Significance
COP1 is an evolutionarily conserved ubiquitin ligase that plays a critical role in regulating the growth patterns of plants in response to light. Light exposure promotes COP1 export from the nucleus, preventing COP1-mediated degradation of transcription factors that regulate light-mediated development (photomorphogenesis). We report that, similar to its regulation in plants, mammalian COP1 localization is modulated by environment signals. Inactivation of a key mitogen-activated protein kinase (MAPK) signaling pathway (e.g., via exposure to anthrax lethal toxin) induces the rapid transit of COP1 from the nuclear envelope to the nucleoplasm. This activity leads to degradation of COP1’s nuclear transcription factor substrates, which are critical for cell survival and proliferation, allowing mammalian cells to adapt rapidly to changing extracellular cues.
Keywords: anthrax lethal toxin, Erk1/2, ubiquitin E3 ligase, COP1, c-Jun
Abstract
Anthrax lethal toxin (LT) is a protease virulence factor produced by Bacillus anthracis that is required for its pathogenicity. LT treatment causes a rapid degradation of c-Jun protein that follows inactivation of the MEK1/2-Erk1/2 signaling pathway. Here we identify COP1 as the ubiquitin E3 ligase that is essential for LT-induced c-Jun degradation. COP1 knockdown using siRNA prevents degradation of c-Jun, ETV4, and ETV5 in cells treated with either LT or the MEK1/2 inhibitor, U0126. Immunofluorescence staining reveals that COP1 preferentially localizes to the nuclear envelope, but it is released from the nuclear envelope into the nucleoplasm following Erk1/2 inactivation. At baseline, COP1 attaches to the nuclear envelope via interaction with translocated promoter region (TPR), a component of the nuclear pore complex. Disruption of this COP1–TPR interaction, through Erk1/2 inactivation or TPR knockdown, leads to rapid COP1 release from the nuclear envelope into the nucleoplasm where it degrades COP1 substrates. COP1-mediated degradation of c-Jun protein, combined with LT-mediated blockade of the JNK1/2 signaling pathway, inhibits cellular proliferation. This effect on proliferation is reversed by COP1 knockdown and ectopic expression of an LT-resistant MKK7-4 fusion protein. Taken together, this study reveals that the nuclear envelope acts as a reservoir, maintaining COP1 poised for action. Upon Erk1/2 inactivation, COP1 is rapidly released from the nuclear envelope, promoting the degradation of its nuclear substrates, including c-Jun, a critical transcription factor that promotes cellular proliferation. This regulation allows mammalian cells to respond rapidly to changes in extracellular cues and mediates pathogenic mechanisms in disease states.
Anthrax lethal toxin (LT) is composed of lethal factor (LF) and the receptor-binding protective antigen (PA), which are encoded on the pXO1 virulence plasmid of Bacillus anthracis (1–4). LF is a zinc-dependent metalloprotease with specific activity against certain mitogen-activated protein kinase kinases (MKKs) (5). The MKKs lie in the middle of the three-tiered mitogen-activated protein kinase kinase kinase (MKKK)–MKK–mitogen-activated protein kinase (MAPK) signaling cascades (6, 7). Extracellular stimuli such as growth factors or cytokines initiate activation of MKKKs that subsequently phosphorylate MKKs, which in turn phosphorylate MAPKs. Activated MAPKs catalyze the phosphorylation of their cytoplasmic and nuclear substrates, which then participate in the regulation of a large variety of cellular processes. LF cleavage of MKKs at their docking sites (D-sites) disrupts the activation of MAPKs, including the extracellular signal-regulated kinases (Erk1/2), p38 MAPKs, and Jun kinases (JNKs), which are activated by MKK1/MKK2, MKK3/MKK6, and MKK4/MKK7, respectively (5, 8–11).
Studies from our laboratory have revealed that LT reduces levels of the c-Jun transcription factor protein by promoting its degradation via inactivation of MKK1/2-Erk1/2 signaling and blocking its gene transcription via inactivation of the MKK4-JNK1/2 signaling pathway (12). c-Jun is a key member of the AP-1 transcription factor family, which regulates a myriad of cellular activities, including cellular proliferation, differentiation, survival, death, and tumorigenesis (13, 14). The level of c-Jun protein is tightly controlled by a process that involves rapid turnover by ubiquitination and degradation. Ubiquitination of c-Jun has been shown to be carried out by several ubiquitin E3 ligases, including Itchy E3 ubiquitin protein ligase (ITCH) (15), F-box, and WD repeat domain containing 7 (FBW7) (16), cullin 4 (CUL4) (17), Sensitive to Apoptosis Gene/RING-box protein 2 (SAG/RBX2) (18), MKKK1 (19), and Constitutive Photomorphogenic1 (COP1) (17, 20).
COP1 was originally identified in the study of the COP/DET/FUS loci in plants and characterized as a key regulator of light-mediated plant development (21, 22), acting to repress photomorphogenesis by promoting the degradation of positive signaling regulators, including photoreceptors and downstream transcription factors such as HY5, HYH, LAF1, and HFR1 (23, 24). Cop1-mutated seedlings undergo photomorphogenic development even in the absence of light (21, 22). In mammals, COP1 serves as a ubiquitin E3 ligase for c-Jun (17, 20), ETS transcription variants (ETV) (25–27), p53 (28), and COP1 itself (29). COP1 protein is evolutionary conserved, comprising three domains: an N-terminal RING-finger motif, a coiled-coil domain, and seven WD40 repeats at its C terminus (30). The RING-finger domain mediates COP1’s interactions with Ub-conjugating enzymes (E2s). The coiled-coil domain is required for formation of COP1 homodimers or heterodimers with other proteins. A COP1 transcript variant lacking its exon 7 and a part of exon 4 produces a protein lacking 24 amino acids (COP1Δ24) in the coiled-coil domain that has no function and cannot mediate c-Jun degradation (17). The WD40 repeats mediate COP1 interaction with its substrates. Although COP1 possesses intrinsic E3 ligase activity and may ubiquitinate some targets on its own, COP1 forms a super protein complex with other proteins in vivo that has a molecular weight over 700 kDa and may preferentially function as a protein complex (31). In mammalian cells, COP1 has been reported to form a complex with De-etiolated-1 (DET1), DNA damage-binding protein 1 (DDB1), cullin 4 (CUL4), and RING-box protein 1 (RBX1) (CRL4COP1-DET1), collectively promoting degradation of its substrates such as c-Jun and ETVs (17, 25).
In plants, COP1 activity is regulated by its subcellular distribution (24, 32). In darkness, COP1 is mainly localized to the nucleus, where it promotes the ubiquitination and degradation of photomorphogenesis-promoting transcription factors. Upon exposure to light, COP1 levels are drastically reduced in the nucleus, allowing reaccumulation of COP1-targeted nuclear-localized transcription factors and subsequent expression of downstream genes required for plant photomorphogenesis. In mammalian cells, COP1 has been shown to be located both in the cytoplasm and nucleus (31), but whether COP1 activity is regulated by alteration of its subcellular distribution was previously unknown. In this study, we observed that at baseline a large fraction of COP1 is located on the nuclear envelope, but Erk1/2 inactivation leads to its rapid redistribution into the nucleoplasm. COP1 tethers to the nuclear envelope via interaction with the nuclear pore protein TPR, and Erk1/2 inactivation disrupts the interaction between COP1 and TPR. Subsequent COP1 translocation to the nucleus promotes the degradation of its nuclear substrates. These findings demonstrate that mammalian COP1 plays a critical role in transducing external signals into the nucleus by sensing Erk1/2 activity and regulating the stability of transcription factors, enabling a rapid response for mammalian cells to adapt to extracellular cues.
Results
Erk1/2 Inactivation Induces Activation of COP1 Ubiquitin E3 Ligase.
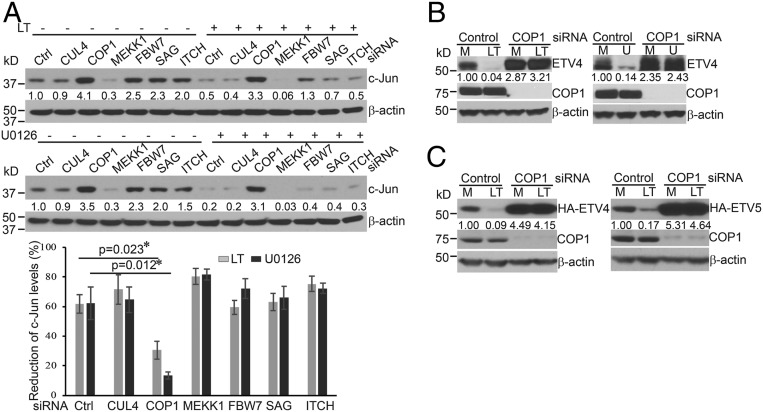
Our previous study showed that Erk1/2 inactivation following treatment with anthrax LT or the MEK1/2 inhibitor, U0126, induces a rapid degradation of c-Jun protein (12). To identify the ubiquitin E3 ligase that mediates Erk1/2 inactivation-induced c-Jun degradation, we investigated a panel of E3 ligases, previously reported to target c-Jun for proteasomal degradation. To this end, Hepa1c1c7 cells were transfected with siRNA targeting CUL4, COP1, MEKK1, FBW7, SAG/RBX2, or ITCH and subsequently treated with LT or U0126. As shown in Fig. 1A, knockdown of COP1, but not other E3 ligases, prevented c-Jun degradation in cells treated with LT or U0126. Prevention of c-Jun degradation by COP1 siRNA transfection was also observed in Hepa1c1c7 cells over a wide range of LT and U0126 concentrations (SI Appendix, Fig. S1). In addition, transfection of COP1 siRNA restored the levels of c-Jun protein in HepG2 and primary murine embryo fibroblast (MEF) cells that were treated with LT or U0126 (SI Appendix, Fig. S2 A and B). These data suggest that inactivation of Erk1/2 activates COP1 and subsequently promotes degradation of COP1 substrates. In addition to c-Jun, ETS transcription variants (ETV1, ETV4, and ETV5) are also known COP1 substrates. We next investigated whether these ETV proteins are degraded in LT-treated cells in a COP1-dependent manner. As expected, treatment with LT or U0126 induced a rapid reduction of endogenous ETV4 and ETV5 in HepG2 cells, which was prevented by pretreatment with the proteasome inhibitor, MG132 (SI Appendix, Fig. S3 A and B). Knockdown of COP1 restored the levels of endogenous ETV4 and ETV5, and HA-tagged ETV4 and ETV5 in Hepa1c1c7 and HepG2 cells that were treated with LT or U0126 (Fig. 1 B and C and SI Appendix, Fig. S3 C and D). Collectively, these data indicate that MKK1/2-Erk1/2 blockade via U0126 or LT treatment activates COP1, leading to a rapid degradation of its substrates.
Fig. 1.
Knockdown of COP1 ubiquitin E3 ligase prevents degradation of c-Jun, ETV4, and ETV5 in cells treated with anthrax LT or U0126. (A) Hepa1c1c7 cells were transfected with control (Ctrl) siRNA or siRNA targeting CUL4, COP1, MEKK1, FBW7, SAG, or ITCH ubiquitin E3 ligases. Two days following transfection, the cells were cultured with or without LT for 2 h (Upper) or U0126 for 1 h (Lower) and then extracted with NuPAGE LDS sample buffer. The expression levels of c-Jun protein in the cell lysates were assessed by Western blotting. Reductions in c-Jun levels were statistically analyzed using data from three independent experiments using an unpaired, two-tailed t test and presented as means ± SE (Bottom). Asterisks depict statistically significant differences between the two groups (P < 0.05). (B and C) Hepa1c1c7 cells (B) and Hepa1c1c7 cells stably expressing HA-ETV4 (C, Left) or HA-ETV5 (C, Right) were transfected with control or COP1 siRNA. Two days following transfection, the cells were cultured with medium alone (M) or LT for 2 h or U0126 (U) for 1 h and then extracted with NuPAGE LDS sample buffer. The expression levels of endogenous ETV-4 (B), HA-ETV4 and HA-ETV5 (C) proteins in the cell lysates were assessed by Western blotting. β-Actin levels were used as loading controls. Band intensities were quantified and normalized using β-actin levels. The normalized amounts are shown under each band. Data shown are representative of at least two independent experiments.
The CRL4COP1-Det1 Complex Is Not Essential for Erk1/2 Inactivation-Induced Degradation of COP1 Substrates.
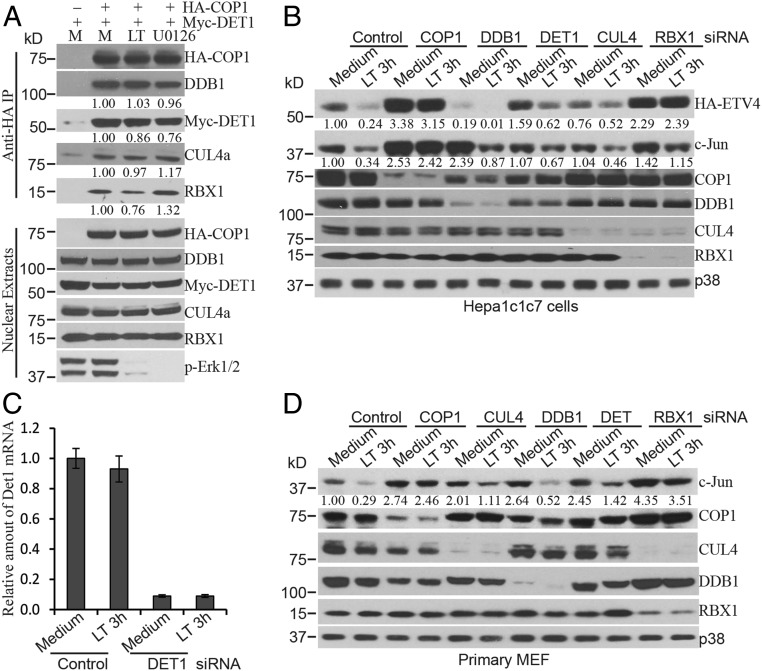
Although COP1 possesses intrinsic E3 ligase activity and may ubiquitinate some targets on its own, it has been reported that COP1 forms a complex with DET1, DDB1, CUL4, and RBX1 (CRL4COP1-Det1) to promote degradation of c-Jun and ETVs (17, 25). We next investigated the effect of Erk1/2 inactivation on the formation of CRL4COP1-Det1 complex. Hepa1c1c7 cells stably expressing HA-COP1 and Myc-DET1 or control cells expressing only Myc-DET1 were cultured with or without LT for 2 h or U0126 for 1 h. Nuclear extracts from these cells were incubated with anti-HA magnetic beads to immunoprecipitate the CRL4COP1-Det1 complex. Western blotting analysis showed that treatment with LT or U0126 did not affect the formation of the CRL4COP1-Det1 complex (Fig. 2A). We then transfected cells with siRNA, targeting the components of CRL4COP1-Det1 complex and examined their role in Erk1/2 inactivation-induced degradation of COP1 substrates. Knockdown of DET1, DDB1, and CUL4 increased the levels of c-Jun at steady state, but surprisingly, knockdown of these three components neither consistently blocked the degradation of c-Jun in LT-treated Hepa1c1c7, primary MEF, or HepG2 cells (Fig. 2 B–D and SI Appendix, Fig. S4), nor prevented the reduction in stably transfected HA-ETV4 and HA-ETV5 protein levels in LT-treated Hepa1c1c7 and HepG2 cells (Fig. 2B and SI Appendix, Fig. S4). In contrast, the knockdown of COP1 substantially reversed the degradation of these proteins. Together, these data suggest that degradation of COP1 substrates following Erk1/2 inactivation may involve an alternative pathway(s) independent of the CRL4COP1-Det1 complex. Of note, knockdown of RBX1 markedly reduced the levels of CUL4 and restored the levels of HA-ETV4 and c-Jun in LT-treated cells (Fig. 2 B and D and SI Appendix, Fig. S4).
Fig. 2.
COP1 functions independently of the CRL4COP1-DET1 complex following LT or U0126 treatment. (A) Hepa1c1c7 cells stably expressing Myc-DET1 alone (used as an immunoprecipitation [IP] control) or with HA-COP1 were cultured with (LT) or without (M) LT for 2 h or U0126 for 1 h. Nuclear extracts prepared from the cells were incubated with anti-HA magnetic beads for IP. The IP complexes were dissolved in NuPAGE LDS sample buffer, and the levels of indicated proteins in the IP complex and nuclear extracts were analyzed by Western blotting. (B–D) Hepa1c1c7 (B and C) and primary MEF (D) were transfected with the indicated siRNA. Two days following transfection, the cells were cultured with or without LT for 2 h and then extracted with NuPAGE LDS sample buffer. The expression levels of indicated proteins in the cell lysates were assessed by Western blotting. Knockdown of target proteins by siRNAs were confirmed at the protein level by Western blotting or at the mRNA level by quantitative PCR (Det1) (C). p38 levels were used as loading controls. Band intensities were quantified and normalized using HA-COP1 (A) or p38 levels (B and D). The normalized protein levels are shown under each band. Data shown are representative of at least two independent experiments.
Erk1/2 Inactivation Promotes Relocation of COP1 from the Nuclear Envelope to Nucleoplasm.
COP1 activity in plant cells is regulated by its subcellular distribution (32). In mammalian cells, COP1 has been shown to be located in both the cytoplasm and the nucleus (31), and we reasoned that its activity may also be regulated by alteration of its subcellular distribution. To investigate this possibility, we initially used a GFP-fusion protein as a tool to visualize COP1 and examine its subcellular distribution. Surprisingly, the GFP-COP1 fusion protein was mostly located in the cytoplasm (SI Appendix, Fig. S5A). In line with its cytosolic distribution, GFP-COP1 was unable to promote the degradation of nuclear c-Jun protein in LT-treated Hepa1c1c7 cells (SI Appendix, Fig. S5B). The abnormal distribution of GFP-COP1 may be caused by the relatively large size of the GFP tag. To overcome the lack of a commercial anti-COP1 antibody that could be applied for immunofluorescent staining in our hands, we generated stable cell lines expressing HA-COP1 and Myc-COP1 fusion proteins and examined their subcellular distribution by immunofluorescence. Both HA-COP1 and Myc-COP1 were predominantly located in the nucleus with enrichment at the nuclear periphery that is likely the nuclear envelope (SI Appendix, Fig. S5C). Western blotting analysis of the cytosol and nuclear extracts also showed that HA-COP1 and Myc-COP1 fusion proteins were mainly located in the nuclear extracts, which matched the distribution of endogenous COP1 (SI Appendix, Fig. S5D). In addition, HA-COP1 but not the nonfunctional HA-COP1Δ24 and ligase-dead HA-COP1-RINGmut promoted c-Jun degradation in cells that were transfected with siRNA specifically targeting endogenous COP1 and then treated with LT or U0126 (SI Appendix, Fig. S5 E and F), demonstrating that HA tag did not affect COP1 function. Of note, transient transfection of 293T cells with HA-COP1 showed increased cytosolic distribution, indicating that high levels of exogenous COP1 may alter cellular distribution (SI Appendix, Fig. S5G).
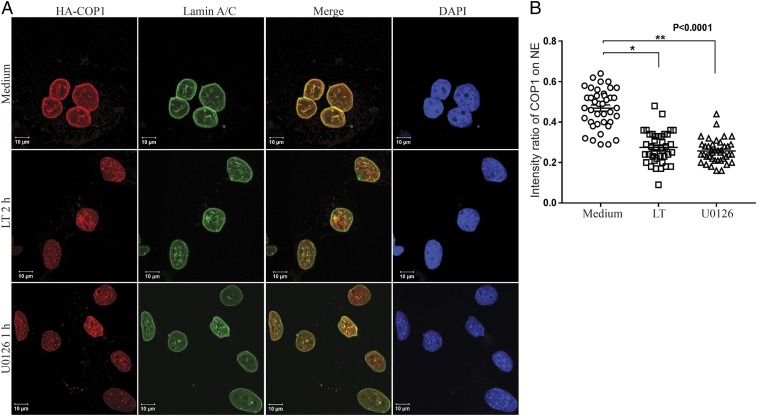
Based on these findings, we chose to use cells stably expressing HA-COP1 or Myc-COP1 to investigate the effect of Erk1/2 inactivation on COP1 subcellular distribution. Hepa1c1c7 and HepG2 cells expressing HA-COP1 were cultured with or without LT (2 h) or U0126 (1 h), and then used for immunofluorescence staining of HA-COP1 and nuclear envelope marker Lamin A/C with anti-HA and anti-Lamin A/C. To render COP1 on the nuclear envelope more visible in immunofluorescent staining, the cells were treated with 1% Triton for 10 s and washed with cold PBS two times before fixation as previously described (31). In untreated cells, COP1 was preferentially located on the nuclear envelope, overlapping with Lamin A/C (Fig. 3 and SI Appendix, Fig. S6). Treatment with LT and U0126 drastically reduced the levels of COP1 on the nuclear envelope and increased its distribution in the nucleoplasm (Fig. 3 and SI Appendix, Fig. S6).
Fig. 3.
Anthrax LT and U0126 treatment promotes a rapid relocation of COP1 from the nuclear envelope (NE) to nucleoplasm. (A) Hepa1c1c7 cells stably expressing HA-COP1 were cultured with or without LT for 2 h or U0126 for 1 h. The cells were then used for immunofluorescence staining with mouse anti-Lamin A/C and rabbit anti-HA primary antibodies followed by goat anti-mouse IgG-Alexa Fluor 633 and goat anti-mouse IgG-Alexa Fluor 568 secondary antibodies. The locations of Lamin A/C, HA-COP1, and the nucleus were assessed by LSM 880 confocal microscope. (B) Fluorescence intensities were quantified using the Bitplane Imaris image analysis software. The intensity ratios of COP1 located on the NE to that in the nucleoplasm were calculated using data from 40 nuclei, from 9 to 13 randomly selected image files obtained from two or three independently performed experiments. Data were statistically analyzed using an unpaired, two-tailed t test at the 95% confidence interval using GraphPad Prism software and presented as means ± SE. P < 0.05 was considered statistically significant.
COP1 Is Attached to the Nuclear Envelope by Interaction with TPR.
We next investigated how Erk1/2 inactivation promotes COP1 redistribution from the nuclear envelope to nucleoplasm. It has been reported that the vertebrate-specific N-terminal extension of COP1 is required for its location to the nuclear envelope (31). Using Hepa1c1c7 cells stably expressing HA-tagged full-length COP1, COP1 lacking part of its coiled-coil domain (COP1Δ24) or COP1 with an N-terminal 70-amino acid deletion (COP1ΔN70), we found that the coiled-coil domain was essential for COP1 localization to the nuclear envelope (SI Appendix, Fig. S7). By Western blotting, we also observed that treatment with 1% Triton for 1 min removed nearly all of the HA-COP1Δ24 but not the HA-COP1ΔN70 protein (SI Appendix, Fig. S7), supporting a dominant role for the coiled-coil domain in promoting COP1 localization to the nuclear envelope.
COP1 is proposed to function in the CRL4COP1-DET1 complex, which degrades c-Jun and ETVs (17, 25). We therefore investigated whether formation of the complex is required for its location to the nuclear envelope. Hepa1c1c7 cells stably expressing Myc-COP1 and HA-DET1 were transfected with siRNA targeting DDB1, or siRNAs targeting both endogenous mouse DET1 and transfected human DET1. Knockdown of DET1 or DDB1 did not affect COP1’s location on the nuclear envelope (SI Appendix, Fig. S8). Interestingly, DDB1 knockdown shifted the predominant distribution of DET1 from the nucleus to the cytoplasm. Moreover, 1% Triton treatment for 1 min removed most of DDB1 and DET1 but not COP1 protein (SI Appendix, Fig. S9), consistent with the conclusion that COP1’s location on the envelope does not require formation of the CRL4COP1-DET1 complex.
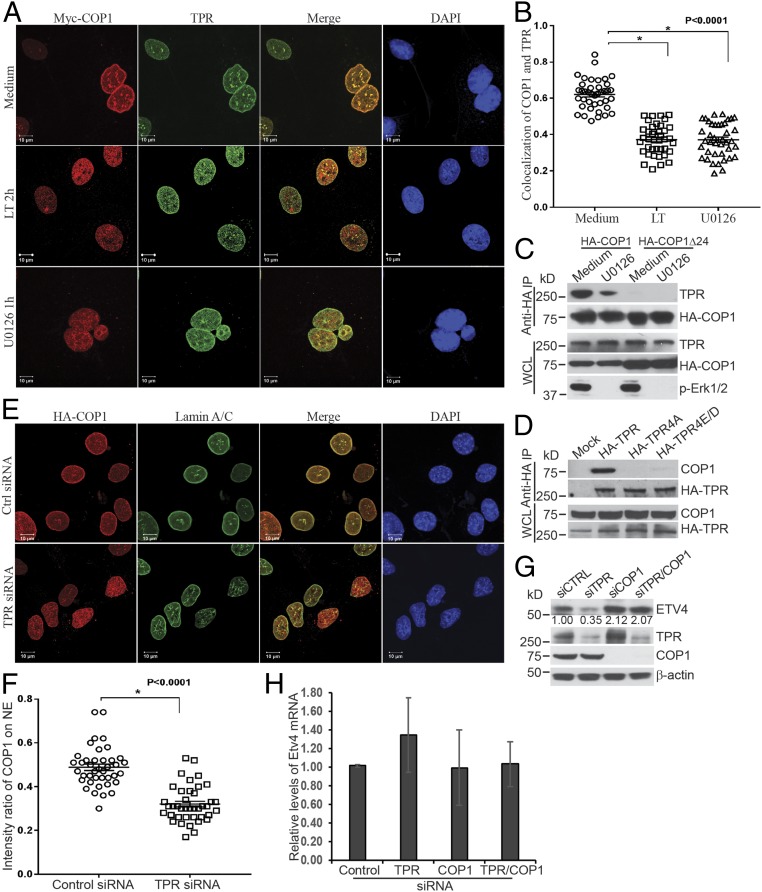
To further elucidate the mechanism through which COP1 attaches to the nuclear envelope in an Erk1/2-dependent manner, immunoprecipitated complexes were prepared from Hepa1c1c7 cells stably expressing HA-COP1 and cultured with MG132 alone or with LT and analyzed using mass spectrometry. TPR was identified as a protein whose abundance in the complex was reduced by treatment with LT (SI Appendix, Fig. S10). The TPR molecule was of particular interest, because it is a component of the nuclear pore complex located in the nuclear envelope basket (33). TPR is also an Erk1/2 substrate, four residues of which can be phosphorylated by Erk1/2 (34). Moreover, TPR was previously shown to coimmunoprecipitate with COP1 (35). By immunofluorescence staining, we observed that COP1 colocalized with TPR on the nuclear envelope. The colocalization of COP1 and TPR was disrupted by treatment with LT or U0126 (Fig. 4 A and B). Western blotting analysis confirmed that TPR coimmunoprecipitates with full-length COP1 but not COP1Δ24, which is in line with the different localization of COP1 and COP1Δ24 (Fig. 4C). Coprecipitation of COP1 and TPR is dependent on Erk1/2 activity, because the interaction of COP1 and TPR was disrupted by U0126 treatment or mutation of the four Erk1/2-phosphorylated residues of TPR (Fig. 4 C and D). As with Erk1/2 inactivation, TPR knockdown caused a redistribution of COP1 from the nuclear envelope to the nucleoplasm (Fig. 4 E and F), leading to COP1-dependent reductions of protein, but not mRNA levels, of endogenous ETV4 and HA-ETV4 (Fig. 4 G and H and SI Appendix, Fig. S11).
Fig. 4.
COP1 attaches to the nuclear envelope via binding to the nucleoporin, TPR. (A) Hepa1c1c7 cells stably expressing Myc-COP1 were cultured with or without LT for 2 h or U0126 for 1 h. The cells were then analyzed by immunofluorescence staining with mouse anti-Myc and rabbit anti-TPR primary antibodies followed by goat anti-mouse IgG-Alexa Fluor 568 and goat anti-mouse IgG-Alexa Fluor 633 secondary antibodies. The location of Myc-COP1, TPR, and the nucleus was assessed by a LSM 880 confocal microscope. (B) Analyses of fluorescence intensity and colocalization of COP1 and TPR were performed using Bitplane Imaris image analysis software. Pearson’s colocalization coefficient was calculated from 40 nuclei, from 9 to 13 randomly selected image files obtained from two or three independently performed experiments. Data were statistically analyzed using an unpaired, two-tailed t test at the 95% confidence interval using GraphPad Prism software and presented as means ± SE. *P < 0.0001. (C) Hepa1c1c7 cells stably expressing HA-COP1 or HA-COP1Δ24 were cultured with or without U0126 for 1 h, and then extracted with xTractor lysis buffer containing protease inhibitors. The cell lysates were then immunoprecipitated with anti-HA magnetic beads. The levels of TPR and HA-COP1 proteins in the immunoprecipitated complex and whole cell lysates (WCLs) were analyzed by Western blotting. (D) 293T cells were transfected with mock, HA-TPR, HA-TPR4A, and HA-TPR4E/D vectors. Two days following the transfection, the cells were extracted with xTractor lysis buffer containing protease inhibitors. The cell lysates were used for immunoprecipitation with anti-HA magnetic beads. The levels of HA-TPR and endogenous COP1 proteins in immunoprecipitated complex and WCLs were analyzed by Western blotting. (E) Hepa1c1c7 cells stably expressing HA-COP1 were transfected with control or TPR siRNA. Two days following transfection, the cells were analyzed by immunofluorescence staining with mouse anti-Lamin A/C and rabbit anti-HA primary antibodies followed by goat anti-mouse IgG-Alexa Fluor 633 and goat anti-rabbit IgG-Alexa Fluor 568 secondary antibodies. The locations of Lamin A/C, HA-COP1, and the nucleus were assessed by LSM 880 confocal microscope. (F) Fluorescence intensities were quantified using the Bitplane Imaris image analysis software. The intensity ratios of COP1 located on the NE to that in the nucleoplasm were calculated using data from 40 nuclei, 9 to 13 randomly selected image files obtained from two or three independently performed experiments. Data were statistically analyzed using an unpaired, two-tailed t test at the 95% confidence interval using GraphPad Prism software and presented as means ± SE. *P < 0.0001. (G and H) Hepa1c1c7 cells were transfected with control or TPR siRNA. Two days after the transfection, the cells were extracted with NuPAGE LDS sample buffer (G) or TRIzol (H). The levels of ETV4 protein (G) and mRNA (H) were measured by Western blotting (G) and quantitative PCR (H). β-Actin levels were used as loading controls. Data shown are representative of three independent experiments (G) or mean ± SE from three independent experiments (H). Intensities of Western blotting bands were quantified, normalized using β-actin levels, and are shown under each band (G).
COP1 Deletion Increases Cellular Proliferation in Suboptimal Conditions.
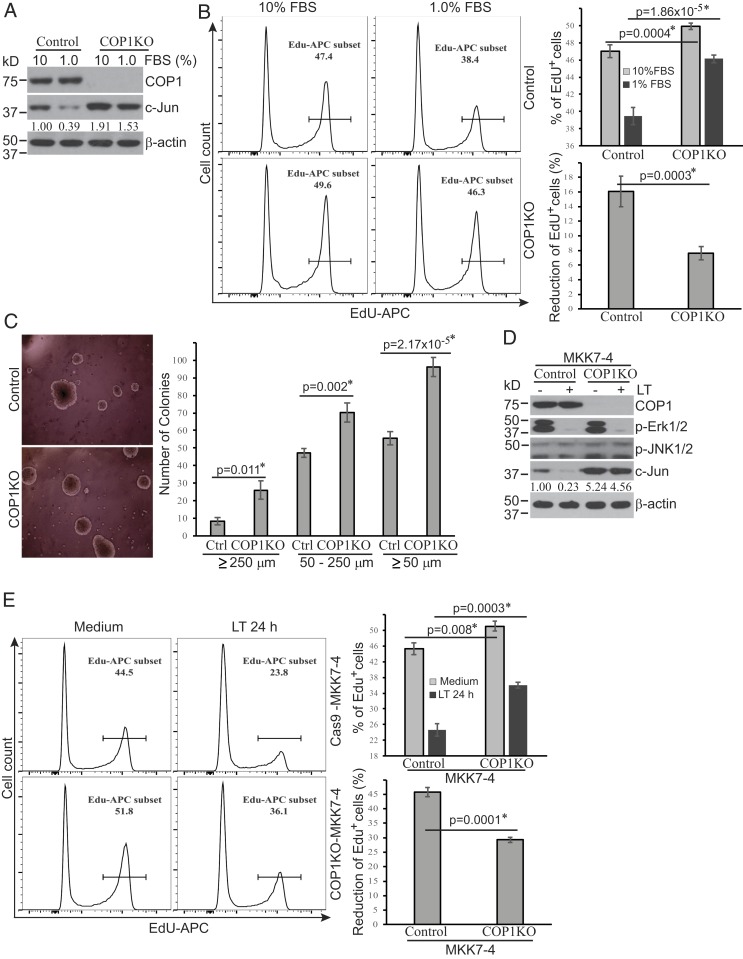
The c-Jun transcription factor has an essential role in promoting cellular proliferation. We next investigated the impact of COP1 deletion on cellular proliferation under suboptimal culture conditions. Hepa1c1c7 cells with COP1 deleted using CRISPR-Cas9 technology maintained higher levels of c-Jun protein than the Cas9-transfected control cells following 24-h culture in medium containing 1% FBS (Fig. 5A). Moreover, COP1-deficient cells exhibited higher proliferation (EdU incorporation) than control cells when cultured in either 10% or 1% FBS (Fig. 5 B, Left and Upper Right panels). The reduction in the percentage of EdU+ cells following culture in 1% FBS medium was markedly smaller for COP1-deficient cells compared to control cells (7.62 ± 0.93% vs. 16.06 ± 2.10%) (Fig. 5 B, Bottom Right), suggesting that the proliferation decline observed in cells cultured in this suboptimal culture condition is primarily COP1 dependent. To address this further, we cultured COP-deficient and control cells in soft agar to assess the effect of COP1 deletion on anchor-independent cell growth. Following 2 wk of culture in soft agar, COP1-deficient cells generated significantly more colonies of all sizes (≥50 µm, 50 to 250 µm, or ≥250 µm) (Fig. 5C), supporting a role for COP1 in restraining cell growth in suboptimal conditions.
Fig. 5.
COP1-dependent pathways regulate cellular proliferation in suboptimal culture conditions. (A and D) COP1-deficient (COP1KO) and control Hepa1c1c7 cells (A) were cultured in media supplemented with 10% or 1% FBS for 24 h. COP1KO and control Hepa1c1c7 cells expressing MKK7-4 (D) were treated with LT for 2 h. Treated cells were analyzed for the expression levels of indicated proteins by Western blotting. c-Jun bands were quantified and normalized using β-actin. The relative amounts are shown under each band. Data shown are representative of two independent experiments. (B and E) Cellular proliferation following culture in media containing 10% or 1% FBS for 24 h (B) or following treatment with or without LT for 24 h (E) was measured using the click-iT EdU cell proliferation kit and analyzed using flow cytometry and FlowJo software. Cell proliferation (EdU+) and proliferation reduction percentages were statistically analyzed using data from four (B) or three (E) independent experiments by an unpaired, two-tailed Student’s t test and presented as mean ± SE. Statistically significant differences were determined by P < 0.01. (C) COP1KO and Ctrl Hepa1c1c7 cells were cultured in soft agar for 2 wk. Colony photos shown are representative of three independent experiments (Left), and colony numbers presented represent mean ± SE from three independent experiments (Right). Statistical analysis was performed using an unpaired, two-tailed Student’s t test and significant difference was determined by P < 0.01.
In contrast to conditions in which MKK4/MKK7-JNK-c-Jun signaling is fully intact, LT cleaves MKK4, presenting a challenge to predicting the functional role of the Erk1/2-TPR-COP1-c-Jun pathway during anthrax infection. To overcome this hurdle, we used Hepa1c1c7 cells ectopically expressing an LT-resistant MKK7-4 fusion protein, designed to preserve JNK1/2 signaling in LT-treated Hepa1c1c7 cells (12). Consistent with our previous report, ectopic expression of this MKK7-4 fusion protein in LT-treated cells maintained activation of the JNK1/2 signaling pathway following LT treatment, and cells with concomitant COP1 deletion preserved higher levels of c-Jun protein at baseline and following LT treatment (Fig. 5D). This was accompanied by higher levels of proliferation in COP1-deficient cells compared to control cells at baseline (51.03 ± 1.24% vs. 45.30 ± 1.56%) and following treatment with LT for 24 h (36.07 ± 0.75% vs. 24.57 ± 1.59%) (Fig. 5E). The percentage of proliferating COP1-deficient cells was 46.8% higher than the percentage in control cells following LT treatment, highlighting the contribution of the Erk1/2-TPR-COP1 pathway in mediating the pathogenic effects of anthrax LT on cellular proliferation.
Discussion
We previously reported that anthrax LT treatment causes a rapid degradation of c-Jun protein via inactivation of the MKK1/2-Erk1/2 signal pathway (12), but the underlying mechanisms were unknown. c-Jun, a basic leucine zip (bZIP) transcription factor, is known to be degraded by the proteasome following ubiquitination mediated by ubiquitin E3 ligases including FBW7 (16), ITCH (15), MEKK1 (19), SAG/ROC2 (18), and COP1 (17, 20, 29). Here we identify COP1, but not other E3 ligases, as the mediator of c-Jun degradation induced by Erk1/2 inactivation. This evolutionarily ancient protein was originally discovered and characterized as a master regulator of light signaling in plants (24). In mammals, COP1 serves as a ubiquitin E3 ligase for itself (29), c-Jun (17, 20), ETVs (25–27), Pea3 (36), and p53 (28) transcription factors. Our findings that ETV4 and ETV5 proteins are degraded in LT- or U0126-treated cells further support a generalized role for Erk1/2 signaling in the regulation of COP1 activity.
Protein ubiquitination is a process catalyzed by a cascade involving three major enzyme components: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). Although Ubc5Hb has been shown to act as an E2 for COP1-mediated ubiquitination in cell-free systems in vitro (29, 37), it is not clear whether this E2 is required for LT-induced c-Jun degradation in vivo. Identification of the E2 that is involved in LT-induced and COP1-mediated c-Jun degradation warrants future investigation.
Although the COP1 RING-finger domain possesses intrinsic E3 ligase activity and may ubiquitinate some targets on its own, COP1 has been reported to function preferentially as a component in the CRL4COP1-DET1 complex with Det1, Ddb1, Cul4, and Rbx1 (17, 25). Zhang et al. recently reported that Erk1/2 inhibition abrogates membrane depolarization-induced accumulation of ETV4 protein in β-cells (36), destabilizing ETV proteins by inhibiting the CRL4COP1/DET1 complex via phosphorylation of DET1 protein at Ser458 (26). However, the authors also showed that interaction between COP1 and DET1 is very weak, and only a very small fraction of COP1 protein is recruited to the DDB1-DET1-CUL4-RBX1 complex (26). Moreover, the mechanism by which DET1 phosphorylation at Ser458 regulates the CRL4COP1/DET1 complex is unknown.
In our study, we observed that DDB1 and DET1 readily coimmunoprecipitate each other, whereas only small fractions of these proteins coimmunoprecipitated with COP1. Triton X-100 treatment removes most of the DDB1, DET1, and RBX1 proteins from the complex, but not COP1. Taken together, these findings suggest that in physiological conditions, the majority of COP1 may not exist in the same complex with DET1 and DDB1. In addition, Erk1/2 inactivation does not affect the formation of the CRL4COP1-DET1 complex. As the degradation of COP1 substrates occurs almost immediately following Erk1/2 inactivation, it seems unlikely that Erk1/2 inactivation activates COP1 by regulating the formation and/or function of the CRL4COP1/DET1 complex in our experimental conditions. Moreover, knockdown of COP1 but not DDB1, DET1, or CUL4 prevents the degradation of COP1 substrates in LT- or U0126-treated cells, suggesting that COP1 may function independently from the CRL4COP1/DET1 complex following Erk1/2 inactivation.
Of note, knockdown of DDB1, DET1, and CUL4 increased the levels of c-Jun but not ETV proteins, suggesting the existence of subtle substrate-specific regulation during the degradation of COP1 substrates. Interestingly, knockdown of RBX1 rendered a more profound restoration of the levels of COP1 substrates in LT-treated cells as compared to knockdown of DDB1, DET1, and CUL4. Nonetheless, this does not necessarily imply that RBX1 acts via the CRL4COP1/DET1 complex; RBX1 has a global effect by recruiting ubiquitin E2 enzymes to a variety of ubiquitin E3 ligases (38). In contrast to the previously proposed model in which Erk1/2 regulates COP1 activity via the CRL4COP1-DET1 complex, our data support an alternative pathway that centers around Erk1/2-dependent translocation of COP1 from a TPR-bound nuclear envelope store, a process that occurs independently from key components of the CRL4COP1-DET1 complex, including DET1, DDB1, and CUL4.
In this study, we observed that GFP fusion and transient overexpression affect the subcellular location of COP1, promoting a more cytoplasmic distribution. COP1 distribution is also known to be regulated by its nuclear import/export (31, 39). The inconsistency of our finding of the intracellular location of GFP-COP1 with previous reports may be due to the use of different cell lines, which may have different expression levels of COP1 import/export machinery proteins. Nevertheless, the increased cytosolic distribution of GFP-tagged and overexpressed COP1 may warrant caution when interpreting data generated from experiments using these overexpression systems. In contrast, the HA-COP1 and Myc-COP1 protein used as tools in our studies predominantly localized to the nucleus, with enrichment at the nuclear periphery. Colocalization of COP1 and Lamin A/C indicates that a large portion of COP1 is located to the nuclear envelope, which is in line with findings from other groups that immunofluorescent staining signals of COP1 are enriched at the periphery of nuclei (29, 39). In line with our results that COP1 functions independently of the CRL4COP1-DET1 complex in the context of Erk1/2 inhibition, COP1 localization to the nuclear envelope is not dependent upon DET1 and DDB1. This contrasts with DET1, which requires DDB1 for its nuclear localization.
Instead, we find that COP1 localization to the nuclear envelope is mediated by TPR, a large protein component of the nuclear pore complex, located at the nuclear basket. The N terminus of TPR contains a large domain dominated by the heptad repeats typical for the coiled-coil-forming proteins (33). The C terminus of TPR contains four residues that can be phosphorylated by Erk1/2 (34). Inactivation of Erk1/2 or mutation of the four Erk1/2-phosporylated residues disrupted the interaction between COP1 and TPR. Moreover, the coiled-coil domain-deficient COP1Δ24 fails to interact with TPR, indicating that the coiled-coil domain of COP1 and Erk1/2-mediated phosphorylation of TPR regulate the TPR/COP1 interaction at the nuclear envelope. It remains to be determined whether COP1 directly or indirectly interacts with TPR.
As the COP1 ubiquitin E3 ligase mediates degradation of transcription factors that are critical for fundamental cell functions, including cellular proliferation, survival, and differentiation, its activity would be predicted to be tightly controlled. In plants, COP1 activity is regulated by its subcellular distribution in plant cells. Upon light exposure, COP1 is transported from the nucleus to the cytoplasm, where it no longer degrades its nuclear transcription factor substrates (32). In this study, we found that inactivation of the MEK1/2-Erk1/2 signaling pathway induces a rapid translocation of COP1 protein from the nuclear envelope to nucleoplasm, where it promotes the degradation of COP1 substrates. The MEK1/2-Erk1/2 signaling pathway is activated by extracellular cues such as growth factors, thereby functioning as a link between extracellular stimulation and nuclear responses by directly phosphorylating numerous substrates, including transcription factors (40). Our findings reveal an additional mechanism through which MEK1/2-Erk1/2 signaling regulates the stability of transcription factors. When cells are in growth-promoting conditions, MEK1/2-Erk1/2 signaling is activated. As a result, COP1 is sequestered on the nuclear envelope and kept in a functionally poised status, allowing accumulation of transcription factors that are required for cellular proliferation. Upon depletion of growth factors, the MEK1/2-Erk1/2 signaling is inactivated, leading to rapid COP1 translocation to the nucleoplasm and subsequent degradation of COP1-targeted transcription factors, leading to cell growth arrest. In parallel to its role in plants, mammalian COP1 protein serves to transduce external signals into the nucleus by altering its subcellular location and thereby modulating the stability of key transcription factors. This regulation pathway provides mammalian cells with the capacity to adapt rapidly to changing extracellular cues and is a target for pathogenic bacteria such as B. anthracis.
The c-Jun protein, originally identified as an oncoprotein, is a key member of the AP-1 transcription factor family (41), which serves a critical role in regulating cellular proliferation, cell survival, and tumorigenesis. In this study, we observed that COP1 deletion promotes anchor-independent cell growth, supporting a previous report that COP1 functions as a tumor suppressor (20). Dissecting the downstream mechanisms associated with the blockade of MKK1/2-Erk1/2 signaling by LT has important clinical implications, especially with respect to the potential use of LT as an antitumor therapeutic reagent for human cancers with mutations in BRAF, which display constitutive activation of the MKK1/2-Erk1/2 pathway (42–44). The survival of BRAF-mutated cells is known to be highly dependent upon MKK1/2-Erk1/2 signaling, which inhibits mitochondria-dependent apoptosis (45, 46). Our data indicate that COP1 is central to the majority of the proliferation inhibition observed in nutrient-deficient conditions (52.6 ± 10.1%), as well as a large component of the antiproliferative effect of LT on Hepa1c1c7 cells (36.6 ± 2.3%), which survive well following 24-h LT treatment. This contrasts with the nonprotective role of COP1 deletion in BRAF-mutated A375 cells (47), which, despite COP1 deletion, die following exposure to LT or MEK1/2 inhibitors such as U0126, SCH772984, and CI-1040 (SI Appendix, Fig. S12). Given that Erk1/2 has numerous substrates and regulates multiple signaling pathways, and a variety of signal pathways are also disrupted when cells are cultured under suboptimal conditions, it is not surprising that COP1 deletion cannot fully restore cellular proliferation of cells that are cultured in nutrient-deficient conditions or following LT treatment. Taken together, COP1 plays a targeted role in regulating cellular proliferation by promoting c-Jun degradation and likely does not mediate all Erk1/2-dependent pathways known to control the survival of certain cancer cells.
Our findings are consistent with a role for COP1-mediated c-Jun degradation as a pathogenic mechanism during anthrax infection. Anthrax LT induces cell cycle arrest and inhibits the proliferation of vascular endothelial cells, intestinal progenitor cells, T cells, and B cells (48–51), contributing to vascular vessel leakage, as well as breakdown of the intestinal barrier and adaptive immune system during anthrax infection. In addition, c-Jun is essential for cytokine production by T cells, which is pivotal for regulating protective adaptive immune responses. Inhibition of IL-2 production in T cells further disrupts protective host immune responses during infection (48). Although the exact contribution of the disruption of the Erk1/2-TPR-COP1-c-Jun pathway in LT-induced pathology is unknown, identification of this pathway has promising implications as a therapeutic target for anthrax infection or a variety of other clinical scenarios in which c-Jun plays a role in pathogenic mechanisms.
Materials and Methods
Cells and Reagents.
Murine liver hepatoma Hepa1c1c7, primary MEF, human hepatocellular carcinoma HepG2, and human embryonic kidney 293T cells were used in this study. COP1-deficient and control Hepa1c1c7 cell lines were generated by Synthego using standard CRISP-Cas9 technology. Hepa1c1c7 cells were cultured with Alpha Minimum Essential Medium (MEM Alpha); HepG2 cells were cultured with Eagle's Minimum Essential Medium; MEF and 293T cells were cultured with Dulbecco’s Modified Eagle’s Medium. All media were supplemented with 10% FBS, 100 IU/mL penicillin, 100 µg/mL streptomycin, and 2 mM l-glutamine. Cultures were conducted at 37 °C in an incubator with 5% CO2. Lyophilized recombinant PA and LF were purchased from List Biological Laboratories, Inc. and reconstituted in sterile water to make stock solutions with final concentrations of 1 mg/mL MG132 (inhibitor of 26 S proteasome) and U0126 (inhibitor of MKK1/2), purchased from EMD Millipore. Unless otherwise specified, LT, U0126, and MG132 were used to treat cells at the concentrations of 1.0 µg/mL, 10 µM, and 10 µM, respectively. xTractor lysis buffer was purchased from Takara (cat. no. 635625). NuPAGE LDS sample buffer (4×) was obtained from Thermo Fisher Scientific (cat. no. NP0007). Taqman gene expression assay primers and probes (see below) were purchased from Thermo Fisher Scientific. The MTS cellular proliferation colorimetric assay kit was purchased from BioVision (cat. no. K300-2500).
Plasmids and Cell Transfection.
cDNA sequences encoding HA- and Myc-tagged human COP1, HA-COP1Δ24, and Myc-tagged human DET1 were synthesized by GenScript and cloned into the retroviral vector, pMIG-w. cDNA sequences encoding GFP and human COP1 were generated by PCR using the high-fidelity PrimeSTAR Max DNA Polymerase (Takara) and cloned into the TA vector pDRIVE (Qiagen). The GFP-COP1 fragment was then released and inserted into pMIG-w vector without the IRES-GFP fragment. cDNA sequences encoding human COP1 with deletion of N-terminal 70 amino acids and mouse ETV4 and ETV5 were generated by PCR amplification and cloned into the retroviral pMIG-w vector. The ligase-dead COP1 mutant (C136A/C139A) was generated using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit from Agilent and then cloned into the retroviral pMIG-w vector. pCDNA3-FLAG-human TPR plasmid was purchased from the Addgene and used as template to generate cDNA sequences encoding three consecutive TPR fragments: TPR-I: 1 to 2,324 containing the NheI restriction enzyme site at its 3′ end; TPR-II: 2,318 to 6,208 containing the NheI and XhoI sites at the 5′ and 3′ ends, respectively; and TPR-III: 6,202 to 7,084 containing XhoI and SalI sites at the 5′ and 3′ ends, respectively. TPR-I and TPR-III were generated by PCR using the high-fidelity PrimeSTAR Max DNA polymerase, and TPR-II was released from pCDNA3-FLAG-human TPR by digestion with NheI and XhoI restriction enzymes. TPR-III was used to generate mutants in which Thr-2102, Thr-2123, and Thr-2200 were mutated to Ala or Glu, and Ser-2141 was mutated to Ala or Asp. The three cDNA fragments were ligated together by inserting them into the TA vector pDRIVE, and the cDNA encoding the full-length TPR was released by digestion with HpaI and SalI restriction enzymes and subcloned into the retroviral vector pMIG-w that was digested with HpaI and XhoI restriction enzymes. All DNA sequences were confirmed by DNA sequencing after cloning. To produce pseudoretrovirus, retroviral plasmids were transfected into 293T cells together with packaging plasmids using the standard calcium phosphate method. Supernatants were harvested 48 h following the transfection, filtered through 0.45-µm filters (Millipore), and then used to transduce cells. siRNA was transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen) following the manufacturer’s suggested protocol. siRNAs were all purchased from Thermo Fisher Scientific and the ID number of the siRNAs used in this study are listed as follows: negative control (4390843, 4390846), mouse Itch (501103, 501104), mouse Fbw7 (86123, 186285), mouse Sag (64316, 150674), mouse Cop1 (70637, 70448), mouse Det1 (s94262, s94264), mouse Ddb1 (s64880, s64881), mouse Cul4a (90859, 176073), mouse Cul4b (s91128, s91129), mouse Rbx1 (s80568, s80569), mouse Tpr (s99370), human COP1 (s34634, s59691), human DET1 (s30103, s30104), human DDB1 (s3979, s3980), human CUL4a (s16046, s14067), human CUL4b (s16042, s16043), human RBX1 (s19386, s19387), and human TPR (s14353, s14355). Two siRNA targeting the same gene were mixed equally to ensure efficient knockdown.
Mass Spectrometry.
Hepa1c1c7 stably expressing HA-COP1 were cultured with or without LT for 2 h or U0126 for 1 h. Nuclear extracts from the cells were isolated using the Nuclear Complex Co-IP Kit from Active Motif. The nuclear extracts were incubated with anti-HA magnetic beads following the manufacturer’s instructions (Thermo Fisher Scientific, cat. no. 88837) to precipitate the COP1 complex. The immune-precipitated complex was then analyzed by mass spectrometry at the Mass Spectrometry and Proteomics Facility of The Ohio State University.
Western Blotting.
Cells were lysed with NuPAGE LDS sample buffer (Invitrogen). Cell lysates were then separated on 4 to 12% NuPAGE BisTris gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad). The membranes were probed with antibodies of interest using standard Western blotting techniques as described previously (11). The following antibodies were used for Western blotting in this study: COP1 (Bethyl Laboratories, cat. no. A300-894A), DDB1 (Bethyl Laboratories, cat. no. A300-462A), CUL4a (Bethyl Laboratories, cat. no. A300-739A), CUL4b (Bethyl Laboratories, cat. no. A303-864A), c-Jun (Cell Signaling, cat. no. 9165), Rbx1 (Cell Signaling, cat. no. 11922), p-Erk1/2 (Cell Signaling, cat. no. 4370), p-JNK1/2 (Cell Signaling, cat. no. 9251), p38 (Cell Signaling, cat. no. 8690), Lamin A/C (Cell Signaling, cat. no. 2032), HA tag (Cell Signaling, cat. no. 3724), Myc tag (Cell Signaling, cat. no. 2278), ETV4 (Proteintech, cat. no. 10684-1-AP), ETV5 (Millipore, cat. no. MABN683), and β-actin (Sigma, cat. no. A5441). The intensities of protein bands were quantified using the Image Studio software supplied with the Odyssey system and normalized to the amounts of β-actin or p38 that were used as loading controls. The reductions in c-Jun protein levels induced by treatment were calculated using the following formula: (normalized c-Jun intensity of the untreated group − normalized c-Jun intensity of the treated group)/normalized c-Jun intensity of the untreated group × 100%. Statistical analyses were performed using the two-tailed Student's t test.
Quantitative PCR.
Total RNA was extracted from cells with TRIzol (Invitrogen) following the manufacturer's suggested protocol. RNA was then reverse transcribed to cDNA using the Omniscript RT kit manufactured by Qiagen. The levels of Etv4 and Det1 mRNA were measured by quantitative PCR performed using standard techniques with a TaqMan probe (Mm00476689_g1 for Etv4 and Mm00547111_m1 for Det1) purchased from Applied Biosystems and normalized to the levels of Gapdh (Mm99999915_g1).
Immunofluorescent Staining.
Cells were plated on fibronectin (Sigma-Aldrich, cat. no. F1141) precoated glass coverslips (10 µg/mL, overnight at 4 °C) in 12-well plates and cultured overnight. Cells were cultured with or without LT for 2 h or U0126 for 1 h, and then incubated with or without 1% Triton X-100 for 10 s followed by two washes with cold PBS. Cells were then fixed in PBS containing 4% paraformaldehyde (Electron Microscopy Sciences, cat. no. 15710) for 15 min and permeabilized with 0.3% Triton X-100/TBS for 15 min. Immunofluorescent staining was performed following a protocol provided by Cell Signaling. Briefly, cells were incubated with blocking buffer containing 5% normal goat serum (Thermo Fisher, cat. no. 500622) at room temperature for 1 h, then incubated with primary antibodies overnight at 4 °C. After three washes with Tris buffered saline with Tween 20 (TBST), the cells were incubated with fluorescent dye-labeled secondary antibodies (Thermo Fisher, 1:200) for 1 h followed by another three washes with TBST. ProLong Gold antifade reagent with DAPI (Invitrogen, cat. no. P36931) was used for mounting specimens on glass slides and nuclear staining. Images were captured using an LSM 880 confocal microscope (Carl Zeiss Microscopy). The following primary antibodies were used: rabbit anti-HA tag (C29F4, Cell Signaling, cat. no. 3724, 1:1,500), mouse anti-Myc tag (9B11, Cell Signaling, no. 2276, 1:3,000), mouse anti-Lamin A/C (4C11, Cell Signaling, cat. no. 4777, 1:100), and rabbit anti-TPR (Novus Biologicals, cat. no. NB100-2866, 1:300). The following secondary antibodies used for staining were purchased from Thermo Fisher Scientific: Alexa Fluor 568-conjugated goat anti-rabbit IgG (cat. no. A21069), Alexa Fluor 633-conjugated goat anti-rabbit IgG (cat. no. A21070), Alexa Fluor 568-conjugated goat anti-mouse IgG (cat. no. A11031), and Alexa Fluor 633-conjugated goat anti-mouse IgG (cat. no. A21053). Fluorescence intensity and colocalization analyses were performed using Bitplane Imaris image analysis software version 9.2.1 (Bitplane, Inc.). Briefly, total fluorescence intensities were measured from the manually identified regions of interest (i.e., nuclear envelop and nucleoplasm of nuclei from each group). Pearson’s colocalization coefficient was calculated from 40 nuclei, randomly selected from 9 to 13 image files obtained from two or three independently performed experiments. The ratio of the intensity of the nuclear envelope divided by the intensity of nucleoplasm was also calculated. Statistical analyses were performed using GraphPad Prism software, version 7.03 (GraphPad Software). The results are presented as means ± SE. The data were analyzed by unpaired, two-tailed t test at the 95% confidence interval, with P < 0.05 considered statistically significant.
Cellular Proliferation Analysis.
COP1-deficient and control Hepa1c1c7 cells (0.15 million cells per well in 12-well plates) were cultured in medium supplemented with 10% or 1% FBS for 24 h. COP1-deficient or control Hepa1c1c7 cells expressing MKK7-4 (0.15 million cells per well in 12-well plates) were treated with or without LT (1 µg/mL) for 24 h. Treated cells were assessed for DNA replication using the click-iT EdU cell proliferation kit (Invitrogen) and the manufacturer’s suggested protocol. EdU incorporation was analyzed using flow cytometry (BD LSRFortessa X-20) and FlowJo software (Tree Star). Proliferation reduction percentages following culture in 1% FBS or LT were calculated using the following formula: proliferation reduction (%) = (% EdU+ cells of untreated group − % EdU+ cells in treated groups)/% EdU+ cells in untreated group × 100%.
Soft Agar Assay.
To assess anchorage-independent growth, soft agar medium (lower layer) was added to the wells of six-well plates (2.0 mL/well, 0.5% agar in MEM Alpha supplemented with 10% FBS). Cells (104/well) were diluted in 0.33% agar in MEM Alpha and layered on top of the 0.5% agar layer. Soft agar cultures were maintained at 37 °C in a 5% CO2 incubator for 2 wk. The colonies that formed in each well were counted and measured using a microscope with a micrometer ruler (Olympus SZX7). Colony photos were obtained using the EVOS microscopy system (4× magnification, Life Technologies).
Supplementary Material
Acknowledgments
We thank Drs. Brian Roelofs, Massimo Gadina, Montserrat Puig, and Ying-Xin Fan for thoughtful review of the manuscript; Dr. Yukinori Endo for his help with confocal microscopy; and Yi Shen for her help with the soft agar assay. This work was supported by US Food and Drug Administration internal funding.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913698117/-/DCSupplemental.
References
- 1.Bhatnagar R., Batra S., Anthrax toxin. Crit. Rev. Microbiol. 27, 167–200 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Moayeri M., Leppla S. H., Vrentas C., Pomerantsev A. P., Liu S., Anthrax pathogenesis. Annu. Rev. Microbiol. 69, 185–208 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Liu S., Moayeri M., Leppla S. H., Anthrax lethal and edema toxins in anthrax pathogenesis. Trends Microbiol. 22, 317–325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu L., Frucht D. M., Bacillus anthracis: A multi-faceted role for anthrax lethal toxin in thwarting host immune defenses. Int. J. Biochem. Cell Biol. 39, 20–24 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Bardwell A. J., Abdollahi M., Bardwell L., Anthrax lethal factor-cleavage products of MAPK (mitogen-activated protein kinase) kinases exhibit reduced binding to their cognate MAPKs. Biochem. J. 378, 569–577 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang L., Karin M., Mammalian MAP kinase signalling cascades. Nature 410, 37–40 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Yang S. H., Sharrocks A. D., Whitmarsh A. J., MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Chopra A. P., Boone S. A., Liang X., Duesbery N. S., Anthrax lethal factor proteolysis and inactivation of MAPK kinase. J. Biol. Chem. 278, 9402–9406 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Tonello F., Montecucco C., The anthrax lethal factor and its MAPK kinase-specific metalloprotease activity. Mol. Aspects Med. 30, 431–438 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Duesbery N. S., et al. , Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280, 734–737 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Ouyang W., Torigoe C., Fang H., Xie T., Frucht D. M., Anthrax lethal toxin inhibits translation of hypoxia-inducible factor 1α and causes decreased tolerance to hypoxic stress. J. Biol. Chem. 289, 4180–4190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang W., Guo P., Fang H., Frucht D. M., Anthrax lethal toxin rapidly reduces c-Jun levels by inhibiting c-Jun gene transcription and promoting c-Jun protein degradation. J. Biol. Chem. 292, 17919–17927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karin M., Liu Zg., Zandi E., AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Kappelmann M., Bosserhoff A., Kuphal S., AP-1/c-Jun transcription factors: Regulation and function in malignant melanoma. Eur. J. Cell Biol. 93, 76–81 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Gao M., et al. , Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Nateri A. S., Riera-Sans L., Da Costa C., Behrens A., The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Wertz I. E., et al. , Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303, 1371–1374 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Gu Q., Bowden G. T., Normolle D., Sun Y., SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage-dependent targeting of c-Jun/AP1 and IkappaB-alpha/NF-kappaB. J. Cell Biol. 178, 1009–1023 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y., et al. , MEKK1 mediates the ubiquitination and degradation of c-Jun in response to osmotic stress. Mol. Cell. Biol. 27, 510–517 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migliorini D., et al. , Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Invest. 121, 1329–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X. W., Caspar T., Quail P. H., cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5, 1172–1182 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Deng X. W., et al. , COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71, 791–801 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Lau O. S., Deng X. W., The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Yi C., Deng X. W., COP1–From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Vitari A. C., et al. , COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., et al. , E3 ubiquitin ligase RFWD2 controls lung branching through protein-level regulation of ETV transcription factors. Proc. Natl. Acad. Sci. U.S.A. 113, 7557–7562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton K., et al. , Ubiquitin ligase COP1 coordinates transcriptional programs that control cell type specification in the developing mouse brain. Proc. Natl. Acad. Sci. U.S.A. 115, 11244–11249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dornan D., et al. , The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Bianchi E., et al. , Characterization of human constitutive photomorphogenesis protein 1, a RING finger ubiquitin ligase that interacts with Jun transcription factors and modulates their transcriptional activity. J. Biol. Chem. 278, 19682–19690 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Marine J. C., Spotlight on the role of COP1 in tumorigenesis. Nat. Rev. Cancer 12, 455–464 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Yi C., Wang H., Wei N., Deng X. W., An initial biochemical and cell biological characterization of the mammalian homologue of a central plant developmental switch, COP1. BMC Cell Biol. 3, 30 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Arnim A. G., Deng X. W., Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Krull S., Thyberg J., Björkroth B., Rackwitz H. R., Cordes V. C., Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 15, 4261–4277 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vomastek T., et al. , Extracellular signal-regulated kinase 2 (ERK2) phosphorylation sites and docking domain on the nuclear pore complex protein Tpr cooperatively regulate ERK2-Tpr interaction. Mol. Cell. Biol. 28, 6954–6966 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi C., Li S., Wang J., Wei N., Deng X. W., Affinity purification reveals the association of WD40 protein constitutive photomorphogenic 1 with the hetero-oligomeric TCP-1 chaperonin complex in mammalian cells. Int. J. Biochem. Cell Biol. 38, 1076–1083 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Suriben R., et al. , β-cell insulin secretion requires the ubiquitin ligase COP1. Cell 163, 1457–1467 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Dornan D., et al. , ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313, 1122–1126 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Wei D., Sun Y., Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: The role in cancer and as cancer targets. Genes Cancer 1, 700–707 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kung J. E., Jura N., The pseudokinase TRIB1 toggles an intramolecular switch to regulate COP1 nuclear export. EMBO J. 38, e99708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roskoski R., Jr, ERK1/2 MAP kinases: Structure, function, and regulation. Pharmacol. Res. 66, 105–143 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Maki Y., Bos T. J., Davis C., Starbuck M., Vogt P. K., Avian sarcoma virus 17 carries the jun oncogene. Proc. Natl. Acad. Sci. U.S.A. 84, 2848–2852 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S., Bugge T. H., Leppla S. H., Targeting of tumor cells by cell surface urokinase plasminogen activator-dependent anthrax toxin. J. Biol. Chem. 276, 17976–17984 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Liu S., et al. , Matrix metalloproteinase-activated anthrax lethal toxin demonstrates high potency in targeting tumor vasculature. J. Biol. Chem. 283, 529–540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samatar A. A., Poulikakos P. I., Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 13, 928–942 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Wang Y. F., et al. , Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, and Mcl-1. Clin. Cancer Res. 13, 4934–4942 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Cook S. J., Stuart K., Gilley R., Sale M. J., Control of cell death and mitochondrial fission by ERK1/2 MAP kinase signalling. FEBS J. 284, 4177–4195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lito P., et al. , Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang H., Cordoba-Rodriguez R., Lankford C. S., Frucht D. M., Anthrax lethal toxin blocks MAPK kinase-dependent IL-2 production in CD4+ T cells. J. Immunol. 174, 4966–4971 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Fang H., Xu L., Chen T. Y., Cyr J. M., Frucht D. M., Anthrax lethal toxin has direct and potent inhibitory effects on B cell proliferation and immunoglobulin production. J. Immunol. 176, 6155–6161 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Depeille P., et al. , Anthrax lethal toxin inhibits growth of and vascular endothelial growth factor release from endothelial cells expressing the human herpes virus 8 viral G protein coupled receptor. Clin. Cancer Res. 13, 5926–5934 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Sun C., et al. , Anthrax lethal toxin disrupts intestinal barrier function and causes systemic infections with enteric bacteria. PLoS One 7, e33583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.