Significance
There is growing interest in pharmacological reactivation of the inactive X chromosome (Xi) as a method of treating X-linked disorders. Indeed, unsilencing of the normal gene copies on the Xi could potentially restore expression of the missing protein for therapeutic benefit. Because general Xi reactivation would be expected to increase expression of some other X-linked genes, the in vivo consequences of any dosage imbalance must be investigated with care. Here, we use multiple mouse models to investigate systemic effects of Xi reactivation and observe a surprising tolerance to loss of Xist RNA and partial Xi reactivation. However, we also observe that Xist and XCI are protective to females during chronic stress in the gut.
Keywords: Xist, Rett syndrome, X reactivation, X inactivation, polyps
Abstract
Long thought to be dispensable after establishing X chromosome inactivation (XCI), Xist RNA is now known to also maintain the inactive X (Xi). To what extent somatic X reactivation causes physiological abnormalities is an active area of inquiry. Here, we use multiple mouse models to investigate in vivo consequences. First, when Xist is deleted systemically in post-XCI embryonic cells using the Meox2-Cre driver, female pups exhibit no morbidity or mortality despite partial X reactivation. Second, when Xist is conditionally deleted in epithelial cells using Keratin14-Cre or in B cells using CD19-Cre, female mice have a normal life span without obvious illness. Third, when Xist is deleted in gut using Villin-Cre, female mice remain healthy despite significant X–autosome dosage imbalance. Finally, when the gut is acutely stressed by azoxymethane/dextran sulfate (AOM/DSS) exposure, both Xist-deleted and wild-type mice develop gastrointestinal tumors. Intriguingly, however, under prolonged stress, mutant mice develop larger tumors and have a higher tumor burden. The effect is female specific. Altogether, these observations reveal a surprising systemic tolerance to Xist loss but importantly reveal that Xist and XCI are protective to females during chronic stress.
In eutherian mammals, dosages of X-linked genes are equalized between XX females and XY males during early development by an epigenetic silencing process known as X chromosome inactivation (XCI) (1–3). XCI ensures not only a dosage balance between XX and XY individuals, but also a critical balance between sex chromosomes and autosomes—the so-called “X-to-autosome ratio.” XCI is mediated by Xist, a long noncoding RNA (lncRNA) transcribed from one of two X chromosomes, which spreads across the chromosome in cis (4–6) and establishes global silencing by repelling activators (7, 8) and recruiting silencers (9–11). XCI and proper dosage compensation are critical for mammalian development, as mouse embryos lacking Xist perish shortly after implantation (12) and rarely survive to birth in an epiblast-specific deletion model for Xist loss (13).
While there is general agreement that Xist is essential for the initiation and establishment of XCI (12, 14–16), to what extent Xist remains necessary in the post-XCI embryonic state and into adulthood continues to be intensively studied. The question impacts not only our general understanding of how epigenetic mechanisms are maintained but also a growing interest in pharmacological inactive X (Xi) reactivation as a method of treating X-linked neurodevelopmental disorders (17–21). In Rett syndrome and CDKL5 syndrome, for example, unsilencing of the wild-type alleles of MECP2 and CDKL5 (respectively) on the Xi could potentially restore expression of the missing protein for therapeutic benefit. Because general Xi reactivation would be expected to increase expression of some other X-linked genes, the in vivo consequences of any dosage imbalance—no matter how small—must be investigated with care.
Although high-level Xist expression persists throughout female life, Xist was once thought to be dispensable after XCI is established in the early embryo, as the Xi appeared to be largely unperturbed in cell culture when Xist is removed (22–24). With the availability of more sensitive and quantitative assays, however, more recent studies have documented the occurrence of partial Xi reactivation when Xist is deleted in post-XCI mouse and human cells (13, 17, 18, 20, 25–27). The partial Xi reactivation results in a shift in X-to-autosome dosage balance (13, 18, 20). Significant physiological consequences have been observed in some cases. For instance, female human induced pluripotent stem cells (hiPSCs) that lose XIST expression and fail to undergo XCI cannot differentiate properly in an in vivo teratoma assay, and instead continue to proliferate as undifferentiated cells (26). In another case, female mice succumb to blood cancer when Xist is deleted using the Vav-Cre driver in hematopoietic stem cells (HSCs) (27). Partial up-regulation of X-linked genes was observed in both the hiPSCs and the mouse HSCs. These observations argue that Xist/XIST expression and maintenance of dosage compensation can be important during development and through adulthood.
These conclusions are, however, at odds with other studies demonstrating that losing Xist expression has no negative consequence. In two independent studies, Xist was conditionally deleted in the brain and shown to have no impact on female mice (18, 20). The female mice lived a normal life span and did not display a predisposition to brain tumors or any other obvious central nervous system disorders, despite partial Xi reactivation. In another study, Sox2-Cre was used to delete Xist around the time of de novo XCI in the periimplantation embryo, and female pups showed no evidence of hematologic cancer or any other tumor (13). Yet another study showed that mice lacking Stc1, a presumptive XCI regulator, live a normal life span (17). From the standpoint of pharmacological safety, these findings are encouraging. However, caution must still be exercised until additional studies address the following critical questions: 1) Is there a tissue-specific sensitivity to Xist deletion? and 2) Are there physiological states in which an organism becomes less able to tolerate Xi reactivation? Here, we investigate these questions by carrying out additional tissue-specific deletions of Xist in mice. Our observations demonstrate a surprising tolerance for Xi reactivation in multiple tissues, but also reveal that chronic stress can unmask a sensitivity to Xist loss in the gut.
Results
Multiorgan Xist Deletion in Postimplantation Embryos Using Meox2-Cre.
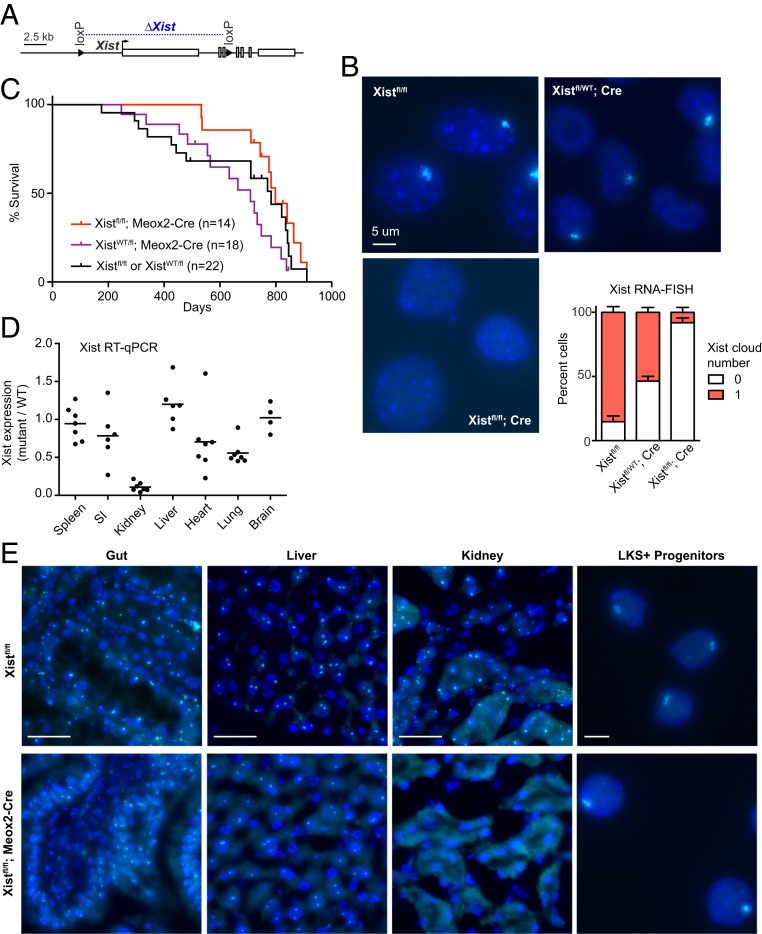
We first generated female mice in which Xist ablation occurs after the establishment of XCI. XCI is normally established by embryonic day 5.5 (E5.5) in mice (28–30). With the goal of eliminating Xist immediately after XCI is established in all somatic tissues, we used the mesenchyme homeobox 2 (Meox2)-Cre driver, which is expressed after uterine implantation in the embryo proper (31–33). We crossed males carrying the Meox2-Cre driver to female mice bearing the conditional Xistfl/fl allele (23) (Fig. 1A) to induce Xist excision specifically in the embryo proper, leaving extraembryonic lineages unaffected by Meox2-Cre. In this setup, Xist expression was expected to be lost in 50% of cells, as the floxed allele would be carried on the Xi in only one-half of cells, due to the random nature of XCI. Xist RNA fluorescent in situ hybridization (RNA-FISH) in mouse embryonic fibroblasts (MEFs) from Xistfl/WT; Meox2-Cre embryos indeed showed loss of the Xist RNA cluster (“cloud”) in ∼50% of cells (Fig. 1B), consistent with the idea that Xist excision occurred in post-XCI in cells of the early embryo. To generate homozygous Xist mutants, we crossed Xistfl/Y males to Meox-Cre; Xistfl/fl females and confirmed loss of Xist expression in >90% of fibroblast cells in the Xist-null female progeny (Fig. 1B).
Fig. 1.
Conditionally deleting Xist shortly after implantation (Meox2-Cre) and establishment of XCI does not affect overall female survival. (A) Schematic of the conditional Xist allele with region of deletion shown in blue. (B) Representative Xist RNA-FISH images of fibroblasts of control (Xistfl/WT), heterozygous mutants (Xistfl/WT; Meox2-Cre), and homozygous mutants (Xistfl/fl; Meox2-Cre). At least 100 nuclei were counted per sample. Xistfl/WT, n = 2 animals; Xistfl/WT; Meox2-Cre, n = 6; Xistfl/fl; Meox2-Cre, n = 2. Error bars denote mean ± SEM. (Scale bar, 5 μm.) (C) Kaplan–Meier survival curve for mouse crosses as indicated. (D) Xist RT-qPCR for panel of tissues from Xist homozygous mutants. The average expression of at least four mutant animals per tissue type is indicated; normalization was to Xist expression in corresponding tissues from four to six control animals. (E) Representative Xist RNA-FISH for tissue sections from control and homozygous mutant animals. LKS+; Lineage− cKit+ Sca1+ hematopoietic progenitors. (Scale bars: 50 μm; for LKS+ hematopoietic progenitors, 5 μm.)
We followed the Xist mutants over 3 y and found that the animals were healthy, fertile, and lived a normal life span (Fig. 1C). Quantitative RT-PCR and Xist RNA-FISH demonstrated variable degrees of Xist RNA depletion in different organs of homozygous Xist mutants (Fig. 1 D and E). In the kidney, Xist expression was nearly completely ablated, suggesting that the kidney tolerates Xist loss well. Wider variation was seen in the heart, lung, and small intestine, suggesting a greater sensitivity to Xist loss. On the other hand, Xist expression was largely preserved in the hematopoietic compartment (spleen), liver, and brain, suggesting that these tissues might not tolerate Xist ablation so well. These findings were surprising. While the hematopoietic compartment is already known to be sensitive to Xist loss (27), the brain has been shown in two independent studies to tolerate Xist loss and Xi reactivation (20, 21). This raised the question of whether Cre was efficiently expressed in the mice. Meox2-Cre is known to be robustly expressed in all tissues and was confirmed here to be active in multiple tissues following excision of the lox-stop-lox (lsl) cassette, enabling Cre-dependent YFP protein expression (SI Appendix, Figs. S1 and S2). We therefore believe that the mosaicism seen in different tissues may arise from selection against Xist-null cells because of their relative survival disadvantage in vivo.
Such mosaicism and presumptive selection against Xist-null cells were also observed in our previous Sox2-Cre Xist deletion model (13). While Sox2-Cre and Meox2-Cre alleles were both used for effecting LoxP recombination in all cells of the epiblast, the alleles were used to answer different biological questions. Xist ablation occurs predominantly before the onset of XCI in the Sox2-Cre model, enabling the study of effects during the de novo establishment phase of random XCI. The Meox2-Cre model, on the other hand, initiates Xist ablation after the establishment phase, enabling the study of Xist during early maintenance phase. Our studies showed that there is strong selective pressure against cells that have lost Xist in both systems, but the outcomes are manifested differently. In the case of Sox2-Cre embryos, most of the female embryos perished during development; surviving embryos were subjected to strong pressure to become XO, an aneuploid state that would obviate the need for dosage compensation (13). In the case of Meox2-Cre embryos, embryos preserved overall organismal fitness by favoring cells that did not delete Xist. The persistence of cells with Xist expression despite efficient recombination (SI Appendix, Fig. S1) suggests strong selective pressure against Xist-null cells. We anticipate that these Xist+ cells would rapidly outpopulate mutant cells in the embryo.
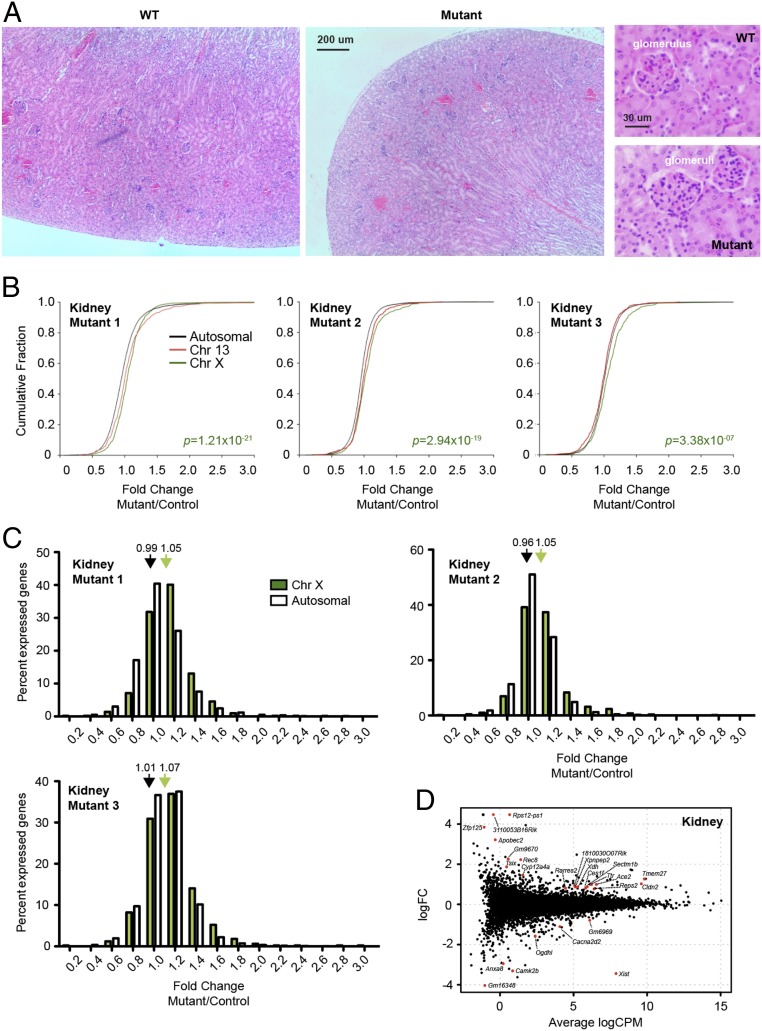
Given that the kidney was the only organ with near-complete depletion of Xist RNA, we continued analysis using kidney tissue. Gross examination did not detect any abnormalities, and histological analysis of glomeruli, proximal and distal convoluted tubules, and collecting ducts was unremarkable, with no evidence of disease (Fig. 2A). To determine whether there is an X–autosomal dosage imbalance resulting from Xist loss, we performed RNA-sequencing (RNA-seq) for three control kidneys and three Xistfl/fl; Meox2-Cre mutant kidney samples. When we looked at the distribution of gene expression fold changes (mutant/wild type) as a whole for genes on the X chromosome, we observed that they were more likely to be up-regulated (by Wilcoxon rank sum test) compared to chromosome 13, a representative autosome with a similar number of genes (Fig. 2B, red curves). There was also a significant increase when X-linked expression was compared to the rest of the genome (Fig. 2B, black curves). The X–autosome imbalances were evident as a right shift in the cumulative distribution plots of fold changes for individual mutants (Fig. 2B), as well as in the combined plot (SI Appendix, Fig. S3A). Notably, the overall fold change for X-linked genes was 1.05 to 1.07 (Fig. 2B and SI Appendix, Fig. S3B), indicating a 5 to 7% increase in X chromosome dosage. This may be an underestimate, considering that this is an averaged value across the entire organ. Although there was a near-complete loss of Xist expression in the kidney, cells that retained Xist may mask the actual degree of reactivation in mutant cells.
Fig. 2.
Xist-deleted organ of Meox2-Cre females show mild X–autosome dosage imbalance. (A) H&E stain of wild-type and mutant kidneys at 40× and 400×. (B) Cumulative distribution plot for fold changes of genes on chromosome 13 (red), chromosome X (green), and all autosomes (black). Three biological replicates (three female mutants; three controls) are shown. P value, Wilcoxon’s rank sum test. (C) Distribution of X-linked (green) vs. autosomal (white) fold changes in three Xist-null kidneys relative to three controls. Average fold changes of X-linked or autosomal genes are indicated. **P < 0.01; ***P < 10−8, Fisher’s exact test. Data represent mean ± SEM. (D) Genome-wide differential expression in the kidney of WT vs. Xist-mutant females. Log fold change in mutant vs. WT shown. Differentially expressed genes (FDR < 0.05) are in red.
While there was a small degree of X reactivation across the whole chromosome, differential gene expression analysis (DE-seq) indicated a small number of up-regulated (19) and down-regulated (7) genes with a false-discovery rate (FDR) < 0.05 by the Benjamini–Hochberg analysis (Fig. 2D). Approximately one-half of all up-regulated genes were X-linked (9 of 19). On the other hand, Xist was the only down-regulated X-linked gene. This suggests that, while multiple X-linked genes might be up-regulated (Fig. 2 B–D), only a few showed a large enough change to be above the threshold for detection by DE-seq. The relatively small number of genes identified by DE-seq in the kidney contrasts with the hundreds of genes showing expression changes in the hematopoietic compartment (27), thereby providing one possible explanation for the kidney’s tolerance of Xist deletion. Altogether, these analyses indicate that the kidney tolerates Xist deletion well despite a small degree of X reactivation across the entire chromosome.
Conditional Xist Deletion in Epithelial Cells Using Keratin14-Cre.
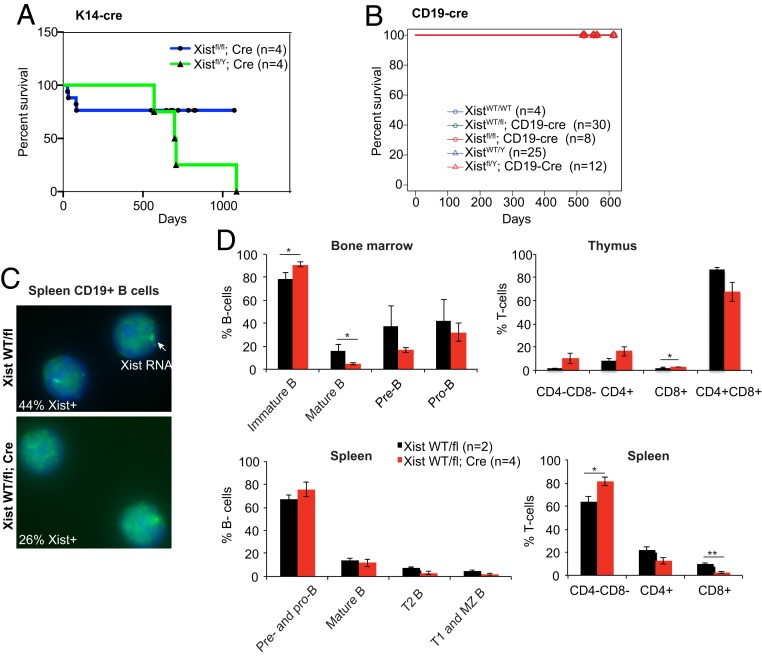
We reasoned that the surprising overall fitness of the female mice exposed to Meox2-Cre likely arose from tissue mosaicism, for the Xist deletion. In an effort to circumvent this, we turned to more tissue-restricted conditional knockouts that act at later stages of development. To delete Xist specifically in epithelial tissues, we used the Keratin14 (K14)-Cre driver (34) that is known to be active in ectoderm-derived tissues including skin (SI Appendix, Fig. S4A). We crossed Xistfl/Y males to Xistfl/fl; K14-Cre females and confirmed the Xist deletion in skin. Despite the absence of Xist, Xist-null females and males lived a normal life span (Fig. 3A) and did not exhibit any obvious skin disorders throughout life. Given the absence of any morbidity and mortality, we did not further characterize the K14-Cre animals. We conclude that deleting Xist in the skin has no phenotypic consequence for either female or male mice.
Fig. 3.
No evident morbidity and mortality in female mice conditionally deleted for Xist in epithelial (K14-Cre) and B cell (CD19-Cre) lineages. (A) Kaplan–Meier kill curves plotted over 1,000 d for K14-cre animals. There were no significance differences between Xistfl/fl; K14-cre or Xistfl/Y; K14-cre males. (B) Kaplan–Meier kill curves plotted over 600 d for CD19-cre animals. There were no differences between control groups and XistWT/fl; CD19-cre or Xistfl/fl; CD19-cre females, or Xistfl/Y; CD19-cre males. Control groups contained XistWT/fl, XistWT/WT, CD19-cre, Xist Xist+/+ females and Xistfl/Y, Xist+/Y, and Xist+/Y; CD19-cre males. All curves are superimposed at 100% survival. (C) Representative Xist RNA-FISH for spleen B cells from XistWT/fl vs. XistWT/fl; CD19-cre females. (D) FACS analyses of B and T cell percentages in bone marrow, thymus, and spleen of XistWT/fl vs. XistWT/fl ; CD19-cre females. See also FACS data in SI Appendix, Fig. S4. B cells in bone marrow were categorized based on expression of B220, IgM, and CD43; Immature B cells (B220−, IgM−), mature B cells (B220+, IgM+), pre-B cells (B220+, IgM−, CD43−), pro-B cells (B220+, IgM−, CD43+). B cells in spleen were categorized based on expression of IgM and IgD; pre- and pro-B cells (IgM−, IgD−), mature B cells (IgDhigh, IgMlow), T2 B cells (IgDhigh, IgMhigh), and T1 and marginal zone (MZ) B cells (IgD−, IgMhigh). T cells were categorized based on expression of CD4 and CD8. Percentage of cells are means ± SEM. Significance is calculated using t test. *P < 0.05; **P < 0.01.
Conditional Xist Deletion in B Cells Using CD19-Cre.
Next, we turned to the blood compartment where we previously observed multilineage tumors upon deleting Xist in HSCs (27). In light of this striking phenotype, we asked whether deleting Xist in a more restricted blood lineage would recapitulate cancer. To this end, we used the CD19-Cre driver, which drives Cre expression throughout B lymphocyte development and differentiation (35). We generated both heterozygous and homozygous Xist-deleted mutant females (SI Appendix, Fig. S4B). However, neither male nor female mice exhibited any excess morbidity nor mortality associated with Xist loss and Kaplan–Meier analysis of survival across a 2-y timeframe showed a normal life span for all Xist genotypes (Fig. 3B).
Because the absence of a B cell disorder might seem surprising, given the multilineage cancer seen in the Vav-Cre-driven Xist deletions in HSCs (27), we performed fluorescence-activated cell sorting (FACS) analysis to examine B cell development and differentiation in the CD19-Cre–deleted heterozygous animals. There was clear Xist loss in B cell lineages, with Xist RNA expressed in only one-half the normal number of cells, consistent with the animals being heterozygous (Fig. 3C). Interestingly, although there was a decrease in the number of Pro-B cells in the bone marrow (SI Appendix, Fig. S4 C–E), the overall percentage was not significantly different (Fig. 3D). There was also a decrease in the percentage of mature B cells (IgM+B220+) (Fig. 3D), but the overall number was not significantly different (SI Appendix, Fig. S4 C–E). No striking changes were seen in other hematologic compartments as well, including thymus and spleen (Fig. 3D and SI Appendix, Fig. S4 C–E). We therefore conclude that deleting Xist in a more restricted blood lineage does not recapitulate the systemwide dysfunction and cancers seen in the HSC model.
Gut-specific Xist Deletion Using Villin-Cre.
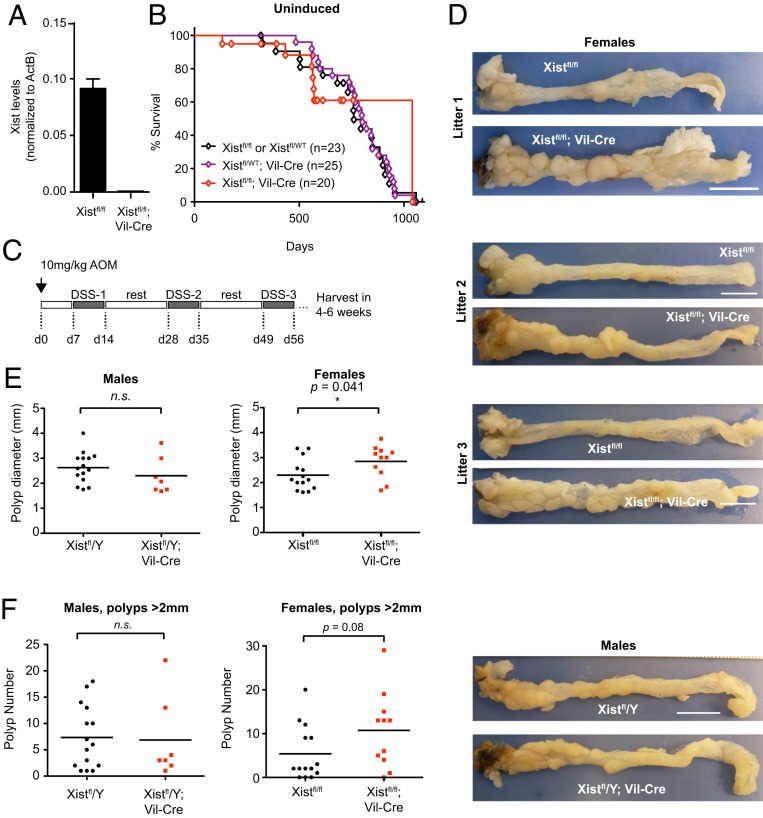
We next turned to the gut—specifically, the gastrointestinal (GI) epithelium, a tissue in which the rate of cell turnover and proliferation is higher than that of any other tissue examined to date. Because renewal of the entire gut epithelia occurs every 3 to 5 d (36), if an Xist phenotype depended on high cell turnover, we would most likely observe it in the gut, in addition to the HSC model shown previously (27). Here, we used the Villin-Cre driver, which is active beginning E12.5 in the epithelial lining of villi and crypts along the entire length of the gut (37). By crossing Xistfl/fl females to Xistfl/Y; Villin-Cre animals, we derived homozygous mutants with Xist loss specifically in the intestinal epithelia (Fig. 4A). Despite a near-complete loss of Xist in the gut epithelium (Fig. 4A), the Xist-null females were surprisingly normal. The females did not show distress, weight loss, or shortened life span relative to control male littermates (Fig. 4B).
Fig. 4.
Chronic stress leads to heavier tumor burden in gut-deleted Xist-mutant females. (A) Xist RT-qPCR for colon epithelial samples; n = 2. Data represent mean ± SEM. (B) Kaplan–Meier survival curve for unstressed animals (not treated with AOM/DSS). (C) AOM/DSS chemical induction protocol. (D) Whole colon from control and Xistfl/fl; Villin-Cre females and Xistfl/Y; Villin-Cre males. (Scale bar, 1 cm.) Three age- and litter-matched biological replicates (mutant female mice) shown. (E) Average polyp diameter in colons of control vs. mutant animals. *P < 0.05 (Student’s t test); n.s, not significant. (F) Average number of polyps in colons of control vs. mutant animals.
A Stress Phenotype: Gut Lacking Xist Shows a Heavy Tumor Burden.
Because development of colorectal cancer typically has a long latency period, tumors might not present clinically during the short (2 to 3 y) life span of a mouse and could therefore go undetected during the study period. Because stress is known to accelerate tumor development in mice, we primed animals using the well-established azoxymethane/dextran sodium sulfate (AOM/DSS) chemical induction method for the study of colorectal cancer development (38). The carcinogen, AOM, reliably induces adenomatous polyps in rodents, but when combined with the inflammatory agent, DSS, considerably shortens the latency time to growth of multiple GI tumors per mouse. We administered all chemical agents in drinking water, introducing AOM at 8 wk of age and following 1 wk later with weeklong exposures to DSS, with 2-wk rest periods in between each DSS exposure for a total of three exposures (Fig. 4C).
As expected, AOM/DSS treatment resulted in formation of multiple colonic polyps in control Xistfl/fl animals of both sexes (Fig. 4 D and E). Xist-null mice also developed tumors. Intriguingly, however, tumor burden was greater in the mutant females relative to controls (Fig. 4 D and E). Increased tumor burden was characterized by greater polyp size (P = 0.041, Student’s t test). This difference was female-specific (Fig. 4 D and E). There was also a tendency toward more numerous tumors in Xist-null females, although the effect size was small and was just below the threshold of statistical significance (Fig. 4F, P = 0.08). However, we believe that the actual number in the mutants was underestimated, due to their microscopic size, which may escape detection during scoring. Taken together, these measurements indicate that female Xist mutants have a heavier tumor burden, at least in terms of tumor size and possibly also in terms of polyp number.
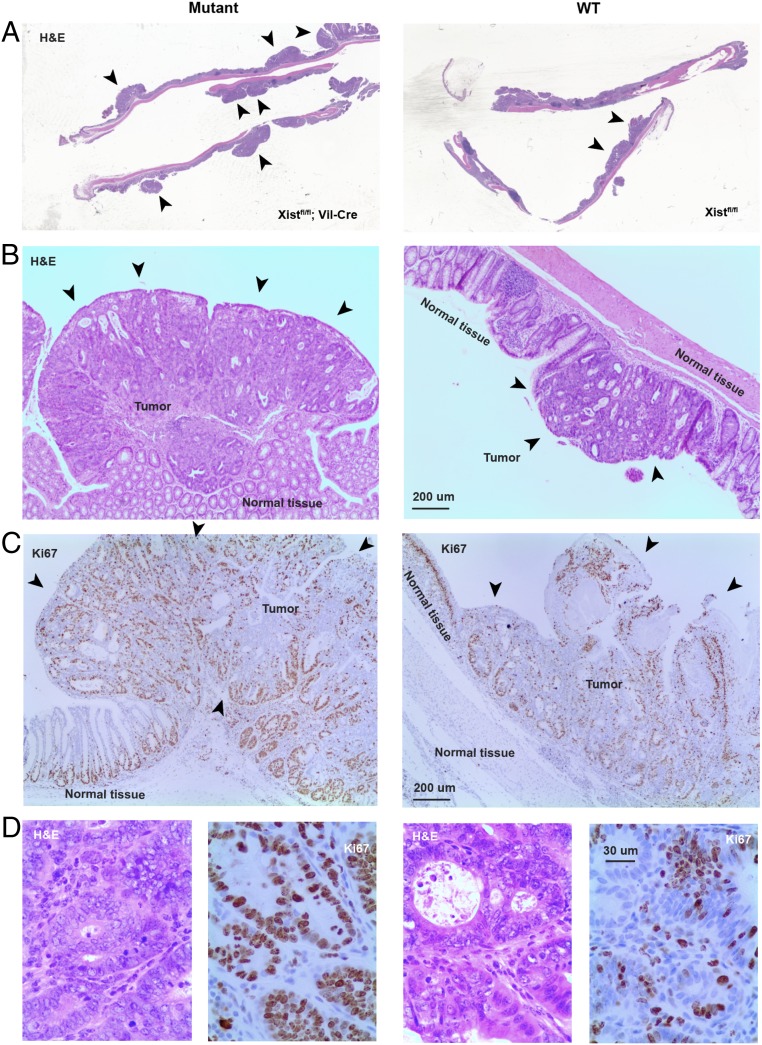
We performed gross and histopathological analyses following necropsy of mutant females and control animals between 4 and 6 wk after the conclusion of DSS treatment. Gross examination of the GI tract revealed findings consistent with greater overall mass in mutant females (Fig. 4D). Hematoxylin and eosin (H&E) staining of longitudinal sections of colon also demonstrated larger polyps in the mutant females in general (Fig. 5A). Histologic sections showed that polyps consisted of tubular adenomas with high-grade dysplasia (Fig. 5 B and D). Severe dysplasia was manifest by hyperchromatic nuclei with increased nuclear to cytoplasmic ratios, prominent nucleoli with clear, open chromatin; increased mitotic rate, and loss of nuclear polarity, nuclear pseudostratification, and nuclear crowding (Fig. 5D and SI Appendix, Fig. S5A). There was also conspicuous loss of goblet and Paneth cell differentiation throughout. Architectural atypia was also prominent with budding, glandular crowding, cribiforming, and focal areas of necrosis (Fig. 5D and SI Appendix, Fig. S5A). Adenomas were marked by Ki67 staining throughout the glandular epithelium vs. confinement to crypts in adjacent normal epithelium, potentially with more Ki67+ cells in the mutant adenomas, although this was difficult to quantify (Fig. 5C and SI Appendix, Fig. S5B). Adenomas in both wild-type and mutant animals manifest the same range of histological findings; however, adenomas were larger in the latter (Figs. 4 D and F and 5B and SI Appendix, Fig. S5). In both cases, there was abrupt transition from normal to dysplastic adenomatous epithelium (Fig. 5B), consistent with clonal adenomatous rather than reactive changes. There was no evidence of invasion into or through the muscularis mucosae as yet, but presence of severe dysplasia suggests that the polyps may be premalignant tumors.
Fig. 5.
Histologic analysis of mutant and control guts following AOM/DSS induction. (A) H&E staining of distal half of colon. The arrowheads point to polyps. Note larger and more polyps in the Xist-null mice relative to control Xist-positive littermates. (B) The 40× magnifications of a single polyp/tumor in mutant vs. control mice. H&E stained. (C) The 40× magnification of immunohistochemical staining for the cell proliferation marker, Ki67 (brown), in mutant and control gut sections. Note that normal tissue shows organized cell proliferation at the base of the crypts only. In tumors, the tissue is disorganized and proliferation is seen throughout the tumor mass. (D) H&E (Left) and Ki67 staining (Right) of mutant and control polyps at 100× magnification.
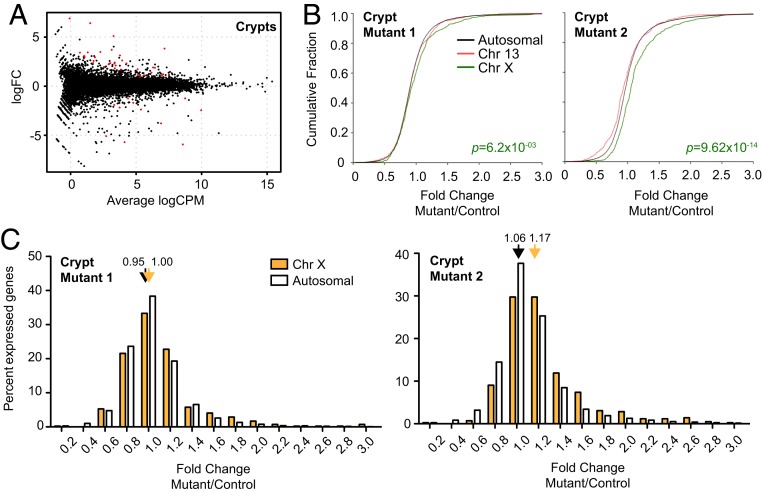
We extracted RNA from the gut of mutant females and control littermates, separately sampling polyps and surrounding normal gut epithelium (crypts) for each animal. In an attempt to understand the basis for a heavier tumor burden in Xist-null females, we compared transcriptomic differences between the clonal tumors (polyps) and surrounding normal tissue from the intestinal crypts. First, we performed differential expression analysis on healthy crypts of Xist-null vs. wild-type animals (Fig. 6). In two biological replicates, we observed 33 significantly up-regulated genes, of which 7 were from the X chromosome (Fig. 6A and SI Appendix, Table S1; Benjamini–Hochberg FDR < 0.05). There were 11 down-regulated genes, none of which was X-linked—except for Xist, the gene deleted in the mutant crypt. Thus, the net cis-effect of Xist deletion is an up-regulation of X-linked genes. Cumulative frequency plots (CFPs) also demonstrated a significant increase in X-linked gene expression relative to Chr13, a representative autosome, or all autosomes in both biological replicates of normal crypts (Fig. 6B; an average of the two mutant samples is shown in SI Appendix, Fig. S6A). Specifically, we noted increased numbers of up-regulated X-linked genes, with range of 1.2- to 1.4-, 2.0- to 2.2-, and 2.6- to 2.8-fold up-regulation (Fig. 6C and SI Appendix, Fig. S6B). Together, these data demonstrate that gut epithelia can remain healthy despite Xi reactivation and increased X-to-autosome gene dosage in the Xist-null females.
Fig. 6.
Transcriptomic analysis of healthy crypts in Xist-deleted and control gut epithelium. (A) Genome-wide differential expression for healthy crypts surrounding polyps. Log2 fold change shown for mutant vs. wild-type crypts. Differentially expressed genes (FDR < 0.05) are in red. (B) CFPs for fold changes of genes on chromosome 13 (red), chromosome X (green), and all autosomes (black). Two biological replicates shown (two pairs of mutant and control females). P value, Wilcoxon’s rank sum test. (C) Distribution of X-linked (blue, polyps; cyan, crypts) vs. autosomal (white) fold changes in Xist-null polyps/crypts relative to corresponding controls. Two biological replicates shown (two pairs of mutant and control females). Average fold changes of X-linked or autosomal genes are indicated. *P < 0.05; **P < 0.01; ***P < 10−8, Fisher’s exact test. Data represent mean ± SEM; n = 2 (crypts), n = 4 (polyps).
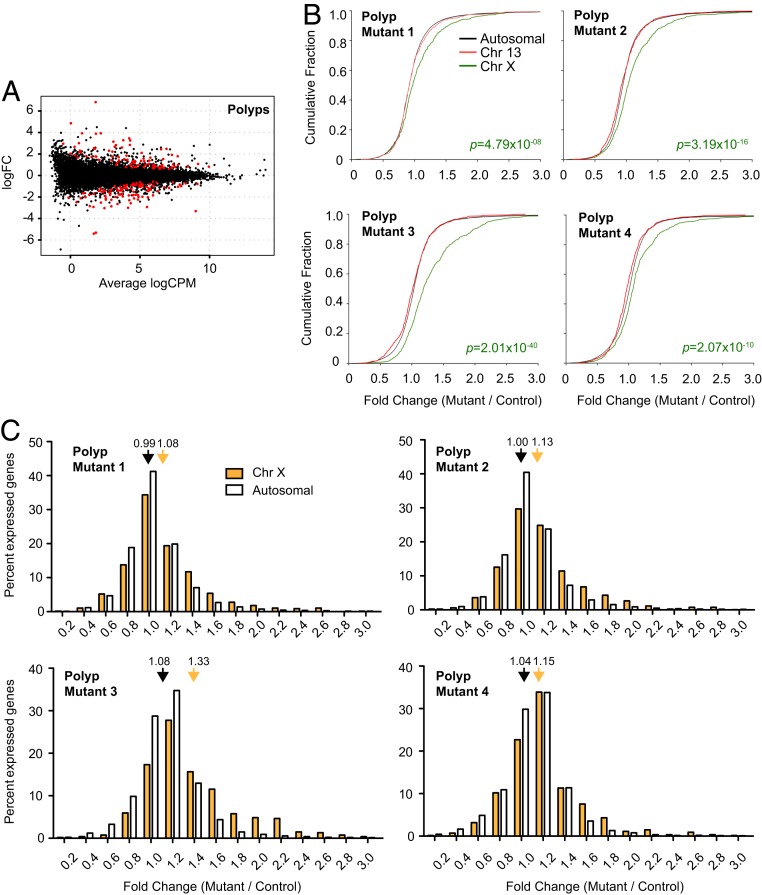
Finally, we turned to transcriptomic analysis of polyps. Four biological replicates (independent clonal tumors from four animals) were examined. Differential gene expression analysis comparing mutant vs. wild-type polyps identified 90 up-regulated genes, of which 24 were from the X chromosome (Benjamini–Hochberg FDR < 0.05; SI Appendix, Table S2 and Fig. 7A). We also identified 102 down-regulated genes (SI Appendix, Table S3), of which none—apart from Xist, which is deleted in the mutant—was X-linked. Thus, there was a major enrichment of up-regulated X-linked genes in the mutant polyps. This enrichment was seen not only when comparing mutant to wild-type polyps but also in comparing mutant polyps to “normal” mutant crypts.
Fig. 7.
Transcriptomic analysis of polyps in Xist-deleted and control gut. (A) Genome-wide differential expression for polyps. Log2 fold change shown for mutant vs. wild type shown. Differentially expressed genes (FDR < 0.05) are in red. (B) CFPs for fold changes of genes on chromosome 13 (red), chromosome X (green), and all autosomes (black). Four biological replicates shown (four pairs of mutant and control females). P value, Wilcoxon’s rank sum test. (C) Distribution of X-linked (blue, polyps; cyan, crypts) vs. autosomal (white) fold changes in Xist-null polyps/crypts relative to corresponding controls. Four biological replicates shown (four pairs of mutant and control females). Average fold changes of X-linked or autosomal genes are indicated. *P < 0.05; **P < 0.01; ***P < 10−8, Fisher’s exact test. Data represent mean ± SEM; n = 2 (crypts), n = 4 (polyps).
CFP analysis confirmed that X-linked genes were indeed more often up-regulated relative to autosomes in all four biological replicates of Xist-null polyps, as evidenced by the shift in the gene expression fold change distribution (Fig. 7B; combined data shown in SI Appendix, Fig. S6A). X-linked genes showed an average fold change of 1.15×, higher than that observed in the crypts, or for the kidney in the Meox2-Cre model (Figs. 2C and 6C). X-linked genes showed an average of 1.2- to 2.0-fold up-regulation relative to autosomes (P < 10−8, Fisher’s exact test). The extent of up-regulation (an average of 1.15-fold; SI Appendix, Fig. S6B) was also much greater than that observed for the kidney (Fig. 2 and SI Appendix, Fig. S3). Given that our quantification of Xi reactivation represents a population average across the tissue, there may be much greater reactivation in individual cells that might be revealed only by single-cell RNA-seq analysis. Such stochastic differences at the single-cell level could determine which cells go on to oncogenic transformation, giving rise to the variable number and size of polyps observed in mutant animals.
Interestingly, among down-regulated genes in the polyps, there was enrichment for genes involved in inflammation and immune response (SI Appendix, Table S4), including the Nlrp family of genes, which are components of the inflammasome complex. This was a paradoxical response, given that the mice were exposed to the inflammatory agent, DSS. Notably, there was no such enrichment for immune response genes in the crypts of mutants relative to those of wild-type animals (SI Appendix, Table S1). These differences suggested that an impaired immune or inflammatory response might aid polyp growth in the mutant gut. We conclude that Xist-null colonic polyps show a dramatic up-regulation of X chromosome expression, that the degree of up-regulation exceeds that observed in surrounding normal gut epithelia, and that inflammatory and immune response genes are enriched among the aberrantly down-regulated genes.
Discussion
Xist was once thought to be dispensable for maintaining the Xi once XCI has been established. Given several works showing that loss of Xist expression leads to a partial reactivation of the Xi, here we have used four mouse models to investigate in vivo consequences of deleting Xist in various organ systems. In recent years, this question has become especially important to answer, because of emerging interest in using Xi reactivation as a strategy to treat X-linked diseases in women. In our present study, when Xist is deleted in the whole body in post-XCI embryonic cells, female pups show no phenotype despite an X-to-autosome dosage imbalance. When Xist is deleted in a more tissue-restricted way—in either skin or B cells—the female mice also live a normal life span without obvious distress. Furthermore, when Xist is deleted in gut epithelia, all female mice are healthy. Taken together with previous work demonstrating an absence of deleterious effects in brain (18, 20), our study supports a systemic tolerance to Xist loss and Xi reactivation in mice.
However, our study also reveals a sensitivity to Xist loss when the gut is subjected to chronic stress. This effect is female specific. When the gut epithelium is repeatedly challenged with a carcinogen (AOM) and inflammatory agent (DSS), tumors develop in both Xist-null and wild-type mice, but the tumor burden is significantly greater in mutant mice. Paradoxically, even though the animals are challenged by the DSS inflammatory agent, inflammatory and immune response genes are down-regulated in the mutant polyps, whereas this difference is not observed in nontumorous crypts. This difference suggests that an abnormal immune or inflammatory response in Xist-deficient gut might drive the higher tumor burden, although further work is required to understand the cause–effect relationship. Thus, Xist and XCI have a protective effect on the gut during prolonged inflammatory stress in female mice. The enablement of a proper inflammatory or immune response might be one of their protective effects during tumorigenesis.
Intriguingly, the degree of X-to-autosomal imbalance differs between the mutant polyps and nontumorous crypts (Figs. 6 and 7). The tumors show a much higher degree of X reactivation than the crypts. This difference could imply that tumors are more likely to arise from cells that have more X–autosomal imbalance in the first place. Alternatively, tumorigenesis could amplify the X reactivation and those cells with greater X–autosomal imbalance could have a survival advantage when under chronic stress. The two possibilities are not mutually exclusive and could in fact both fuel the higher tumor burden in Xist-null mice. Importantly, the propensity toward a higher tumor burden occurs only when animals are chronically stressed (Fig. 4). The unstressed Xist-null gut did not show any greater predisposition toward tumors than wild-type mice.
Work in Drosophila has demonstrated that, when there is chromosomal instability (CIN) such as that induced by depletion of spindle assembly checkpoint proteins, the dosage compensation machinery resets in response to changes in the copy number of X-linked genes (39). Female cells that lose one X chromosome activate the MSL dosage compensation complex, which are normally repressed in females. MSL activation increases gene transcription from the remaining X. In contrast, XY males that gain an X chromosome repress their normally active MSL complex to avoid excessive gene dosage. Given the importance of CIN in tumorigenesis (40), we wonder whether Xist may be called upon to restore X–autosomal balance in CIN scenarios. The inability to suppress effects of the CIN phenotype, at least for X-linked genes in Xist-null polyps, may be one possible explanation for the severe X–autosome imbalance seen in mutant polyps. This may also account for the heavier tumor burden in Xist-deleted guts, especially in light of the high incidence of CIN in colorectal cancers (41). Moreover, Xist itself has been implicated in the maintenance of stability of the X chromosomes (42), and its absence could therefore have a direct effect on tumorigenesis and polyp size.
When viewed in the light of previous Xist deletion models, our findings enable further conclusions regarding how Xist loss impacts organism health. Clearly, if Xist is deleted prior to the establishment of XCI, female embryos either do not survive past the periimplantation period (12) or die later during embryogenesis, with only a rare number surviving to birth when XO cellular mosaicism is present (13). These results speak to the absolutely essential nature of Xist for the establishment of XCI, which in turn is critical for female survival and fitness. During the maintenance phase, female mice that lose Xist function in HSCs develop a fulminant blood cancer with nearly full penetrance (27). On the other hand, two independent brain-specific Xist deletion models show no phenotype of any kind (20, 21). This difference has led to speculation that tumorigenesis may result from an Xist deletion only in cell types with high proliferation rates (21, 27).
However, our current work indicates that a high proliferation rate alone is not sufficient for Xist-related morbidities. Skin and gut are two of the most highly proliferative tissues, with gut cells turning over every 3 to 5 d and matching cell division rates typically seen in the bone marrow. However, the K14-Cre and Villin-Cre experiments did not reveal any morbidity or mortality associated with the respective Xist deletions. The gut-specific deletion showed a phenotypic difference only under prolonged stress. Thus, we suggest that a high proliferation rate alone is not sufficient to drive disease when Xist function is lost. Chronic stress may also be required, at least in the gut. Additionally, activation of specific X-linked oncogenes may be necessary. For example, HSCs could be extra sensitive to Xist loss due to the fact that the X chromosome houses Gata1, a master regulator of hematopoiesis, which is known to be oncogenic when overexpressed (41). For the skin, B cells, or kidney, perhaps the lack of tissue- and/or stage-specific oncogenes on the X chromosome may be more of a barrier to cancer transformation. Tumorigenesis often requires multiple initiation events, such as misregulation or mutations of several oncogenes or tumor suppressor genes. In the gut, for example, mutations in Wnt signaling genes induced by AOM administration is required for polyp formation in the established cancer model (38). Last, there is precedence for the idea of Xist hypersensitivity in the hematopoietic system. Full-body induction of Xist expression on one or both X chromosomes in 4-wk-old male and female mice resulted in bone marrow hypocellularity, anemia, and defects in both myeloid and erythroid differentiation (43). As such, unique properties of the hematopoietic system could also represent this as a special, tissue-specific case for how somatic cells respond to Xist RNA loss.
In summary, our present work cautions against premature conclusions that an lncRNA lacks function when it has only been challenged by a limited set of environmental conditions. Xist RNA was once thought to have no further function for Xi silencing after XCI is established. The example shown here demonstrate unequivocally that Xist plays a crucial role during maintenance of Xi silencing, that Xist expression is required to maintain cellular homeostasis, and that it is protective for females during chronic stress. We predict that functions of other lncRNAs whose deletions have not yielded strong phenotypes would ultimately be revealed by specific but as-yet-undefined physiological perturbations.
Materials and Methods
Animal Studies.
Xistfl/fl mice (129Sv/Jae background) were a gift from R. Jaenisch, Whitehead Institute, Cambridge, MA. B6.129S4-Meox2tm1(cre)Sor/J mice (JAX 003755), B6.Cg-Tg(Vil1-cre)997Gum/J mice (JAX 004586), and B6.129P2(C)-Cd19tm1(cre)Cgn/J mice (JAX 006785) were procured from The Jackson Laboratory. Cd19 Cre genotyping were done using two primer sets that recognize Cre containing locus (1084, 5′- GCG GTC TGG CAG TAA AAA CTA TC-3′; 1085, 5′- GTG AAA CAG CAT TGC TGT CAC TT-3′) or wild-type locus (1589, 5′-CCT CTC CCT GTC TCC TTC CT-3′; 1590, 5′-TGG TCT GAG ACA TTG ACA ATC A-3′). Genotyping for Xist 2lox or wild-type allele was done using the following primer sets: Xint3F: 5′-GGC CAG TTT CTG ACA CCC TA-3′; Xint3R: 5′-CAC TGG CAA GGT GAA TAG CA-3′; 370: 5′-GTG CCATAT CAGTGA GCT CTC -3′; 371: 5′-TGT CAC CTA CCA ATG AGA GAT CC-3′; 389: 5′-AAC CAA GGT TGA GAG AGC AAA-3′. All progeny were maintained on a 129Sv/Jae, C57BL/6 mixed genetic background. For timed matings, the morning of copulation plug detection was considered 0.5 dpc. Embryos were harvested at E14.5 for MEF isolation. Mouse husbandry and experiments were carried out as stipulated by the Massachusetts General Hospital (MGH) Institutional Animal Care and Use Committee (IACUC).
RNA-FISH.
Cultured fibroblasts were either seeded onto coverslips or cytospun to glass slides at 1,000 rpm for 10 min (Shandon Cytospin). Fixation was performed using 4% paraformaldehyde for 10 min at room temperature before permeabilization with 0.5% Triton-X/PBS for 5 min on ice. For preparation of tissue cryosections, tissue was harvested fresh, embedded in Tissue-Plus OCT (Fisher Healthcare), and snap-frozen in a dry ice/isopentane slurry. Tissues were cut to 12-μm-thick sections. Fixation and permeabilization for tissue sections were carried out as for fibroblasts. RNA-FISH was performed as previously described (25, 44).
Reverse Transcription and Quantitative PCR.
Fresh tissue was collected into RNALater (Qiagen). Tissue homogenization was performed with the Tissue Lyzer II (Qiagen) using 2-mm stainless-steel beads, in TRIzol reagent. For reverse transcription, cDNA was prepared using SuperScript III (Thermo Fisher Scientific) with random hexamers (Promega). Quantitative PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad); relative gene expression was calculated by the standard curve method.
Isolation of Bone Marrow, Spleen, and Thymus Cells and FACS.
To isolate bone marrow cells, both femurs and tibias were harvested from each animal, and bone marrow cells were flushed with 3 mL of 1× PBS/5% fetal calf serum (FCS) into a 50-mL tube (Falcon) using a 22.5-gauge needle. The cells were then passed through a 100-μ strainer to collect single cells. To collect spleen and thymus cells, organs were dissected and passed through a 100-μ strainer by crushing with forceps and collecting cells in 3 mL of cold 1× PBS/5% FCS. Next, cell suspensions were spun down, and the cell pellet was resuspended in 2 mL of ACK Lysis buffer (Thermo Fisher Scientific) and incubated for 2 min (bone marrow, thymus) or 10 min (spleen) at 4 °C to lyse red blood cells. Cells were resuspended in 2 mL of 1× PBS/5% FCS, passed through a 70-μ strainer and counted using a cell counter (Nexalom). For RNA-FISH analyses, B cells were isolated using MagniSort B cell enrichment kit (Invitrogen). For FACS analyses, cells were stained using the following antibodies. B cell markers were as follows: anti-B220 (RA3-6B2) (BD), anti-IgM (eB121-15F9) (Thermo Fisher Scientific), anti-CD43 (S7) (Biolegend), anti-CD21/CD35 (7G6) (BD), anti-CD23 (B3B4) (BD), and anti-IgD (11-26c.2a) (BD). T cell markers were as follows: anti-CD4 (RM4-5) (BD) and anti-CD8 (53-6.7) (BD). Propidium iodide (Invitrogen) was used to determine cell viability. B cell subpopulations were discriminated based on earlier reports (45). FACS was done using the BD FACSAria II (BD Bioscience); data analysis was performed using FlowJo, version 10.
Induction of Colorectal Polyps by AOM/DSS.
Eight- to 10-wk-old mice were intraperitoneally injected with AOM (Sigma Aldrich) diluted to 1 mg/mL in saline using a 25-gauge needle. A 10 mg/kg body weight dose was used; animals were weighed right before injection. The 2% (wt/vol) dextran sulfate (40 to 50 kDa; Affymetrix) was dissolved in water and administered for 7-d cycles. Normal drinking water ad libitum was provided in between DSS treatment cycles. Animals were housed in groups no bigger than three during duration of chemical induction protocol. Mouse husbandry and experiments were carried out as stipulated by the MGH IACUC.
Histopathology.
Tissues were fixed in 10% neutral buffered formalin (Fisher Scientific) overnight and stored in 70% ethanol for further processing. Antibodies for immunohistochemistry were as follows: β-catenin, Santa-Cruz, 1496, at 1:100; YFP, Abcam, ab6556, at 1:400. Images were acquired using the Nikon Eclipse 90i microscope with Q-imaging MicroPublisher RTV color camera.
Isolation of Intestinal Epithelia.
Mouse colons were harvested, opened lengthwise, and rinsed twice in cold PBS. For extraction of the crypt fraction, a modified protocol from Sato and Clevers (46) was used. Whole colon was first transferred to dissociation buffer 1 (30 mM EDTA, 1.5 mM DTT in PBS) and incubated for 20 min at 4 °C with rotation. Tissue was then cut into 1-mm segments and transferred to dissociation buffer 2 (30 mM EDTA in PBS) and incubated at 37 °C for 10 min. Mechanical dissociation was performed to dislodge crypts from tissue; this was repeated five to seven times with fresh 0.1% BSA/PBS. The crypt suspension was filtered through 100-μm strainer to remove debris and then spun down to cell pellet at 200 × g, 4 °C.
Strand-Specific RNA-Seq.
Strand-specific RNA-seq was performed as described (13). Briefly, 4 μg of total RNA (RNA integrity number [RIN] value ≥ 8, except for crypt fractions where RIN ≥ 7) from mouse tissues or isolated crypts was depleted of ribosomal RNA (RiboMinus Eukaryote Kit, version 2; Thermo Fisher Scientific) as per manufacturer’s recommendations, DNase-treated for 30 min at room temperature, and purified using RNeasy MinElute Cleanup kit (Qiagen). Eluted RNA was then quantified (Quant-iT RiboGreen RNA Assay; Thermo Fisher Scientific) and 30 ng of RNA was fragmented in 6 mM MgCl2 buffer (10 min; 94 °C) before first-strand (Superscript III; Thermo Fisher Scientific) and second-strand (NEBNext Ultra Directional RNA Second Strand Synthesis Module; NEB) cDNA synthesis. Adaptor oligos were ligated to the cDNA library (NEBNext Chip-Seq Library Master Mix; NEB). Libraries were amplified with multiplexing barcodes (NEBNext Multiplex Oligos for Illumina; NEB) and quantified (KAPA Library Quantification kit; Kapa Biosystems) before 50-nt paired-end sequencing was performed on the Illumina HiSeq 2000 or HiSeq 2500.
RNA-Seq Analysis.
Read quality was verified by FastQC before alignment with Tophat2 (47) to the mouse reference genome (NCBI37/mm9). Read counting was performed with HTseq-count (48) against all ENSEMBL transcripts (ENSEMBL release 67). Normalization and differential expression analysis was performed using the EdgeR package (49). For analyzing distributions of gene expression fold changes on Chr X and autosomes, only genes with cpm ≥ 1 are considered.
Data Availability.
All sequencing data have been deposited in the European Bioinformatics Institute’s repository under ArrayExpress accession number E-MTAB-8161. All other data and materials will be shared upon request.
Supplementary Material
Acknowledgments
We thank R. Mostoslavsky, K. Hochedlinger, W. Bender, B. Payer, and all members of the J.T.L. laboratory for stimulating discussions and valuable advice. We thank R. Jaenisch for Xistfl/fl mice, and W. Press for mouse colony maintenance. This work was supported by grant funding from the Agency for Science, Technology and Research, Singapore (to L.Y.); and the Rett Syndrome Research Trust, Rettsyndrome.org, and the Howard Hughes Medical Institute (to J.T.L.).
Footnotes
Competing interest statement: J.T.L. is a cofounder of Translate Bio and Fulcrum Therapeutics, and an advisor to Skyhawk Therapeutics. To our knowledge, none of the companies is presently working on Xi reactivation.
Data deposition: All sequencing data have been deposited in the European Bioinformatics Institute’s repository (ArrayExpress accession no. E-MTAB-8161).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917203117/-/DCSupplemental.
References
- 1.Starmer J., Magnuson T., A new model for random X chromosome inactivation. Development 136, 1–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disteche C. M., Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46, 537–560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jégu T., Aeby E., Lee J. T., The X chromosome in space. Nat. Rev. Genet. 18, 377–389 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Brockdorff N., et al. , The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Brown C. J., et al. , The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542 (1992). [DOI] [PubMed] [Google Scholar]
- 6.Clemson C. M., McNeil J. A., Willard H. F., Lawrence J. B., XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minajigi A., et al. , Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349, aab2276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jégu T., et al. , Xist RNA antagonizes the SWI/SNF chromatin remodeler BRG1 on the inactive X chromosome. Nat. Struct. Mol. Biol. 26, 96–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J., et al. , Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat. Genet. 28, 371–375 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Kohlmaier A., et al. , A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2, E171 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J., Sun B. K., Erwin J. A., Song J.-J., Lee J. T., Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R., Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Yang L., Kirby J. E., Sunwoo H., Lee J. T., Female mice lacking Xist RNA show partial dosage compensation and survive to term. Genes Dev. 30, 1747–1760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penny G. D., Kay G. F., Sheardown S. A., Rastan S., Brockdorff N., Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Kalantry S., Purushothaman S., Bowen R. B., Starmer J., Magnuson T., Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 460, 647–651 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Namekawa S. H., Payer B., Huynh K. D., Jaenisch R., Lee J. T., Two-step imprinted X inactivation: Repeat versus genic silencing in the mouse. Mol. Cell Biol. 30, 3187–3205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatnagar S., et al. , Genetic and pharmacological reactivation of the mammalian inactive X chromosome. Proc. Natl. Acad. Sci. U.S.A. 111, 12591–12598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrette L. L. G., et al. , A mixed modality approach towards Xi reactivation for Rett syndrome and other X-linked disorders. Proc. Natl. Acad. Sci. U.S.A. 115, E668–E675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sripathy S., et al. , Screen for reactivation of MeCP2 on the inactive X chromosome identifies the BMP/TGF-β superfamily as a regulator of XIST expression. Proc. Natl. Acad. Sci. U.S.A. 114, 1619–1624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adrianse R. L., et al. , Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion. Epigenetics Chromatin 11, 50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrette L. L. G., Blum R., Ma W., Kelleher R. J. 3rd, Lee J. T., Tsix-Mecp2 female mouse model for Rett syndrome reveals that low-level MECP2 expression extends life and improves neuromotor function. Proc. Natl. Acad. Sci. U.S.A. 115, 8185–8190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown C. J., Willard H. F., The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature 368, 154–156 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Csankovszki G., Panning B., Bates B., Pehrson J. R., Jaenisch R., Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 22, 323–324 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Csankovszki G., Nagy A., Jaenisch R., Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153, 773–784 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L. F., Huynh K. D., Lee J. T., Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell 129, 693–706 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Anguera M. C., et al. , Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11, 75–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yildirim E., et al. , Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727–742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monk M., Harper M. I., Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281, 311–313 (1979). [DOI] [PubMed] [Google Scholar]
- 29.Mak W., et al. , Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Payer B., et al. , Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol. Cell 52, 805–818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tallquist M. D., Soriano P., Epiblast-restricted Cre expression in MORE mice: A tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26, 113–115 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Wu L., et al. , Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421, 942–947 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Yagi R., et al. , Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development 134, 3827–3836 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Dassule H. R., Lewis P., Bei M., Maas R., McMahon A. P., Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775–4785 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Rickert R. C., Roes J., Rajewsky K., B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25, 1317–1318 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barker N., Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Madison B. B., et al. , Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 277, 33275–33283 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Neufert C., Becker C., Neurath M. F., An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2, 1998–2004 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Clemente-Ruiz M., et al. , Gene dosage imbalance contributes to chromosomal instability-induced tumorigenesis. Dev. Cell 36, 290–302 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Whyatt D., et al. , An intrinsic but cell-nonautonomous defect in GATA-1-overexpressing mouse erythroid cells. Nature 406, 519–524 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Torres E. M., Williams B. R., Amon A., Aneuploidy: Cells losing their balance. Genetics 179, 737–746 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savarese F., Flahndorfer K., Jaenisch R., Busslinger M., Wutz A., Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol. Cell Biol. 26, 7167–7177 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunwoo H., Wu J. Y., Lee J. T., The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl. Acad. Sci. U.S.A. 112, E4216–E4225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carsetti R., Characterization of B-cell maturation in the peripheral immune system. Methods Mol. Biol. 271, 25–35 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Sato T., Clevers H., Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–328 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Kim D., et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S., Pyl P. T., Huber W., HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited in the European Bioinformatics Institute’s repository under ArrayExpress accession number E-MTAB-8161. All other data and materials will be shared upon request.