coiled-coil domain of angiopoietin-like protein 4 confers its inhibitory effects on lipoprotein lipase by disrupting the enzyme dimerization. J. Biol. Chem. 284, 11942–11952 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferraro D. M., Lazo N., Robertson A. D., EX1 hydrogen exchange and protein folding. Biochemistry 43, 587–594 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Nimonkar A. V., et al. , A lipoprotein lipase –GPI-anchored high density lipoprotein binding protein 1 fusion lowers triglycerides in mice: Implications for managing familial chylomicronemia syndrome. J. Biol. Chem. 10.1074/jbc.RA119.011079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Péterfy M., Lipase maturation factor 1: A lipase chaperone involved in lipid metabolism. Biochim. Biophys. Acta 1821, 790–794 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang L., Lookene A., Wu G., Olivecrona G., Calcium triggers folding of lipoprotein lipase into active dimers. J. Biol. Chem. 280, 42580–42591 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Koerner C. M., Roberts B. S., Neher S. B., Endoplasmic reticulum quality control in lipoprotein metabolism. Mol. Cell. Endocrinol. 498, 110547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundberg E. L., Deng Y., Burd C. G., Syndecan-1 mediates sorting of soluble lipoprotein lipase with sphingomyelin-rich membrane in the Golgi apparatus. Dev. Cell 51, 387–398.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robal T., Larsson M., Martin M., Olivecrona G., Lookene A., Fatty acids bind tightly to the N-terminal domain of angiopoietin-like protein 4 and modulate its interaction with lipoprotein lipase. J. Biol. Chem. 287, 29739–29752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leth J. M., Mertens H. D. T., Leth-Espensen K. Z., Jørgensen T. J. D., Ploug M., Did evolution create a flexible ligand-binding cavity in the urokinase receptor through deletion of a plesiotypic disulfide bond? J. Biol. Chem. 294, 7403–7418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guttman M., Weis D. D., Engen J. R., Lee K. K., Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J. Am. Soc. Mass Spectrom. 24, 1906–1912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SAXS data and rigid modeling for the GPIHBP1•LPL•Fab5D2 complex have been deposited at the Small Angle Scattering Biological Data Bank, https://www.sasbdb.org/ (accession no. SASDHF4).