Significance
Understanding coevolution in crop–parasite systems is critical to management of myriad pests and pathogens confronting modern agriculture. In contrast to wild plant communities, parasites in agricultural ecosystems are usually expected to gain the upper hand in coevolutionary “arms races” due to limited genetic diversity of host crops in cultivation. Here, we develop a framework to characterize associations between genome variants in global landraces (traditional varieties) of the staple crop sorghum with the distribution of the devastating parasitic weed Striga hermonthica. We find long-term maintenance of diversity in genes related to parasite resistance, highlighting an important role of host adaptation for coevolutionary dynamics in smallholder agroecosystems.
Keywords: species distribution modeling, genotype–environment association analysis, environmental niche modeling
Abstract
Host–parasite coevolution can maintain high levels of genetic diversity in traits involved in species interactions. In many systems, host traits exploited by parasites are constrained by use in other functions, leading to complex selective pressures across space and time. Here, we study genome-wide variation in the staple crop Sorghum bicolor (L.) Moench and its association with the parasitic weed Striga hermonthica (Delile) Benth., a major constraint to food security in Africa. We hypothesize that geographic selection mosaics across gradients of parasite occurrence maintain genetic diversity in sorghum landrace resistance. Suggesting a role in local adaptation to parasite pressure, multiple independent loss-of-function alleles at sorghum LOW GERMINATION STIMULANT 1 (LGS1) are broadly distributed among African landraces and geographically associated with S. hermonthica occurrence. However, low frequency of these alleles within S. hermonthica-prone regions and their absence elsewhere implicate potential trade-offs restricting their fixation. LGS1 is thought to cause resistance by changing stereochemistry of strigolactones, hormones that control plant architecture and below-ground signaling to mycorrhizae and are required to stimulate parasite germination. Consistent with trade-offs, we find signatures of balancing selection surrounding LGS1 and other candidates from analysis of genome-wide associations with parasite distribution. Experiments with CRISPR–Cas9-edited sorghum further indicate that the benefit of LGS1-mediated resistance strongly depends on parasite genotype and abiotic environment and comes at the cost of reduced photosystem gene expression. Our study demonstrates long-term maintenance of diversity in host resistance genes across smallholder agroecosystems, providing a valuable comparison to both industrial farming systems and natural communities.
Host–parasite interactions can be powerful and dynamic selective forces maintaining genetic variation in natural populations (1). In wild-plant pathosystems, long-term balancing selection often maintains diverse resistance alleles in host populations (2–4). When rare alleles provide a selective advantage, negative frequency-dependent selection drives cycling of resistance and virulence alleles [i.e., fluctuating Red Queen dynamics sensu (5)] (2, 6). Fitness costs of resistance and spatiotemporal changes in selection can also maintain diversity across gradients of parasite pressure (7).
In contrast to wild-plant communities, where fluctuating Red Queen dynamics have frequently been observed, low host diversity in agricultural settings is often assumed to permit runaway “arms races” in fast-evolving parasites (4, 8). Relative to smallholder farms, however, industrial-scale farming accounts for a fraction of global production for many food crops (9). The dynamics of host adaptation to parasites across diverse smallholder agricultural systems remains poorly known, despite relevance for identifying novel resistance alleles and managing crop genetic resources (e.g., preserving germplasm both ex situ and in situ; ref. 10). Are coevolutionary dynamics in smallholder farming systems more similar to natural plant pathosystems, where high connectivity among genetically diverse patches can help promote evolution of host resistance (11)?
Current approaches for identifying and studying the evolution of resistance alleles often involve scoring large panels of diverse individuals (in genome-wide association studies [GWAS]) or many recombinant individuals deriving from controlled crosses (in linkage mapping) and using DNA-sequence information to identify genomic regions associated with parasite susceptibility. These mapping studies have revealed numerous insights to mechanisms of plant–pathogen dynamics (12), but require extensive phenotyping and genotyping effort for adequate statistical power. By contrast, if traditional local crop or livestock varieties (known as landraces) are locally adapted to regions of high parasite prevalence (13–16), then a different “bottom-up” approach may be used. Compared to modern improved varieties, which may have lost resistance alleles due to bottlenecks and selection in optimal environments (17), landraces and wild relatives may be a rich source of resistance alleles to sympatric parasites. Specifically, to map loci underlying local adaptation, one can identify loci where allele frequency is associated with environmental conditions, known as genotype–environment associations (GEAs). GEAs of georeferenced landraces have been a powerful strategy for understanding the genetic basis of local adaptation to gradients of abiotic stressors (14, 15). Furthermore, landraces can be studied to test the hypothesis that a putatively environmentally adapted allele, identified from a limited set of experimental environments and genetic backgrounds, is indeed adaptive across a wide range of similar environments and across diverse genetic backgrounds (14).
In this study, we extend GEAs to biotic stress gradients (16) to evaluate the frequency and distribution of alleles in genomes of sorghum that confer resistance to the African witchweed Striga hermonthica (Delile) Benth., a root hemiparasite of the broomrape family (Orobanchaceae). Sorghum bicolor (L.) Moench is the world’s fifth most important crop and was domesticated from the wild progenitor Sorghum arundinaceum (Desv.) Stapf in Africa more than 5,000 y ago (18). Sorghum is particularly important due to its tolerance of marginal environments compared to maize and rice. While plant responses to environment are influenced by many processes, in recent years, the role of strigolactones (SLs), hormones that regulate shoot branching (19), root architecture (20), and response to abiotic stress (21), have received increasing attention. SLs exuded from roots, particularly under nutrient (22) and water (23) limitation, promote interactions with beneficial arbuscular mycorrhizal fungi (24) and also function as germination stimulants for many root-parasitic plants. S. hermonthica attacks cereal crops and is one of the greatest biotic threats to food security in Africa, causing billions of US dollars in crop losses annually (25). Several resistance mechanisms have been reported among cultivated and wild sorghums (26).
Here, we evaluate the hypothesis that a geographical selection mosaic across gradients of biotic interactions maintains genetic diversity in sorghum landrace resistance to S. hermonthica (27, 28). To identify genomic signatures of host adaptation to parasites, we first developed species distribution models (SDMs) for S. hermonthica and searched for statistical associations between sorghum genotype and modeled parasite prevalence at the location of origin for each sorghum landrace. We validated our approach by characterizing diversity and geographic distribution of loss-of-function alleles at the putative sorghum resistance gene LOW GERMINATION STIMULANT 1 (LGS1) (29). Loss-of-function mutations at LGS1, a gene of unknown function with a sulfotransferase domain, is thought to underlie a quantitative trait locus (QTL) that alters stereochemistry of the dominant SL in sorghum root exudates, from 5-deoxystrigol to orobanchol (29). Orobanchol is considered a weaker stimulant of S. hermonthica germination, conferring resistance (29, 30). Our ecological genetic analyses of diverse sorghum landraces suggest that LGS1 loss-of-function mutations are adaptive across a large region of high S. hermonthica prevalence in Africa. However, using comparisons of CRISPR–Cas9-edited sorghum, we also present evidence supporting potential trade-offs for LGS1 loss-of-function due to high sensitivity of some S. hermonthica genotypes to orobanchol and subtle impacts on host fitness. In addition to focused analyses on LGS1, we performed genome-wide tests of association with parasite distribution. We investigated patterns of polymorphism surrounding candidate resistance genes with evidence of locally adaptive natural variation to determine whether balancing selection has maintained diversity in S. hermonthica resistance over evolutionary time.
Results
S. hermonthica Distribution Model.
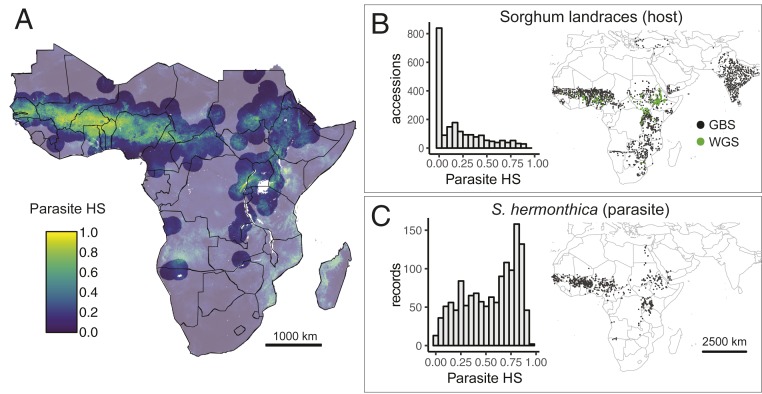
We predicted that host alleles conferring resistance or tolerance would be strongly associated with the geographic distribution of Striga parasites. To identify regions of likely S. hermonthica occurrence in the absence of continent-wide surveys, we built MaxEnt SDMs (31). This approach uses locations of known species occurrences and multivariate environmental data to generate predictions of suitable habitat across a landscape. The optimal model showed good ability to predict occurrence with an area under the receiver operating characteristic curve (AUC) value of 0.86 (Fig. 1). A high degree of overlap was observed between models generated using all S. hermonthica records (n = 1,050) and a subset of 262 occurrences that were observed specifically in fields of sorghum (Schoener’s D = 0.82; I = 0.97). Annual rainfall and total soil nitrogen (N) were the most informative variables for predicting S. hermonthica occurrence (SI Appendix, Table S1). Compared to all grid cells in the study background, distributions of environmental values for locations with high habitat suitability (HS) were generally restricted to a narrower range of intermediate values of precipitation and soil quality (SI Appendix, Figs. S1 and S2). Locations with the highest HS scores exhibited mean annual rainfall ranging from ∼500 to 1,300 mm/y and soil N ranging from ∼400 to 1,000 g/kg (10th to 90th percentiles for all cells with HS > 0.5; SI Appendix, Table S1). Soil clay content also contributed substantially to the sorghum-only model, and clay content in locations with the highest HS scores ranged from 12 to 29% (10th to 90th percentile, HS > 0.5, all-occurrence model) or up to 36% (sorghum-only model; SI Appendix, Table S1).
Fig. 1.
A subset of global sorghum landraces are from parasite-prone areas. (A) S. hermonthica HS scores across Africa based on MaxEnt SDM. To account for areas with suitable habitat where parasites have not been recorded, HS for accessions more than 200 km from any S. hermonthica record was set to zero; transparent colors demarcate areas of this HS masking. (B) Geographic distribution and frequency histogram of HS scores at locations of all georeferenced and genotyped sorghum landrace accessions. Landraces with available GBS (n = 2,070) and WGS (n = 143) data are shown. (C) Geographic distribution and frequency histogram of HS scores at locations of 1,369 S. hermonthica occurrence records. HS, habitat suitability; SDM, species distribution model; GBS, genotyping-by-sequencing; WGS, whole genome sequencing.
LGS1 Associations with S. hermonthica Occurrence.
We predicted that sorghum resistance alleles would be more common in parasite-prone regions. Evaluating this prediction also allowed us to validate our SDM-based GEA approach by characterizing associations between S. hermonthica distribution and genetic variation at LGS1 (Sobic.005G213600). LGS1 is thought to cause a known QTL for resistance to Striga species (29).
Using whole-genome sequencing data (∼25× coverage) from 143 sorghum landraces, we found evidence for three naturally occurring mutations resulting in LGS1 loss-of-function (Fig. 2). Two ∼30-kb deletions were identified between positions 69,958,377 to 69,986,892 (n = 5 accessions) and 69,981,502 to 70,011,149 (n = 4 accessions; SI Appendix, Table S2 and Fig. 2A). These deletions appeared to be identical to two previously described as resistance alleles, lgs1-2 and lgs1-3 (29), although breakpoint positions reported by our structural variant caller differed slightly. No single nucleotide polymorphism (SNPs) in a separate genotyping-by-sequencing (GBS) dataset of >2,000 sorghum landraces tagged lgs1-2 or lgs1-3 (SI Appendix, Fig. S3), and so we imputed large deletions identified from the whole-genome sequencing (WGS) dataset to the GBS dataset based on patterns of missing data (Methods). Deletion imputations were validated by testing root exudate from a subset of sorghum accessions for their ability to induce S. hermonthica germination (SI Appendix, Fig. S4). We tested four genotypes with likely deletion alleles, and these genotypes stimulated significantly fewer Striga seeds to germinate compared to eight other genotypes that did not show strong evidence for lgs1-2 or lgs1-3 deletions (linear mixed-effects model with genotype random effect; deletion genotypes stimulated germination of 8.43 fewer seeds out of 75 per replicate; Wald 95% CI = 1.49, 15.36).
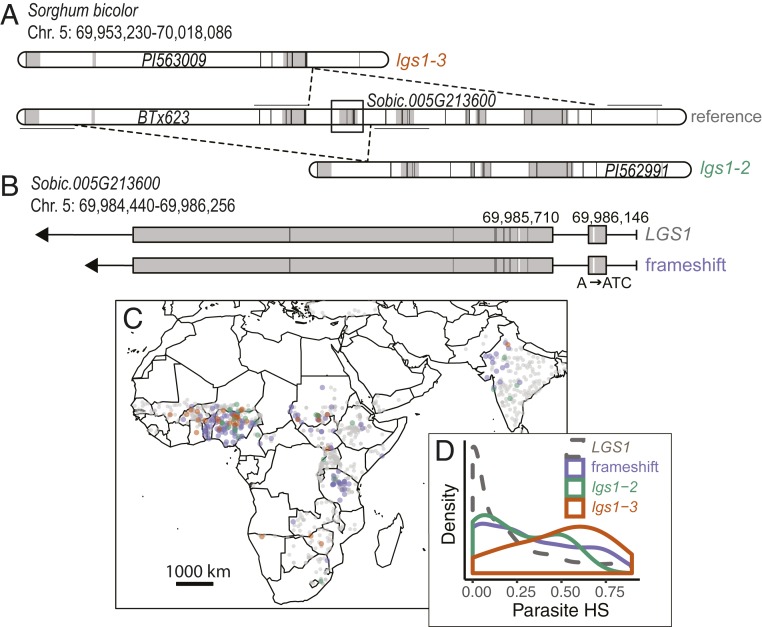
Fig. 2.
LGS1 loss-of-function alleles are broadly distributed within parasite-prone regions. (A and B) Schematic of large deletion variants (A) and frameshift mutation (B) impacting sorghum LGS1, a locus involved in resistance to S. hermonthica. Gray shading indicates position of gene models (A) or coding regions (B). Vertical black bars indicate the position of SNPs in the GBS dataset, and horizontal black lines denote 5-kb flanking regions used to impute deletion calls. In B, vertical white bars show the frameshift mutation (position 69,986,146) and the SNP at position 69,985,710 that tags the frameshift in the GBS dataset. Chr., chromosome. (C) Geographic distribution of LGS1 alleles in sorghum landraces. (D) Distribution of parasite HS scores at locations of sorghum accessions with lgs1-2 (n = 25), lgs1-3 (n = 34), frameshift (n = 131), or intact LGS1 (n = 785). HS, habitat suitability.
In addition to lgs1-2 and lgs1-3, we identified a previously unknown 2-bp insertion predicted to cause a frameshift variant in the beginning of the LGS1 coding region (position 69,986,146; allele frequency in WGS dataset = 8%). The frameshift was linked to an SNP genotyped in the GBS dataset (Fig. 2B; T/A at position 69,985,710; D′ = 0.93, r2 = 0.84). All nine accessions with the frameshift in the WGS dataset also shared a 315-bp deletion (positions 69,984,268 to 69,984,583) overlapping 143 bp of the 3′ untranslated region in the 1,580-bp second exon of LGS1.
Three of the six total independent LGS1 putative loss-of-function alleles characterized here and elsewhere (29) were present at low frequency in the GBS panel. Among accessions with SNP calls in LGS1, 7.0% of accessions exhibited SNP calls consistent with homozygous lgs1-2 and lgs1-3 in the GBS dataset, and the SNP tagging the frameshift was present at an allele frequency of 15% (Dataset S1). LGS1 loss-of-function alleles were found in diverse genetic backgrounds and geographic regions (SI Appendix, Fig. S5 and Table S2 and Dataset S1), suggesting that these mutations have had time to spread, that their benefit is not strongly masked by epistasis, and that costs of resistance are not strong enough to prevent their spread. Most landraces with LGS1 loss-of-function alleles and known botanical race [which largely correspond to genetic clusters (3)] from the GBS dataset were guinea (71/211 accessions), durra-caudatum (30/83 accessions), or caudatum (20/165 accessions; Dataset S1; 298 lacked assignments).
LGS1 loss-of-function alleles were more common among landraces with high parasite HS scores (Fig. 2 C and D). However, correlations with population structure reduced power to detect associations with these resistance alleles after accounting for kinship (SI Appendix, Fig. S5). The median S. hermonthica HS score was 0.20 for accessions homozygous for lgs1-2, 0.54 for accessions homozygous for lgs1-3, 0.25 for accessions with the frameshift, and 0.09 for accessions without evidence for LGS1 loss-of-function. The difference in S. hermonthica HS score between lgs1-3 and LGS1 intact accessions was statistically significant before (P < 0.001, Wilcoxon rank-sum test), but not after, accounting for relatedness (P = 0.10, mixed-linear model [MLM]). Frameshift associations with HS were also statistically significant prior to correction for relatedness (P < 0.001, Wilcoxon rank sum; P = 0.69, MLM). We observed modest support for associations between lgs1-2 and S. hermonthica HS before correcting for relatedness (P = 0.06, Wilcoxon rank-sum test; P = 0.53, MLM). Results were similar considering a subset of just African accessions (Plgs1-3 < 0.001, Pframeshift < 0.001, and Plgs1-2 = 0.07; Wilcoxon rank-sum test). For sorghum landraces within parasite-prone regions (<200 km from an occurrence record), naturally occurring LGS1 loss-of-function alleles were more common among landraces from low-nutrient (particularly low N) environments (PN < 0.001, PP = 0.004; Kolmogorov–Smirnov test comparing distribution of soil N or P for LGS1 intact vs. LGS1 loss-of-function landraces), but evenly distributed across precipitation gradients (P = 0.4; Kolmogorov–Smirnov test).
Parasite-Associated SNPs Across the Sorghum Genome.
We performed genome-wide tests of association with predicted parasite HS score. A scan of 317,294 SNPs across 2,070 sorghum landraces revealed 97 genomic regions exhibiting significant associations with S. hermonthica distribution at a false discovery rate (FDR) of 5% (Fig. 3A and SI Appendix, Table S4). Of SNPs exceeding the threshold for significance, 45 were present within 1 kb of a predicted gene model (SI Appendix, Table S4). Three SNPs exceeding this threshold were in QTL previously associated with Striga resistance (32), including one intron variant in the uncharacterized gene model Sobic.001G227800 (SI Appendix, Table S4). Another SNP was present in an intron of Sobic.007G090900, a gene model with high homology to SMAX1/D53, which is degraded in an SL-dependent manner to control downstream SL signaling and is associated with tillering and height in rice (33). SNPs among those with the strongest associations to parasite occurrence were also found in genes related to suberin and wax ester biosynthesis (Sobic.007G091200, synonymous variant), including phenylalanine ammonia–lyase (Sobic.006G148800, intron variant). Phenylalanine ammonia–lyase is highly up-regulated in the resistant rice line Nipponbare compared to a susceptible line during infection with S. hermonthica (34) and is associated with increased lignin deposition and postattachment resistance (35).
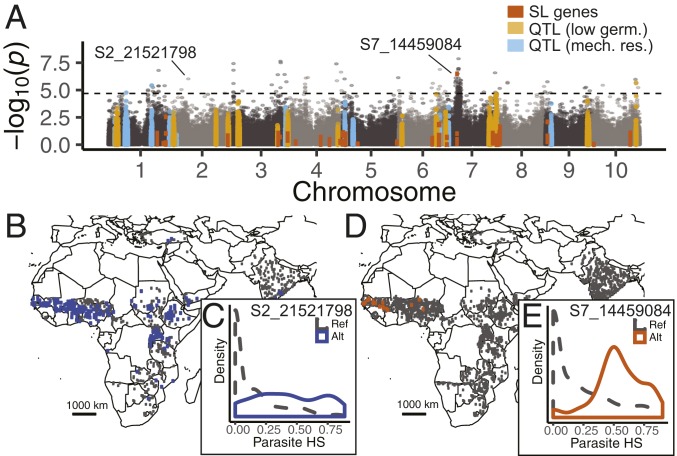
Fig. 3.
Sorghum genome-wide associations with parasite distribution implicate cell-wall and SL-signaling genes. (A) Genome-wide association with parasite HS score, based on 317,294 SNPs with MAF > 0.01 in 2,070 sorghum landraces. SNPs in genomic regions linked to SL biosynthesis/signaling (red), resistance through formation of a mechanical barrier (light blue), or low S. hermonthica germination (orange) are indicated. Germ., germination; mech. res., mechanical resistance. The dashed line represents significance threshold at a FDR of 5%. (B) Geographic distribution of reference and alternate alleles for an SNP (S2_21521798) in a pectinesterase gene (MAF = 0.275). (C) Distribution of parasite HS scores for sorghum accessions segregating for S2_21521798. (D) Geographic distribution of reference and alternate alleles for an SNP (S7_14459084) in a gene homologous to SMAX1 (MAF = 0.014). (E) Distribution of parasite HS scores for sorghum accessions segregating for S7_14459084. Alt, alternate; Ref, reference.
Across all gene models tagged by SNPs in the genome-wide analysis (i.e., within 1 kb), no Gene Ontology (GO) term met the threshold for significance after correction for multiple comparisons. The strongest enrichment scores were in genes with GO terms related to cell-wall organization (GO:0071555; corrected P = 0.13, mean p score for 48 genes = 0.27), cell wall (GO:0005618; corrected P = 0.17, mean p score for 70 genes = 0.29), and pectinesterase activity (GO:0030599; corrected P = 0.22, mean p score for 39 genes = 0.26). The strongest SNP association to parasite occurrence in a pectinesterase gene model was in Sobic.002G138400 (SNP S2_21521798, P = 0.01; SI Appendix, Table S4). Although the allele associated with parasite occurrence at S2_21521798 was not predicted to cause an amino acid change, it tagged a haplotype block encompassing complex structural variation 204.4 kb upstream of the gene model, suggesting a potential regulatory variant or nearby presence/absence variation. Overall, SNPs in genes related to SL biosynthesis and signaling (SI Appendix, Table S3) did not show a significant enrichment for associations with S. hermonthica distribution (uncorrected P = 0.09).
Signatures of Balancing Selection in Candidate Regions.
We further investigated three candidate genes with polymorphism that exhibited distinct geographic patterns and had known or potential roles in S. hermonthica resistance. Elevated Tajima’s D values can indicate an excess of shared polymorphism at a locus, expected for regions of the genome under balancing selection, whereas strongly negative values can indicate an excess of low-frequency polymorphism, expected under either purifying or positive selection. Two 5-kb genomic regions, spanning SNPs in LGS1 (SNP S5_69986146 in gene model Sobic.G005G213600) and a pectinesterase gene (SNP S2_21521798 in gene model Sobic.002G138400; minor allele frequency [MAF] = 0.275) exhibited elevated values of Tajima’s D compared to 1,000 randomly sampled 5-kb windows containing or overlapping gene models (P = 0.02, Sobic.G005G213600; P = 0.05, Sobic.002G138400; SI Appendix, Fig. S6). Regions of elevated Tajima’s D were localized to relatively small windows centered on SNPs associated with S. hermonthica HS, and larger window sizes produced weaker signals. We looked for these alleles in several genomes of wild relatives of sorghum, but found no reads mapping to the pectinesterase gene and no evidence for the LGS1 loss-of-function alleles characterized using data from previously sequenced accessions of S. propinquum (Kunth) Hitchc. (n = 2) and S. arundinaceum (as synonym S. bicolor subsp. Verticilliflorum [Steud.] de Wet ex Wiersema & J.Dahlb. [n = 2]) (36).
We did not observe strong departures from the neutral expectation of Tajima’s D for the region surrounding a gene with homology to SMAX1 (gene model Sobic.007G090900 tagged by SNP S7_14459084; P = 0.6). The minor allele at S7_14459084 was at low frequency in the GBS dataset (MAF = 0.014) and most common in West Africa (Fig. 3C), which was not well sampled in the WGS dataset. The signal of association with S. hermonthica occurrence extended more than 7.5 Mb on chromosome 7 (Fig. 3A), but we did not have sufficient sampling to suggest that S7_14459084 tags an incomplete or soft sweep in either the GBS or WGS datasets, according to the haplotype-based statistic nSL (37).
LGS1-Mediated Resistance Depends on Parasite Population and Environment.
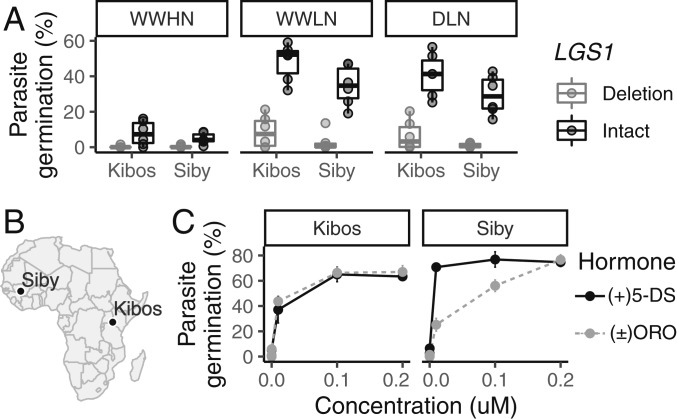
We further characterized the effects of loss-of-function variation at LGS1 by comparing multiple aspects of performance of newly generated lines of sorghum with CRISPR–Cas9 deletions of LGS1. Root exudate from these LGS1 deletion lines induced substantially lower S. hermonthica germination compared to control lines (P < 0.001, likelihood ratio test for fixed effect of deletion; Fig. 4A). However, the benefit of LGS1 deletion was conditional on the specific parasite population (P = 0.005, likelihood ratio test). We observed <1% germination in response to exudate from LGS1 deletion lines in an S. hermonthica population from Siby, Mali (95% Wald CI: 0.0, 4.0% germination) under the most stressful treatment (drought and low nutrient), but 6.1% germination in an S. hermonthica population from Kibos, Kenya (95% Wald CI: 0.2, 12.4% germination). In contrast, exudate from wild-type Macia grown under the same conditions induced mean germination of 40% (Siby; 78% germination in response to 0.2 μM GR24) or 29% (Kibos; 66% germination in response to 0.2 μM GR24). The Kenyan S. hermonthica similarly showed higher germination sensitivity to an orobanchol standard compared to the Malian S. hermonthica (Fig. 4C). The fact that Kenyan S. hermonthica showed similar germination response to 5-deoxystrigol and orobanchol standards, while germinating at a higher rate on intact LGS1 alleles vs. deletions, suggests that LGS1 deletion has effects on SLs beyond changing the ratio of 5-deoxystrigol to orobanchol in exudate.
Fig. 4.
Efficacy of LGS1 loss-of-function varies across abiotic and biotic gradients. (A) Germination of two S. hermonthica populations in response to root exudate from 3-wk-old wild-type Macia and CRISPR–Cas9-edited sorghum seedlings with lgs1 deletion alleles. Plants were grown under well-watered high-nutrition (WWHN), well-watered low-nutrition (WWLN), or drought low-nutrition (DLN) conditions. Germination was 0% in response to diH2O and 66% (Kibos) or 78% (Siby) in response to 0.2 μM GR24. (B) Origin of two tested S. hermonthica populations. (C) Germination of two S. hermonthica populations in response to synthetic SLs, (+)5-deoxystrigol (5-DS) or (±)orobanchol (ORO).
LGS1 Loss-of-Function Reduces Expression of Photosystem Genes.
Changes in SLs (and, indeed, any hormone) are potentially pleiotropic, given their many functions, suggesting that trade-offs may be associated with LGS1 variation. In CRISPR–Cas9-edited sorghum, we found 505 differentially expressed genes in roots (244 down-regulated in LGS1 knockout lines and 261 up-regulated) or 2,167 differentially expressed in shoot (917 down-regulated in LGS1 knockout lines and 1,250 up-regulated) at 6 wk after planting compared to wild-type Macia. Of all differentially expressed genes, 185 were differentially regulated in both root and shoot, including a transcription factor most highly expressed in sorghum panicles during floral initiation (Sobic.010G180200; down-regulated in knockout) and a GRAS family transcription factor homologous to RGA, which in Arabidopsis thaliana represses giberellic acid-induced vegetative growth and floral initiation (Sobic.008G168400; up-regulated in knockout) (38). In shoots, photosystem II genes were most enriched among differentially expressed genes (GO:0009523, P < 0.001; 11 of 36 genes with corrected P < 0.05; Dataset S2). Only GO terms related to photosystem I (GO:0009522, P < 0.001; 8 of 10 genes with corrected P < 0.1) and photosynthetic light harvesting (GO:0009765, P < 0.001; 12 of 18 genes with corrected P < 0.05) were enriched among differentially expressed genes in roots. All differentially expressed photosystem I, photosystem II, and light-harvesting genes were down-regulated in LGS1 knockout lines, with lower expression in roots than shoots, and 1.7 to 7.2× higher expression in Macia wild type (log2 fold-change ratios ranging from 0.77 to 2.84; Dataset S2).
Six of the light-harvesting or photosystem I genes down-regulated in roots of LGS1 knockout lines were also differentially expressed in nutrient-stressed roots of a resistant sorghum line (SRN39), which produces high levels of orobanchol, compared to a susceptible sorghum line (Shanqui Red) that produces mainly 5-deoxystrigol (29) (Dataset S2). SL biosynthesis genes were up-regulated in the LGS1-deficient sorghum line SRN39 compared to the susceptible line, including two genes with homology to NSP1 (Sobic.001G341400, P < 0.001; and Sobic.002G372100, P = 0.03), CCD7 (Sobic.006G170300, P < 0.001), and LBO (Sobic.003G418000, P = 0.01; SI Appendix, Fig. S7 and Table S3). Of these four genes, only LBO was also significantly up-regulated in the CRISPR LGS1 knockout line (in shoots), suggesting that the natural low-germination stimulating line carries additional alleles altering SL profiles.
In addition to reduced expression of photosynthesis genes, we found evidence for subtle effects of LGS1 loss-of-function on sorghum growth. Total leaf area of 65-d-old plants was not significantly different among three independent CRISPR knockout lines grown in nutrient-rich soil (SI Appendix, Fig. S8). In contrast, deletion lines had smaller leaf area relative to wild-type control and two event-null lines (P < 0.05, Scheffé’s method; SI Appendix, Fig. S8). However, no significant differences were seen in a separate experiment on younger plants grown under nutrient limitation, in which we did not detect a significant effect of LGS1 deletion on dry biomass, specific leaf area (SLA), single-photon avalanche diode (SPAD)-based measurement of leaf chlorophyll, or root number. Consistent with differential expression of transcription factors involved in growth and floral initiation (e.g., Sobic.008G168400), it may be that phenotypic differences are only detectable later in development or in some environments.
Discussion
Pests, pathogens, and parasites threaten human health and food security in a changing world, but understanding mechanisms of resistance across diverse taxa remains challenging. Here, we evaluated the hypothesis that geographic selection mosaics maintain genetic diversity in host resistance alleles across gradients of parasite occurrence in smallholder farming systems. We extended GEA analyses to biotic environmental gradients using SDMs to model high-resolution variation in parasite occurrence at continent scales. We report strong associations with parasite occurrence for candidate resistance loci in the sorghum–S. hermonthica pathosystem and characterize diverse loss-of-function mutations in the sorghum resistance gene LGS1. Geographic distribution of loss-of-function alleles suggests that LGS1-conferred resistance is stable across some range of environments and genetic backgrounds. However, the low frequency and paucity of LGS1 loss-of-function alleles outside of parasite-prone areas, combined with the germination sensitivity of some parasite populations to orobanchol and impacts on host photosynthesis regulation, suggest there may be trade-offs associated with LGS1 loss-of-function.
Our results support the hypothesis that spatial variation in selective pressures controls geographic clines in the frequency of host resistance alleles. The patterns we characterized are likely representative of long-term averaged conditions as opposed to a snapshot of coevolution because our parasite-distribution model used occurrence records spanning more than 150 y, and the landraces we studied were collected across the last several decades. In addition to temporal genetic variation (for example, due to negative frequency-dependent selection), many parasites like S. hermonthica are highly genetically diverse across space (26), so that host resistance phenotype depends on local parasite genotypes (39). Depending on the parasite genotype used in experiments, this host by parasite genotype interaction might obscure GWAS for host resistance (40). By capturing variation at coarse scales, the spatial perspective of parasite-associated host genomic variation presented here could facilitate complementary, inexpensive detection of genomic regions contributing to resistance across diverse parasite populations.
Our study revealed evidence for locally adaptive, natural variation in genes related to cell-wall modification. Cell-wall-modifying enzymes, including pectinesterases, are highly expressed in the developing haustorium of parasitic plants and in the host–parasite interface (41–44). Pectinesterases de-esterify pectin in plant cell walls, making it accessible to other cell-wall-degrading enzymes and loosening cell walls. However, some studies have suggested that in the presence of Ca2+, de-esterified pectin forms egg-box structures and, instead, stiffens cell walls (45). Rigidification of sorghum cell walls by their own pectinesterases (such as Sobic.002G138400; SI Appendix, Table S4) or reduced activity could help defend against parasitic invaders. Notably, Yang et al. (43) reported haustorium-specific expression and positive selection on pectinesterase inhibitors in parasitic plant lineages, and a pectinesterase inhibitor showed exceptionally high host-species-associated expression in field populations of S. hermonthica (46). Parasite inhibitors could interact with host pectinesterases or help maintain integrity of parasite cell walls in the face of high pectinesterase expression during haustorial invasion.
Our results also suggest that LGS1 loss-of-function alleles may be adaptive in S. hermonthica-prone regions, but that costs of resistance may limit their distribution. Loss-of-function alleles are relatively uncommon, but higher in frequency and broadly distributed where parasites occur (Fig. 2C). Associations for the putative resistance locus LGS1 were not statistically significant after correcting for kinship, likely due to covariation with genomic background (SI Appendix, Fig. S5), which has been shown to substantially reduce power to detect causal loci in locally adapted sorghum landraces (14) and in simulations (47). Experiments with genome-edited sorghum, however, supported an adaptive role of LGS1 loss-of-function, particularly in marginal environments (Fig. 4A). The diversity of loss-of-function variants reported here (Fig. 2 A and B) and elsewhere (29), their wide geographic distribution (Fig. 2C), and an excess of high-frequency polymorphism localized to LGS1 (SI Appendix, Fig. S6) are further consistent with long-term maintenance of LGS1 diversity under balancing selection. The underlying mechanisms could include negative frequency-dependent selection or spatiotemporally variable selection favoring different alleles in different environments, depending on the relative costs of resistance.
Costs of resistance linked to impacts of SL structural changes on host fitness could include impaired signaling to arbuscular mycorrhizal fungi, susceptibility to S. hermonthica genotypes sensitive to orobanchol, or impacts on endogenous SL signaling. Consistent with the second hypothesis, we found higher sensitivity to root exudates from CRISPR-edited LGS1 deletion lines for S. hermonthica from Kenya compared to S. hermonthica from Mali (Fig. 4). Our findings indicate the existence of S. hermonthica alleles that promote germination on orobanchol, alleles that could increase in frequency following an increase in cultivation of LGS1 deletion sorghum. Consistent with the third hypothesis, we found evidence of systemic down-regulation of photosystem-related genes in LGS1 knockout lines, corresponding to a previously identified role of SLs as positive regulators of light harvesting (48). Perturbations of photosynthesis regulatory networks due to changes in SL profiles could be buffered by other components of the system, ultimately resulting in subtle, but detectable, impacts on host fitness (SI Appendix, Fig. S8).
Taken together, this study provides evidence of locally adaptive natural variation in sorghum parasite-resistance genes across African smallholder farming systems. We describe long-term maintenance of diversity in known and novel candidates implicated in preattachment and postattachment resistance to the parasitic plant S. hermonthica. However, the possibility of trade-offs and the existence of orobanchol-sensitive S. hermonthica populations suggest potential pitfalls with widespread deployment of the LGS1 loss-of-function allele in sorghum cultivation. Our findings highlight the complexity of interacting abiotic, biotic, and human pressures shaping genome polymorphism across environments in cultivated species.
Methods
SDMs.
Genome–environment association analyses are designed to identify putatively locally adaptive genetic loci where allelic variation is strongly associated with home environments (49). To employ this approach with biotic gradients, we required information on local parasite pressure for each sorghum landrace. We used SDMs to estimate the HS of S. hermonthica at the location of each georeferenced sorghum landrace, under the assumption that modeled HS scores are a reasonable proxy of parasite success averaged over the long term and in comparison with sites where the parasite never occurs.
S. hermonthica SDMs were constructed with MaxEnt, a machine learning tool for predicting HS for a species of interest given a set of environmental variables and presence-only data (31). We compiled 1,369 occurrence records for S. hermonthica (Dataset S3; additional details are in SI Appendix). We also created “sorghum-only” models based on a subset of S. hermonthica records (n = 262) that were annotated as occurring specifically on sorghum. Environmental variables were chosen based on prior knowledge of the ecology of S. hermonthica (50). We included bioclimatic and topographic variables (annual rainfall, mean temperature of the wettest quarter, isothermality, potential evapotranspiration [PET], and topographic wetness index) from the CHELSA (51) and ENVIREM datasets (52). Soil variables (clay content, N, and P) were based on continental and global-scale soil-property maps (53, 54). SDMs were implemented and evaluated with ENMeval, using the “checkerboard2” method for data partitioning, which is designed to reduce spatial autocorrelation between testing and training records (55).
Sorghum LGS1 Loss-of-Function Alleles.
Fine-scale natural variation in sorghum LGS1 was characterized by using WGS data from a set of 143 georeferenced landraces from the Sorghum Bioenergy Association Panel (BAP) (56). The BAP includes both sweet and biomass sorghum types; accessions from the five major sorghum botanical races (durra, caudatum, bicolor, guinea, and kafir); and accessions from Africa, Asia, and the Americas. The BAP accessions were sequenced to ∼25× coverage and genotyped as part of the TERRA-REF (Terraphenotyping Reference Platform) project (https://www.terraref.org/). This WGS-derived dataset is referred to throughout the manuscript as the “WGS dataset” to distinguish it from the GBS dataset used for GEA.
We characterized three loss-of-function alleles in LGS1 using data from the WGS dataset. Frameshift and nonsense mutations were identified using SnpEff (Version 4.3t) for SNP calls and small indels in Sobic.005G213600 (57). To characterize large deletion variants, we quality-trimmed reads with BBDuk (https://sourceforge.net/projects/bbmap/; qtrim = rl, trimq = 20) and aligned to the S. bicolor (Version 3.1) reference genome (US Department of Energy [DOE] Joint Genome Institute; https://phytozome.jgi.doe.gov/) with BWA MEM (Version 0.7.17) (58). Duplicates were removed with SAMBLASTER (Version 0.1.24) (59), and structural variants were called for each landrace with LUMPY (Version 0.2.13) (60). SVTYPER (Version 0.6.0) was used to call genotypes for structural variants ≤1 Mb spanning Sobic.005G213600 (61).
After characterizing the LGS1 deletion breakpoints using the WGS dataset, we imputed deletion calls to the GBS dataset. We considered the LGS1 region to be deleted if at least one SNP was called in the 5-kb-region flanking positions of deletion breakpoints, but all data were missing between breakpoints. We considered the LGS1 region to be present if at least one SNP was called within the Sobic.005G213600 gene model. Fifteen low-coverage samples, with missing data extending 5 kb into flanking regions, were excluded.
Experimental Validation of LGS1 Deletion and Frameshift Alleles.
LGS1 loss-of-function alleles were validated by testing 12 accessions from the Sorghum Association Panel (SAP) (62) for their ability to stimulate S. hermonthica germination (SI Appendix, Table S5). Seed was obtained from the US National Plant Germplasm System through the Germplasm Resources Information Network (US Department of Agriculture [USDA], Agricultural Research Service, Plant Genetic Resources Conservation Unit). Two accessions were reported to be resistant to S. hermonthica due to low germination stimulation; two accessions were susceptible; and eight accessions had unknown resistance (29, 63). Root exudates were harvested 43 d after planting (DAP) and used for S. hermonthica germination assays (see SI Appendix for a detailed description of plant growth conditions and germination assays).
GEAs.
We performed a genome-wide scan for SNPs in the sorghum genome strongly associated with values of HS estimated by our S. hermonthica distribution model. Sorghum genotypic information was extracted from a public dataset of accessions genotyped by using GBS (14, 64–66). This dataset comprises a diverse set of worldwide accessions including germplasm from the SAP (62), the Mini-Core Collection (67), and the BAP (56). Beagle (Version 4.1) was used to impute missing data based on the Li and Stephens (68) haplotype frequency model (69). The average missing rate in the nonimputed dataset is 0.39 (66). After excluding sorghum accessions with missing coordinates and SNPs with MAF less than 0.01, the dataset, referred to throughout the manuscript as the “GBS dataset,” included 1,547 African landraces among 2,070 georeferenced accessions total genotyped at 317,294 SNPs.
At each location of a georeferenced accession in the GBS dataset, we extracted logistic output from the S. hermonthica distribution model (71) as the environment. To account for regions where predicted HS was high but S. hermonthica had not been recorded, we cropped model predictions to within 200 km from any occurrence record and set values outside of this range to zero to derive for each grid cell an S. hermonthica occurrence score ranging from zero to one; more than half of sorghum accessions were from locations with parasite HS scores greater than zero (Fig. 1B). Genome-wide associations for each SNP with S. hermonthica occurrence were computed for the GBS dataset by using an MLM fit with GEMMA (Version 0.94) (70). To take into account relatedness among individuals, we used a centered kinship matrix (-gk 1) generated from all 317,294 SNPs before calculation of p score statistics (-lmm 3). P scores were adjusted for multiple comparisons by using the Benjamini and Hochberg (72) procedure (FDR = 0.05). To visualize genomic regions previously implicated in resistance to S. hermonthica, locations of QTL from the linkage mapping study of Haussman et al. (32) in the S. bicolor (Version 3.0) genome were downloaded from the Sorghum QTL Atlas (https://aussorgm.org.au/) (73). We tested for associations with LGS1 loss-of-function mutations using the same procedure and kinship matrix as for the genome-wide association analysis.
We identified gene functions enriched for associations with parasite distribution using the gene-score resampling method in ErmineJ (74). This method places higher value on gene scores than their relative ranking and does not require choice of a threshold for significance. For each gene model, we used the lowest p score from GEMMA of any SNP within 1 kb of gene-model boundaries, and enrichment analyses were performed by using the mean of all gene scores in the gene set. Gene sets were created by using GO terms for all gene models in the S. bicolor (Version 3.0) genome (annotation version 3.1). We also created a custom gene set comprising 30 gene models implicated in SL biosynthesis and signaling (SI Appendix, Table S3). Enrichment analysis was performed with 200,000 iterations, excluding gene sets with less than 5 or more than 200 genes.
Signatures of Selection in Candidate Genomic Regions.
We performed scans for selection in 1-Mb regions surrounding focal SNPs. Linkage disequilibrium between sites was determined with vcftools (Version 0.1.15) (–geno-r2 parameter). To identify regions under balancing selection among a subset of African landraces, we used Tajima’s D calculated with vcftools in nonoverlapping 5-kb windows, excluding SNPs with more than 70% missing data. P values for candidate regions under selection were calculated based on the empirical distribution of Tajima’s D for 1,000 randomly sampled 5-kb windows that overlapped or fully encompassed gene models. We searched for sweeps using the nSL statistic with selscan (Version 1.2.0a) (75).
Performance of CRISPR–Cas9-Edited Sorghum.
We evaluated potential trade-offs to LGS1 loss-of-function by testing S. hermonthica germination response and growth of genome-edited sorghum. LGS1 was deleted in the sorghum line Macia by using the CRISPR–Cas9 system to produce three independent genome-edited lines, with sequence confirmation using Southern-by-Sequencing (SI Appendix).
We measured plant growth rate in a greenhouse at Corteva Agriscience using a hyperspectal imaging system to determine total leaf area of each plant (day temperature: 26 °C; night temperature: 20 °C; 16-h photoperiod). We compared three independent CRISPR-edited lines (lgs1-d lines 1 to 3), the wild-type Macia line, and two event-null transformation lines that had intact LGS1 but SNPs at the two cutting sites flanking LGS1. Experimental design was unbalanced incomplete block with replications of 10 to 33 plants. Images were collected weekly on days 23 to 65 after planting. Total leaf area per plant was analyzed by using one-way ANOVA followed by Scheffé’s method.
In a separate experiment at Pennsylvania State University (Penn State), we compared the same three CRISPR-edited lines and two independently bulked seed batches of wild-type Macia under three conditions: well-watered with high nutrition (WWHN), well-watered with low nutrition (WWLN), and drought with low nutrition (DLN). Plants (n = 19 per line and treatment) were grown in autoclaved sand in cone-tainers under natural-light conditions in a greenhouse (day temperature: 27 °C; night temperature: 24 °C). For the WWHN treatment, plants were fertilized every other day with 15 mL of 1/10th strength Miracle-Gro Plant Food (24% N, 8% P, and 16% potassium) and watered with 15 mL of tap water on days without fertilization. Plants were watered every day with 15 mL of tap water (WWLN treatment) or every 4 d with 15 mL water (DLN treatment). Three plants per line and treatment were harvested at 21 DAP for collection of root exudates as described for validation of natural deletion alleles (SI Appendix). Germination assays testing root exudate in 12-well plates (three technical replicates per biological replicate, ∼60 seeds per well) were performed in the USDA quarantine facility at Penn State, using S. hermonthica seeds collected in 2016 from a sorghum field in Kibos, Kenya (0°40′S; 34°49′E) and in 2018 from a sorghum field in Siby, Mali (12°23′ N; 8°20′ W). Germination assays used a preconditioning temperature of 29 °C for 11 d in 1 mL of deionized water (diH2O) prior to addition of 1.5 mL of root exudate. A complementary assay tested the response of the same S. hermonthica populations to five concentrations (0, 1 × 10−10, 1 × 10−8, 1 × 10−7, and 2 × 10−7 M) of (±)orobanchol and (+)5-deoxystrigol (Chemical Abstracts Service nos. 220493-64-1 and 151716-18-6; Olchemim).
To compare performance of wild-type and CRISPR-edited sorghum, we also measured biomass (shoot and root dried weight), chlorophyll concentration (measured via SPAD meter), root architecture traits (number of crown, adventitious [shoot-borne], and seminal roots), shoot architecture traits (plant height, leaf number, total shoot length, and leaf internode lengths), and SLA at 41 DAP. We tested significance of deletion alleles using generalized linear mixed models for count data (root number) or linear mixed effects models (all other traits) in the R package lme4 (76), where deletion and treatment were fixed effects and sorghum lines were random effects. For germination rate data, fixed effects of S. hermonthica population (Kibos or Siby) and root weight were also included.
LGS1 and Expression of Genes in SL Synthesis and Signaling Pathways.
To assess the impact of putatively locally adaptive variation at LGS1, we studied transcriptomes for roots and shoots (excluding leaf blade) of CRISPR knockout (lgs1-d line 3) and wild-type Macia lines at 30 DAP under WWLN, the treatment under which plants exuded the most SLs (Fig. 4). RNA extractions were performed with the NucleoSpin RNA Plant kit (Machery-Nagel). We used the 3′-TagSeq approach (77), which quantifies messenger RNA based on the 3′ end of transcripts. Complementary DNA libraries were sequenced on a shared lane of an Illumina Nova-Seq at the Genomic Sequencing and Analysis Facility at the University of Texas at Austin. We recovered between 10 million and 15 million raw single-end 100-bp reads per sample and compared expression differences using the TagSeq (Version 2.0) pipeline (https://github.com/Eli-Meyer/TagSeq_utilities/). We also generated TagSeq libraries using root tissue from five replicate individuals each of sorghum lines Shanqui Red (Plant Introduction [PI] no. 656025) and SRN39 (PI 656027) grown under nutrient-deficient conditions (SI Appendix). Count data were analyzed in DESeq2 with FDR correction (ɑ = 0.05) (78). Enrichment analysis was performed as for genome-wide association analysis, except that only the set of gene models with nonzero expression were used as the background.
Data Availability.
Raw reads generated for the TagSeq study have been deposited in the National Center for Biotechnology Information Sequence Read Archive database under BioProject accession no. PRJNA542394. S. hermonthica occurrence records are available in SI Appendix. Environmental niche models and additional datasets are available from Penn State ScholarSphere (https://doi.org/10.26207/bfct-ca95) (71).
Supplementary Material
Acknowledgments
We thank the many collectors, volunteers, and herbarium curators who made this work possible and are particularly grateful to Marie-Hélène Weech and the staff of the Royal Botanic Gardens Kew and the Muséum National d’Histoire Naturelle (MNHN), Paris. Historical data from French herbaria were obtained thanks to “Les Herbonautes” (MNHN/Tela Botanica), part of Infrastructure Nationale e-ReColNat (ANR-11-INBS-0004). We thank Alice MacQueen for comments that improved the manuscript; Ping Che for assistance with tissue culture; Meizhu Yang for molecular analysis of CRISPR mutants; Eric Schultz for hyperspectral phenotyping; and the dean of Eberly College and head of the Department of Biology for their support in construction of the USDA–Animal and Plant Health Inspection Service-certified Parasitic Plant Containment Laboratory at Penn State. WGS data used here are from the TERRA-REF project, funded by the Advanced Research Projects Agency-Energy, US DOE, under Award DE-AR0000594. This study is based on work supported by an NSF Postdoctoral Research Fellowship in Biology (to E.S.B.) under Grant 1711950. The views and opinions of authors expressed herein do not necessarily state or reflect those of the US government or any agency thereof.
Footnotes
Competing interest statement: H.G. and N.D.C. are employees of Corteva Agriscience.
This article is a PNAS Direct Submission. J.N.T. is a guest editor invited by the Editorial Board.
Data deposition: Raw TagSeq reads generated for this study have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database, https://www.ncbi.nlm.nih.gov/sra (BioProject accession no. PRJNA542394). Environmental niche models and additional datasets are available from Penn State ScholarSphere (https://doi.org/10.26207/bfct-ca95).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908707117/-/DCSupplemental.
References
- 1.Haldane J. B. S., Disease and evolution. Ric. Sci. 19, 68–76 (1949). [Google Scholar]
- 2.Stahl E. A., Dwyer G., Mauricio R., Kreitman M., Bergelson J., Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400, 667–671 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Tack A. J. M., Thrall P. H., Barrett L. G., Burdon J. J., Laine A.-L., Variation in infectivity and aggressiveness in space and time in wild host-pathogen systems: Causes and consequences. J. Evol. Biol. 25, 1918–1936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karasov T. L., Horton M. W., Bergelson J., Genomic variability as a driver of plant-pathogen coevolution? Curr. Opin. Plant Biol. 18, 24–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockhurst M. A., et al. , Running with the Red Queen: The role of biotic conflicts in evolution. Proc. Biol. Sci. 281, 20141382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergelson J., Dwyer G., Emerson J. J., Models and data on plant-enemy coevolution. Annu. Rev. Genet. 35, 469–499 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Agrawal A., Lively C. M., Infection genetics: Gene-for-gene versus matching- alleles models and all points in between. Evol. Ecol. Res. 4, 79–90 (2002). [Google Scholar]
- 8.Möller M., Stukenbrock E. H., Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 15, 756–771 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Samberg L. H., Gerber J. S., Ramankutty N., Herrero M., West P. C., Subnational distribution of average farm size and smallholder contributions to global food production. Environ. Res. Lett. 11, 124010 (2016). [Google Scholar]
- 10.Jensen H. R., Dreiseitl A., Sadiki M., Schoen D. J., The Red Queen and the seed bank: Pathogen resistance of ex situ and in situ conserved barley. Evol. Appl. 5, 353–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jousimo J., et al. , Disease ecology. Ecological and evolutionary effects of fragmentation on infectious disease dynamics. Science 344, 1289–1293 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Bartoli C., Roux F., Genome-wide association studies in plant pathosystems: Toward an ecological genomics approach. Front. Plant Sci. 8, 763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer R. S., Purugganan M. D., Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Lasky J. R., et al. , Genome-environment associations in sorghum landraces predict adaptive traits. Sci. Adv. 1, e1400218 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romero Navarro J. A., et al. , A study of allelic diversity underlying flowering-time adaptation in maize landraces. Nat. Genet. 49, 476–480 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Vajana E., et al. , Combining landscape genomics and ecological modelling to investigate local adaptation of indigenous Ugandan Cattle to East Coast fever. Front. Genet. 9, 385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaillard M. D. P., Glauser G., Robert C. A. M., Turlings T. C. J., Fine-tuning the ‘plant domestication-reduced defense’ hypothesis: Specialist vs generalist herbivores. New Phytol. 217, 355–366 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Winchell F., Stevens C. J., Murphy C., Champion L., Fuller D. Q., Evidence for sorghum domestication in fourth millennium BC eastern Sudan: Spikelet morphology from ceramic impressions of the Butana group. Curr. Anthropol. 58, 673–683 (2017). [Google Scholar]
- 19.Gomez-Roldan V., et al. , Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Ruyter-Spira C., et al. , Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 155, 721–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha C. V., et al. , Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. U.S.A. 111, 851–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoneyama K., et al. , Nitrogen deficiency as well as phosphorus deficiency in sorghum promotes the production and exudation of 5-deoxystrigol, the host recognition signal for arbuscular mycorrhizal fungi and root parasites. Planta 227, 125–132 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ruiz-Lozano J. M., et al. , Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 39, 441–452 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Akiyama K., Matsuzaki K., Hayashi H., Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Spallek T., Mutuku M., Shirasu K., The genus Striga: A witch profile. Mol. Plant Pathol. 14, 861–869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timko M. P., Huang K., Lis K. E., Host resistance and parasite virulence in Striga–host plant interactions: A shifting balance of power. Weed Sci. 60, 307–315 (2012). [Google Scholar]
- 27.Laine A.-L., Resistance variation within and among host populations in a plant–pathogen metapopulation: Implications for regional pathogen dynamics. J. Ecol. 92, 990–1000 (2004). [Google Scholar]
- 28.Thompson J. N., Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 153, S1–S14 (1999). [Google Scholar]
- 29.Gobena D., et al. , Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl. Acad. Sci. U.S.A. 114, 4471–4476 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohemed N., et al. , Genetic variation in Sorghum bicolor strigolactones and their role in resistance against Striga hermonthica. J. Exp. Bot. 69, 2415–2430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips S. J., Anderson R. P., Schapire R. E., Maximum entropy modeling of species geographic distributions. Ecol. Modell. 190, 231–259 (2006). [Google Scholar]
- 32.Haussmann B. I. G., et al. , Genomic regions influencing resistance to the parasitic weed Striga hermonthica in two recombinant inbred populations of sorghum. Theor. Appl. Genet. 109, 1005–1016 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Jiang L., et al. , DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swarbrick P. J., et al. , Global patterns of gene expression in rice cultivars undergoing a susceptible or resistant interaction with the parasitic plant Striga hermonthica. New Phytol. 179, 515–529 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Mutuku J. M., et al. , The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiol. 179, 1796–1809 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mace E. S., et al. , Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4, 2320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrer-Admetlla A., Liang M., Korneliussen T., Nielsen R., On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 31, 1275–1291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverstone A. L., Ciampaglio C. N., Sun T., The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltis N. E., et al. , Interactions of tomato and Botrytis cinerea genetic diversity: Parsing the contributions of host differentiation, domestication, and pathogen variation. Plant Cell 31, 502–519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacPherson A., Otto S. P., Nuismer S. L., Keeping pace with the Red Queen: Identifying the genetic basis of susceptibility to infectious disease. Genetics 208, 779–789 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Losner-Goshen D., Portnoy V. H., Mayer A. M., Joel D. M., Pectolytic activity by the haustorium of the parasitic plant Orobanche L. (Orobanchaceae) in host roots. Ann. Bot. 81, 319–326 (1998). [Google Scholar]
- 42.Honaas L. A., et al. , Functional genomics of a generalist parasitic plant: Laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 13, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z., et al. , Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol. Biol. Evol. 32, 767–790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun G., et al. , Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat. Commun. 9, 2683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hocq L., Pelloux J., Lefebvre V., Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 22, 20–29 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Lopez L., et al. , Transcriptomics of host-specific interactions in natural populations of the parasitic plant purple witchweed (Striga hermonthica). Weed Sci. 67, 397–411 (2019). [Google Scholar]
- 47.Forester B. R., Lasky J. R., Wagner H. H., Urban D. L., Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations. Mol. Ecol. 27, 2215–2233 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Mayzlish-Gati E., et al. , Strigolactones are positive regulators of light-harvesting genes in tomato. J. Exp. Bot. 61, 3129–3136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rellstab C., Gugerli F., Eckert A. J., Hancock A. M., Holderegger R., A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 24, 4348–4370 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Mohamed K. I., Musselman L. J., Riches C. R., The genus Striga (Scrophulariaceae) in Africa. Ann. Mo. Bot. Gard. 88, 60–103 (2001). [Google Scholar]
- 51.Karger D. N., et al. , Climatologies at high resolution for the earth’s land surface areas. Sci. Data 4, 170122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Title P. O., Bemmels J. B., ENVIREM: An expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography 41, 291–307 (2018). [Google Scholar]
- 53.Hengl T., et al. , SoilGrids250m: Global gridded soil information based on machine learning. PLoS One 12, e0169748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hengl T., et al. , Soil nutrient maps of Sub-Saharan Africa: Assessment of soil nutrient content at 250 m spatial resolution using machine learning. Nutr. Cycl. Agroecosyst. 109, 77–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muscarella R., et al. , ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 5, 1198–1205 (2014). [Google Scholar]
- 56.Brenton Z. W., et al. , A genomic resource for the development, improvement, and exploitation of sorghum for bioenergy. Genetics 204, 21–33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faust G. G., Hall I. M., SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Layer R. M., Chiang C., Quinlan A. R., Hall I. M., LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 15, R84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang C., et al. , SpeedSeq: Ultra-fast personal genome analysis and interpretation. Nat. Methods 12, 966–968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casa A. M., et al. , Community resources and strategies for association mapping in sorghum. Crop Sci. 48, 30–40 (2008). [Google Scholar]
- 63.Hess D. E., Ejeta G., Butler L. G., Selecting sorghum genotypes expressing a quantitative biosynthetic trait that confers resistance to Striga. Phytochemistry 31, 493–497 (1992). [Google Scholar]
- 64.Elshire R. J., et al. , A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morris G. P., et al. , Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. U.S.A. 110, 453–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu Z., Olatoye M. O., Marla S., Morris G. P., An integrated genotyping-by-sequencing polymorphism map for over 10,000 sorghum genotypes. Plant Genome 12, 180044 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Upadhyaya H. D., et al. , Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 49, 1769–1780 (2009). [Google Scholar]
- 68.Li N., Stephens M., Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browning B. L., Browning S. R., Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98, 116–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou X., Stephens M., Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellis E., Striga hermonthica environmental niche models. Penn State ScholarSphere. 10.26207/bfct-ca95. Deposited 24 January 2020. [DOI]
- 72.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 57, 289–300 (1995). [Google Scholar]
- 73.Mace E., et al. , The Sorghum QTL Atlas: A powerful tool for trait dissection, comparative genomics and crop improvement. Theor. Appl. Genet. 132, 751–766 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Gillis J., Mistry M., Pavlidis P., Gene function analysis in complex data sets using ErmineJ. Nat. Protoc. 5, 1148–1159 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Szpiech Z. A., Hernandez R. D., selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31, 2824–2827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 77.Meyer E., Aglyamova G. V., Matz M. V., Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol. Ecol. 20, 3599–3616 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads generated for the TagSeq study have been deposited in the National Center for Biotechnology Information Sequence Read Archive database under BioProject accession no. PRJNA542394. S. hermonthica occurrence records are available in SI Appendix. Environmental niche models and additional datasets are available from Penn State ScholarSphere (https://doi.org/10.26207/bfct-ca95) (71).