Significance
Mitochondria are dynamic double-membraned organelles, which undergo cycles of division and fusion mediated by dynamin-related GTPases. How dynamins mediate mitochondrial inner membrane fusion is not understood. In humans, defective inner membrane fusion causes a degenerative form of blindness termed dominant optic atrophy. Here, we solve the structure of a trimer assembly of an inner membrane fusion dynamin. Our observations underscore the structural plasticity of dynamins and suggest a previously unidentified mode of how they might mediate membrane fusion events.
Keywords: membrane fusion, mitochondria, dynamin, inner membrane, structure
Abstract
The fusion of inner mitochondrial membranes requires dynamin-like GTPases, Mgm1 in yeast and OPA1 in mammals, but how they mediate membrane fusion is poorly understood. Here, we determined the crystal structure of Saccharomyces cerevisiae short Mgm1 (s-Mgm1) in complex with GDP. It revealed an N-terminal GTPase (G) domain followed by two helix bundles (HB1 and HB2) and a unique C-terminal lipid-interacting stalk (LIS). Dimers can form through antiparallel HB interactions. Head-to-tail trimers are built by intermolecular interactions between the G domain and HB2-LIS. Biochemical and in vivo analyses support the idea that the assembly interfaces observed here are native and critical for Mgm1 function. We also found that s-Mgm1 interacts with negatively charged lipids via both the G domain and LIS. Based on these observations, we propose that membrane targeting via the G domain and LIS facilitates the in cis assembly of Mgm1, potentially generating a highly curved membrane tip to allow inner membrane fusion.
The fusion and fission of mitochondria are mediated by dynamin-like proteins (DLPs) (1–5). The fission process requires dynamin-related protein 1 (Drp1) in mammals (6), which is recruited to the surface of mitochondria by membrane-bound receptors and forms helical oligomers similar to dynamin-1, which produces force for membrane scission in a GTP hydrolysis-dependent manner (7–9). The fusion process also relies on two distinct classes of related dynamin GTPases that separately control the fusion of the outer and inner membranes (10). Mitofusins (MFNs) in mammals (11) and Fzo1 in yeast (12) mediate outer mitochondrial membrane (OMM) fusion, whereas inner mitochondrial membrane (IMM) fusion is mediated by optic atrophy 1 (OPA1) in mammals (13) and mitochondrial genome maintenance 1 (Mgm1) in yeast (10). Mutations in human OPA1 cause dominant optic atrophy (14, 15), whereas deleterious mutations or deletion of Mgm1 in yeast results in mitochondrial fragmentation, aberrant cristae structure, and loss of mitochondrial DNA and respiration (16–21). These observations underscore the physiological importance of IMM fusion, but the molecular mechanisms underlying this event are largely unknown.
Mitochondrial homotypic membrane fusion mediated by DLPs is mechanistically different from that of viral fusogens or SNARE-mediated membrane fusion events (22–24). The best studied homotypic fusion by DLPs is homotypic ER fusion mediated by the integral membrane protein atlastin (ATL) (25, 26). Structural and biochemical analyses have revealed that ATL-mediated membrane fusion is driven by GTP-dependent dimerization of the GTPase (G) domain, which tethers opposing membranes. Within docked dimers, GTP hydrolysis drives a conformational change that locks the helix bundle (HB) associated with its G domain into a parallel position, bringing the ATL-attached bilayers together to promote lipid mixing in a manner that is dependent on a C-terminal amphipathic helix (27, 28). ATL undergoes multiple rounds of GTP hydrolysis to maintain membrane tethering (29, 30). Recent studies of membrane-bound MFN suggest that OMM fusion is likely mechanistically similar to ATL, with trans interactions mediated by G domains harnessed to mediate the tethering and fusion of the OMM. Specifically, the minimal GTPase domain (MGD) of MFN consists of a typical G domain and an associated HB (31, 32). The G domain of MFN forms a nucleotide-dependent dimer, MFN-MGD–mediated tethering is GTP-dependent, and the HB undergoes a very similar inward swinging conformational change upon GTP hydrolysis (33). Finally, a functionally analogous amphipathic helix has been identified in the C terminus of MFN (34).
The mechanism underlying DLP-mediated IMM fusion is distinct from that used by ATL and MFN. Mgm1 and OPA1 exist in both long, membrane-bound and short, soluble forms, which act together to mediate fusion (35–40). An N-terminal transmembrane (TM) domain proximal to the G domain anchors Mgm1 and OPA1 to the inner membrane, with the remainder of the protein facing the intermembrane space. Fusion is regulated by different intramitochondrial proteases in yeast and humans that mediate proteolysis at sites between the TM and G domains to generate the short forms of Mgm1 and OPA1 (41–44).
Mgm1 and OPA1 have been shown to deform liposomes into tubules by forming helical oligomers (45, 46), which is a characteristic of fission DLPs. The structural basis for the helical assembly of short isoform of Chaetomium thermophilum Mgm1 (CtMgm1) was reported recently using both X-ray analysis of crystals and electron cryotomography analysis of membrane associated structures (47). Similar to fission DLPs, a stalk-mediated tetramer was used as a CtMgm1 building block to form helical filaments on both positively and negatively curved membranes (47). Based on these structures and their functional interrogation, the helical assemblies of Mgm1 were proposed to be used for fusion and cristae formation, respectively.
To address how IMM DLPs mediate membrane fusion, we determined the X-ray structure of Saccharomyces cerevisiae short-Mgm1 (s-Mgm1) in complex with GDP. As previously described, the s-Mgm1 monomer possesses a similar configuration as scission DLPs, such as dynamin-1 or DRP1. However, in crystals, s-Mgm1 exists as a stacked trimer assembly distinct from the tetrameric assemblies recently reported in helical assemblies of CtMgm1 and by scission DLPs (47). Biochemical and cellular analyses indicate that trimers exist in solution and that amino acids comprising the trimer interfaces are critical for mitochondria fusion in vivo. These observations underscore the plasticity of s-Mgm1 assemblies and suggest a model in which the IMM DLP functions to mediate fusion via a mechanism that is distinct from both DLP-mediated membrane scission and DLP-mediated homotypic membrane fusion.
Results
Crystal Structure of s-Mgm1.
To investigate the mechanisms underlying Mgm1-mediated fusion, we determined the structure of S. cerevisiae s-Mgm1 (ScMgm1). The region between the site of proteolysis (residue 91) and the G domain (residue 184) is predicted to be unstructured (Fig. 1A and SI Appendix, Fig. S1), and the full-length s-Mgm1 (residues 92–881), which includes this region, was largely insoluble. Thus, we expressed in Escherichia coli a truncated version (residues 184–881) lacking this unstructured region and purified and crystallized it in the presence of GDP and BeF3−. Given that this construct possesses nearly all key elements of short, soluble Mgm1, we refer to it as s-Mgm1. The structure was determined at 3.2-Å resolution by single anomalous diffraction (SI Appendix, Table S1). Three Mgm1 molecules were present in the asymmetric unit, with nearly identical overall structures (SI Appendix, Fig. S2A).
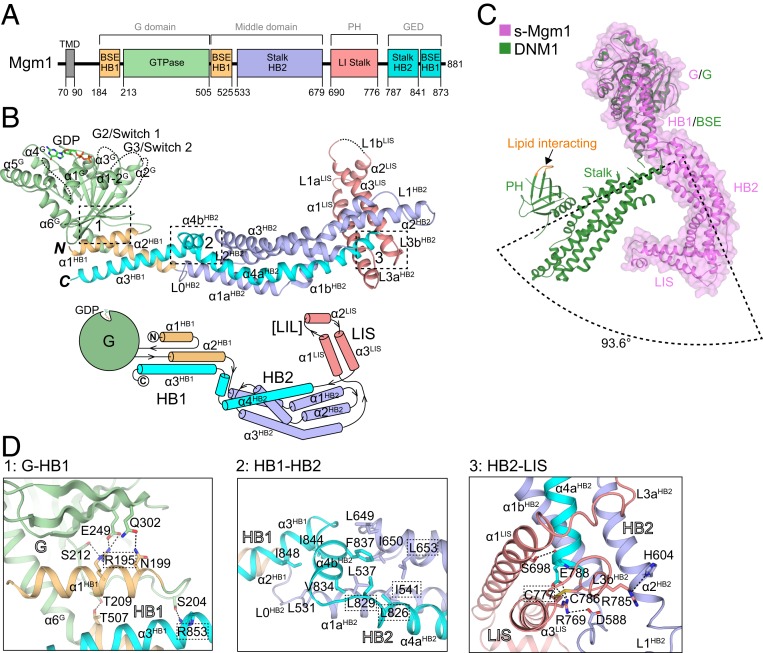
Fig. 1.
Crystal structure of s-Mgm1. (A) Scheme showing the domains of yeast Mgm1. Domains of s-Mgm1 and their corresponding domains in dynamin-1 are given in boxes. Previously used domain names in dynamin are listed above. Regions of s-Mgm1 are colored with residues numbered: HB1-forming helices that are connected to the GTPase are colored in yellow, the GTPase in green, HB2 in light blue, lipid-interacting stalk (LIS) in pink, and the C-terminal helices that complement HB1 an HB2 in cyan. BSE, bundle signaling element; HB, helix bundle; TMD, transmembrane domain. (B) Structure of the GDP-bound form of s-Mgm1. Regions of s-Mgm1 are colored as in A. Major helices and loops are labeled and numbered according to domain. A topology plot is shown below with the same coloring scheme. LIL, lipid-interacting loop. (C) Superposition of s-Mgm1 in magenta and dynamin-1 in green (PDB ID code: 3SNH) with the G domains aligned. Major domains are labeled. A surface representation of s-Mgm1 is shown. The orientation difference between the Mgm1-HB2 and DNM1-stalk is indicated. (D) Zoom-in views of boxes indicated in B. Key domains, secondary structures, and key residues are labeled. Residues that caused severe defects in yeast cells when mutated are highlighted.
The overall structural configuration of s-Mgm1 is similar to dynamin-1 (Fig. 1 A and C); the s-Mgm1 structure consists of an N-terminal G domain, followed by two HBs (HB1 and HB2), and a helical hairpin at the distal end, termed the lipid interacting stalk (LIS, residues 687–777), as it has been implicated in lipid binding with several key lysine residues (48, 49) (Fig. 1B). HB1 corresponds to the bundle signaling element (BSE) of dynamin-1, the longer HB2 corresponds to the stalk domain of dynamin-1, and LIS corresponds to the dynamin pleckstrin-homology (PH) domain. As in dynamin-1, the amino acid chain following the LIS domain forms helices in the HBs, positioning the N and C termini in close proximity.
Key intramolecular interactions that play roles in maintaining the overall shape of s-Mgm1 were identified (Fig. 1D). The HB1 domain is closely associated with the G domain, as R195 in α1 of Mgm1-HB1 (α1HB1) interacts with S212 and E249 of the central β-sheet, and N199 in the same helix engages Q302 (Fig. 1D, Left). In addition, T507 in α2HB1 and R853 in α3HB1 interact with T209 and S204, respectively (Fig. 1D, Left). Compared to the dynamin-1 G domain/BSE dimer that forms via a G-G interface, the position of HB1 in the s-Mgm1 structure resembles that of the cognate dynamin-1 BSE domain in the semiclosed state (50), in which the two BSE domains are proximal. Superimposition of the G domains of Mgm1 and nucleotide-free dynamin-1 indicates that the HB1 of Mgm1 is similarly positioned relative to the BSE in dynamin-1 (Fig. 1C, with an rmsd of 2.387). In addition, HB2 of Mgm1 is nearly perpendicular to the corresponding dynamin-1 stalk (Fig. 1C). When overlaid, the first half of the HB2 is close to the dynamin-1 stalk, but the second half differs (SI Appendix, Fig. S3A, with an rmsd of 4.567). The connecting regions between HB1 and HB2, including loop 0 in HB2 (L0HB2) and α4bHB2, form a zigzag conformation that is stabilized mostly by three clusters of hydrophobic interactions (Fig. 1 D, Center and SI Appendix, Fig. S3B, box 1): I844-I848-L531, V834-L829-L537-L826-I541, and F837-L649-I650-L653. Finally, a majority of the LIS domain is well-ordered (SI Appendix, Fig. S3B, box 2) and oriented perpendicular to HB2, with R769 in α3LIS forming salt bridges with D588 and E788 in HB2 (Fig. 1 D, Right). A disulphide bond (C777-C786) was observed in the base of the LIS (SI Appendix, Fig. S3B, box 3), likely ensuring the orientation of the LIS relative to HB2.
Nucleotide Binding and Hydrolysis.
Although GDP and phosphate-mimicking BeF3− were added during crystallization, only GDP was observed in the electron density map (Fig. 2A). GDP is shallowly associated with the G domain, interacting mainly with the G1/P loop and G4 motifs of the GTPase, where the side chain of S224 and main chain atoms of S220 and G222 bind to the two phosphates of GDP. K386 and D388 in G4 contact the guanosine of the nucleotide (Fig. 2B), facilitated by T415 in the neighboring area, which is unique to Mgm1.
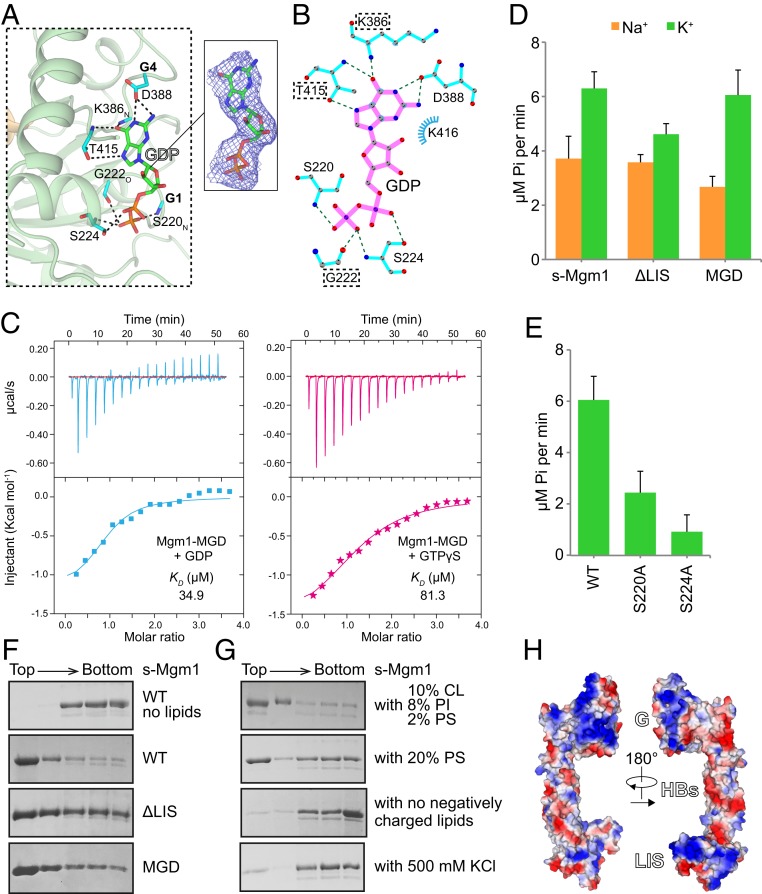
Fig. 2.
GTP hydrolysis and lipid interactions of Mgm1. (A) The active site of Mgm1 is shown in stick form with key residues highlighted. The 2Fo − Fc electron density maps (1.0σ contour) of GDP are shown as wire mesh (blue). (B) Interaction map of the catalytic core generated by LIGPLOT. (C) Binding affinity of GDP or GTPγS for Mgm1-MGD as measured by ITC. A 2 mM nucleotide solution was titrated stepwise into 0.1 mM protein. The dissociation constant (KD) is given in Inset. The data are representative of at least three repetitions. (D) The GTPase activity of various Mgm1 constructs was measured in the presence of 500 mM NaCl/KCl and 5 mM MgCl2. A total of 5 μM of protein was used for each sample. GTP hydrolysis was measured by phosphate release at saturating GTP concentrations (0.5 mM). Data are presented as the mean ± SD of nine measurements from three independent experiments. (E) As in D, but with WT or mutant s-Mgm1 in KCl-containing buffer. (F) Liposome flotation assay with various Mgm1 constructs. Liposomes (2 mM, POPC:POPE:CL:PI:DOPS:Rhodamine-PE = 55:23.5:10:8:2:1.5) were mixed with 2 μM purified protein at room temperature for 30 min. Fractions collected after centrifugation were analyzed by SDS/PAGE and Coomassie blue staining. The data are representative of at least three repetitions. (G) As in F, but with liposomes of different lipid compositions or different buffer conditions. (H) Electrostatic analysis of s-Mgm1. Negatively charged surfaces are colored in red and positively charged surfaces in blue. The s-Mgm1 molecule is viewed from both sides.
To characterize the nucleotide binding and hydrolysis properties of the G domain, we constructed a version of the protein containing only the G and HB1 domains (Mgm1-MGD) by directly attaching the last helix of s-Mgm1, α3HB1, to α2HB1 via a flexible linker. As predicted by the structure, Mgm1-MGD exhibited a weak association with GDP or a poorly hydrolysable GTP analog GTPγS when measured by isothermal calorimetry (ITC) (Fig. 2C). However, both s-Mgm1 and Mgm1-MGD possessed a capacity for GTP hydrolysis (Fig. 2D) that was enhanced by using K+ in place of Na+-containing buffers. We also observed that removal of the LIS domain from s-Mgm1 did not significantly change GTPase activity (Fig. 2D), but GTP hydrolysis was reduced when the nucleotide-interacting S220 and S224 of G1 were individually mutated to Ala (Fig. 2E).
Lipid Interactions.
Previously, s-Mgm1 was shown to interact with membranes containing the negatively charged inner membrane lipid cardiolipin (CL) that promoted s-Mgm1 assembly and GTPase activity, implying a role for CL in Mgm1-mediated fusion (51). Consistently, we observed an interaction of s-Mgm1 with membranes containing CL using membrane flotation assays, where a majority of purified s-Mgm1 floated to the top of the density gradient in a CL-containing liposome-dependent manner (55% 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine [POPC], 25% 1-palmitoyl-2-oleoyl-glycero-3-phosphoethanolamine [POPE], 10% CL, 8% glycero-3-phosphoinositol [PI], and 2% 1,2-dioleoyl-glycero-3-phosphoserine [DOPS]; Fig. 2F). We examined CL specificity by testing negatively charged phosphatidylserine (PS) in liposomes and observed that these liposomes were also able to recruit s-Mgm1, as evidenced by its efficient flotation (Fig. 2G). In contrast, s-Mgm1 was not observed to interact with liposomes containing only a mixture of PC and PE (Fig. 2F). The presence of a high salt concentration (500 mM KClCl) interfered with the interaction between s-Mgm1 and negatively charged liposomes (Fig. 2G), whereas varied membrane curvature when using liposomes of various diameters (50 nm vs. 400 nm) had no detectable impact on s-Mgm1 liposome binding (SI Appendix, Fig. S4B). These results indicate that s-Mgm1 binds membranes via negatively charged lipids.
Based on previously published work, we tested whether the LIS, which is in the cognate position of the lipid-interacting PH domain in dynamin-1, plays an essential role in membrane association. The lipid-interacting activity of s-Mgm1 ΔLIS was similar to that observed for s-Mgm1 in flotation assays (Fig. 2F), suggesting that Mgm1 possesses additional membrane-interacting regions.
Consistently, liposome flotation of Mgm1-MGD indicated that the G domain also possesses lipid-binding activity (Fig. 2F). This observation was supported using liposome sedimentation assays, which also demonstrated that only the Mgm1-MGD and LIS, but not HB2, cosediment with CL-containing liposomes (SI Appendix, Fig. S4A). Notably, s-Mgm1 and s-Mgm1 domain interactions were enhanced when the liposome CL concentration was increased from 10 to 20% (SI Appendix, Fig. S4A). Taken together, these observations suggest that s-Mgm1 interacts with negatively charged membranes via both its MGD and LIS.
The preference of s-Mgm1 for negative charges and the salt sensitivity of s-Mgm1 membrane interaction suggest that positive surfaces in s-Mgm1 are membrane-binding determinants. Electrostatic analysis revealed two positive clusters, one in the G domain and the other in the LIS (Fig. 2H), which is consistent with the membrane-binding activity mapped to these regions. The substitution of Lys/Arg with Ala in the G domain did not substantially affect the s-Mgm1–lipid interactions (SI Appendix, Fig. S4C). When Lys/Arg clusters were analyzed in the context of MGD, mutations at two sites, one in the back of the G domain (5K/R to Ala; K486/K487/R489/K491/K494) and one in the front (3K/R to Ala; K277/K280/R281), resulted in significantly reduced coflotation with liposomes (SI Appendix, Fig. S4D). However, the GTPase activity of the 5K/R and 3K/R mutants was not different as compared to the WT protein (SI Appendix, Fig. S4F). Similarly, mutation of the two exposed Lys residues (K740/K747) near the lipid-interacting loop region to Ala abrogated the binding of the LIS domain to negatively charged liposomes (SI Appendix, Fig. S4E). The 5K/R cluster located on the back of the G domain is likely involved in self-assembly, as discussed below. However, the front located 3K/R cluster on the G domain site and the K740/K747 residues on the LIS are oriented on the same side of s-Mgm1, suggesting that they cooperatively function to mediate the interaction of s-Mgm1 with CL-containing membranes.
Assembly of Mgm1.
The asymmetric structural unit was composed of three s-Mgm1 molecules in a “head-to-tail” trimer configuration with a threefold noncrystallographic symmetry axis, where the head is the G domain and the tail is the HB2/LIS region (Fig. 3A).
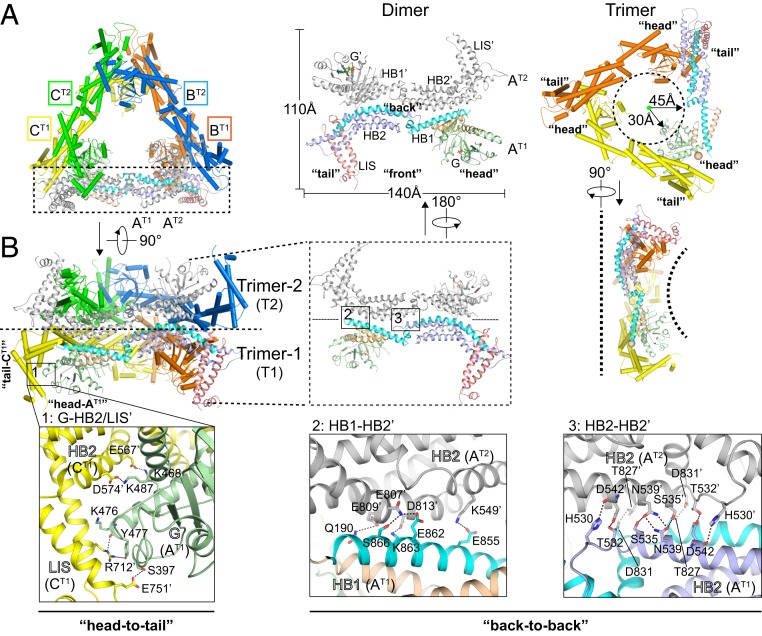
Fig. 3.
Structural assembly of s-Mgm1. (A) Crystallographic assembly of s-Mgm1. The dimer and trimer are shown from different angles. Key dimensions and major domains are labeled. Components from the second protomer in the dimer are tagged with prime (′). Three molecules in the trimer are designated A, B, and C, and two stacking trimers are termed T1 and T2. (B) As in A, but with zoomed-in views of the head-to-tail interface (G-HB2′/LIS′, box 1) and back-to-back interface (HB1-HB2′, box 2; HB2-HB2′, box 3).
Two sets of hydrophilic interactions between G and HB2/LIS are present at the interfaces, which bury a total area of 1,237.9 Å2 (Fig. 3 B, Left). In HB2 of one protomer, E567 and D574 form salt bridges with K468 and K487 in the G domain of the neighboring protomer, respectively. In addition, interactions were observed between the LIS and G domain in which the LIS R712 interacts with a backbone oxygen and Y477 in the neighboring G domain, and LIS E751 forms a hydrogen bond with S397 in the G domain.
Symmetry operation revealed a hexamer formed by an additional dimeric assembly between two trimers with a crystallographic twofold symmetry axis (Fig. 3B). The “back-to-back” stacking face of the trimer was relatively flat, with a buried area of 1,947.3 Å2, whereas the other side was concave with an inward-tilting wall formed by the three G-LIS domain interfaces (Fig. 3 A, Right, dotted lines). The dimer interface between trimers is generated by antiparallel stacking of the HB domains of the two protomers. In the center of the stack, the two HB2 domains form an intertwined hydrogen-bonding network (Fig. 3B, Right). In this hydrogen-bonding network, N539 has a homotypic interaction and also interacts with S535, D542 engages H530 and T532 at both side chain and main chain atoms, and T827 is paired with D831. At the periphery of the stack, multiple hydrophilic interactions occur (Fig. 3 B, Center) in which E807 in HB2 interacts with S866 and E862 in the neighboring HB1. In addition, K549 bridges to E855, E809 reaches Q190, and D813 reaches K863.
To test whether the oligomerization state of s-Mgm1 observed in the crystal reflects Mgm1 oligomerization in solution, we performed analytical ultracentrifugation (AUC). As previously reported, s-Mgm1 was mostly monomer in the absence of nucleotide (51), but we also observed s-Mgm1 dimer (∼20%) and trimer (∼10%) species with no detection of a tetramer (Fig. 4A and SI Appendix, Fig. S6A). The trimer species was reduced in an s-Mgm1 R712A/E751A mutant predicted, based on the structure, to compromise the head-to-tail interface (Fig. 4A and SI Appendix, Fig. S6A). Consistent with the crystallization conditions, the addition of GDP/BeF3− or GDP slightly shifted the dimer/trimer peaks assessed by AUC, but it did not alter the oligomerization pattern (SI Appendix, Fig. S6B). Interestingly, the addition of GTPγS resulted in the detection of a s-Mgm1 oligomeric species that migrated by AUC between dimer and trimer (SI Appendix, Fig. S6B). These results indicate that s-Mgm1 forms nucleotide-independent trimers in solution and suggest that GTP binding may inhibit the head-to-tail association.
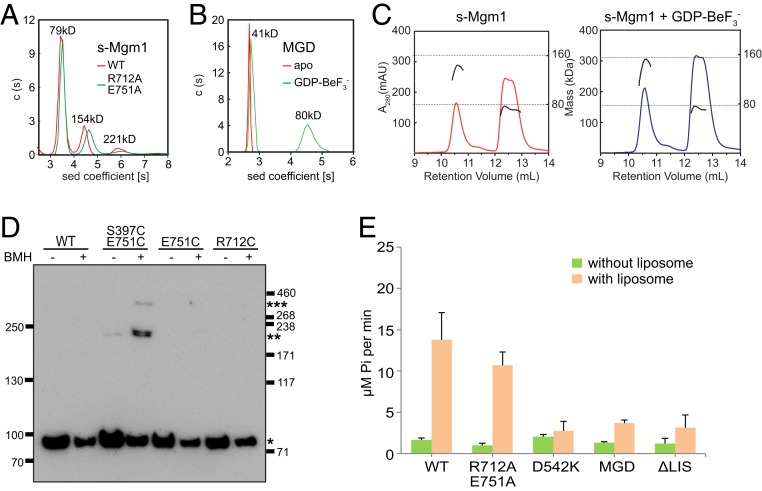
Fig. 4.
Oligomerization of s-Mgm1. (A) The size of s-Mgm1 (theoretical molecular mass 80.0 kDa), both WT and R712A/E751A mutant, was determined at 25 μM by AUC in a buffer containing 500 mM KCl. The estimated molecular masses are given above the peaks (in kilodaltons, kDa). The data are representative of at least three repetitions. (B) As in A, but with Mgm1-MGD (theoretical molecular mass 43.2 kDa) in the absence or presence of 0.5 mM GDP and 2.5 mM BeF3− in a buffer containing 500 mM KCl. (C) The size of s-Mgm1 (theoretical molecular mass 80.0 kDa) was determined by MALS coupled with gel filtration in the absence or presence of 0.5 mM GDP and 2.5 mM BeF3− in a buffer containing 500 mM KCl. The estimated molecular masses are shown by the right axis. The data are representative of at least three repetitions. (D) Purified HA-tagged Mgm1 (0.6 μM), WT, or indicated mutant was treated by 5 μM BMH at room temperature for 20 min. Samples were quenched and analyzed by SDS/PAGE and immunoblotting (IB) with anti-HA antibodies. Different oligomeric states are indicated by asterisks (*). Molecular markers are shown at the side (in kilodaltons). The data are representative of at least three repetitions. (E) The GTPase activity of various Mgm1 constructs was measured in the absence or presence of liposomes (1 mM, POPC:POPE:CL:PI:DOPS:Rhodamine-PE = 39:28.5:20:8:4:1.5) in a buffer with 100 mM KCl and 5 mM MgCl2. A total of 1 μM protein was used for each sample. GTP hydrolysis was measured by phosphate release at saturating GTP concentrations (0.5 mM). Data are presented as the mean ± SD of nine measurements from three independent experiments.
We also performed analytical gel filtration in combination with multiangle light scattering (MALS). Similar to AUC, s-Mgm1 was present as monomers and dimers in the absence of nucleotide, and this oligomeric state was not altered in the presence of GDP and BeF3− (Fig. 4C). However, in contrast to AUC, no trimer was detected, possibly due to relatively low protein concentrations during gel filtration. We hypothesized that nucleotide-independent s-Mgm1 dimerization is mediated by the HB-HB interface in the crystal utilized for hexamer formation. Consistently, mutant proteins containing changes at residues in the HB-HB interface regions, including D542A, were compromised for dimer formation in SEC-MALS in the absence of nucleotides (SI Appendix, Fig. S5). Therefore, these results are consistent with the formation of a nucleotide-independent, back-to-back s-Mgm1 dimer via the HB-HB interface.
Based on the s-Mgm1 structure, the MGD construct, which lacks HB2 and the LIS, is predicted to not support HB-HB interface dimer or head-to-tail trimer assembly. Consistently, Mgm1-MGD behaved as a monomer in AUC (Fig. 4B). However, similar to other DLP MGDs, dimer formation was observed in the presence of GDP and BeF3−. Thus, this MGD nucleotide-dependent dimer likely forms through a G-G interface.
To further test whether the “head-to-tail” interface exists in solution, we performed Cys-based cross-linking. Four surface-exposed Cys (C341, C676, C709, and C763) were mutated to Ser in s-Mgm1 and additional Cys were introduced at the “head-to-tail” interface. When a bifunctional Cys cross-linker bismaleimidohexane (BMH) was added, we detected dimers and trimers with the S397C/E751C mutant by SDS/PAGE analysis (Fig. 4D). Higher molecular mass s-Mgm1 species were not observed in the presence of BMH with either WT protein (with 4CS mutations) or mutants E751C or R712C (Fig. 4D). These results support the existence of the s-Mgm1 head-to-tail interface, consistent with the possible formation of an s-Mgm1 trimer.
Membrane-associated assembly of DLPs, including that of Mgm1, has been reported to stimulate GTP hydrolysis. No stimulation was observed when s-Mgm1 was incubated with liposomes containing 20% PS (SI Appendix, Fig. S6C), which efficiently recruited s-Mgm1 based on flotation assays. In contrast, liposomes containing 10% or 20% CL stimulated s-Mgm1 GTPase activity fivefold and eightfold, respectively (SI Appendix, Fig. S6C and Fig. 4E). These results suggest CL selectively promotes s-Mgm1 GTP hydrolysis.
Next, we tested a role for s-Mgm1 interfaces identified in the crystal structure in CL-dependent GTPase activity of s-Mgm1. The D542K mutation, predicted to disrupt the back-to-back s-Mgm1 interface, was severely deficient in CL- stimulated GTP hydrolysis (Fig. 4E). In addition, the MGD and ΔLIS mutant were also deficient in CL-stimulated GTP hydrolysis, consistent with defective nucleotide-independent assembly and membrane association, respectively. In contrast, the s-Mgm1 R712A/E751A mutant predicted to compromise the head-to-tail interface was not significantly affected in CL- stimulated GTP hydrolysis in comparison to s-Mgm1 (Fig. 4E). These results suggest that the s-Mgm1 back-to-back interface plays a role in Mgm1 assembly, which functions to stimulate GTP hydrolysis.
In Vivo Function of Mgm1 Mutants.
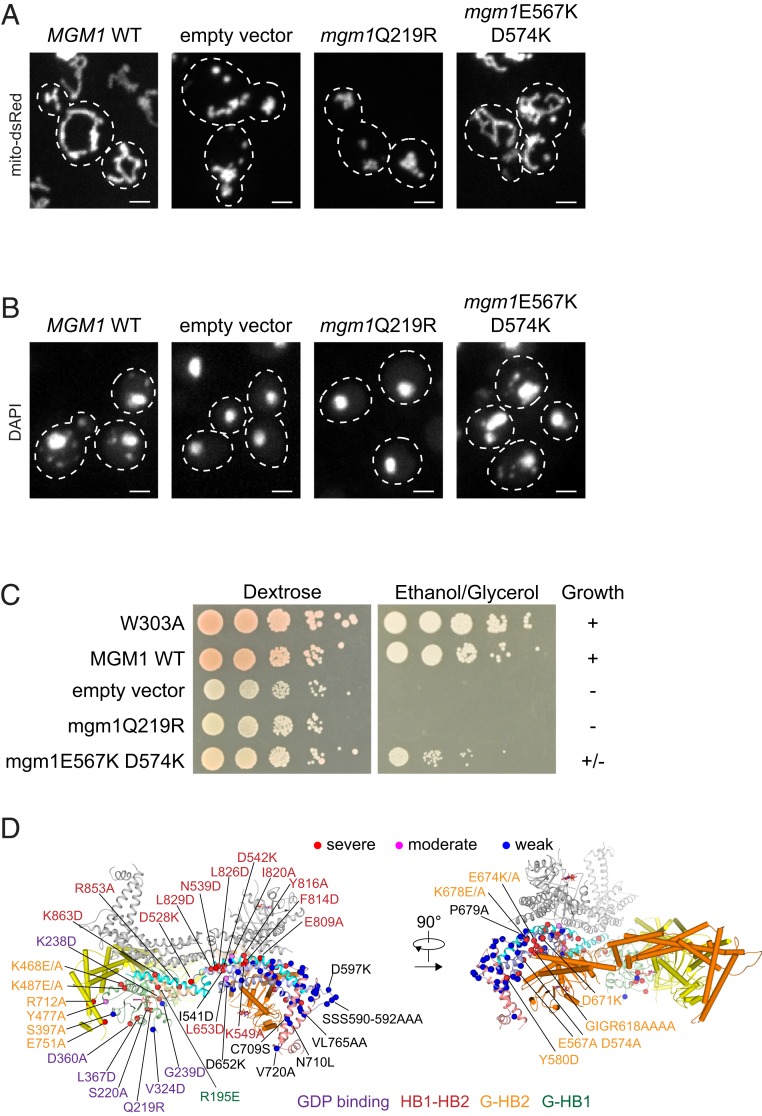
Our observations indicate that s-Mgm1 forms trimers and dimers of trimers in vitro. To test whether these Mgm1 structures function in vivo, we generated mutations predicted to disrupt both intramolecular and intermolecular interfaces involved in creating these structures and tested them for their ability to complement the loss of Mgm1 function (Figs. 1 and 5). In yeast cells, Mgm1-dependent IMM fusion is required for the maintenance of mitochondrial tubular networks and mitochondrial DNA (mtDNA) and, consequently, the ability to respire (18–20). Mitochondrial morphology was assessed in Δmgm1 cells co expressing empty vector, WT MGM1, or mutant mgm1 with mitochondrial-targeted DsRed (mito-DsRed) (Fig. 5A and SI Appendix, Table S2). Cells were also stained with DAPI and imaged to visualize mtDNA to assess mtDNA maintenance (Fig. 5B and SI Appendix, Table S2). The respiratory function of mgm1 mutant alleles was assessed by growth on the nonfermentable carbon sources ethanol and glycerol (Fig. 5C and SI Appendix, Table S2). Mutants were categorized as severe, moderate, or weak based on the combination of a reduced percentage of cells containing tubular network-like mitochondrial morphology and mtDNA, and by the severity of growth on a nonfermentable carbon source (Fig. 5D and SI Appendix, Table S2).
Fig. 5.
Functional studies of Mgm1 in yeast cells. (A) Mitochondrial morphology was visualized in ∆mgm1 cells expressing mitochondrial targeted-dsRed with indicated mgm1 plasmids. Cell boundaries are highlighted by dashed lines. (B) mtDNA was visualized using DAPI as described in cells with indicated mgm1 plasmids. (C) Growth analysis of indicated strains by serial dilution on media containing dextrose (Left) and nonfermentable carbon sources, ethanol and glycerol (Right). Growth (+), no growth (−), and partial growth (+/−). (D) Schematic representation of mutation mapped onto s-Mgm1 trimer viewed from two angles (listed in SI Appendix, Table S2). Phenotypes of mitochondrial morphology, mtDNA maintenance, and growth on ethanol/glycerol from mutations are categorized into “severe” (red dots), “moderate” (magenta dots), and “weak” (blue dots). Other color-coded residues include purple for nucleotide binding, red for the HB1-HB2 interface in antiparallel “back-to-back” stacking, green for intramolecular G-HB1 association, and yellow for G-HB2 interactions in “head-to-tail” assembly. (Scale bars, 2 μm.)
Mutations that affected Mgm1 function moderately or severely in vivo (Fig. 5D, magenta and red dots, respectively) were in residues predicted to be required for GDP binding (Fig. 5D, mutations in purple), head-to-tail interface/trimer formation (Fig. 5D, G-HB2 mutations in yellow), and back-to-back interface/dimer formation (Fig. 5D, HB1-HB2 mutations in red; SI Appendix, Table S2). In contrast, individual mutations in the LIS (residues 690–776) did not severely affect Mgm1 function in cells, as they exhibited phenotypes similar to wild type (Fig. 5D, blue dots; SI Appendix, Table S2), which is consistent with in vitro data indicating that redundant residues in both the G domain and LIS serve to target Mgm1 to the membrane. Along these lines, mutations in HB2 that are not near observed assembly interfaces retained Mgm1 function (Fig. 5D, blue dots; SI Appendix, Table S2). Thus, this functional analysis is consistent with a mechanism of Mgm1 forming head-to-tail and back-to-back assemblies.
Discussion
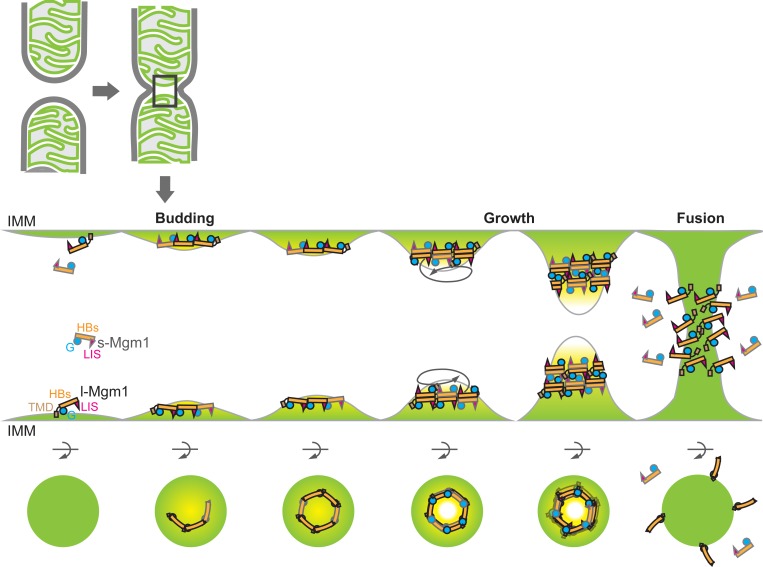
Recently, crystal and electron cryotomography structures of s-Mgm1 from C. thermophilum were reported and demonstrate the formation of stalk-mediated s-Mgm1 tetramers, which form membrane associated helical filaments on both negatively and positively curved membranes via the LIS domain (47). Such helical assemblies are proposed to remodel membranes to promote both inner membrane fusion and cristae biogenesis. The stacking interface of CtMgm1 is conserved and corresponds to the “back-to-back” interface of ScMgm1 reported here (SI Appendix, Fig. S1), and the different utilization of this interface underscores the possible plasticity of Mgm1 structures. In contrast, the “head-to-tail” interface we observe in the trimer is not conserved in CtMgm1. Our structural and biochemical analyses suggest that this trimer would associate with membranes via both the G domain and LIS (Fig. 6). In our model, s-Mgm1 would be recruited to the IMM surface and associate via its concave face to create a head-to-tail trimeric assembly. We speculate that these concave assemblies could promote the formation of a curved bilayer, in a manner analogous to a BAR domain, to generate an unstable membrane tip to promote fusion. Our proposed model represents a previously unidentified way to harness the ability of DLPs to interact with membranes and self-assemble for membrane fusion.
Fig. 6.
A model of Mgm1-mediated fusion. See Discussion for details.
Both the long and short forms of Mgm1 are required for efficient fusion (36, 37). Given the hydrophilic nature of the interaction between s-Mgm1 and lipids, it is likely that s-Mgm1 is not stably anchored to membranes. To promote membrane curvature, the stable membrane association of s-Mgm1 may be mediated via its association with l-Mgm1, which possesses a TM domain. Indeed, the requirement for l-Mgm1 may explain why both s-Mgm1 and l-Mgm1 exist as monomers, in contrast to the obligate dimers observed for the structurally similar scission dynamins (38). l-Mgm1 may not be capable of forming homooligomeric assemblies, as the addition of the long form is capable of disrupting the assembly of s-Mgm1 (51).
The membrane association of the CtMgm1 helical assemblies is mediated by the LIS. Our biochemical data also demonstrate an interaction of the ScLIS with membranes containing CL. CL is made within mitochondria, and the IMM possesses the highest percentage of this lipid in the cell. In many IMM complexes, it is a critical and highly specific cofactor/effector, and our data are consistent with a cofactor role for Mgm1-mediated function. However, we observe that in addition to the LIS, the G domain possesses the ability to interact with CL membranes. Although we were unable to determine key residues among the abundant Lys/Arg residues on the surface of the G domain, our data suggest that monomeric s-Mgm1 may bind to the membrane in multiple orientations and use these positively charged residues redundantly for membrane targeting. However, when locked in a head-to-tail arrangement, our structure predicts that the 5K/R cluster would be buried and the only available positive membrane-interacting patch in s-Mgm1 would be formed by the 3K/R (K277/K280/R281, see SI Appendix, Fig. S4 C and D). In combination with K740/K747 in the LIS, the tandem two-point interacting face on the concave side of the Mgm1 assembly would ensure efficient membrane-dependent scaffolding and membrane bending.
As a fusogenic DLP, our results suggest that Mgm1 assembly may not be harnessed for fusion in a manner similar to ATL and MFN. Although we observed a GG dimer-like interface between two s-Mgm1 molecules in the crystal packing (SI Appendix, Fig. S2B), no substantial interactions occurred between them. In addition, although the HB1/BSE domain adopts a relatively closed conformation, the HB2 domains point in opposite directions and cause the ends of the molecules to localize far from one another, making it unlikely that this structure represents a postfusion state, as seen with ATL and MFN. Similarly, when two s-Mgm1 molecules are aligned in the OPA1-MGD dimer structure, the ends of the molecules in the dimer are distant (SI Appendix, Fig. S2C). Collectively, it is possible that the potential GG dimer interface does not directly contribute to the fusion reaction, but it could regulate the formation of the actual fusogenic assembly.
In summary, we have provided data indicating that the inner membrane fusion dynamin ScMgm1 is capable of forming a concave membrane associated trimeric structure. This unique ScMgm1 trimer suggests a mechanism of Mgm1-mediated fusion.
Methods
Protein Preparation.
s-Mgm1 (residues 184–881) was cloned into pET-28a vector with an N-terminal 6×His and thrombin cleavage site. Transformed E. coli BL21 (DE3) cells were cultured at 37 °C in Luria–Bertani broth media containing 100 mg/L kanamycin. After the OD600 reached 0.6, the culture was cooled to 4 °C and supplemented with 0.25 mM isopropyl β-d-thiogalactoside. Cells were induced at 16 °C for 18 h, harvested, resuspended in lysis buffer (20 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, 4 mM MgCl2), and homogenized using an ultrahigh-pressure cell disrupter (JNBIO) at 4 °C. Cell lysates were clarified by centrifugation and recombinant protein isolated by Ni-NTA affinity chromatography and eluted with elution buffer (20 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, 4 mM MgCl2, 250 mM imidazole). The protein was then further purified by passage through a heparin column (GE Healthcare). Purified s-Mgm1 was concentrated to 3.5 mg/mL and stored at −80 °C. The selenomethionine (SeMet)-substituted protein was expressed as previously described (33) and purified by the same procedure as described for the native protein.
Crystallization, Data Collection, and Structure Determination.
Crystals were obtained by mixing 2 μL of protein solution with an equal volume of reservoir solution with equilibration against 400 μL of reservoir solution. The nucleotide mixture (GDP and BeF3−) was prepared with 2 mM GDP in 20 mM Tris, 150 mM NaCl, 4 mM MgCl2, 2 mM BeSO4, and 20 mM NaF. The same recipe was used for the biochemical assays, except in some conditions, KCl and KF were used instead of the Na equivalents. The crystals for s-Mgm1/GDP were obtained in the reservoir solution containing 100 mM KI, 8% PEG 3350, 250 mM NaCl, 10 mM Tris (pH 8.0), and 2 mM MgCl2. The crystals for SeMet sMgm1/GDP were obtained by microseeding with native crystal, and SeMet crystal was collected to 3.2 Å on a beamline BL17 at Shanghai Synchrotron Radiation Facility. Data were processed and scaled in the XDS program suite (52). Se-atom searching, initial phase calculations, and density modifications were performed with PHENIX (53). The model was built manually with COOT (54) and subsequently refined with PHENIX. A summary of the final refinement statistics is shown in SI Appendix, Table S1. Structural figures were prepared by the program PyMOL (https://pymol.org/2/).
ITC.
ITC was performed with a MicroCal iTC200 instrument (GE Healthcare) at 16 °C as described previously (33). In brief, 2 mM indicated nucleotides were injected in a stepwise manner into ∼100 μM Mgm1-MGD. Acquired data were analyzed by the program Origin 7.0 (GE Healthcare) with a “One Set of Binding Sites” fitting model.
Multiangle Light Scattering.
MALS was performed at 25 °C with an 18-angle static light scattering detector (DAWN HELEOS II, Wyatt) in conjunction with an analytical size exclusion chromatography column (Superdex200 10/300 or Superdex75 10/300, for s-Mgm1 and Mgm1-MGD, respectively) as described previously. For each run, 100 μL of 2 mg/mL purified sMgm1p/Mgm1-MGD (WT or mutant) was applied after incubating with or without 0.5 mM of the corresponding ligand overnight on ice. The column was equilibrated with 20 mM Tris 8.0, 250 mM NaCl/250 mM KCl, 4 mM MgCl2, and 2 mM dithiothreitol (DTT). MALS results were analyzed using the provided ASTRA software.
GTPase Activity Assay.
GTPase assays were performed using the Enzchek phosphate assay kit (Invitrogen). Reactions were performed in a 100-μL volume with 20 μL of 5× reaction buffer (100 mM Hepes, pH 7.4, 2.5 M KCl or NaCl, 25 mM MgCl2), 200 μM 2-amino-6-mercapto-7-methylpurine riboside, 0.1 U purine nucleoside phosphorylase (PNP), and 5 μM protein, and incubated for 20 min at 30 °C in a 96-well plate (Corning). Reactions were initiated by the addition of 0.5 mM GTP (Jena Bioscience). The absorbance was measured at 360 nm every 30 s over 30 min at 30 °C using a microplate reader (Synergy 4, BioTek). The rate of phosphate release was then calculated based on a standard curve.
Flotation Assays.
The lipids (Avanti Polar Lipids; 55:23.5:10:8:2:1.5 mol % POPC:POPE:CL:soy-PI:DOPS:Rhod-PE) were dried to a film, hydrated with 25 mM Hepes (pH 7.4) containing 100 mM KCl, and extruded through polycarbonate filters with a pore size of 50 nm, 100 nm, or 400 nm. The liposomes (2 mM) were mixed with 2 μM purified protein and incubated at room temperature for 30 min. The 30 μL mixture of proteins and liposomes was mixed with 100 μL of 1.9 M sucrose and overlaid with 100 μL of 1.25 M sucrose and 20 μL of 0.25 M sucrose, all in 25 mM Hepes (pH 7.4) and 100 mM KCl. The samples were centrifuged in a Beckman TLS 55 rotor at 174,000 × g at 4 °C for 60 min. The gradient was fractionated into five 50-μL fractions and analyzed by SDS/PAGE and Coomassie blue staining.
Liposome Pelleting Assays.
For the pelleting assays, Mgm1 fragments were cloned, expressed, and purified with a 6xHIS tag. The MGD-like construct contains residues 170–530 and 842–872 of ScMgm1. The HB2 construct contains residues 520–839. The LIS construct contains residues 689–775. IMM-like liposomes were prepared (51) and pelleting assays performed as described previously (55). Briefly, liposomes (final concentration 1.2 mg/mL) were incubated with 1.2 μM recombinant proteins at room temperature for 90 min. Following incubation, liposomes and recombinant proteins were centrifuged in a TLA-100 rotor in Beckman TLA-100 Ultracentrifuge at 40,000 × g for 30 min at 4 °C. Supernatant and pellet were separated and the pellet resuspended in an equivalent volume of 20 mM Tris⋅HCl (pH 8) and 150 mM NaCl as supernatant. Laemmli (6×) was added to the pellet and supernatant and the samples analyzed by SDS/PAGE and Coomassie blue staining.
Cys Cross-linking Assays.
C-terminal HA-tagged s-Mgm1 was PCR amplified and inserted into pET28a. To avoid nonspecific cross-linking, four exposed Cys (C341, C676, C709, C763) were mutated into Ser. All other point mutants, including E751C, S397C, R712C, were also generated by site-directed mutagenesis from s-Mgm1. The proteins were purified from E. coli BL21 (DE3) cells. The harvested cells were resuspended in lysis buffer (40 mM Hepes-KOH, pH 7.4, 250 mM NaCl, 250 mM KCl, 1 mM DTT) and lysed by sonication at 4 °C. Cell lysates were clarified by centrifugation at 9,000 × g for 10 min, and the supernatant was collected. The protein was isolated by Ni-NTA chromatography (GE Healthcare) and eluted with elution buffer (20 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 300 mM imidazole). The protein was then further purified by HiTrap Heparin HP affinity column (GE Healthcare) and gel filtration chromatography (Superdex-200; GE Healthcare). Purified protein (0.6 μM) was incubated with 5 μM BMH (Thermo Scientific) at room temperature for 20 min in 25 mM Hepes, pH 7.4, 250 mM KCl, 250 mM NaCl. The samples were analyzed by 6% Tris-Acetate SDS/PAGE and immunoblotted with mouse anti-HA antibody (Sigma-Aldrich). HiMark LC5699 (Invitrogen) and 26619 (Thermo Scientific) were used as PAGE Rulers.
Strains, Media, and Plasmids.
Media preparations and genetic techniques were performed as described previously (56). All yeast strains were constructed by direct transformation using the lithium acetate method, and transformants were selected on minimal media (SD) with appropriate selection. All ∆mgm1 strains expressing WT or mutant MGM1 alleles were generated by plasmid shuffle with pRS316-dnm1G385D as described previously (51). To construct the pRS425-MGM1 alleles, MGM1 (including ∼400 bp 5′ of the start codon and 30 bp after the stop codon) was amplified by PCR with SacII and SacI sites at the 5′ and 3′ ends, respectively. The Mgm1 fragment was ligated into pRS425 and sequenced to confirm. The Mgm1 point mutations were created by site-directed mutagenesis of pRS425-MGM1 wild type or cloned into linearized pRS425 using a Gibson isothermal assembly reagent according to the manufacturer’s instructions. Internal deletions were made using a PCR-based method with complementary primers 20–30 nt in length on either side of the altered nucleotides. Mitochondria were marked fluorescently with mitochondria-targeting DsRed (57, 58).
Yeast Growth Assays.
Cells were grown to log phase in appropriate synthetic medium to select for plasmids, pelleted, and resuspended in water at a concentration of 0.5 OD600/mL. Two microliters of 10-fold serial dilutions were plated on yeast extract peptone dextrose (YPD) and yeast extract peptone ethanol/glycerol (YPEG) plates. Cells were grown for ∼48 h (YPD) or 48–60 h (YPEG) at 30 °C.
Staining of mtDNA.
To monitor mtDNA foci in vivo, cells were grown at 30 °C to log phase in the appropriate synthetic medium to select for plasmids and fixed in 70% ethanol with 100 ng/mL DAPI for 2–3 min. Cells were washed in PBS and imaged by fluorescence microscopy. The mtDNA was quantified by counting the number of cells containing at least two mtDNA foci. A minimum of 50 cells were counted for each experiment.
Fluorescence Microscopy.
For all fluorescence microscopy experiments, cells were grown at 30 °C to log phase in the appropriate synthetic medium to select for plasmids. Cells were mounted and imaged on glass bottom SensoPlates (Greiner 655892) pretreated with Con A (Sigma-Aldrich C2010, 0.25 mg/mL) for 1 h, followed by drying, or directly mounted and imaged on MICRO slides (VWR 48300-025). Cells were imaged at 25 °C. For fluorescence microscopy, an Olympus IX70 Deltavision Microscope with a 60× 1.4 N.A. objective (Olympus) and 100 W mercury lamp (Applied Precision, Inc.) was used to assess the mitochondrial morphology. Two- and three-dimensional light microscopy data were collected using an integrated, cooled, CCD-based Princeton Micromax Camera equipped with a Sony Interline Chip. Z-series images were taken with a step size of 0.2 µm. Three-dimensional datasets were processed using DeltaVision’s iterative, constrained, 3D deconvolution method to remove out-of-focus light. Deconvolved images were analyzed in SoftWorx (Applied Precision). For confocal microscopy, Z-series images (step size 0.2 μm) of cells were collected using the spinning-disk module of a Marianas SDC Real Time 3D Confocal-TIRF microscope (Intelligent Imaging Innovations) fitted with a 100× 1.46 N.A. objective and Photometrics QuantEM EMCCD camera. Images were captured using SlideBook (Intelligent Imaging Innovations) and linear adjustments made using ImageJ. Morphology was assessed manually by categorization based on the tubular or fragmented characteristics of the mitochondria, counting a minimum of 50 cells per experiment with at least three fields of view.
Data Availability.
Atomic coordinates and structure factors of GDP-bound s-Mgm1 have been deposited in the Protein Data Bank, http://www.rcsb.org under the ID code 6JSJ. All other data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Alicia M. Prater for proofreading, Dr. Sha Sun for help with model illustration, the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for analytical support, Dr. Jaclyn Bonner from the University of California, Davis for technical assistance, and the staff at beamline BL17U of Shanghai Synchrotron Radiation Facility for help with diffraction data collection. J.H. is supported by National Key Research and Development Program Grant 2016YFA0500201, National Natural Science Foundation of China Grants 91854202 and 31630020, and Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB39000000. L.Y. is a Tsinghua-Peking Center for Life Sciences Postdoctoral Fellow and is supported by National Key Research and Development Program Grant 2017YFC0840302 and National Natural Science Foundation of China Grant 3170040488. Z.R. is supported by National Key Research and Development Program Grant 2017YFC0840300. J.N. is supported by NIH Grants R37GM097432 and R01GM126081.
Footnotes
The authors declare no competing interest.
Data deposition: Atomic coordinates and structure factors of GDP-bound s-Mgm1 have been deposited in the Protein Data Bank, http://www.rcsb.org (PDB ID code 6JSJ).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919116117/-/DCSupplemental.
References
- 1.Lackner L. L., Nunnari J. M., The molecular mechanism and cellular functions of mitochondrial division. Biochim. Biophys. Acta 1792, 1138–1144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoppins S., Nunnari J., The molecular mechanism of mitochondrial fusion. Biochim. Biophys. Acta 1793, 20–26 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Mishra P., Chan D. C., Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pernas L., Scorrano L., Mito-morphosis: Mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78, 505–531 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Youle R. J., van der Bliek A. M., Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoppins S., Lackner L., Nunnari J., The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Otsuga D., et al. , The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ingerman E., et al. , Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramachandran R., et al. , The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 26, 559–566 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meeusen S., et al. , Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127, 383–395 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Hales K. G., Fuller M. T., Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Hermann G. J., et al. , Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L., OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 101, 15927–15932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander C., et al. , OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Delettre C., et al. , Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Jones B. A., Fangman W. L., Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 6, 380–389 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Guan K., Farh L., Marshall T. K., Deschenes R. J., Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet. 24, 141–148 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Shepard K. A., Yaffe M. P., The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 144, 711–720 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong E. D., et al. , The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160, 303–311 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong E. D., et al. , The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341–352 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sesaki H., Southard S. M., Yaffe M. P., Jensen R. E., Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell 14, 2342–2356 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahn R., Scheller R. H., SNAREs–engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Wickner W., Schekman R., Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Südhof T. C., Rothman J. E., Membrane fusion: Grappling with SNARE and SM proteins. Science 323, 474–477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J., et al. , A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orso G., et al. , Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Bian X., et al. , Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 3976–3981 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T. Y., et al. , Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. U.S.A. 109, E2146–E2154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu T. Y., et al. , Cis and trans interactions between atlastin molecules during membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 112, E1851–E1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J., Rapoport T. A., Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin. Cell Dev. Biol. 60, 105–111 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Qi Y., et al. , Structures of human mitofusin 1 provide insight into mitochondrial tethering. J. Cell Biol. 215, 621–629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y. L., et al. , MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542, 372–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan L., et al. , Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion. Nat. Struct. Mol. Biol. 25, 233–243 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Huang X., et al. , Sequences flanking the transmembrane segments facilitate mitochondrial localization and membrane fusion by mitofusin. Proc. Natl. Acad. Sci. U.S.A. 114, E9863–E9872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McQuibban G. A., Saurya S., Freeman M., Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S., Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sesaki H., Southard S. M., Hobbs A. E. A., Jensen R. E., Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem. Biophys. Res. Commun. 308, 276–283 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Ishihara N., Fujita Y., Oka T., Mihara K., Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25, 2966–2977 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griparic L., Kanazawa T., van der Bliek A. M., Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178, 757–764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Z., Chen H., Fiket M., Alexander C., Chan D. C., OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esser K., Tursun B., Ingenhoven M., Michaelis G., Pratje E., A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 323, 835–843 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S., Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Duvezin-Caubet S., et al. , Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281, 37972–37979 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Del Dotto V., Fogazza M., Carelli V., Rugolo M., Zanna C., Eight human OPA1 isoforms, long and short: What are they for? Biochim. Biophys. Acta Bioenerg. 1859, 263–269 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Ban T., Heymann J. A., Song Z., Hinshaw J. E., Chan D. C., OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19, 2113–2122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rujiviphat J., et al. , Mitochondrial genome maintenance 1 (Mgm1) protein alters membrane topology and promotes local membrane bending. J. Mol. Biol. 427, 2599–2609 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Faelber K., et al. , Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1. Nature 571, 429–433 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meglei G., McQuibban G. A., The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion. Biochemistry 48, 1774–1784 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Rujiviphat J., Meglei G., Rubinstein J. L., McQuibban G. A., Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284, 28682–28686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faelber K., et al. , Crystal structure of nucleotide-free dynamin. Nature 477, 556–560 (2011). [DOI] [PubMed] [Google Scholar]
- 51.DeVay R. M., et al. , Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine P. V., et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Besprozvannaya M., et al. , GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. eLife 7, e31019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guthrie C., Fink G., Guide to Yeast Genetics and Molecular Biology (Academic Press, San Diego, 1991). [Google Scholar]
- 57.Westermann B., Neupert W., Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Naylor K., et al. , Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281, 2177–2183 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors of GDP-bound s-Mgm1 have been deposited in the Protein Data Bank, http://www.rcsb.org under the ID code 6JSJ. All other data are included in the manuscript and SI Appendix.