Significance
X-ray crystallography often fails to determine the positions of hydrogen atoms, which play crucial roles in enzymatic reactions. Despite many X-ray crystallographic studies, the reaction mechanism of copper-containing nitrite reductases (CuNIRs), which reduce nitrite using two protons, has been controversial. The high-resolution neutron structure of a CuNIR reveals the protonation states of catalytic residues and key water molecules, thus providing insights into the catalytic mechanism. The catalytic Cu is shown to be coordinated by a hydroxide ion and not water. Furthermore, the hydrogen-deuterium exchange ratio suggests that intramolecular electron transfer is involved in a hydrogen-bond jump. These observations are consistent with previous computational chemistry; therefore, our study forms a bridge between the structural biology and quantum chemistry of CuNIRs.
Keywords: neutron crystallography, copper-containing nitrite reductase, structural biology
Abstract
Copper-containing nitrite reductases (CuNIRs) transform nitrite to gaseous nitric oxide, which is a key process in the global nitrogen cycle. The catalytic mechanism has been extensively studied to ultimately achieve rational control of this important geobiochemical reaction. However, accumulated structural biology data show discrepancies with spectroscopic and computational studies; hence, the reaction mechanism is still controversial. In particular, the details of the proton transfer involved in it are largely unknown. This situation arises from the failure of determining positions of hydrogen atoms and protons, which play essential roles at the catalytic site of CuNIRs, even with atomic resolution X-ray crystallography. Here, we determined the 1.50 Å resolution neutron structure of a CuNIR from Geobacillus thermodenitrificans (trimer molecular mass of ∼106 kDa) in its resting state at low pH. Our neutron structure reveals the protonation states of catalytic residues (deprotonated aspartate and protonated histidine), thus providing insights into the catalytic mechanism. We found that a hydroxide ion can exist as a ligand to the catalytic Cu atom in the resting state even at a low pH. This OH-bound Cu site is unexpected from previously given X-ray structures but consistent with a reaction intermediate suggested by computational chemistry. Furthermore, the hydrogen-deuterium exchange ratio in our neutron structure suggests that the intramolecular electron transfer pathway has a hydrogen-bond jump, which is proposed by quantum chemistry. Our study can seamlessly link the structural biology to the computational chemistry of CuNIRs, boosting our understanding of the enzymes at the atomic and electronic levels.
The denitrification process in the global nitrogen cycle is an anaerobic respiratory system of microorganisms through which electron acceptor nitrogen oxide (NOx) molecules are reduced in a stepwise manner to gaseous dinitrogen and released into the atmosphere (1). Copper-containing nitrite reductases (CuNIRs) are involved in the key step of denitrification: one-electron reduction of nitrite to nitric oxide (NO2− + 2H+ + e− → NO + H2O), which generates gaseous NO and results in the removal of terrestrial fertilizer. Although CuNIRs are divided into class I and class II types depending on the lengths of loop regions (2), both types usually show commonly shared homotrimeric structures. Each monomeric unit of CuNIRs consists of tandem cupredoxin domains and contains one type 1 copper (T1Cu) and one type 2 copper (T2Cu) sites (3–6) (Fig. 1A). T1Cu absorbs light at around 600 nm because of ligand-to-metal charge transfer from the sulfur atom of cysteine (Cys) to T1Cu and shows blueish colors (7). This Cu site, also known as blue copper, is involved in accepting an electron from physiological electron donor proteins. The T1Cu sites in CuNIRs generally have a ligand set composed of two histidine (His), one methionine (Met), and one Cys residues. The Cys residue is juxtaposed to one of the His ligands at the T2Cu site to form a Cys-His bridge. The electron acquired by T1Cu is efficiently transferred through this bridge to the catalytic T2Cu site, which is coordinated by three equatorial His residues and axial water. A quantum chemical analysis suggests that the intramolecular electron transfer (ET) pathway between T1Cu and T2Cu in CuNIR has a hydrogen-bond jump between the Cys amide O atom and the imidazole Nδ atom of T2Cu ligand His (route A in Fig. 1A) (8), although experimental evidence for this has not yet been presented. There are two conserved catalytic residues above the T2Cu site, aspartate (AspCAT) and His (HisCAT), both of which assist proton transfer (PT) to the substrate (9–12). AspCAT and HisCAT are connected via one water molecule that is designated as bridging water (BW) and considered to be involved in PT. Because the PT reaction is coupled with intramolecular ET (13, 14), the CuNIR reaction is one of the natural examples of proton coupled electron transfer (PCET).
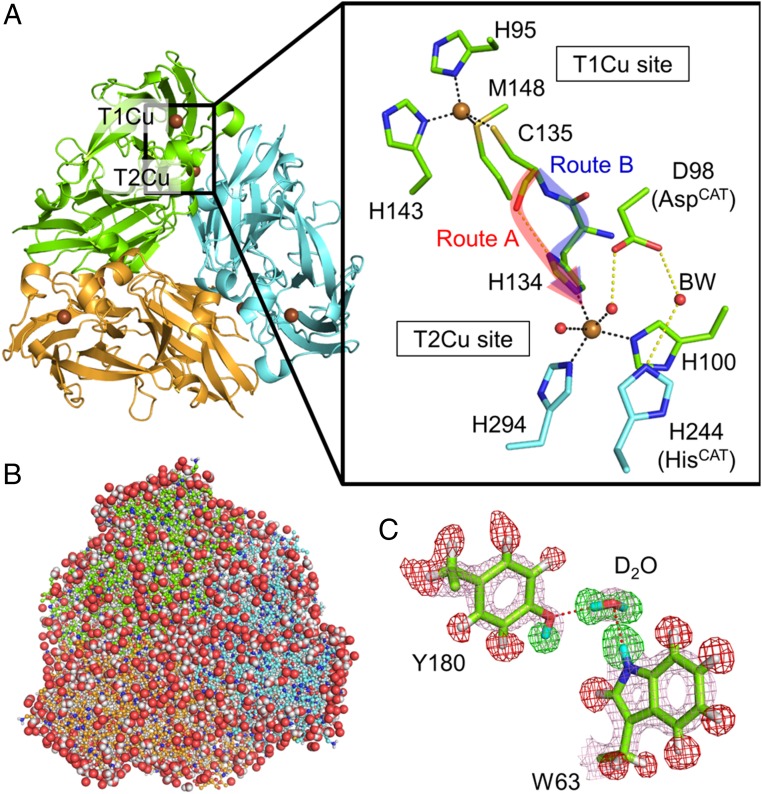
Fig. 1.
Structure of GtNIR. (A) Overall structure and copper sites of GtNIR. Two possible intramolecular ET routes, route A (with a hydrogen-bond jump) and route B (with only covalent bonds), are indicated by red and blue arrows, respectively. This figure is generated from an oxidized GtNIR structure determined by SFX (PDB ID code 4YSA). (B) Neutron structure of GtNIR trimer. The protein structure is drawn by balls and sticks. Water molecules are shown by sphere representation. Both H and D atoms are colored in white for the clarity. (C) An example of sigma-A–weighted neutron scattering length density maps. The 2Fo–Fc map (courted at 2.0 σ) is shown by pink meshes. The positive (+4.0 σ) and negative (−4.0 σ) Fo–Fc maps are shown by green and red meshes, respectively. H and D atoms are colored in white and cyan, respectively.
Conventional X-ray crystallography has contributed to the understanding of the mechanisms of ET and nitrite reduction in CuNIRs (3, 9, 14–18). However, hydrated electrons generated by X-ray radiation, especially by synchrotron X-ray radiation, can easily cause reduction of metal centers in metalloproteins, which leads to local structural changes (19, 20). This phenomenon, known as photoreduction, complicates the interpretation of X-ray crystallographic data. Spectroscopic and crystallographic analyses confirm that the metal centers of CuNIRs are reduced by X-ray radiation (21–23). Along this line of thinking, newly developed techniques such as serial femtosecond crystallography (SFX) (23, 24) and serial femtosecond rotational crystallography (SF-ROX) (25, 26) by using X-ray free electron lasers (XFELs) have been applied to damage-free structure determination of resting and substrate-bound states of several CuNIRs. Alternatively, the multiple structures serially from one crystal (MSOX) method uses X-ray radiation as a trigger of the reduction reaction to visualize an in crystallo catalytic process of CuNIR (27, 28). MSOX shows a side-on nitrosyl product bound to T2Cu as is observed in NO-soaked CuNIR crystals (15, 18). However, several computational studies have suggested that direct release of NO without forming nitrosyl copper species is energetically favorable and hydroxyl T2Cu remains as a stable product (29–32). The calculated activation energy shows good agreement with an activation energy value determined experimentally. Moreover, spectroscopic data are inconsistent with the presence of the side-on NO species (33). Thus, some researchers have supposed that nitrosyl T2Cu is an experimental artifact (31).
Among the steps of the CuNIR reaction, the PT mechanism is most ambiguous because of the difficulty in detecting the positions of hydrogen (H) atoms and protons (H+) only using X-ray diffraction (XRD) analysis. Because the amplitude of X-ray scattering is in proportion to the number of electrons by which X-rays are diffracted, the signals from light atoms are weak and often buried under those from their adjacent C, O, and N atoms. Consequently, H atoms and H+ are difficult to observe with XRD even at an atomic resolution showing minimum Bragg spacing (dmin) better than 1.2 Å at which individual atoms can be separately observed (34). Nakamura et al. recently described an extreme representative of “hidden H atoms” at an atomic resolution, in which a subångström (0.64 Å) resolution XRD data from a cellulase crystal fails to visualize the positions of H atoms and H+ on catalytically important residues (35). Neutron diffraction (ND) crystallography is a powerful tool to determine the positions of H atoms and H+. Because neutrons are diffracted by atomic nuclei, the amplitude of neutron scattering depends only on the structures of atomic nuclei. Accordingly, H atoms and atoms of its isotope deuterium (D) contribute to neutron scattering to the same extent as C, N, and O atoms. Moreover, neutron beams with much weaker energy than X-ray ones are thought to have an advantage in determining intact metalloprotein structures without photoreduction. In fact, ND analyses have been applied to iron-containing cytochrome c peroxidase (36) and ascorbate peroxidase (37) to directly observe protonation states of reaction intermediates that are unresolved by spectroscopic and XRD analyses. A neutron crystal structure of a class I CuNIR from Achromobacter cycloclastes (AcNIR) in the resting state has recently been determined at a resolution of 1.8 Å with a quasi-Laue neutron diffractometer (26). Although this structure has provided insights into the protonation state around T2Cu, deprotonated HisCAT at pD5.4 (pH5.0) found in it is inconsistent with previous experimental (11, 12, 38) and computational (29) results. Since CuNIRs are found in a broad spectrum of organisms, investigation of other CuNIRs is needed to confirm that deprotonated HisCAT in the resting state is a universal phenomenon among CuNIRs.
Here, we performed time-of-flight (TOF) neutron diffraction data collection (39) to obtain high quality ND data from a large crystal of a class II CuNIR from Geobacillus thermodenitrificans (GtNIR). Our data record near-atomic resolution showing dmin of 1.50 Å, which is comparable to covalent bond distances dominant in proteins. We carried out joint refinement (40) against the ND data with XRD data collected from the same crystal to visualize the protonation state of the catalytic site of GtNIR in its resting state. Furthermore, because we used a hydrogen-deuterium (H-D) exchange method, we could obtain information about the H-D exchange ratio of each residue, which provides a clue to protein dynamics.
Results
Quality of the Neutron Structure.
We collected ND data from a single GtNIR crystal that diffracted neutrons beyond 1.40 Å resolution at 100 K (SI Appendix, Fig. S1). The data were processed by a profile fitting method (41) using reflections up to 1.50 Å resolution. The highest resolution shell shows a signal-to-noise ratio of 2.7 and completeness of 99.7% (SI Appendix, Table S1). Then, we performed joint neutron and X-ray (NX) refinement, which gives better models than the refinement done only with ND data (40). Although we collected X-ray data up to 1.03 Å resolution, NX joint refinement with X-ray data up to 1.30 Å resolution yielded better R values and less noise for both the electron and neutron scattering length density maps compared to when we used full X-ray data. The final Rwork/Rfree values for the ND (1.50 Å resolution) and XRD (1.30 Å resolution) data were 14.1/15.8 and 9.73/11.2%, respectively, indicating good accordance of the constructed model with the observed structure factors. Our data clearly visualized the locations of H/D atoms in GtNIR, and a total of 3,043 H/D atoms and 285 hydrating water positions were identified (Fig. 1 B and C). The data quality was sufficiently high to show that some amide H atoms deviated from their Cα-N-C planes. For example, the strong negative peak in the neutron scattering length mFo–DFc difference Fourier (Fo–Fc neutron) map showed the deviated position of the amide H atom of Thr293 (SI Appendix, Fig. S2). This is consistent with the observation that the dihedral angle of the peptide bond between Val292 and Thr293 was refined to be 144.9°, showing a highly distorted peptide bond.
In addition to T1Cu and T2Cu, the X-ray data showed the electron density of extra Cu ions (Cu3 and Cu4) binding to GtNIR. Cu3, which almost fully occupied the site, is coordinated by surface residues, His42, His83, and Glu53 from the adjacent trimer (SI Appendix, Fig. S3A). Cu4, which is ligated by His244 (HisCAT in GtNIR), showed occupancy of 0.1 (SI Appendix, Fig. S3B). These Cu ions were observed and confirmed by anomalous dispersion signals in a previous study (23). They are thought to be artifacts caused by the presence of highly concentrated Cu ions in the crystallization solution. The presence of Cu4 has been a problem for several years because it binds to HisCAT and impedes the understanding of the catalytic mechanism. However, Cu4 did not contribute so much to the neutron-scattering length density map compared with the case for the electron density map (SI Appendix, Fig. S3C). This is because the Cu atom only diffracts neutrons to the same extent as C, O, N, and D atoms, whereas it strongly does X-rays even when present at a trace amount. Consequently, Cu4 with very low occupancy is almost negligible in the ND data. We found another advantage of complementary analysis with XRD and ND when we refined the T2Cu site. A sodium (Na) ion having occupancy of 0.16 was observed in the electron density map as is found in a previous SFX structure of GtNIR (23). Although the X-ray scattering of a Na atom is comparable to those of C, O, and N atoms, its neutron scattering is significantly weaker than theirs. Therefore, low occupancy Na is negligible in the ND data (SI Appendix, Fig. S3C), supporting our previous speculation that the site is occupied by Na not a water molecule. Because the Na ion has been only observed in the damage-free XFEL structure, its presence indicates that we could collect the present X-ray data with low radiation damages. Furthermore, our data did not show the presence of a diatomic foreign ligand on the T2Cu atom, which is found in aerobically manipulated GtNIR after X-ray photoreduction (22, 42). Therefore, the T2Cu site observed is considered to be in the oxidized state.
Protonation States Around Catalytic T2Cu.
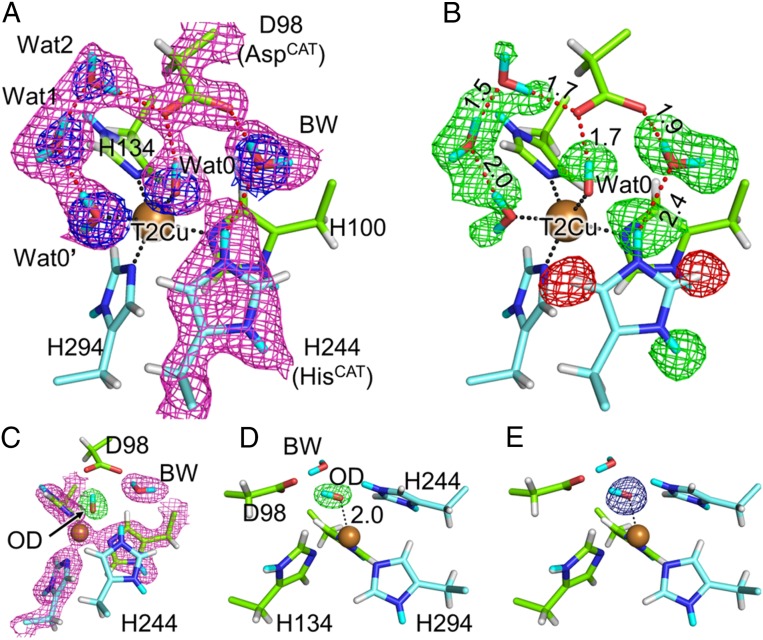
Previous high-resolution XRD analyses of GtNIR depict five water molecules located around the T2Cu site (Wat0, Wat0′, Wat1, Wat2, and BW) (23, 43). Among them are ligands to the T2Cu atom (Wat0 and Wat0′). We could observe all of these water molecules in the present XRD study, although Wat0′ was partially replaced with a chloride ion having occupancy of 0.3 as is found in a previous XRD study (43). The ND data also showed the presence of these five solvent molecules (Fig. 2A). The electron density and the neutron-scattering length density maps of Wat2 were slightly weak probably due to its flexibility as is observed in the XFEL experiment (23). The occupancy of Wat2 was refined to be 0.73. The Fo–Fc neutron map can detect the position of H and D atoms (Fig. 2B). Negative and positive peaks in the H/D omit Fo–Fc neutron map indicate the presence of H and D atoms, respectively. We observed a positive peak between the Oδ atom of AspCAT and Wat2, which formed a hydrogen bond with AspCAT. There was another positive peak between the Oδ atom of AspCAT and Wat0. Because these peaks were respectively closer to Wat2 and Wat0 than to AspCAT, we concluded that AspCAT was deprotonated in the resting state, which is consistent with an earlier result from Fourier transform infrared spectroscopy (44) and computational studies (29). Contrary to the case of AspCAT, the Fo–Fc neutron map showed positive density at around the Nδ and Nε atoms of HisCAT. Additionally, the neutron scattering length 2mFo–DFc difference Fourier (2Fo–Fc neutron) map showed the presence of the D atoms (note that the omit map of Fig. 2B was calculated with the model including Cu4 and that Cu4 did not fit the neutron map; SI Appendix, Fig. S3C). Therefore, HisCAT was basically protonated in the resting state. This observation accords with the earlier density functional theory (DFT) calculation showing that protonated HisCAT is stabilized when AspCAT is deprotonated (29). According to the occupancy of Cu4, we conveniently refined the D atom on the Nε atom in HisCAT with occupancy of 0.9. Because the Nε atom in HisCAT was too close to Cu4 (1.7 Å), there should be another conformation of HisCAT that was invisible in our data. BW showed positive electron density around it in the Fo–Fc neutron map and its shape of the 2Fo–Fc neutron map looked triangular corresponding to the molecular structure of water. Owing to such features found in the neutron maps, we could construct a complete hydrogen bond network around the catalytic residues (Fig. 2B).
Fig. 2.
Protonation state of the T2Cu site. (A) Sigma-A–weighted 2Fo–Fc maps for X-ray (blue, contoured at 1.5 σ) and neutron (magenta, contoured at 1.0 σ) data. (B) Positions of H and D atoms. Sigma-A–weighted Fo–Fc neutron maps are shown by green (+3.0 σ) and red (−3.0 σ) meshes. Hydrogen bonds are illustrated by dashed red lines, and their distances are shown in ångströms. Dashed black lines indicate coordination bonds. H and D atoms are colored by white and cyan, respectively. Na, Cl, and Cu4 atoms are eliminated for the clarity. (C) Sigma-A–weighted 2Fo–Fc neutron (magenta, contoured at 1.5 σ) and Fo–Fc OD-omit neutron (green, contoured at +6.0 σ) maps. (D) The side view of C. The coordination bond between OD and T2Cu (dashed black line) is shown in Å unit. (E) Sigma-A–weighted Fo–Fc OD-omit electron density map contoured at +6.0 σ, which is viewed from the same direction as D.
Direct Observation of a Hydroxide Ligand on the T2Cu Atom.
There were two water ligands on the T2Cu atom, Wat0 and Wat0′. Previous crystallographic reports on CuNIRs from Achromobacter xylosoxidans (AxNIR) (45) and Rhodobacter sphaeroides (RsNIR) (46) revealed that the binding sites of ligand water can change depending on the pH. Furthermore, pH is temperature-dependent; however, the presence and positions of Wat0 and Wat0′ were not temperature/pH-dependent in our experimental system because their positions in the present data collected at 100 K were almost the same as those in the room temperature SFX structure (SI Appendix, Fig. S4). While the Fo–Fc neutron map indicated that one of the D atoms in Wat0 directed AspCAT, we could not observe the positive peak of the other D atom. All other water molecules at the T2Cu site including Wat0′ showed two positive peaks around them, being neutral water molecules (D2O) (Fig. 2B). Refinement with models such as one D2O molecule resulted in significant negative peaks in the Fo–Fc neutron map, while a deuterated hydroxide model (OD−) having full occupancy showed good agreement with the observed data (Fig. 2 C and D and SI Appendix, Fig. S5). Such OH-type molecules are often observed in neutron crystallography and are usually interpreted as being a result of rotational disorder of water molecules along one of two O-H bond axes (47, 48). However, ligand water molecules are usually immobilized on metal atoms as was observed in the water ligands to Cu3 at the molecular surface, which clearly showed two positive peaks of D atoms in the Fo–Fc neutron map and were refined as D2O (SI Appendix, Fig. S6). Wat0 also appeared to be fixed on the T2Cu atom with a low B-factor value of 15 Å2 for the O atom and its rotational motion was geometrically restricted. Comparison between neutron and X-ray data ruled out the possibility that a diatomic molecule such as O2 or NO binds to T2Cu (Fig. 2 A, D, and E). Therefore, we interpreted that a hydroxide ion existed on the T2Cu atom even at pD 5.3 (pH 4.9) in the preparation of the crystal. Although direct measurement of the pH in our cryogenic experiment was impossible, protonation states of proton-acceptable residues in our structure indicated that pH in the experimental crystal was not as high as that at room temperature; i.e., four of seven free His residues (typical pKa of His: ∼6) were protonated and a glutamate residue Glu34 (typical pKa of Glu: ∼4) positioned at the protein surface was protonated (SI Appendix, Fig. S7). Thus, the observed hydroxide species at low pH might not be the same as a hydroxide-bound state of T2Cu that is proposed to be present in high pH environments (38, 45, 49). The negatively charged hydroxide ion may be stabilized by positive charges of T2Cu and protonated HisCAT. A DFT study shows that the T2Cu site of GtNIR energetically prefers a [Cu(OH)]+ state after the nitrite reduction reaction and direct release of NO (30).
Proton Channel.
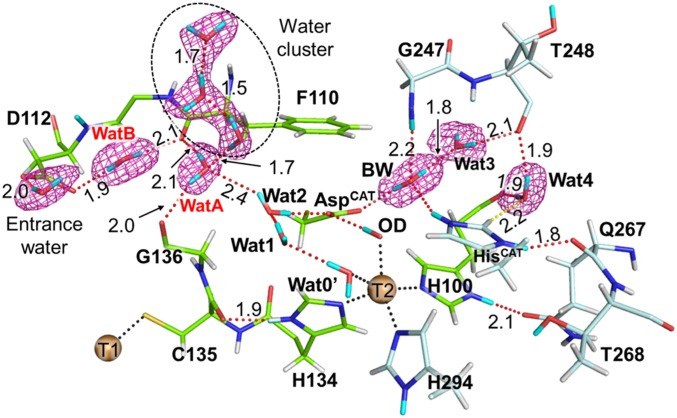
Wat2 next to AspCAT was a terminal water molecule in the major proton channel (MPC), which lined up water molecules from the molecular surface to AspCAT. Asp112 was located at the other end of the MPC and formed a hydrogen bond with one of the bulk water molecules (entrance water) (Fig. 3 and SI Appendix, Fig. S8). Two water molecules, WatA and WatB, constituted a hydrogen bond network between Asp112 and Wat2. These two water molecules did not directly form a hydrogen bond with each other. Instead, the carbonyl O atom of Phe110 mediated the proton channel. The carbonyl O atom of Gly136 was another residue that supported the location of WatA, and the carbonyl O atoms of Phe110 and Gly136 fixed the direction of WatA. Although WatA was the closest water molecule to Wat2 in the MPC, the observed distance between them was slightly long (2.4 Å). This long hydrogen bond and the indirect connection between WatA and WatB indicate that PT from bulk water needs thermodynamic fluctuations to rearrange hydrogen bond networks. This is consistent with that protein flexibility coupled with solvent motions controls proton transfer, which precedes the ET reaction in CuNIR (50). A slightly low occupancy of Wat2 implies its flexibility, which may be important to bring proton to the T2Cu site via AspCAT. Moreover, a water cluster floating above WatA may mediate PT to Wat2.
Fig. 3.
Complete hydrogen bond networks in the major proton channel. The 2Fo–Fc neutron map (courted at 1.0 σ) is shown by meshes. Coordination and hydrogen bonds are shown by dotted black and red lines, respectively. D and H atoms are colored by cyan and white, respectively. Na, Cl, and Cu4 atoms are eliminated for the clarity. Only the major conformation of Wat3 is shown here for the clarity.
BW was a hydrogen donor to AspCAT and Wat3 that formed hydrogen bonds with the carbonyl O atom of Thr248. BW was a hydrogen acceptor from the amide group of Gly247. Thr248 interacted with Wat4, which formed a hydrogen bond with a T2Cu ligand, His100. Furthermore, Wat4 had a weak CH-O hydrogen bond interaction with HisCAT as proposed in the previous XRD study (43).
The electron density map revealed the flexibility of Wat3, with it being in two conformations. The H-D exchange ratios of main chain amides also suggest the flexibility around the T2Cu site. The D atom occupancies were higher in the surface loop regions than in the rigid β-barrel cupredoxin cores (SI Appendix, Fig. S9A). The amide H atoms of residues on the interior loop that is located above the T2Cu site were significantly replaced with D atoms (SI Appendix, Fig. S9B). Because the amide moieties of Gly247 and Thr248 formed hydrogen bonds with water molecules (Fig. 3), the amide H-D exchange reaction occurs more easily at these residues than in other nearby residues. The Gly residue, which is conserved among CuNIRs (SI Appendix, Fig. S9C), may be an important factor that provides flexibility to the catalytic site.
T1Cu Site and Intramolecular ET Pathway.
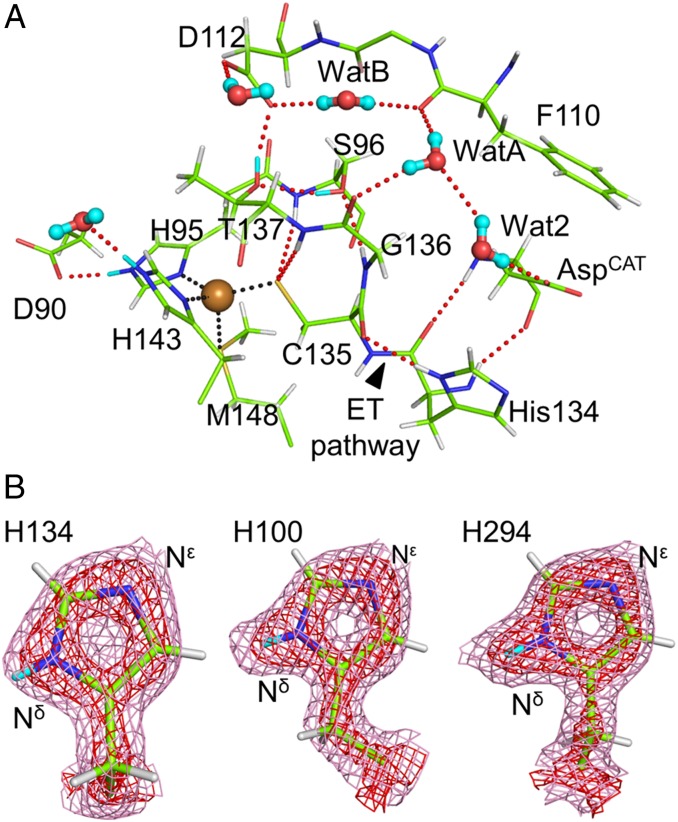
Fig. 4A shows standard hydrogen bonds present around the T1Cu site. The Cys residue is an essential ligand of the T1Cu site because the electronic interaction between the Sγ atom of the Cys ligand and T1Cu dominantly controls the property of the T1Cu site such as color and redox potential (7). The Fo–Fc neutron map indicated that Cys135 in GtNIR is deprotonated (SI Appendix, Fig. S10 A and B), unlike the result of neutron crystallography on amicyanin, a small T1Cu-containig protein, in which the ligand Cys residue was protonated (51). This difference can be explained by the number of hydrogen bonds that the Cys residue forms. Deprotonated Cys in GtNIR was stabilized by two hydrogen bonds between the Sγ atom and the main chain amide groups in Ser96 and Thr137. In amicyanin, the hydrogen bond corresponding to that between Cys135 and Ser96 in GtNIR is conserved, whereas the residue at the position of Thr137 of GtNIR is replaced with Pro and cannot form a hydrogen bond with the Cys ligand (SI Appendix, Fig. S10C). Although the Oγ atoms of Ser96 and Thr137 were close to the Sγ atom of Cys135 (3.7 and 3.4 Å, respectively), the neutron structure did not show their hydrogen bonding to Cys135. The Oγ atom of Thr137 was a hydrogen donor to Asp112, the entrance residue of the MPC, and a hydrogen acceptor from the Oγ atom of Ser96 that is a hydrogen acceptor from the amide moiety of Gly136. The observed hydrogen bond network shows the connection between the T1Cu site and the MPC, which may be involved in PCET accelerated by solvent-slaved protein motions (50).
Fig. 4.
Detailed hydrogen bond networks around the T1Cu site. (A) Standard hydrogen bonds around the T1Cu sites. Water molecules are illustrated by ball-stick models. T1Cu is shown by a brown sphere. (B) Low H-D exchange ratio on the Nδ atom of H134. Sigma-A–weighted 2Fo–Fc neutron maps contoured at 1.0 (pink) and 3.5 σ (red) show low neutron scattering length density at the D atom of H134 compared to those at other T2Cu ligands, H100 and H294. Coordination and hydrogen bonds are shown by dotted black and red lines, respectively. D and H atoms are colored by cyan and white, respectively.
A previous computational study suggests that the intramolecular ET pathway is route A (hydrogen-bond jump) described in Fig. 1A and not route B (covalent peptide bonds). Note that neutron scattering length density around the D atom on the Nδ atom in His134 was less clear than those of other two His ligands to T2Cu. Because the X-ray data shows the presence of the H/D atom on the Nδ atom of His134 (SI Appendix, Fig. S11), we considered that the neutron scattering from this atom position is canceled by positive and negative scattering from partially occupied D and H atoms, respectively. Occupancy refinement resulted in that only 31% of the H atom on the Nδ atom in His134 was replaced with the D atom while the other two His ligands to T2Cu showed 100% H-D exchange (Fig. 4B). This observation indicates that route A has a tight hydrogen bond, which suppresses H-D exchange reactions. The hydrogen bond distance between the carbonyl O atom of Cys135 and the H atom on the Nδ atom of His134 was 1.9 Å. In SI Appendix, Supplementary Results and Discussion, we provided the results and discussion on the network of weak hydrogen bonds around T1Cu, which was revealed by neutron crystallography.
Discussion
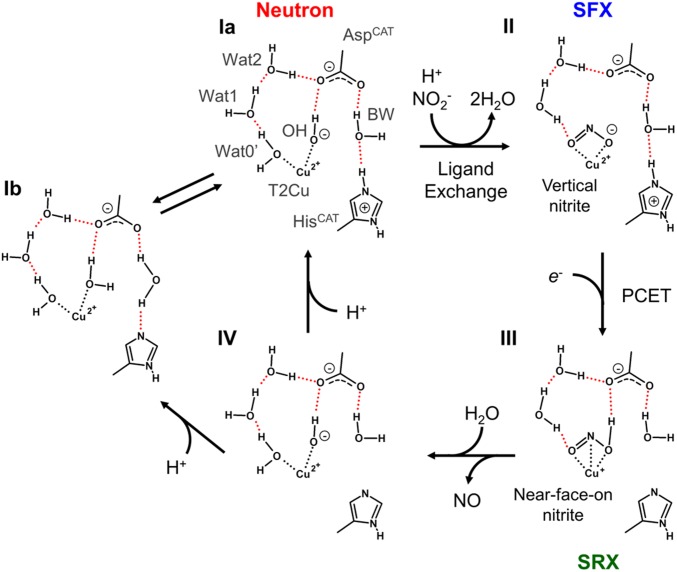
We determined the high-quality and high-resolution neutron structure of GtNIR in its resting state. This structure provides insights into the catalytic mechanism of CuNIRs because it directly visualizes protonation states of catalytic residues, deprotonated AspCAT and protonated HisCAT, which are in good agreement with the results from computational chemistry and spectroscopic experiments. However, this state is inconsistent with the observation in the AcNIR neutron structure showing both deprotonated AspCAT and HisCAT at pD5.5. There are two external ligands to T2Cu in our structure, one of which is OH, not a water molecule, while the AcNIR neutron structure shows only one axial ligand to T2Cu, which can be refined as a water molecule*. This discrepancy may be explained by the equilibrium of the protonation states; that is, a proton can be easily exchanged between HisCAT and the axial ligand to T2Cu. Although Halsted et al. proposed that HisCAT is kept deprotonated during the enzymatic turnover (26), our structure at similar pD casts doubt on this view. Our observation of the hydroxide on T2Cu reminds of the possible OH-bound state in the course of nitrite reduction, which is proposed by DFT calculation using a GtNIR structure (30, 52). A DFT study using another CuNIR, RsNIR, also shows that the OH-bound state is the most stable intermediate after nitrite reduction (29). Our structure is direct evidence that the OH-bound state is stably present in the real CuNIR even at low pH, suggesting the nitrite reduction mechanism without forming Cu-nitrosyl coordination. In fact, side-on NO coordination to T2Cu observed in several crystal structures has been suspected from spectroscopic and computational viewpoints (31) although the recent time-resolved MSOX study captures the Cu-nitrosyl intermediate after NO2− reduction (28). Therefore, the Cu-nitrosyl species is thought to be stabilized by environments in CuNIR crystals such as the presence of a slightly bulky residue above the T2Cu site (53). The combination of the present study and previous experimental and computational studies supports the enzymatic cycle shown in Fig. 5. The computational chemistry shows that the OH-bound state (species Ia) energetically prefers protonation of the hydroxide ion before ligand exchange by nitrite (52). The nitrite ion on the oxidized T2Cu atom shows an η2-O,O vertical conformation that is observed by the damage-free XFEL experiment (species II) (24). Our neutron structure suggests that one of the axial ligand binding sites tends to be occupied by a negative charge. Therefore, the nitrite ion at this moment may be in a negatively-charged and electron-localized state. The vertical conformation of nitrite reduces electronic repulsion with negatively charged AspCAT. The electron-localized state may have an advantage in the following N-O bond cleavage. After the T2Cu site receives an electron from the T1Cu site, the proton on HisCAT is transferred by a redox-coupled proton switch mechanism to nitrite through BW (species III) (24). Now that protonated nitrite can form a hydrogen bond with AspCAT, this state prefers the near-face-on (or near-side-on) manner in which the N atom of nitrite directly interacts with the T2Cu atom, which is observed in photoreduced synchrotron XRD structures (24, 54). The electron moves from the T2Cu atom to nitrite to form NO, which is released without forming the Cu-nitrosyl species. The resulting state (species IV) requires another proton to reconstitute the resting state Ia. Considering that another ND experiment observes species Ib at low pH, there should be an equilibration between Ia and Ib, which implies that proton transfer between the Cu ligand and HisCAT can easily occur via BW.
Fig. 5.
Proposed reaction mechanism of nitrite reduction in CuNIR. Coordination and hydrogen bonds are shown by dotted black and red lines, respectively. Species I, II, and III are observed in the present neutron structure, our previous SFX structure, and our previous structures determined by synchrotron radiation crystallography (SRX), respectively.
The H-D exchange ratios observed in neutron crystallography are related to protein dynamics (55, 56). The high and low D atom occupancies indicate flexible and rigid structures, respectively. The D atom occupancy at the Nδ atom of His134, with which the carbonyl O atom of Cys135 forms a hydrogen bond, was significantly lower than those of other T2Cu ligand His residues. This observation shows that His134 and Cys135 maintain a relatively tight hydrogen bond and keep the local structure rigid. The DFT calculation proposes the corresponding hydrogen bond in AcNIR is a part of the intramolecular ET pathway (8), although it has not been experimentally confirmed. The rigid hydrogen bond observed in our neutron structure is experimental supporting evidence for the presence of the hydrogen-bond jump because CuNIRs must prefer a rigid ET pathway that can reduce reorganization energy in the ET reaction.
The combination of high-quality TOF neutron diffraction data collection with NX joint refinement enables neutron structural determination at near-atomic resolution even for relatively large proteins such as GtNIR with a molecular mass of ∼106 kDa. In several aspects, our neutron structure of GtNIR shows good agreement with computational results, which have not been obvious from earlier XRD studies. We can conclude that high-resolution neutron crystallography is so powerful that it can smoothly connect experimental and computational studies.
Materials and Methods
GtNIR was expressed in Escherichia coli BL21 (DE3) and purified by column chromatography. A large GtNIR crystal was obtained by a combination of micro- and macro-seeding methods at 20 °C. The crystal was placed in the deuterated solution for 1 wk. Complete methods for protein expression, purification, and crystallization are described in SI Appendix, Materials and Methods. The crystal was flash-cooled in a nitrogen-gas stream and then data collection was performed at a beamline BL03 iBIX in the Materials and Life Sciences Experimental Facility of the Japan Proton Accelerator Research Complex (J-PARC) (57, 58). X-ray data were collected from the same crystal at a beamline AR-NE3A of Photon Factory Advanced Ring (PF-AR) (59) at 100 K using a Pilatus 2M-F detector (DECTRIS). Processing of data and structural analysis were performed as described in SI Appendix, Materials and Methods. The coordinate file and the structure factor file are deposited in the Protein Data Bank (PDB ID code 6L46). The raw data are available at the Integrated Resource for Reproducibility in Macromolecular Crystallography (https://proteindiffraction.org).
Supplementary Material
Acknowledgments
This study was partly supported by Grant-in-Aid for Young Scientists (B) 17K17862 (to Y.F.) and Grant-in-Aid for Scientific Research (B) 18H02004 (to T.I.). We appreciate the student fellowship of the Japan Atomic Energy Agency (JAEA) in 2010 (to Y.F.) because our neutron crystallography project for CuNIR started at JAEA in 2010 when we recorded the first neutron diffraction from a large CuNIR crystal at Japan research reactor-3 (JRR-3) (Proposal 2010-urgent-8). We thank Dr. K. Kurihara and Dr. T. Ohhara for their support at JRR-3. Neutron data were collected at BL03 of J-PARC under the project for Ibaraki prefectural local government beamline (Proposal 2018PX0010). X-ray data were collected at AR-NE3A of PF-AR (Proposal 2018RP-04). We acknowledge supports from beamline staffs. We also thank Dr. S. Suzuki and Dr. M. Nojiri who guided to the study of CuNIRs; Dr. H. Koteishi and Dr. M. Lintuluoto for discussion; Ms. J. Kim for checking the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinate file and the structure factor file are deposited in the Protein Data Bank (PDB ID code 6L46). The raw data collected in this study are available at the Integrated Resource for Reproducibility in Macromolecular Crystallography (https://proteindiffraction.org).
*Omitting D atoms on the ligand water molecule, we re-refined the AcNIR neutron structure (PDB ID code 6GTJ) with the deposited coordinates and the structure factor. We could not observe any positive Fo–Fc peaks around it, implying the ambiguity of its protonation state.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918125117/-/DCSupplemental.
References
- 1.Zumft W. G., Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulanger M. J., Murphy M. E. P., Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: A new class of copper-containing nitrite reductases. J. Mol. Biol. 315, 1111–1127 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Godden J. W., et al. , The 2.3 angstrom X-ray structure of nitrite reductase from Achromobacter cycloclastes. Science 253, 438–442 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Kukimoto M., et al. , X-ray structure and site-directed mutagenesis of a nitrite reductase from Alcaligenes faecalis S-6: Roles of two copper atoms in nitrite reduction. Biochemistry 33, 5246–5252 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Dodd F. E., Van Beeumen J., Eady R. R., Hasnain S. S., X-ray structure of a blue-copper nitrite reductase in two crystal forms. The nature of the copper sites, mode of substrate binding and recognition by redox partner. J. Mol. Biol. 282, 369–382 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Inoue T., et al. , Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 A resolution: Comparison of blue and green nitrite reductases. J. Biochem. 124, 876–879 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Solomon E. I., Szilagyi R. K., DeBeer George S., Basumallick L., Electronic structures of metal sites in proteins and models: Contributions to function in blue copper proteins. Chem. Rev. 104, 419–458 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Hadt R. G., Gorelsky S. I., Solomon E. I., Anisotropic covalency contributions to superexchange pathways in type one copper active sites. J. Am. Chem. Soc. 136, 15034–15045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy M. E. P., Turley S., Adman E. T., Structure of nitrite bound to copper-containing nitrite reductase from Alcaligenes faecalis. Mechanistic implications. J. Biol. Chem. 272, 28455–28460 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Boulanger M. J., Kukimoto M., Nishiyama M., Horinouchi S., Murphy M. E., Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J. Biol. Chem. 275, 23957–23964 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Kataoka K., Furusawa H., Takagi K., Yamaguchi K., Suzuki S., Functional analysis of conserved aspartate and histidine residues located around the type 2 copper site of copper-containing nitrite reductase. J. Biochem. 127, 345–350 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi K., Tagawa S., Deligeer, Suzuki S., The pH-dependent changes of intramolecular electron transfer on copper-containing nitrite reductase. J. Biochem. 126, 408–412 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Brenner S., et al. , Demonstration of proton-coupled electron transfer in the copper-containing nitrite reductases. J. Biol. Chem. 284, 25973–25983 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leferink N. G., et al. , Proton-coupled electron transfer in the catalytic cycle of Alcaligenes xylosoxidans copper-dependent nitrite reductase. Biochemistry 50, 4121–4131 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Tocheva E. I., Rosell F. I., Mauk A. G., Murphy M. E., Side-on copper-nitrosyl coordination by nitrite reductase. Science 304, 867–870 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Antonyuk S. V., Strange R. W., Sawers G., Eady R. R., Hasnain S. S., Atomic resolution structures of resting-state, substrate- and product-complexed Cu-nitrite reductase provide insight into catalytic mechanism. Proc. Natl. Acad. Sci. U.S.A. 102, 12041–12046 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nojiri M., et al. , Structural basis of inter-protein electron transfer for nitrite reduction in denitrification. Nature 462, 117–120 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Tocheva E. I., Rosell F. I., Mauk A. G., Murphy M. E. P., Stable copper-nitrosyl formation by nitrite reductase in either oxidation state. Biochemistry 46, 12366–12374 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Schlichting I., et al. , The catalytic pathway of cytochrome p450cam at atomic resolution. Science 287, 1615–1622 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Yano J., et al. , X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. U.S.A. 102, 12047–12052 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hough M. A., Antonyuk S. V., Strange R. W., Eady R. R., Hasnain S. S., Crystallography with online optical and X-ray absorption spectroscopies demonstrates an ordered mechanism in copper nitrite reductase. J. Mol. Biol. 378, 353–361 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Fukuda Y., et al. , Insights into unknown foreign ligand in copper nitrite reductase. Biochem. Biophys. Res. Commun. 464, 622–628 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Fukuda Y., et al. , Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography. J. Biochem. 159, 527–538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda Y., et al. , Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. Proc. Natl. Acad. Sci. U.S.A. 113, 2928–2933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halsted T. P., et al. , An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase. IUCrJ 5, 22–31 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halsted T. P., et al. , Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography. IUCrJ 6, 761–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horrell S., et al. , Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal. IUCrJ 3, 271–281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horrell S., et al. , Enzyme catalysis captured using multiple structures from one crystal at varying temperatures. IUCrJ 5, 283–292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh S., Dey A., Sun Y., Scholes C. P., Solomon E. I., Spectroscopic and computational studies of nitrite reductase: Proton induced electron transfer and backbonding contributions to reactivity. J. Am. Chem. Soc. 131, 277–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lintuluoto M., Lintuluoto J. M., DFT study on nitrite reduction mechanism in copper-containing nitrite reductase. Biochemistry 55, 210–223 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Solomon E. I., et al. , Copper active sites in biology. Chem. Rev. 114, 3659–3853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin X., Deng L., Hu C., Li L., Chen X., Copper-containing nitrite reductase employing proton-coupled spin-exchanged electron-transfer and multiproton synchronized transfer to reduce nitrite. Chemistry 23, 14900–14910 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S., et al. , Resolution of the spectroscopy versus crystallography issue for NO intermediates of nitrite reductase from Rhodobacter sphaeroides. J. Am. Chem. Soc. 129, 10310–10311 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheldrick G. M., Phase annealing in SHELX-90: Direct methods for larger structures. Acta Crystallogr. A 46, 467–473 (1990). [Google Scholar]
- 35.Nakamura A., et al. , “Newton’s cradle” proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography. Sci. Adv. 1, e1500263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadei C. M., et al. , Heme enzymes. Neutron cryo-crystallography captures the protonation state of ferryl heme in a peroxidase. Science 345, 193–197 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Kwon H., et al. , Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme. Nat. Commun. 7, 13445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y., et al. , Catalytic function and local proton structure at the type 2 copper of nitrite reductase: The correlation of enzymatic pH dependence, conserved residues, and proton hyperfine structure. Biochemistry 41, 7464–7474 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Langan P., Greenea G., Schoenborn B. P., Protein crystallography with spallation neutrons: The user facility at Los Alamos neutron science center. J. Appl. Cryst. 37, 24–31 (2004). [Google Scholar]
- 40.Adams P. D., Mustyakimov M., Afonine P. V., Langan P., Generalized X-ray and neutron crystallographic analysis: More accurate and complete structures for biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 65, 567–573 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano N., et al. , Application of profile fitting method to neutron time-of-flight protein single crystal diffraction data collected at the iBIX. Sci. Rep. 6, 36628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda Y., et al. , Crystallographic study of dioxygen chemistry in a copper-containing nitrite reductase from Geobacillus thermodenitrificans. Acta Crystallogr. D Struct. Biol. 74, 769–777 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Fukuda Y., et al. , Structural insights into the function of a thermostable copper-containing nitrite reductase. J. Biochem. 155, 123–135 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Zhang H., Boulanger M. J., Mauk A. G., Murphy M. E. P., Carbon Monoxide binding to copper-containing nitrite reductase from Alcaligenes faecalis. J. Phys. Chem. B 104, 10738–10742 (2000). [Google Scholar]
- 45.Ellis M. J., et al. , X-ray structure of a blue copper nitrite reductase at high pH and in copper-free form at 1.9 A resolution. Acta Crystallogr. D Biol. Crystallogr. 57, 1110–1118 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Jacobson F., et al. , pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase. J. Biol. Chem. 282, 6347–6355 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Habash J., et al. , Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography. Acta Crystallogr. D Biol. Crystallogr. 56, 541–550 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Chatake T., Ostermann A., Kurihara K., Parak F. G., Niimura N., Hydration in proteins observed by high-resolution neutron crystallography. Proteins 50, 516–523 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Wijma H. J., Jeuken L. J., Verbeet M. P., Armstrong F. A., Canters G. W., A random-sequential mechanism for nitrite binding and active site reduction in copper-containing nitrite reductase. J. Biol. Chem. 281, 16340–16346 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Hedison T. M., Heyes D. J., Shanmugam M., Iorgu A. I., Scrutton N. S., Solvent-slaved protein motions accompany proton coupled electron transfer reactions catalysed by copper nitrite reductase. Chem. Commun. (Camb.) 55, 5863–5866 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Sukumar N., Mathews F. S., Langan P., Davidson V. L., A joint x-ray and neutron study on amicyanin reveals the role of protein dynamics in electron transfer. Proc. Natl. Acad. Sci. U.S.A. 107, 6817–6822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lintuluoto M., Lintuluoto J. M., DFT study on enzyme turnover including proton and electron transfers of copper-containing nitrite reductase. Biochemistry 55, 4697–4707 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Merkle A. C., Lehnert N., The side-on copper(I) nitrosyl geometry in copper nitrite reductase is due to steric interactions with isoleucine-257. Inorg. Chem. 48, 11504–11506 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Fukuda Y., Inoue T., High-temperature and high-resolution crystallography of thermostable copper nitrite reductase. Chem. Commun. (Camb.) 51, 6532–6535 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Kossiakoff A. A., Protein dynamics investigated by the neutron diffraction-hydrogen exchange technique. Nature 296, 713–721 (1982). [DOI] [PubMed] [Google Scholar]
- 56.Bennett B., et al. , Neutron diffraction studies of Escherichia coli dihydrofolate reductase complexed with methotrexate. Proc. Natl. Acad. Sci. U.S.A. 103, 18493–18498 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka I., et al. , Neutron structure analysis using the IBARAKI biological crystal diffractometer (iBIX) at J-PARC. Acta Crystallogr. D Biol. Crystallogr. 66, 1194–1197 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Kusaka K., et al. , Evaluation of performance for IBARAKI biological crystal diffractometer iBIX with new detectors. J. Synchrotron Radiat. 20, 994–998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamada Y., et al. , AR-NE3A, a new macromolecular crystallography beamline for pharmaceutical applications at the Photon Factory. AIP Conf. Proc. 1234, 415–418 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.