Significance
We present high-resolution crystal structures of the bacterial heliorhodopsin 48C12, a representative of the recently discovered family of microbial rhodopsins. In opposite to all other rhodopsins, heliorhodopsins face the cytoplasm of the cells with their N termini. The structures of two different states of 48C12 reveal specific features of heliorhodopsins, such as existence of a water-filled cavity in the cytoplasmic side near the retinal Schiff base able to accommodate anions of triangular geometry, such as nitrate, carbonate, or acetate, and completely hydrophobic organization of the inner extracellular part of the protein. Hence, the structure gives important insights into possible functions of 48C12.
Keywords: rhodopsin, membrane protein, X-ray crystallography, crystal structure, retinal
Abstract
Rhodopsins are the most abundant light-harvesting proteins. A new family of rhodopsins, heliorhodopsins (HeRs), has recently been discovered. Unlike in the known rhodopsins, in HeRs the N termini face the cytoplasm. The function of HeRs remains unknown. We present the structures of the bacterial HeR-48C12 in two states at the resolution of 1.5 Å, which highlight its remarkable difference from all known rhodopsins. The interior of HeR’s extracellular part is completely hydrophobic, while the cytoplasmic part comprises a cavity (Schiff base cavity [SBC]) surrounded by charged amino acids and containing a cluster of water molecules, presumably being a primary proton acceptor from the Schiff base. At acidic pH, a planar triangular molecule (acetate) is present in the SBC. Structure-based bioinformatic analysis identified 10 subfamilies of HeRs, suggesting their diverse biological functions. The structures and available data suggest an enzymatic activity of HeR-48C12 subfamily and their possible involvement in fundamental redox biological processes.
Microbial and animal visual rhodopsins (classified into types 1 and 2 rhodopsins, respectively) comprise an abundant family of seven-helical transmembrane proteins that contain a covalently attached retinal cofactor (1–3). On absorption of a photon, the retinal isomerizes, triggering a series of conformational transformations correlating with functional and spectral states known as the photocycle (4–6). Microbial rhodopsins are universal and the most abundant light-harvesting proteins on Earth. Before 1999, only rhodopsins from halophilic archaea had been known. About 30 y after the discovery of the first rhodopsin (bacteriorhodopsin [bR]) (2), the first nonhaloarchaeal rhodopsin was reported (Neurospora rhodopsin) (7). Soon after that, metagenomics studies by Beja et al. (8) led to the discovery in 2000 of a rhodopsin gene in marine Proteobacteria that was named accordingly proteorhodopsin (pR). Since then, 7,000 microbial rhodopsins were identified. They are present in all of the three domains of life (bacteria, archaea, and eukaryotes) as well as in giant viruses (4). The discovery of channel rhodopsins (9) led to development of optogenetics, the revolutionary method for controlling cell behavior in vivo in which microbial rhodopsins play the key role (10–13).
Several rhodopsins with new functions have recently been discovered and characterized. Among the members of the rhodopsin family are light-driven proton, anion and cation pumps, light-gated anion and cation channels, and photoreceptors (3, 5, 14, 15). Genomic and metagenomic studies dramatically expanded the world of rhodopsin sequences, some of which were found in unexpected organisms and habitats: for example, sodium-pumping rhodopsins in Flavobacteria (16, 17) and the rhodopsins from giant viruses (18–20). The widely spread presence and importance of pR-based phototrophy in the marine environment (21) were identified. Recently, rhodopsins that function as inward proton pumps were discovered (22, 23).
Despite diversity of their functions and differences in the structures, all of these rhodopsins are oriented in the membranes in the same way. Their N termini always face the outside of the cells. In 2018, Pushkarev et al. (24) discovered a new large family of rhodopsins, named heliorhodopsins (HeRs), facing the cytoplasmic space of the cell with their N termini. It was found that they are present in Archaea, Bacteria, Eukarya, and viruses.
The function of HeRs is not yet known (25, 26). Moreover, the structural data on HeRs are limited to the very recently reported model of archaeal HeR (TaHeR) (27). Here, we present two crystallographic structures of the HeR-48C12 discovered in an actinobacterial fosmid from freshwater Lake Kinneret (24) corresponding to two states of the protein, both solved at 1.5-Å resolution. The structures show an astonishingly large difference between the organization of HeRs and other type I rhodopsins. For instance, the protein has a big cavity in the cytoplasmic part containing the cluster of water molecules, which is likely to serve as proton acceptor from the retinal Schiff base (RSB). Ten of 48C12 amino acids are highly conserved within all HeRs, and we believe that its structure and the discussed mechanisms will be a basis for understanding this abundant family and also, the evolution of rhodopsins in general.
Results
Structure of the HeR-48C12 at Neutral pH.
HeR-48C12 was crystallized using the in meso approach similarly to our previous works (28). Rhombic crystals appeared in 2 wk and reached 150 μm in length and width, with the maximum thickness of 20 μm. We have solved the crystal structure of 48C12 at pH 8.8 at 1.5 Å (29). The crystals of P21 symmetry contained two protomers organized in a dimer in the asymmetric unit (SI Appendix, Figs. S1 and S2). The high-resolution structure reveals 233 water molecules and 31 lipid fragments.
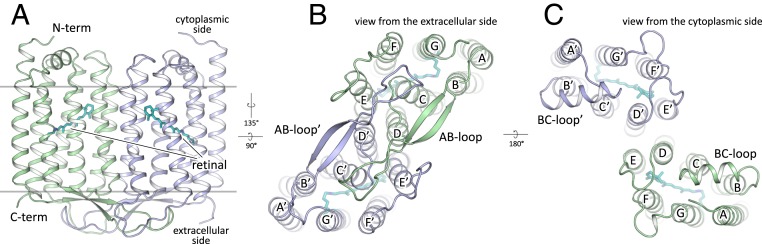
Similarly to other type I rhodopsins, each 48C12 protomer has seven transmembrane α-helices connected by three extracellular and three intracellular loops. However, some of the loops are relatively large and have certain secondary structure (Fig. 1). The extracellular AB loop of 48C12 (residues 34 to 64) is ∼40-Å long and forms a β-sheet with the length of ∼17 Å (Fig. 1B). It extends in the direction of the second protomer of the dimer while remaining parallel to the membrane surface, and thus, it covers the extracellular surface of the nearby molecule (Fig. 1 and SI Appendix, Fig. S1). The intracellular BC loop comprises 14 residues (86 to 98) and forms an α-helix with the length of ∼18 Å (Fig. 1C). Other loops and N and C termini, although not forming regular secondary structures, are well ordered and therefore, are completely resolved. The relative location of the α-helices is also altered in comparison with other microbial rhodopsins with known structure (SI Appendix, Figs. S3 and S4). Particularly, the most notable differences occur in the helices A, D, and E (SI Appendix, Fig. S4).
Fig. 1.
Overall architecture of the HeR-48C12 dimer. (A) Side view of the dimer. Hydrophobic/hydrophilic membrane boundaries are shown with gray lines. (B) View from the extracellular side. (C) View from the cytoplasmic side. Cofactor retinal is colored teal.
Dimerization Interface of 48C12.
The 48C12 protomers interact in the dimer via helices D and E (Fig. 1 B and C and SI Appendix, Fig. S5), with a broad hydrophobic interface in the middle part (inside the membrane) and interactions between polar residues, specifically Asp127 and Tyr179′ at the extracellular and Tyr151 and Asp158′ at the cytoplasmic sides of the membrane. Tyr179′ side chain is additionally connected through a hydrogen bond to the main chain of the AB loop of the neighboring protomer (nitrogen of Thr44). The AB loop itself almost does not interact directly with the neighbor protomer, although it is stabilized by several hydrogen bonds mediated by numerous water molecules located on the extracellular surface of the dimer.
Several well-ordered lipid molecules are present in the structure, surrounding the protein dimer (SI Appendix, Fig. S6). Two of them permeate the HeR between helices E and F near the β-ionone ring of the retinal cofactor with the hydrocarbon tails. Surprisingly, the pocket of the hydrocarbon chain comprises polar amino acids Asn207 and Asn138 and one water molecule. Asn207 is also exposed to the surface of the extracellular part of the 48C12 protomer in the middle of the membrane and is highly conserved within HeRs.
The structures of the protomers within the 48C12 dimer are similar (rmsd between protomers 0.144 Å); however, there are differences in the EF-loop organization and α-helical BC-loop location, and 3-Å displacement of the cytoplasmic end of helix A is observed (SI Appendix, Fig. S7). Consequently, positions of several residues inside the protomers are slightly varied. Since general features of the HeR structure are the same in both molecules, we will describe mostly the protomer A. Nevertheless, we will also describe the differences between the protomers where appropriate.
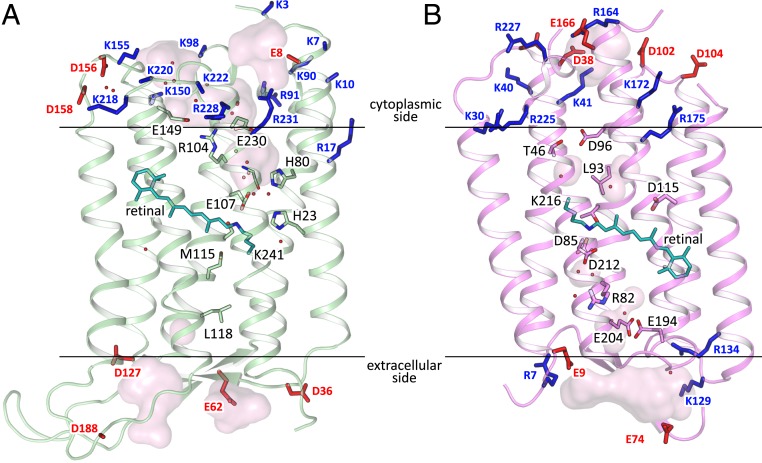
Mechanism of Topological Inversion of HeRs.
It is known that insertion and folding of membrane proteins is guided by the “positive-inside rule” (30). Using the structure of 48C12, we analyzed the location of positively and negatively charged residues in the cytoplasmic and extracellular domains of the protein and compared it with bR (Fig. 2). Notably, in 48C12, all of the positively charged residues are located exclusively at the cytoplasmic side of the protein, which is consistent with the positive-inside rule (30). Importantly, some of these residues, such as Arg91, Lys218, Lys222, and Arg231, are highly conserved in the subfamily of 48C12 (SI Appendix, Fig. S8). In addition, unlike bR, HeR contains only negative amino acids in the extracellular polar part of the proteins, which is characteristic for this subfamily. Thus, we suggest that HeRs follow the “positive-inside and negative-outside rule” rather than just the positive-inside rule.
Fig. 2.
Comparison of 48C12 (green) and bR (purple; PDB ID code 1C3W). (A) Side view of the HeR-48C12. N terminus is at the cytoplasmic side of the membrane. (B) Side view of the bR. N terminus is at the extracellular side of the membrane. Hydrophobic/hydrophilic membrane boundaries are shown with black lines. Positively and negatively charged residues on the protein cytoplasmic and extracellular surfaces are shown in blue and red, respectively.
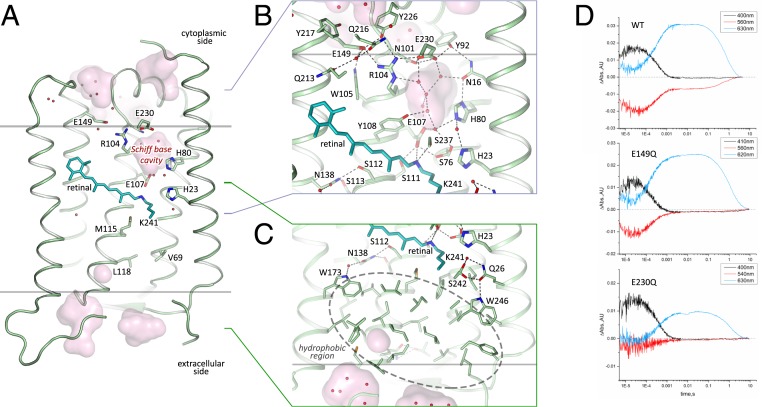
Structure of the Extracellular Region.
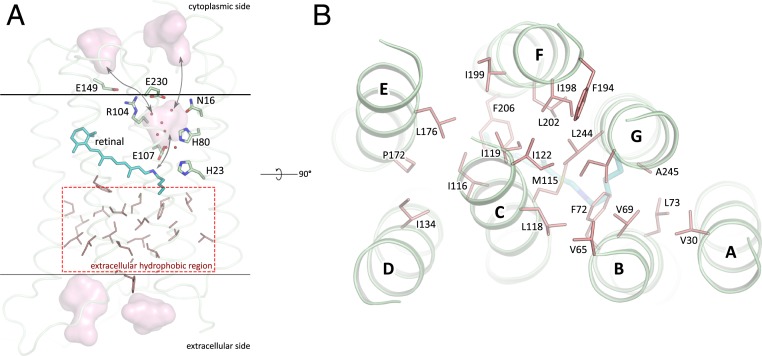
As HeRs are topologically inverted in the membrane relative to other type I rhodopsins, the extracellular part of 48C12 corresponds to the cytoplasmic part of classical microbial rhodopsins, such as bR. However, in 48C12 the internal region, embedded in the extracellular leaflet of the lipid bilayer, is completely hydrophobic and does not comprise any charged or polar amino acids and solvent-accessible cavities (Figs. 3C and 4). Hereafter, we denote this part as the hydrophobic extracellular region. Nevertheless, several clusters of polar amino acids are located at the extracellular half of the protein inside the membrane but on the outer surface of the protein. Helices A and G interact by hydrogen bonding of Gln26 with Ser242 and Trp246, while helices F and G are also connected by a hydrogen bond between Gln247 and Ser201. We suggest that these interactions support the internal hydrophobic configuration at the extracellular side. The absence of any charged or/and polar amino acids inside the region may explain the absence of any proton/ion pumping by 48C12 (24).
Fig. 3.
Structure of the 48C12 protomer. (A) Side view of the protomer in the membrane. (B) Detailed view of the cytoplasmic part. (C) Detailed view of the extracellular side and the hydrophobic region. Cofactor retinal is colored teal. Hydrophobic/hydrophilic membrane boundaries are shown with gray lines. Cavities are calculated with HOLLOW (31) and shown in pink. Charged residues in 48C12 are shown with thicker sticks. Helices F and G are not shown. (D) Time evolution of the transient absorption changes of photo-excited 48C12, wild-type (WT), E230Q, and E149Q mutant forms. The characteristic wavelengths of intermediate states are slightly shifted in the mutants. The O2-state decay is almost two times longer in both 48C12 variants. Abs, absorption; AU, absorption unit.
Fig. 4.
Hydrophobic residues in the extracellular part of 48C12. (A) Side view of the 48C12 protomer. The residues comprising the extracellular hydrophobic region are colored red. The region is embedded in the extracellular half of the lipid bilayer and is contoured with dashed red rectangle. Membrane core boundaries are shown with black lines. Black arrows indicate putative pathways connecting the inner cavity (SBC), cytoplasmic side of the protein, and RSB. Cavities are colored pink. Water molecules in the inner cavity are shown with red spheres. (B) View on the hydrophobic region from the extracellular surface of the protein. Loops are hidden for clarity. Hydrophobic residues in the extracellular internal part of the 48C12 protomer are colored red. Cofactor retinal is colored teal.
Retinal Binding Pocket and Cavity in the RSB Region.
The retinal binding pocket of 48C12 (Figs. 1 and 2) is also different from that of microbial rhodopsins with known structures. Near the retinal molecule, helices C and D are connected by hydrogen bonding of Asn138 (analog of Asp115 in bR and Asp156 in channelrhodopsin-2 [ChR2]) with Ser112 (analog of Thr90 in bR and Thr128 in ChR2) and Ser113. The Asn138 side chain is also stabilized by hydrogen bonding with Trp173 through a well-ordered water molecule (Fig. 3C). In the region of the β-ionone ring of the retinal molecule, only two residues (Met141 and Ile142) are similar to those in bR (SI Appendix, Fig. S9). Although many of the residues of the pocket walls remain aromatic in 48C12, there are notable alterations, such as, for example, Phe206 in the position of Trp182 in bR and Trp105 instead of Tyr83 and Tyr108 in the place of Trp86. All of these residues are highly conserved in HeRs. Interestingly, polar Gln213 (in the position of Trp189 in bR) is located close to the β-ionone ring.
The Schiff base is surrounded by an unusual, for the rhodopsins of type I, set of residues: for example, Ser237 replaces Asp212 (extremely conserved aspartate in type I rhodopsins), Glu107 replaces Asp85, His23 replaces Met20, the bulky Phe72 replaces Val49, Met115 replaces Leu93, and Ser76 replaces Ala53. In this configuration, RSB is hydrogen bonded directly to Glu107 (RSB counterion) and Ser237. The Glu107 side chain is stabilized by two serine residues (Ser76 and Ser111).
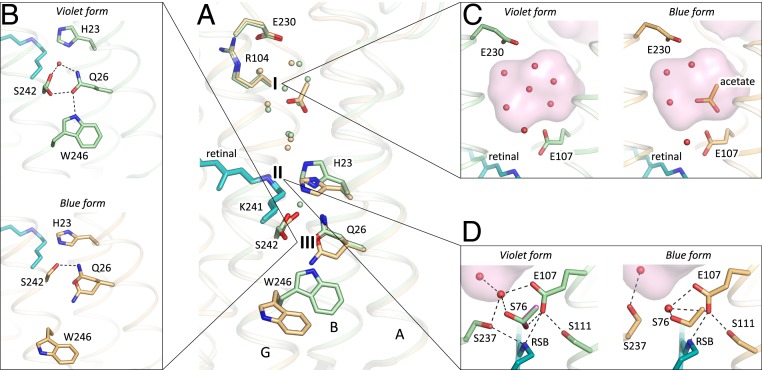
The distinctive feature of HeR-48C12 is the presence of a large hydrophilic cavity in the vicinity of the Schiff base (Schiff base cavity [SBC]) between residues Glu107 and Arg104 (analog of Arg82 in bR). The SBC is separated from the cytoplasmic bulk volume with the only side chain of Asn101 (Fig. 3); is surrounded by polar residues Glu107, His23, His80, Ser237, Glu230, Tyr92, Asn16, Asn101, and Tyr108; and is filled with six water molecules (Fig. 3B). The listed amino acids, together with the water molecules, create a dense hydrogen bonding network, which protrudes from the RSB to Arg104. The Arg104 side chain is pointed toward the cytoplasm and is stabilized by Glu230, Glu149, and also, Tyr226. It should be noted that, in protomer B, there is an alternative conformation of Arg104, Glu230, and Tyr226, which, however, does not affect the shape of the cavity. The Glu149 side chain is additionally stabilized by Trp105 and by a water-mediated hydrogen bond to Gln216 and Gln213. The calculation of the hydrophilic/hydrophobic membrane boundaries shows that Glu149 is located out of the hydrophobic part of the membrane (Fig. 3A) and can be accessed from the cytoplasmic bulk, which is also proved by the cavities calculations using HOLLOW (31). In protomer B, the accessibility of Glu149 from the bulk is lower, mostly because of slight alterations of the helices positions. Importantly, all of the residues mentioned in this paragraph are highly conserved within all of the known HeRs (SI Appendix, Figs. S8 and S10). This fact together with their structural roles points toward their functional importance.
Role of the SBC.
As it was shown in previous studies, His23, His80, and Glu107 do not act as a proton acceptor group from the RSB; however, His23 and His80 are important for proton transfer (24, 25). Our structure shows that the rechargeable amino acids E149 and E230 are connected to the RSB via a continuous network of hydrogen bonds but have not yet been studied. To understand better their roles for the HeR functioning, we produced E149Q and E230Q mutants and studied the properties of their photocycles. First of all, we should stress that these mutants were not stable during purification. Moreover, E230Q degrades quickly on illumination even while remaining in the lipid membranes. It indicates that both amino acids are important for the protein stabilization. We measured the transient absorption of the mutants with the solubilized (not purified) protein (Fig. 3D). Formation of the K/M photocycle intermediate was observed for both mutants; therefore, neither Glu149 nor Glu230 is the proton acceptor. However, the O2-state decay in mutants is more than two times longer than that of the wild-type protein (Fig. 3D), which indicates the involvement of Glu149 and Glu230 in the protein function. Thus, none of charged amino acids, surrounding the SBC, are a proton acceptor. Taking into account all facts [also the absence of charged amino acids in the hydrophobic extracellular internal part of the protein and that the proton is not transiently released to the aqueous phase (24)], we conclude that the only candidate for the proton acceptor is the water cluster in the cavity. Indeed, water molecules were shown to play key roles in functioning of microbial rhodopsins (32). Thus, we suggest that proton is stored in the aqueous phase of the cavity after its release from the RSB and is returned to the RSB in the end of 48C12 photocycle.
To learn more about the movement of the charges inside the protein, we performed time-resolved studies of electrogenic behavior of the protein. On the laser flash illumination (532 nm, 10 ns) of proteoliposomes containing 48C12, a generation of transmembrane electric potential was observed (SI Appendix, Fig. S11). The rise of the membrane potential corresponds to the outward transfer of the positive charge. We observed the major (∼9 µs, ∼70%) and the minor (∼30 µs, ∼30%) parts of the potential increase (SI Appendix, Fig. S11). The characteristic time of the first component coincides with the generation of the M state and corresponds to deprotonation of the RSB in accordance with the photocycle (SI Appendix, Figs. S12 and S13). The minor part may relate to spectroscopically silent conformational relaxation of the proton and charged residues on the M-state formation triggered by deprotonation of the RSB. After that, a drop of membrane potential is observed. The first component of the drop (∼0.5 ms, ∼10 to 20% of the maximum of the potential) coincides with the decay of the M to the O1 states, which corresponds to reprotonation of the RSB in accordance with the photocycle (SI Appendix, Fig. S12) and means the movement of the proton in the opposite direction. The next component of a complete decay of the electric potential (∼500 ms) to zero correlates with the spectroscopic transitions from the O1 to the precursor of the ground state (SI Appendix, Figs. S12 and S13). It is accompanied by the movement of the charged residues to the ground-state positions/states. Remarkably, the movement of the proton in 48C12 is very different from that in all of the known proton pumps. Indeed, in the case of bR (33, 34) and pR (35), similar experiments showed that, on proton translocation, it always moves in one certain direction (inside proteoliposomes in case of bR) during the entire photocycle. In case of 48C12, the direction of the proton movement is reversed on reprotonation of the RSB. The results of this time-resolved study are in favor of our hypothesis that the SBC plays the role of a collective primary proton acceptor.
Structure of 48C12 at Acidic pH.
While the biological function of HeRs remains unknown, thorough study of different 48C12 states is of great potential benefit for elucidating it. To investigate the conformational rearrangements in the HeR associated with pH decrease, we also solved the crystal structure of 48C12 at 1.5 Å using the crystals grown at pH 4.3 (36). Indeed, pH of the surrounding solution affects the functionality and the structure of microbial rhodopsins due to protonation or deprotonation of the key residues (37–39). Moreover, it was shown for bR that the structure of the protein at acidic pH is similar to that of its M state (40). Thus, analysis of HeR structure at low pH may be of high importance for understanding of its biological function and possible rearrangements in protein structure during photocycle. While the crystal packing is the same as in the crystals grown at neutral pH, with one 48C12 dimer in the asymmetric unit, the crystals were colored blue (maximum absorption wavelength of 568 nm) at acidic pH, while at neutral pH, they were violet (maximum absorption wavelength of 552 nm), which corresponds to the color of the wild-type protein in solution under the same conditions (25) (SI Appendix, Fig. S1). We designate these two 48C12 forms as blue and violet, respectively. The color shift is presumably caused by the protonation of the Glu107 residue (24). Key differences between the two structures are shown in Fig. 5. In general, the backbone organization is the same at both acidic and neutral pH values (rmsd between models 0.158 Å); however, the cytoplasmic parts of helices A and B are displaced for 1 to 2 Å, respectively, in the blue form (SI Appendix, Fig. S7).
Fig. 5.
Comparison of the violet (shown in green) and blue (shown in orange) forms of 48C12. (A) Alignment of the two models. The three most notable differences between two structures are 1) the cavity at the cytoplasmic side, 2) rearrangements of the residues near the RSB, and 3) loss of the water molecule between His23 and Ser242 in the blue form and rearrangements of the Gln26 and Trp246 side chains. Water molecules are shown with the spheres and colored green and orange, corresponding to the violet and blue forms of 48C12, respectively. (B) Detailed view of the Ser242-Gln26-Trp246 cluster and His23 in the violet and blue forms. (C) Detailed view of the cavity (active site) at the cytoplasmic side in the violet and blue forms. (D) Detailed view of the RSB and surrounding residues in the violet and blue forms. In violet form, Ser76 assumes two alternative conformations (second is colored magenta for clarity). Cavities are colored pink.
At the cytoplasmic side, the main difference is observed in the organization of the water molecules inside the SBC (Fig. 5C). The hydrogen bonds network propagating from the RSB to Arg104 and Glu230 is present in both models. Interestingly, the difference Fo-Fc electron densities at 1.5-Å resolution indicate the presence of a triangular molecule in the SBC (SI Appendix, Fig. S14F). As the crystallization buffer contained only one molecule of triangular geometry, the acetate anion, the densities were fitted with an acetate (CH3COO−) molecule (Fig. 5 and SI Appendix, Fig. S14E). It fits the density well; however, the acetate can mediate only two hydrogen bonds instead of three bonds necessary to fit the environment of two water molecules 3 and 5 and Glu107 (Fig. 5C and SI Appendix, Fig. S14 C and D). Other molecules that could fit the triangular density and create three hydrogen bonds with water molecules 3 and 5 and Glu107 could be nitric acid (NO3−) or bicarbonate (HCO3−) (SI Appendix, Fig. S14 C and D). This is in line with a very recent publication, where anion binding was shown in the 48C12 mutated at the Glu107 position (41). Particularly, NO3− binding was demonstrated spectroscopically in the E107A and E107Q mutants, which imitate the wild-type protein with the protonated (neutralized) Glu107.
Transient absorption spectroscopy of the 48C12 in the presence of acetate indicated that, at neutral pH, the anion does not affect the kinetics of the protein (SI Appendix, Figs. S12 and S13). On the contrary, at pH 5.0 we observed ∼1.5 times slowdown of the K/M-state decay and ∼2 times acceleration of the O2-state formation (SI Appendix, Figs. S12 and S13). We also observed a slight (1.5-nm) shift of the maximum absorption wavelength of the 48C12 in the presence of acetate at acidic but not at neutral pH (SI Appendix, Fig. S12). The results of the spectroscopy experiments correlate nicely with the obtained model of the 48C12 at acidic pH. Indeed, the anion does not interact directly with the RSB, which may explain only the small shifts of the maximum-absorption wavelength on anion binding. At the same time, acetate is hydrogen bonded to the neutralized Glu107 side chain, which is possible exclusively at low pH values. This explains the absence of any effects of the acetate on the spectra and kinetics of the 48C12 at neutral pH. Therefore, the structure of 48C12 at acidic pH reveals the structural basis of anion binding in the core of the protein and shows the ability of the HeR to bind the molecules of triangular geometry in the SBC.
While at neutral pH, RSB is stabilized through hydrogen bonding to Ser237 and Glu107, at acidic pH it is still bound to Glu107; however, it is slightly shifted toward Ser111, thus weakening the connection to Ser237 (Fig. 5D). Ser76 is in a single conformation at acidic pH and does not stabilize Glu107 anymore. Ser237 flips from the RSB toward the cavity at the cytoplasmic part of the protein (Fig. 5D). At the same time, His23 is reoriented compared with the structure at neutral pH and forms a hydrogen bond with Ser76 in the blue form. The reorientation of His23 may be caused by protonation of Glu107 (24), by protonation of His23 itself, or by the combination of these events. Nevertheless, the reorientation of His23 toward the extracellular side in the blue form of HeRs results in loss of the water molecule, which is coordinated by Gln26 in the purple form (Fig. 5B). The organization of the Ser242-Gln26-Trp246 cluster, located at the extracellular half of the protein surface inside the hydrophobic membrane, is also disturbed (Fig. 5B). Prominently, Trp246, conserved in most HeRs (SI Appendix, Figs. S8 and S10) and exposed to the surrounding lipid bilayer, loses the hydrogen bond to Gln26 and reorients in the blue form of 48C12. Such reorientation might trigger a signal transduction cascade if HeRs are light sensors (24), similarly to sensory rhodopsins II, where there is also an aromatic amino acid in the helix G, Tyr199, which controls the signal transducer protein (42). Alternatively, a latch-like motion of Trp246 might create a defect in the surrounding lipid membrane and open a pathway toward Gln26, His23, and the retinal.
Comparison of the Structures of the Archaeal and Bacterial HeRs.
Very recently, the structure of the archaeal HeR TaHeR at 2.4-Å resolution was reported by Shihoya et al. (27). This allowed us to compare HeRs from different origin.
In general, structures of 48C12 and TaHeR are similar, with the rmsd between models of 0.66 Å (SI Appendix, Fig. S15 A and B). Both proteins form dimers in the lipid bilayer. The most notable differences occur in the AB-loop localization in which β-turn in case of 48C12 is moved closer to the nearby protomer (SI Appendix, Fig. S15C). The DE loop is slightly longer in 48C12 and is displaced by around 9 Å in comparison with TaHeR (SI Appendix, Fig. S15C).
The organization of the inner parts of both proteins is also similar (SI Appendix, Fig. S15F). Side chains of Arg104 (Arg105 in TaHeR), Glu107 (Glu108 in TaHeR), Glu230 (Glu227 in TaHeR), Glu149 (Glu150 in TaHeR), and Tyr217 (Tyr214 in TaHeR) are slightly different in the models; however, the overall configuration is similar (SI Appendix, Fig. S15F). The polar cavity, similar to the SBC of the 48C12, is found in TaHeR near the RSB and filled with water molecules. It should be noted that the number of waters inside the proteins (and particularly, in the cavity) and also, those bound at the protein surface are much higher in the model of 48C12, most probably due to higher resolution of the model.
Surprisingly, the structure of 48C12 revealed similar fenestration on the surface of the protein as that found in TaHeR (SI Appendix, Figs. S6 and S15 D and E). As described in more details in ref. 27 and in Dimerization Interface of 48C12, the fenestration is occluded by the hydrocarbon chain in both proteins. However, in case of the archaeal protein, lipid molecule goes through the fenestration, while in 48C12, the end of the hydrocarbon chain is well ordered in the concavity (SI Appendix, Fig. S6). Thus, the fenestration is tighter in case of 48C12, presumably due to the presence of two bulky hydrophobic residues, Ile170 and Phe203, and the positions of Phe172 and Ala200 of the TaHeR (SI Appendix, Fig. S15 D and E).
Structure-Based Bioinformatic Analysis of HeRs.
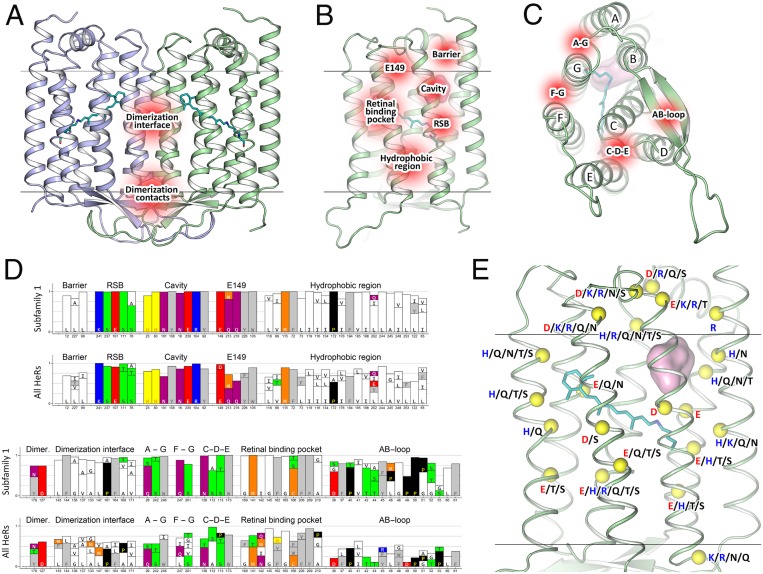
The structures of 48C12 allowed us to identify amino acid residues, comprising the key regions of the HeR (Fig. 6). Based on the comparison of these amino acids in different HeRs, we classified HeRs in 10 subfamilies with potentially different properties. The subfamilies are presented in a phylogenetic tree (SI Appendix, Fig. S16). The groups that contain less than 10 members were merged into “unsorted proteins.”
Fig. 6.
Key regions of 48C12 and HeRs family identified by structure-based bioinformatical analysis. (A) View of the 48C12 dimer with identified regions (shown in red) composed of conservative residues of the dimerization interface (polar, responsible for contacts between protomers and hydrophobic, responsible for hydrophobic interaction inside the membrane). (B) View of the 48C12 protomer with the key regions (shown in red) composed of conservative residues. (C) View of the 48C12 protomer with the polar clusters (shown in red) shown to be conservative among subfamily 1 and most of other subfamilies. (D) Most conservative residues among subfamily 1 and all HeRs, comprising the key regions of 48C12. (E) The location of the residues in 48C12, which are probable analogs of charged or polar residues in other subfamilies of HeRs (selected using a sequence alignment) (SI Appendix, Figs. S16 and S17). The backbone carbon atoms of these residues are shown with yellow spheres. The polar cavity in the cytoplasmic part is shown with a pink surface. Cofactor retinal is colored teal.
The group of 48C12 (subfamily 1) is the largest and comprises 195 proteins of the 479 unique sequences of HeRs currently available (24, 43). The majority of HeRs of subfamily 1 have bacterial origin, with most of them from Actinobacteria. However, representatives of the subfamily are also found in Chloroflexi and Firmicutes of the Terrabacteria group and also, in Proteobacteria and the Planctomycetes, Verrucomicrobia, and Chlamydiae (PVC) group. The host of the unique protein A0A0L0D8K8 is a eukaryote Thecamonastrahens. Importantly, the sequences belong to both gram-positive and gram-negative bacteria, which is inconsistent with the previously made conclusion (26).
Those residues that are conservative in the most of the proteins were identified (SI Appendix, Fig. S10). The alignment of the 10 most distinct HeRs of subfamily 1 is shown in SI Appendix, Fig. S17. Using the structure of 48C12 as a reference, we identified the following regions of protein composed of conserved residues as potentially important for the function of 48C12, correspondingly for the whole 48C12 subfamily, and in some cases, for all HeRs (Fig. 6).
Namely, HeRs have a conservative pattern of the residues that stabilize the RSB (Ser237, Glu107, Ser111, and Ser76) (SI Appendix, Figs. S17 and S18). The SBC and surrounding charged and polar residues (His23, His80, Asn101, Tyr108, Asn16, Glu230, Arg104, Tyr92) together with residues Leu12, Leu96, and Leu227, forming a hydrophobic barrier between the cavity and the cytoplasm, are almost completely conserved in subfamily 1 (SI Appendix, Figs. S17 and S18). The polar region near Glu149 and Arg104 is also conserved (Glu149, Gln216, Tyr226, Trp105, Gln213) (SI Appendix, Figs. S17 and S18). We found that the common feature of HeRs is the hydrophobic organization of the extracellular internal part (Fig. 3 and SI Appendix, Figs. S17 and S18). Indeed, only a few HeR subfamilies have members with charged or polar residues in this region. This fact is very interesting from the functional point of view and will be discussed in the following paragraphs. As it was already mentioned, 48C12 has three clusters composed of polar residues (Gln26/Ser242/Trp246, Gln247/Ser201, and Ser112/Ser113/Asn138/Trp173), which are structurally important presumably for the interactions between the helices and for stabilization of the protein. Importantly, many residues of the characteristic for 48C12 long AB loop and dimerization interface are conserved within subfamily 1. To determine whether the same regions and residues are conserved within other HeR subfamilies, we performed additional bioinformatic analysis of the whole family.
Comparison of HeRs Subfamilies.
The most conservative residues in HeR family are similar to those of subfamily 1 with some variations. The filling of the cytoplasmic part, particularly the RSB region, the SBC, the hydrophobic barrier separating the cavity from the cytoplasm, and the region near Glu149, as well as the hydrophobic extracellular configuration are highly conserved within all HeRs (SI Appendix, Figs. S8 and S10). These regions include such polar and charged residues as Ser237, Glu107, Ser111, Ser76, His23, His80, Asn101, Tyr108, Asn16, Glu230, Arg104, Tyr92, Glu149, Gln216, Tyr226, Trp105, and Gln213, which were shown to be structurally important in 48C12 (SI Appendix, Figs. S8 and S10). Indeed, only a few HeR subfamilies have variations in the listed parts (SI Appendix, Fig. S19). It should be noted that, although Gln213 is almost completely conserved among HeRs of subfamily 1, methionine is an often variant for this position in HeRs. In addition, the analogs of the residues, comprising the clusters at the surface of 48C12 (Gln26/Ser242/Trp246 and Gln247/Ser201), are present in most of the HeRs.
The differences between HeR subfamilies were identified by a comparison of the residues, comprising structurally important regions in 48C12. In general, amino acids responsible for dimerization are not conserved in all HeRs. However, in most cases, analogs of Tyr179 and Asp127 are present (except subfamily 2), but hydrophobic residues of the dimerization interface are different in almost all of the groups. The AB loop is conserved only within some subfamilies but varies notably from group to group in size and amino acid composition. Despite this, residue Pro40 is highly conserved among all HeRs and is part of a β-sheet of the AB loop of 48C12.
Subfamily 2 comprises 19 members and mostly consists of viral proteins, but there are two representatives of Euryarchaeota; the bacterial PVC group and eukaryota are also presented with one protein. We found that this group is the most distinct from all others, especially in the organization of the extracellular part and the retinal binding pocket. Interestingly, one of the members of this group has two Asn residues near the cytoplasmic inner cavity in the positions of His23 and His80 of 48C12. A lot of its members have glutamate in helix F in the position of Leu202 in 48C12, which belongs to its hydrophobic extracellular part. There are no analogs in microbial rhodopsins for Glu202, which thus may be a key determinant of the subfamily 2 protein function. A highly conserved Pro172, which makes a π-bulge in helix E of 48C12, also characteristic only for HeRs, is absent in group 2; however, proline is present in position 168 (helix E) of 48C12 in almost all of its members. This alteration may change the shape of helix E and affect the folding of the protein. The retinal binding pocket in HeRs of group 2 is extremely different from that of other subfamilies, especially due to the presence of positively charged His residues in positions 162 and 166 of the reference protein 48C12. Analogs of Asn138 are also absent in group 2.
Subfamilies 3, 4, and 5 have variations from 48C12 in the retinal binding pocket. Particularly, methionine and asparagine in subfamily 3 are placed in the positions of Gln213 and Ile142 of 48C12, respectively. The same asparagine is present in groups 4 and 5; however, it alternates with asparagine in the position of Asn138, and thus, only the Asn residue is present near the β-ionone ring of the retinal.
Subfamilies 7, 8, and 9 have a very interesting feature of conservative Tyr in position 202 of 48C12. Asn is present in the position of Ile142 of 48C12 in all members of groups 8 and 9 and in some representatives of subfamily 7. Group 9 also has no analogs of Asn138 of 48C12.
Unsorted proteins group includes the most different HeRs (SI Appendix, Fig. S20). These proteins presumably maintain the polar cavity in the cytoplasmic part; however, its surroundings are varied. Most interesting, in subgroup U1, histidine is present in the position of Asn16 in addition to two histidines in the positions of His23 and His80 of 48C12. Moreover, at the extracellular side, two glutamates are present in all members of subfamily U1 in the positions of Leu73 and Ile116 of 48C12. Glutamate is also found in the positions of Pro172 (subfamily U2), Val69, and Leu202 (subfamily U8) of 48C12. Positively charged residues also appear in the extracellular side of the members of subfamilies U1, U6, U7, and U11, such as His residues in the position of Leu73 and Arg and Lys residues in the position of Leu253 of 48C12. The positions of 48C12, with analogs in other HeRs that are occupied by unusual charged or polar residues, and possible variants of those residues are shown in Fig. 6. These charged amino acids, especially located in the extracellular part of the proteins, may be crucial for the functions of those HeRs.
Subfamilies 3, 5, 6, 8, U1, U2, U3, U4, U5, U6, and U12 consist exclusively of bacterial proteins. Subfamilies 4, 7, 9, and U13 represent archaeal HeRs (except one bacterial protein from subfamily 4 and one from subgroup U13), mostly Euryarchaeota, but subfamily 7 also has members of Asgard and TACK groups. Subfamilies U8, U9, and U11 comprise proteins of eukaryotic origin.
Discussion
Molecular Mechanisms and Biological Function(s) of HeRs.
The biggest surprise of the first studies of HeRs (the studies of 48C12) is that the attempts to identify amino acids playing the roles of primary proton acceptor and proton donor to the RSB failed (24, 25). Such amino acids are key functional determinants in all known rhodopsins. Another important fact is that Pushkarev et al. (24) did not observe any translocation of the proton (an ion) through the protein to its polar surfaces. High-resolution crystallographic structures of 48C12 HeR, which represents the most abundant subfamily of HeRs (195 of 479 currently known unique sequences), were solved at 1.5-Å resolution with the crystals obtained at pH 8.8 and 4.3, respectively. The structures correspond to the two different forms of the protein. Both structures show remarkable difference between HeRs and all of the known rhodopsins. The retinal binding pocket and the parts of the cytoplasmic and extracellular regions of the protein, which are determinants of the function of the known rhodopsin, are also different. There is no analog to this protein among other type 1 (microbial) and type 2 (visual) rhodopsins.
In the cytoplasmic part of the protein, a large cavity (SBC), filled with six water molecules at pH 8.8, is located close to the RSB. The SBC is surrounded by highly conservative charged amino acids His23, His80, Arg104, Glu107, and Glu230, the protonated RSB and also by polar residues Asn16, Tyr92, Asn101, Tyr108, and Ser237. The amino acids and the RSB are interconnected by an extensive hydrogen network mediated by the water molecules (Fig. 2). There are two pathways from the cavity to the bulk. From one of the sides, the cavity is separated from the bulk by only the Asn101 residue found on the surface of the protein at the level of the hydrophobic/hydrophilic interface (Fig. 4 and SI Appendix, Fig. S21). From the other side, the cavity is delineated by Arg104, found in almost all rhodopsins as a major gate between the RSB and the bulk.
The major difference between the two structures is that, at lower pH, the SBC comprises a planar triangle-shaped molecule in the cavity. Remarkably, several residues mentioned above (namely His23, His80, Arg104, Glu107, Tyr108, Ser237) were subjected to alanine substitution (25), which in all cases, led to the changes of absorption spectra. This result supports the presence of a strong interaction of the SBC with the RSB. In its turn, this means that isomerization of the retinal modifies the properties of the SBC since the base is directly connected to the cavity through Glu107 via a hydrogen bond (Figs. 3B and 5D).
The structure also suggests why the previous attempts to identify the proton acceptor and the proton donor have failed (24, 25). One of the reasons is that one of the possible amino acid candidates for these roles (for instance, Glu230, which is the key member of the active site) was overlooked in the previous studies because of poor prediction of the protein topology in the membrane (figure 1 in ref. 25). However, additional mutational analysis suggests that the cluster of water molecules in the SBC plays a role of a reservoir for the proton dissociated from the RSB. Importantly, the transfer of the RSB proton to the hydrophobic extracellular part of the protein on isomerization of the retinal seems to be problematic due to high free energy penalty. We suppose that the RSB proton dissociates on isomerization of the retinal but does not leave the SBC during the photocycle. We also suggest that the SBC may play the role of an “active site” for substrate binding inside 48C12. In the latter case, the proton released from RSB during the photocycle might interact with the substrate in the reaction H+ + substrate− → reduced substrate, like in carbon fixation, which is known as one of the most important biosynthetic processes in biology (44).
Since the extracellular part of the protein is highly hydrophobic, the transfer of the RSB proton on isomerization of the retinal to this part of the protein is energetically unfavorable. It means that, in opposite to bR, the RSB proton of 48C12 does not follow the RSB on retinal isomerization, but it dissociates from the base, remains in the cytoplasmic part of the protein, and is temporarily accommodated in the cavity. Then, the proton moves back on reisomerization of the retinal and reprotonates the RSB.
Biological Role of HeR Subfamily 1.
At this point, it is difficult to establish the primary role of HeR, even of the best studied subfamily 1. Pushkarev et al. (24) suggested that HeRs may function as sensory proteins. This conclusion was based on two observations. First, the authors did not detect any ion translocation activity of the protein (under experimental conditions corresponding to pH 8.1). Second, the photocycle of the protein (measured at pH 8.5) was several seconds long, which is characteristic for sensory rhodopsins (3). Along these lines, we noted above that HeR possesses a hydrogen bond-forming aromatic amino acid Trp246 that faces the membrane and that might change its conformation under illumination, similarly to Tyr199 in NpSRII (42, 45). Usually, the genes of sensory rhodopsins have a gene coding for a signal transducer protein located nearby and often cotranscribed (46, 47). At present, two distinct types of sensory rhodopsins are known: SRII-like photoreceptors utilize transmembrane chemoreceptor-like transducer proteins, whereas Anabaena sensory rhodopsin (ASR) utilizes a soluble transducer protein that dissociates from ASR on illumination (3). In case of subfamily 1, however, no conserved proteins, which could potentially be signal transducers, could be detected in the genomic neighborhood. On the other hand, in some microbes (aquatic actinobacteria, such as the marine Candidatus Actinomarina) that most often contain HeRs, their genes are surrounded by two large clusters of Nuo genes (SI Appendix, Fig. S22), the products of which are the key proteins in respiratory chains. At this point, it is unclear what this might mean. However, the ability of HeRs to bind the triangular anions, like carbonate, in the SBC suggests the possibility of its involvement in carbon fixation.
The analysis of the presence/absence of HeRs in monoderm and diderm representatives of the Tara Oceans and 25 freshwater lakes metagenomes led to the conclusion that HeRs were absent in diderms, confirming their absence in cultured Proteobacteria. Judging by a specific semipermeability of outer membranes of diderms, the authors proposed a role of HeRs in light-driven transport of amphiphilic molecules (26). However, the structures of 48С12 do not support such function for HeRs. Moreover, according to the literature data, we conclude that, in fact, there is no clear evidence of the HeRs presence only in monoderms (48). For example, some of the proteins of subfamily 1 are originating from Bacteroidetes, Gemmatimonadetes, and Proteobacteria, all of which are assumed to be diderms. Some HeRs from other subfamilies are found in the phyla Thermotogae and Dictioglomy, which also have diderm cells.
Although at this point, we cannot provide a definitive role for HeR, we would like to advance a hypothesis. In many (if not most) cases, HeRs are found in pelagic microbes living in the photic zone of aquatic habitats (freshwater or marine). They appear in microbes that often also contain a classical rhodopsin, typically a proton pump, providing the cell with unlimited energy as long as there is light. The transfer of the proton from the retinal to the interior of the cell, likely reducing a molecule of carbonate or nitrate, might act like cyanobacterial (or plant) photosystem II, transforming light energy into reducing power to form precursors of cell biomass. This would transform the microbes containing the two kind of rhodopsins in primary producers, like cyanobacteria, and would help explain the extraordinary success of some of them, such as the actinobacteria that are the most abundant microbes in most photic freshwater habitats. Further structure-guided functional studies are necessary to clarify the biological role (roles) of this unusual family of rhodopsins.
Materials and Methods
Protein Expression and Purification.
The gene of HeR-48C12 (UniProt ID no. A0A2R4S913; NIH GenBank accession no. AVZ43932.1) was synthesized de novo and optimized for expression in Escherichia coli with Thermo Fisher Scientific GeneOptimizer service. The optimized gene was introduced into StabyCodon T7 expression plasmid system (Delphi Genetics) via NdeI and XhoI (Thermo Fisher Scientific) that led to the addition of 6×His tag to the C terminus of the gene. The resulting plasmid DNA was sequenced (Eurofins Genomics) and used to transform E. coli C41 strain.
The protein expression procedure is adopted from ref. 49 but slightly modified. The culture was cultivated at 37 °C in the autoinduction media (1% [wt/vol] Trypton, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] glycerol, 0.05% [wt/vol] glucose, 0.2% [wt/vol] lactose, 10 mM (NH4)2SO4, 20 mM KH2PO4, 20 mM Na2HPO4 adjusted pH 7.8) containing 150 µg/mL ampicillin antibiotic to optical density600(OD600) = 0.8. After the cultivation temperature was decreased to 26 °C with subsequent addition of 150 µg/mL ampicillin, 20 µM all-trans Retinal (solubilized in Triton X-100 detergent), and 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), the culture was grown overnight. The concentration of antibiotic after induction was maintained with addition of an extra 150 µg/mL each 2 h.
The cells were then collected and disrupted at 20,000 psi with an M-110P homogenizer (Microfluidics) in the buffer containing 30 mM Tris⋅HCl, pH 8.0, 0.3 M NaCl, 0.04% Triton X-100, 50 mg/L DNase I (Sigma-Aldrich), and cOmplete protease inhibitor mixture (Roche). The total cells’ lysate was ultracentrifugated at 120,000 relative centrifugal force (rcf). Then, membranes were isolated, dispensed in the same buffer without DNase (with addition of 1% [wt/wt] n-dodecyl β-D-maltoside [DDM] detergent and 5 mM all-trans retinal), and stirred overnight at 4 °C.
The nonsoluble fraction was separated by ultracentrifugation at 120,000 rcf for 1 h at 4 °C. The resulting soluble protein mixture was loaded to nickel-charged affinity resin (Ni-NTA) resin (Cube Biotech). The column with loaded resin was washed with 3 column volume (CV) of washing buffer WB1 (30 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 10 mM Imidazole, 0.05% Triton, 0.2% DDM) and washing buffer WB2 (30 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 50 mM Imidazole, 0.05% Triton, 0.2% DDM). Then, HeR was eluted with elution buffer (EB) (30 mM Tris-HCl, pH 7.8, 0.3 M NaCl, 250 mM l-Histidine [AppliChem], 0.05% Triton, 0.1% DDM). The eluted protein mixture was subjected to the size exclusion chromatography column Superdex200 Increased 10/300 GL (GE Health Care Life Sciences) preequilibrated with size-exclusion chromatography (SEC) buffer (30 mM Tris-HCl, 50 mM NaPi, pH 7.8, 300 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 2 mM 6-aminohexanoic acid, 0.075% DDM). The fractions were analyzed; those containing the 48C12 rhodopsin with peak ratio of ∼1.25 and lower were collected, and protein was concentrated to 20 mg/mL with 50-kDa concentration tubes at 5,000 rcf and flash cooled with liquid nitrogen.
Flash Photolysis Setup.
The laser flash photolysis was similar to that described by Chizhov and coworkers (50–52) with minor differences. The excitation system consisted of Nd:YAG laser Q-smart 450 mJ with OPO Rainbow 420- to 680-nm range (Quantel). Samples were placed into a 5 × 5-mm quartz cuvette (Starna Scientific) and thermostabilized via sample holder qpod2e (Quantum Northwest) and Huber Ministat 125 (Huber Kältemaschinenbau AG). The detection system beam emitted by 150-W xenon lamp (Hamamatsu) housed in LSH102 universal housing (LOT Quantum Design) passed through a pair of Czerny–Turner monochromators MSH150 (LOT Quantum Design). The received monochromatic light was detected with photomultiplier tubes (PMT) R12829 (Hamamatsu). The data recording subsystem was represented by a pair of DSOX4022A oscilloscopes (Keysight). The signal offset was measured by one of oscilloscopes, and the PMT voltage was adjusted by Agilent U2351A DAQ (Keysight). The absorption spectra of the samples were measured before and after each experiment on an Avaspec ULS2048CL fiber spectrophotometer paired with AVALIGHT D(H)S Balanced light source.
Preparation of Samples for Flash Photolysis.
The wild-type protein sample for flash photolysis assay was purified in the same manner as for crystallization but increased from 0.3 to 0.6 M NaCl concentration on each purification step. The purified wild-type protein was 100× diluted in buffer containing 30 mM Hepes, pH 7.0, 1 M NaCl, and 2% DDM to concentration of ca. 0.5 mg/mL The measurement was performed in the following way. The 350-μL sample was placed into the 5-mm light path cuvette, and the temperature of the sample was set to 20 °C. Then, the protein sample was exposed to a 6-ns pulse of mean of 3.5 mJ (SD 6% on 1,000 pulses) at 545 nm. The transient absorption changes data were recorded (in 350- to 700-nm light range; step 10 nm) from 1 microsecond (mks) up to 5 s with two oscilloscopes with overlapping ranges (range ratio 1:1,000) and averaged for 20 pulses for each wavelength. The data compression reduced the initial number of data points per trace to ca. 900 points. The samples of E230Q and E149Q mutant proteins were prepared without purification similar to ref. 25 with modification. The E. coli C41 cells were disrupted at 20,000 psi with M-110P homogenizer in buffer containing 30 mM Tris⋅HCl, pH 8.0, 1 M NaCl, and DNase I, and nonsoluble fraction was sedimented at 120,000 rcf. The 5 g of the membranes were then washed and resuspended in 20 mL of the buffer containing 30 mM Hepes, pH 7.0, and 1 M NaCl. After homogenization, the 2% DDM was added to 1.6 mL of suspension, and the sample was incubated for 30 min at 4 °C. Later, samples were applied to the centrifugation (for 10 min at 4 °C, 15,000 rcf), and supernatant was collected for characterization. The flash photolysis measurement of E230Q/E149Q mutant-containing samples was performed at 400 to 610 nm (step 70 nm; each reading averaged for 20 pulses) at 20 °C using 6-ns excitation pulses of 3.5 mJ at 545 nm.
Proteoliposomes Preparation.
Liposomes were produced from asolectin (20 mg/mL; type IVS, 40% [wt/wt] phosphatidylcholine content; Sigma) by sonication (at 22 kHz, 60 μA) for 2 min in 1 mL of 25 mM Hepes-NaOH buffer, pH 7.5. Reconstitution of the protein into liposomes was carried out by mixing the liposomes with protein in 1.5% (wt/vol) octyl β-D-glucopyranoside (OG) at the lipid/protein ratio of 100:1 (wt/wt) for 30 min in the dark. Removal of detergent was performed according to using Bio-Beads SM-2 absorbent (Bio-Rad). The detergent was removed by addition of a 20-fold excess of Bio-Beads (by weight) and stirring the suspension for 3 h at room temperature.
Electrometric Time-Resolved Measurements of the Membrane Potential.
Generation of the transmembrane electric potential difference ΔΨ was studied using a direct electrometric setup with time resolution of 100 ns as described in refs. 33 and 34. This technique includes fusion of the proteoliposomes with the surface of a collodion phospholipid-impregnated film (a membrane) separating two sections of the measuring cell filled with a buffer solution. The membrane should be thin enough and possess large electric capacitance (about 5 nF) for detecting fast charge translocation events. A pulsed Nd-YAG laser (YG-481, λ = 532 nm, pulse half-width 12 ns, flash energy up to 40 mJ; Quantel) was used as a source of flashes. In the process of the light-driven proton transfer, 48C12 creates ΔΨ across the vesicle membrane, which is proportionately divided with the measuring membrane and thus, can be detected by Ag+/AgCl electrodes immersed in a solution at different sides of the membrane. Typically, the measuring membrane has high resistance of 2 to 3 GOhm, and the light-induced ΔΨ decays with a time constant of several seconds generation.
Crystallization.
The crystals were grown with an in meso approach (45, 53), similar to that used in our previous work (17, 28). The solubilized protein in the crystallization buffer was mixed with premelted at 42 °C monoolein (Nu-Chek Prep) to form a lipidicmesophase. The 150-nL aliquots of a protein–mesophase mixture were spotted on a 96-well lipidic cubic phase (LCP) glass sandwich plate (Marienfeld) and overlaid with 500 nL of precipitant solution by means of the NT8 crystallization robot (Formulatrix). The best crystals of the violet form were obtained with a protein concentration of 20 mg/mL and the precipitant solution of 2.0 M ammonium sulfate and 0.1 M Tris⋅HCl, pH 8.8. For the blue form, the best crystals were grown with the same protein concentration of 20 mg/mL and the precipitant solution of 2.0 M ammonium sulfate and 0.1 M sodium acetate, pH 4.3. The crystals were grown at 20 °C to observable size in 2 wk for both types. The rhombic-shaped crystals reached 150 μm in length and width, with maximum thickness of 20 μm. Crystals of both forms were incubated for 5 min in cryoprotectant solution (2.0 M ammonium sulfate, 0.1 M Tris⋅HCl, pH 8.8 for the violet form and 2.0 M ammonium sulfate, 0.1 M sodium acetate, pH 4.3, for the blue form supplied with 20% [wt/vol] glycerol) before harvesting. All crystals were harvested using micromounts (MiTeGen), and they were flash cooled and stored in liquid nitrogen. Absorption spectra from the 48C12 crystals were measured at ID29s beamline of the European Synchrotron Radiation Facility, Grenoble, France, at 300 K (54).
Collection and Treatment of Diffraction Data.
X-ray diffraction data were collected at Proxima-1 beamline of the SOLEIL, Saint-Aubin, France, at 100 K with an EIGER 16M detector and at P14 beamline of the PETRAIII, Hamburg, Germany, at 100 K with an EIGER 16M detector. We processed diffraction images with XDS (55) and scaled the reflection intensities with AIMLESS from the CCP4 suite (56). The crystallographic data statistics are presented in SI Appendix, Table S1. The molecular replacement search model was generated by RaptorX web server (57) based on the exiguobacterium sibiricum rhodopsin (ESR) structure [Protein Data Bank (PDB) ID code 4HYJ (6)]. Initial phases were successfully obtained in P21 space group by the molecular replacement using phenix.mr_rosetta (58) of the PHENIX (59) suite. The initial model was iteratively refined using REFMAC5 (60), PHENIX, and Coot (61). The cavities were calculated using HOLLOW (31). Hydrophobic–hydrophilic boundaries of the membrane were calculated using PPM server (62).
Bioinformatics Analysis.
Multiple amino acid alignment was performed using Clustal Omega algorithm (63). The HeRs database was downloaded from InterPro (44) and merged with the database provided in original article (24). Phylogenetic tree was constructed, and classes were identified using iTOL server software version 4.3.2 (64). For removing those proteins above certain similarity threshold, we used CD-HIT suite (65). Cutoff similarity threshold is always specified. Calculations of conservative amino acids were performed using an in-house C# application written using Visual Studio Community 2017. Most conservative regions were identified, and normalized results were visualized using in-house Wolfram Mathematica Notebooks. Genome sequence of the single-amplified genome AG-333-G23, belonging to the marine Ca. Actinomarinales group, was downloaded from the National Center for Biotechnology Informatio (NCBI) database (biosample no. SAMN08886063). Encoded genes were predicted using Prodigal v2.6 (66). transport RNA (tRNA) and ribosomal RNA (rRNA) genes were predicted using tRNAscan-SE v1.4 (67), ssu-align v0.1.1 (68), and metarna (69). Predicted protein sequences were compared against the NCBI nr database using DIAMOND (70) and against COG (71) and TIGFRAM (72) using HMMscan v3.1b2 (73) for taxonomic and functional annotation. A custom database containing both type I and type III rhodopsins (24) was used to identify putative homologs. Resulted significative genes (HMMscan, E value 1e-15) were then confirmed by determining the secondary structure and the presence of domains with InterPro (43).
Supplementary Material
Acknowledgments
We thank O. Volkov and A. Yuzhakova for technical assistance. We also thank A. Royant (ID29S Cryobench Laboratory, European Synchrotron Radiation Facility) for help with collection of the 48C12 crystal optical absorption spectra. We acknowledge the Structural Biology Groups of the Swiss Light Source (Villigen, Switzerland), SOLEIL (Saint-Aubin, France), and PETRAIII (Hamburg, Germany) for granting access to the synchrotron beamlines. This work was supported by the common program of Agence Nationale de la Recherche (ANR), France and Deutsche Forschungsgemeinschaft, Germany Grants ANR-15-CE11-0029-02/FA 301/11-1 and MA 7525/1-1 and by funding from Frankfurt: Cluster of Excellence Frankfurt Macromolecular Complexes by the Max Planck Society (E.B.) and Commissariat à l’EnergieAtomique et aux Energies Alternatives (Institut de Biologie Structurale)–Helmholtz-Gemeinschaft Deutscher Forschungszentren (Forschungszentrum Jülich) Special Terms and Conditions 5.1 Specific Agreement. This work used the platforms of the Grenoble Instruct-ERIC (European Research Infrastructure Consortium) Center (Integrated Structural Biology Grenoble; UMS 3518 CNRS-CEA-UJF-EMBL) within the Grenoble Partnership for Structural Biology. Platform access was supported by French Infrastructure for Integrated Structural Biology (FRISBI) Grant ANR-10-INBS-05-02 and Grenoble Alliance for Integrated Structural Cell Biology (GRAL), a project of University Grenoble Alpes Graduate School (Ecoles Universitaires de Recherche) CBH-EUR-GS Grant ANR-17-EURE-0003. The reported study (particularly spectroscopy of the protein crystals) was funded by Russian Foundation for Basic Research (RFBR) and CNRS according to Research Project 19-52-15017. Measurements of electrogenic properties were supported by RFBR Grant 18-04-00503a (to S.S.). Bioinformatics search of new HeRs (including viral proteins) was supported by Russian Science Foundation (RSF) Project 19-44-06302. F.R.-V. thanks L’Application des Epreuves Individuelles/ The Spanish Federation of Rare Diseases (AEI/FEDER), European Union (EU) Grant ‘VIREVO’ CGL2016-76273-P (cofunded with FEDER funds).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The crystal structure of the 48C12 heliorhodopsin in the violet form at pH 8.8 reported in this paper has been deposited in the Protein Data Bank (PDB, ID code 6SU3). The crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 reported in this paper has been deposited in the PDB (ID code 6SU4).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915888117/-/DCSupplemental.
References
- 1.Shalaeva D. N., Galperin M. Y., Mulkidjanian A. Y., Eukaryotic G protein-coupled receptors as descendants of prokaryotic sodium-translocating rhodopsins. Biol. Direct 10, 63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oesterhelt D., Stoeckenius W., Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 (1971). [DOI] [PubMed] [Google Scholar]
- 3.Gushchin I., Gordeliy V., “Microbial rhodopsins” in Membrane Protein Complexes: Structure and Function, Harris J. R., Boekema E. J., Eds. (Subcellular Biochemistry, Springer, Singapore, 2018), pp. 19–56. [DOI] [PubMed] [Google Scholar]
- 4.Béjà O., Lanyi J. K., Nature’s toolkit for microbial rhodopsin ion pumps. Proc. Natl. Acad. Sci. U.S.A. 111, 6538–6539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst O. P., et al. , Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gushchin I., et al. , Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 12631–12636 (2013). Correction in: Proc. Natl. Acad. Sci. U.S.A.110, 14813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieszke J. A., Spudich E. N., Scott K. L., Borkovich K. A., Spudich J. L., A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38, 14138–14145 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Beja O., et al. , Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Nagel G., et al. , Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deisseroth K., et al. , Next-generation optical technologies for illuminating genetically targeted brain circuit. J. Neurosci. 26, 10380–10386 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bamann C., Nagel G., Bamberg E., Microbial rhodopsins in the spotlight. Curr. Opin. Neurobiol. 20, 610–616 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Nagel G., et al. , Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15, 2279–2284 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K., Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Govorunova E. G., Sineshchekov O. A., Li H., Spudich J. L., Microbial rhodopsins: Diversity, mechanisms, and optogenetic applications. Annu. Rev. Biochem. 86, 845–872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandori H., Ion-pumping microbial rhodopsins. Front. Mol. Biosci. 2, 52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon S. K., et al. , Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol. Evol. 5, 187–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gushchin I., et al. , Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 22, 390–395 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Yutin N., Koonin E. V., Proteorhodopsin genes in giant viruses. Biol. Direct 7, 34 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bratanov D., et al. , Unique structure and function of viral rhodopsins. Nat. Commun. 10, 4939 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Needham D. M., et al. , A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc. Natl. Acad. Sci. U.S.A. 116, 20574–20583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Béjà O., Spudich E. N., Spudich J. L., Leclerc M., DeLong E. F., Proteorhodopsin phototrophy in the ocean. Nature 411, 786–789 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Inoue K., et al. , A natural light-driven inward proton pump. Nat. Commun. 7, 13415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko V., et al. , Inward H+ pump xenorhodopsin: Mechanism and alternative optogenetic approach. Sci. Adv. 3, e1603187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pushkarev A., et al. , A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558, 595–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh M., Inoue K., Pushkarev A., Béjà O., Kandori H., Mutation study of heliorhodopsin 48C12. Biochemistry 57, 5041–5049 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Flores-Uribe J., et al. , Heliorhodopsins are absent in diderm (Gram-negative) bacteria: Some thoughts and possible implications for activity. Environ. Microbiol. Rep. 11, 419–424 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Shihoya W., et al. , Crystal structure of heliorhodopsin. Nature 574, 132–136 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Volkov O., et al. , Structural insights into ion conduction by channelrhodopsin 2. Science 358, eaan8862 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Kovalev K., Volkov D., Astashkin R., Alekseev A., Gushchin I., Gordeliy V., Crystal structure of the 48C12 heliorhodopsin in the violet form at pH 8.8. Protein Data Bank. https://www.rcsb.org/structure/6su3. Deposited 12 September 2019.
- 30.von Heijne G., Gavel Y., Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174 671–678 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Ho B. K., Gruswitz F., HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerwert K., Freier E., Wolf S., The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta 1837, 606–613 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Drachev L. A., et al. , Direct measurement of electric current generation by cytochrome oxidase, H+-ATPase and bacteriorhodopsin. Nature 249, 321–324 (1974). [DOI] [PubMed] [Google Scholar]
- 34.Drachev L. A., Kaulen A. D., Khitrina L. V., Skulachev V. P., Fast stages of photoelectric processes in biological membranes. I. Bacteriorhodopsin. Eur. J. Biochem. 117, 461–470 (1981). [DOI] [PubMed] [Google Scholar]
- 35.Siletsky S. A., et al. , Electrogenic steps of light-driven proton transport in ESR, a retinal protein from Exiguobacterium Sibiricum. Biochim. Biophys. Acta 1857, 1741–1750 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Kovalev K., Volkov D., Astashkin R., Alekseev A., Gushchin I., Gordeliy V., Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3. Protein Data Bank. https://www.rcsb.org/structure/6su4. Deposited 12 September 2019.
- 37.Kovalev K., et al. , Structure and mechanisms of sodium-pumping KR2 rhodopsin. Sci. Adv. 5, eaav2671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Der A., et al. , Alternative translocation of protons and halide ions by bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 88, 4751–4755 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris A., et al. , A new group of eubacterial light-driven retinal-binding proton pumps with an unusual cytoplasmic proton donor. Biochim. Biophys. Acta 1847, 1518–1529 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Okumura H., Murakami M., Kouyama T., Crystal structures of acid blue and alkaline purple forms of bacteriorhodopsin. J. Mol. Biol. 351, 481–495 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Singh M., Katayama K., Béjà O., Kandori H., Anion binding to mutants of the Schiff base counterion in heliorhodopsin 48C12. Phys. Chem. Chem. Phys. 21, 23663–23671 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Moukhametzianov R., et al. , Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature 440, 115–119 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Mitchell A. L., et al. , InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J., et al. , Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Chang. 2, 106–110 (2012). [Google Scholar]
- 45.Gordeliy V. I., et al. , Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 419, 484–487 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Seidel R., et al. , The primary structure of sensory rhodopsin II: A member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc. Natl. Acad. Sci. U.S.A. 92, 3036–3040 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung K. H., Trivedi V. D., Spudich J. L., Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 47, 1513–1522 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Sutcliffe I. C., New insights into the distribution of WXG100 protein secretion systems. Antonie van Leeuwenhoek 99, 127–131 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Chizhov I., Engelhard M., Temperature and halide dependence of the photocycle of halorhodopsin from Natronobacterium pharaonis. Biophys. J. 81, 1600–1612 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chizhov I., et al. , The photophobic receptor from Natronobacterium pharaonis: Temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75, 999–1009 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chizhov I., et al. , Spectrally silent transitions in the bacteriorhodopsin photocycle. Biophys. J. 71, 2329–2345 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordeliy V. I., et al. , Crystallization in lipidic cubic phases: A case study with bacteriorhodopsin. Methods Mol. Biol. 228, 305–316 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Von Stetten D., et al. , In crystallo optical spectroscopy (icOS) as a complementary tool on the macromolecular crystallography beamlines of the ESRF. Acta Crystallogr. D Biol. Crystallogr. 71, 15–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Källberg M., et al. , Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 7, 1511–1522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terwilliger T. C., et al. , phenix.mr_rosetta: Molecular replacement and model rebuilding with Phenix and Rosetta. J. Struct. Funct. Genomics 13, 81–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chojnacki S., Cowley A., Lee J., Foix A., Lopez R., Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucleic Acids Res. 45, W550–W553 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciccarelli F. D., et al. , Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Huang Y., Niu B., Gao Y., Fu L., Li W., CD-HIT suite: A web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowe T. M., Eddy S. R., TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nawrocki E. P., “Structural RNA homology search and alignment using covariance models,” PhD thesis, Washington University in St. Louis, St. Louis, MO (2009).
- 69.Huang Y., Gilna P., Li W., Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25, 1338–1340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buchfink B., Xie C., Huson D. H., Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Tatusov R. L., et al. , The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haft D. H., et al. , TIGRFAMs: A protein family resource for the functional identification of proteins. Nucleic Acids Res. 29, 41–43 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eddy S. R., Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.