Abstract
Background
Among all cases of nosocomial pneumonia, Staphylococcus aureus is the second most prevalent pathogen (17.8%). In Europe, 29.9% of the isolates are oxacillin-resistant. The changing epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) nosocomial infections and the decreasing susceptibility to first-line antibiotics leave clinicians with few therapeutic options. The objective of our study was to determine the antimicrobial susceptibility, the associated molecular mechanisms of resistance and the epidemiological relatedness of MRSA strains isolated from the endotracheal tubes (ETT) of intubated critically ill patients in the intensive care unit (ICU) with nosocomial pneumonia caused by Staphylococcus aureus.
Methods
The antimicrobial susceptibility to vancomycin, linezolid, ciprofloxacin, clindamycin, erythromycin, chloramphenicol, fusidic acid, gentamicin, quinupristin-dalfopristin, rifampicin, sulfamethoxazole/trimethoprim, and tetracycline were measured. Resistance mechanisms were then analyzed by polymerase chain reaction and sequencing. Molecular epidemiology was carried out by multi-locus sequence typing.
Results
S. aureus isolates were resistant to ciprofloxacin, erythromycin, gentamicin, tetracycline, clindamycin, and fusidic acid. The most frequent mutations in quinolone-resistant S. aureus strains were S84L in the gyrA gene, V511A in the gyrB gene, S144P in the grlA gene, and K401R/E in the grlB gene. Strains resistant to erythromycin carried the ermC, ermA, and msrA genes; the same ermC and ermA genes were detected in strains resistant to clindamycin. The aac(6′)-aph(2″) gene was related to gentamicin resistance, while resistance to tetracycline was related to tetK (efflux pump). The fusB gene was detected in the strain resistant to fusidic acid. The most frequent sequence types were ST22, ST8, and ST217, which were distributed in four clonal complexes (CC5, CC22, CC45, and CC59).
Conclusions
High levels of resistance to second-line antimicrobials threatens the treatment of nosocomial respiratory infections due to methicillin-resistant S. aureus with decreased susceptibility to linezolid and vancomycin. The wide genotypic diversity found reinforces the central role of ICU infection control in preventing nosocomial transmission.
Keywords: Endotracheal tube, Biofilm, Methicillin-resistant Staphylococcus aureus, Respiratory infection, Hospital-acquired pneumonia, Ventilator-associated pneumonia, MLST, Mechanism of resistance, Clonal complexes
Background
Hospital-acquired pneumonia (HAP) and ventilator-associated pneumonia (VAP) are the principal causes of infection among critically ill patients in intensive care units (ICU) [1]. Among all such cases of nosocomial pneumonia, Staphylococcus aureus is the second most prevalent pathogen (17.8%), with 29.9% of the isolates in Europe being oxacillin-resistant [2]. The changing epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) nosocomial infections, the decreasing susceptibility to first-line antibiotics, such as vancomycin-intermediate Staphylococcus aureus (VISA), linezolid resistant MRSA [3], and community-associated MRSA (CA-MRSA) leave clinicians with few therapeutic options. In this context, an accurate description of the mechanisms of antimicrobial resistance in MRSA nosocomial pneumonia in ICU could help in the design of novel therapies. Knowledge of resistance-related phenotypic and genotypic changes is critical for the development of new drugs. When designing a new antibiotic, the previously described resistance mechanisms must be taken into account. The new antimicrobial should be able to overcome the resistance mechanisms, or should be aimed at new targets where the probability that the microorganism has developed resistance is lower [4].
Given that few new antimicrobial agents have been approved in the last 10 years, it is anticipated that the problems associated with resistance will only worsen. Antibiotics currently approved for MRSA nosocomial pneumonia are linezolid (an oxazolidinone), vancomycin (a glycopeptide), ceftobiprole (an extended-spectrum cephalosporin) and Telavancin (a lipoglycopeptide). Tedizolid (a second-generation oxazolidinone) is pending authorization for systemic treatment of HAP [5]. Other secondary options when these agents cannot be used include, either alone or in combination, quinolones (ciprofloxacin or levofloxacin), macrolides (erythromycin), aminoglycosides (gentamicin), tetracyclines, clindamycin (a lincosamide), and fusidic acid.
A wide range of resistance mechanisms have been described for S. aureus including PBP alterations (β-lactam agents), cell wall structure modifications (glycopeptides), point mutations in the quinolone resistance-determining regions of GyrA and GrlA (quinolones), inactivating enzymes (aminoglycosides) ribosome alterations (macrolides, lincosamides, oxazolidones and tetracyclines), efflux pumps (tetracyclines, macrolides, quinolones) or spontaneus mutations in the gene fusA encoding the ribosomal translocase elongation factor G (fusidic acid) [6, 7]. However, little recent information is available on the mechanisms of resistance in S. aureus strains obtained from mechanically ventilated patients, or whether or not these mechanisms are associated with particular circulating S. aureus clones.
The aim of this study was to determine the antimicrobial susceptibility, the associated molecular mechanisms of resistance, and the epidemiological relatedness of MRSA strains isolated from the ETTs of intubated critically ill patients in the intensive care unit (ICU) with nosocomial pneumonia caused by Staphylococcus aureus.
Materials and methods
Study design, sample collection and bacterial isolates
Clinical S. aureus (17 MRSA and three methicillin-susceptible isolates) were collected from ETTs after extubation during a prospective observational study carried out in four European tertiary hospitals from September 2013 to December 2016 [8]. The participating centers were the Hospital Clinic of Barcelona (Spain), the Hospital del Mar (Critical Care Department; Barcelona, Spain), the Hospital Universitario Central de Asturias (Intensive Medicine Service; Oviedo, Spain), and the Fondazione IRCCS Ca′ Granda (Adult Intensive Care; Ospedale Maggiore Policlinico, Milan, Italy). Patients were included if they were older than 18 years, mechanically ventilated (with ≥48 h of orotracheal intubation), had microbiologically confirmed nosocomial MRSA pneumonia, and were treated for ≥48 h with either linezolid or vancomycin.
This study was carried out in compliance with the latest revision of the Declaration of Helsinki (Fortaleza, Brazil, October 2013) and was conducted in accordance with the requirements of Law 14/2007 of July 3, of Biomedical Research. The study was approved by the institution’s Internal Review Board (registry number 2012/7927). Written informed consent was obtained from patients or their next-of-kin.
Antimicrobial susceptibility testing
The minimal inhibitory concentrations of vancomycin and linezolid were determined by E-Test. Antimicrobial susceptibility was performed using the Kirby-Bauer method and the ATCC 25923 strain (S. aureus) as a control [9]. The following antibiotics were tested: ciprofloxacin (5 μg), clindamycin (2 μg), erythromycin (15 μg), chloramphenicol (30 μg), fusidic acid (10 μg), gentamicin (10 μg), quinupristin-dalfopristin (15 μg), rifampicin (5 μg), Sulfamethoxazole/trimethoprim (25 μg), and tetracycline (30 μg). Screening of inducible clindamycin resistance was performed by the D-test for strains resistant to clindamycin. Replicates of each susceptibility test were performed. All results were interpreted according to the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [10].
Mechanisms of resistance
We tested the most common mechanisms of resistance to ciprofloxacin, clindamycin, erythromycin, chloramphenicol, fusidic acid, gentamicin, quinupristin-dalfopristin, rifampicin, sulfamethoxazole/trimethoprim, and tetracycline. Each mechanism was screened by polymerase chain reaction (PCR) with the primers and conditions shown in Table 1 and electrophoresis in agarose gel using the 100-bp DNA ladder as a marker for molecular weight (Invitrogen). The PCR products were sequenced by Sanger methods (Genewiz, Germany), and were analyzed by alignment with the template sequence at GenBank [7].
Table 1.
Primers used in this study
| Primer Pair | Amplified product | Sequence (5′ to 3′) | Amplicom size | Anneling temperature | References |
|---|---|---|---|---|---|
| gyrA-F | gyrA | ATG GCT GAA TTA CCT CAA TC | 398 bp | 55°C | 13 |
| gyrA-R | GTG TGA TTT TAG TCA TAC GC | ||||
| gyrB-F | gyrB | CAGCGTTAGATGTAGCAAGC | 680 bp | 55°C | 17 |
| gyrB-R | CGATTTTGTGATATCTTGCTTTCG | ||||
| grlA-F | grlA | CAG TCG GTG ATG TTA TTG GT | 469 bp | 55°C | 13 |
| grlA-R | CCT TGA ATA ATA CCA CCA GT | ||||
| grlB-F | grlB | GIG AAG CIG CAC GTA A | 363 bp | 50°C | 13 |
| grlB-R | TCI GTA TCI GCA TCA GTC AT | ||||
| ermA-F | erm(A) | TAT CTT ATC GTT GAG AAG GGA TT | 138 bp | 55°C | 5 |
| ermA-R | CTA CAC TTG GCT TAG GAT GAA A | ||||
| ermC-F | erm(C) | CTT GTT GAT CAC GAT AAT TTC C | 189 bp | 55°C | 5 |
| ermCR | ATC TTT TAG CAA ACC CGT ATT C | ||||
| msrA-F | msrA | TCC AAT CAT TGC ACA AAA TC | 162 bp | 55°C | 5 |
| msrA-R | AAT TCC CTC TAT TTG GTG GT | ||||
| aac(6′)-aph(2″)F | aac(6′)-aph(2″) | TTG GGA AGA TGA AGT TTT TAG A | 173 bp | 55°C | 5 |
| aac(6′)-aph(2″)R | CCT TTA CTC CAA TAA TTT GGC T | ||||
| tetK-F | tetK | GTA GCG ACA ATA GGT AAT AGT | 360 bp | 55°C | 6 |
| tetK-R | GTA GTG ACA ATA AAC CTC CTA | ||||
| fusB-F | fusB | ATT CAA TCG GAA AAC TAT AAT GAT A | 292 bp | 60°C | 21 |
| fusB-R | TTA TAT ATT TCC GAT TTG ATG CAA G | ||||
| 16srRNA-F | 16S rRNA | GGA GGA AGG TGG GGA TGA CG | 245 bp | 55°C | 5 |
| 16srRNA-R | ATG GTG TGA CGG GCG GTG TG | ||||
| arcC-F | arcC | TTGATTCACCAGCGCGTATTGTC | 450 bp | 55°C | 10 |
| arcC-R | AGGTATCTGCTTCAATCAGCG | ||||
| aroE-F | aroE | ATCGGAAATCCTATTTCACATTC | 450 bp | 55°C | 10 |
| aroE-R | GGTGTTGTATTAATAACGATATC | ||||
| glpF-F | glpF | CTAGGAACTGCAATCTTAATCC | 450 bp | 55°C | 10 |
| glpF-R | TGGTAAAATCGCATGTCCAATTC | ||||
| gmk-F | gmk | ATCGTTTTATCGGGACCATC | 450 bp | 55°C | 10 |
| gmk-R | TCATTAACTACAACGTAATCGTA | ||||
| pta-F | pta | GTTAAAATCGTATTACCTGAAGG | 450 bp | 55°C | 10 |
| pta-R | GACCCTTTTGTTGAAAAGCTTAA | ||||
| tpi-F | tpi | TCGTTCATTCTGAACGTCGTGAA | 450 bp | 55°C | 10 |
| tpi-R | TTTGCACCTTCTAACAATTGTAC | ||||
| yqiL-F | yqiL | CAGCATACAGGACACCTATTGGC | 450 bp | 55°C | 10 |
| yqiL-R | CGTTGAGGAATCGATACTGGAAC |
Multi-locus sequence typing
Allelic profiles of seven S. aureus housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, yqiL) were analyzed and confirmed in 2% agarose gel. Next, PCR products were sequenced by Gemewiz and sequence alignment was done by the ClustalW software. These genes were linked by the multi-locus sequence typing (MLST) database (https://MLST.net; https://pubmlst.org/saureus/) to assign the sequence type. Phylogenetic analysis was carried out using comparative eBURST V3 software employing the eBURST algorithm (http://www.phyloviz.net/goeburst) [11, 12].
Results
MRSA positive samples
Twenty strains of S. aureus were isolated and characterized. Of these, 17 were methicillin-resistant, as confirmed by the oxacillin E-Test, and three were methicillin- susceptible.
Antimicrobial susceptibility
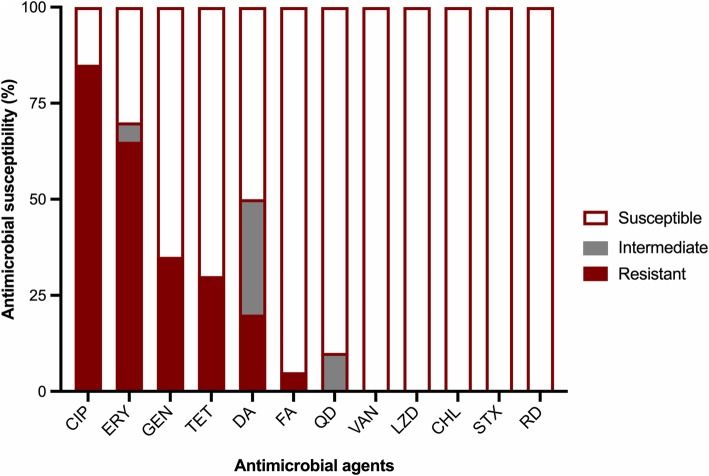
Although there was high susceptibility to linezolid, three strains showed hetero-resistant subpopulations to this antimicrobial agent (strain 1: CC22 (HUCA), strain 2: CC59 (HCP) and strain 8: CC22 (Hospital del Mar) (Table 2). In total, 40% of S. aureus strains were resistant to three or more different antimicrobial agents, with 85% resistant to ciprofloxacin, 65% to erythromycin, 35% to gentamicin, 30% to tetracycline, 20% to clindamycin, and 5% to fusidic acid (Fig. 1). Two strains showed inducible resistance to clindamycin. All the strains were susceptible to vancomycin, linezolid, chloramphenicol, sulfamethoxazole/trimethoprim and rifampicin.
Table 2.
Resistance patterns and mechanisms of resistance
| QRDR Mutations | Resistant genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ETT code | STs | CC | Origin | Resistance patern | gyrA | gyrB | grlA | grlB | ERY- DA | GEN | TET | FA |
| 1 | ST954 | CC22 | HUCA | CIP - ERY- DA | S144P | K401R | ermC | |||||
| 2 | ST87 | CC59 | HCP | ERY- TET | msrA | TeT K | ||||||
| 3 | *ST8 | CC5 | HCP | CIP - GEN - TET | S84L, T129I | S144P | K401R | aac(6´)-aph(2") | TeT K | |||
| 4 | ST217 | CC22 | HCP | CIP - ERY | S84L, S85P | S144P, V82S, Y83V, E84R | K401R, D507E | ermC | ||||
| 5 | *ST8 | CC5 | HCP | ERY - TET | msrA | TeT K | ||||||
| 6 | ST45 | CC45 | HCP | CIP - GEN | E134N, L135T | S144P | K401E | aac(6´)-aph(2") | ||||
| 7 | ST22 | CC22 | Hospital del Mar | CIP - ERY - DA | S84L, T129K | Y500T, H501T | S144P, G78A, E84R | ermC | ||||
| 8 | ST22 | CC22 | Hospital del Mar | CIP - ERY | S84L, S85P, T129I | V511A, V303C | S144P, S81P | ermC | ||||
| 9 | ST8 | CC5 | Hospital del Mar | CIP - ERY - GEN - TET | S84L, T129K, I131S | S144P | K401E | msrA | aac(6´)-aph(2") | TeT K | ||
| 10 | ST22 | CC22 | Hospital del Mar | CIP | S84L, S85P | V511A, R447H | S144P | K401R, L440F | ||||
| 11 | ST22 | CC22 | Hospital del Mar | CIP - GEN - TET | S84L, K130G | V511A, G339D, K312T | S144P | K401E, K400Q, D507E | aac(6´)-aph(2") | TeT K | ||
| 12 | ST22 | CC22 | Policlinic Milan | CIP - ERY | S84L, T129K | S144P | K401R | ermC | ||||
| 13 | ST1535 | - | Policlinic Milan | CIP - GEN - TET - FA | R447L | S144P | K403Q, H478Y, D507E | aac(6´)-aph(2") | TeT K | FusB | ||
| 14 | ST22 | CC22 | Policlinic Milan | CIP - ERY | S84L, T129I | H501T | S144P, S80F | ermC | ||||
| 15 | ST83 | CC5 | Policlinic Milan | CIP - ERY | S84L | H501T | S144P | ermC | ||||
| 16 | ST217 | CC22 | Policlinic Milan | CIP - ERY - GEN - DA | S84L | L418F | S144P | K401E | ermA | aac(6´)-aph(2") | ||
| 17 | ST403 | CC5 | Policlinic Milan | CIP | T129K | V511A, S425G, R447N | S144P | D503E, A504P | ||||
| 18 | ST1221 | CC5 | Policlinic Milan | CIP - ERY | S84L, E88A | S144P, S80F | K401E, D507N | ermC | ||||
| 19 | ST22 | CC22 | Policlinic Milan | CIP - ERY - GEN - DA | S84L | S144P | K403Q, A504P, D507E | ermA | aac(6´)-aph(2") | |||
* ETT collected at different ICU. ETT code 3, 5, and 6 = Methicillin sensitive Staphylococcus aureus
Abbreviations: CC Clonal Complex, CIP Ciprofloxacin, DA Clindamycin, ERY Erythromycin, FA Fusidic acid, GEN Gentamicin, HCP Hospital Clinic (Spain), HUCA Hospital Universitario Central de Asturias (Spain); Policlinic Milan, Fondazione IRCCS Ca′ Granda (Italy), QRDR Quinolone Resistance-Determining Region, ST sequence type; Strain 20, susceptible to all antimicrobial agents tested (ST217, CC22), TET Tetracycline
Fig. 1.
Antimicrobial susceptibility. Abbreviations: CIP, ciprofloxacin; ERY, erythromycin; GEN, gentamicin; TET, teracycline; DA, clindamycin; FA, fusidic acid; QD, quinupristin-dalfopristin; VAN, vancomycin; LZD, linezolid; CHL, cloranphenicol; STX, sulfamethoxazole/trimetoprim; RD, rifampicin
Mechanisms of resistance
Mutations in gyrA, gyrB, grlA, and grlB genes were found in ciprofloxacin-resistant S. aureus strains. The most frequent mutations were S84 L in gyrA (76.5%), V511A in gyrB (23.5%), S144P in grlA (100%), and K401R/E in grlB (58.8%). Erythromycin resistance was related to the ermC (61.5%), ermA (15.4%), and msrA genes (23.1%). Aminoglycoside-resistant strains contained the aac(6′)/aph(2″) gene, while tetracycline-resistant strains contained the tetK gene. In strains that were resistant to clindamycin, the ermC (50%) and ermA (50%) were detected in equal numbers. Finally, the fusB gene was detected in the strain resistant to fusidic acid. The presence of the fusB gene in plasmid pUB101 and plasmid pUB102 was not confirmed.
Phylogenetic analysis
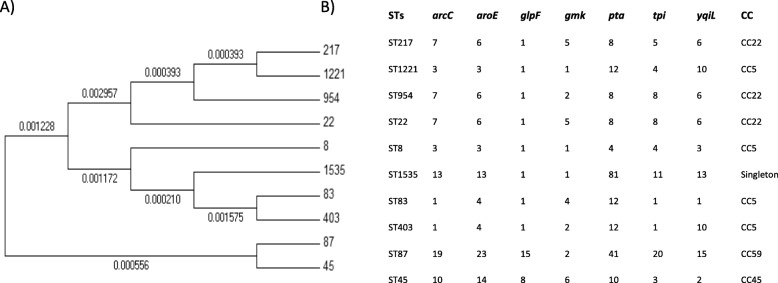
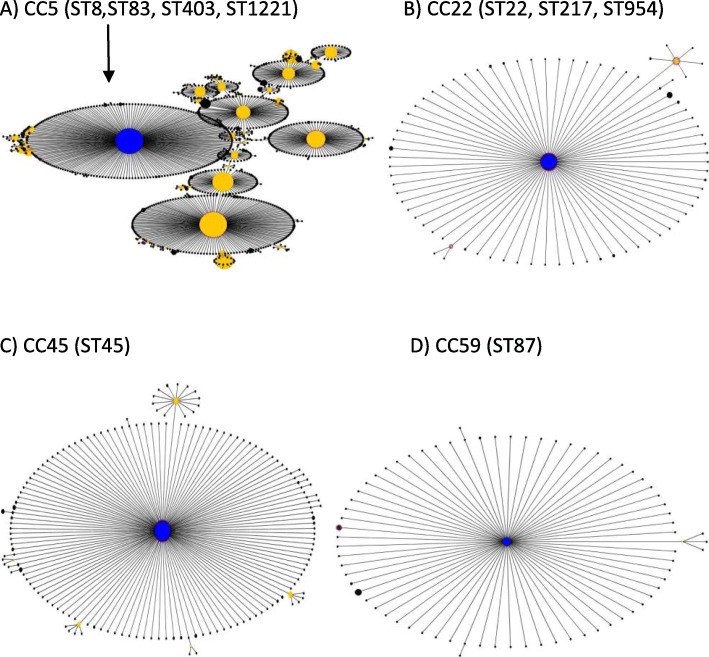
The following S. aureus sequence types (STs) were the most common: ST22 (35%), ST8 (15%), and ST217 (15%). However, ST87, ST83, ST45, ST954, ST403, ST1221, and ST1535 were found with a frequency of 5%. The hospitals where these were collected are shown in Table 2, and the phylogenetic tree shows the genetic proximity (Fig. 2). The allelic profile of each sequence type and clonal complex is also shown in Fig. 2. Our sequence types were distributed in four clonal complexes: CC5 included ST8, ST83, ST403 and ST1221; CC22 included ST22, ST217, and ST954; CC45 included only ST45; and CC59 included only ST87 (Fig. 3). In addition, ST1535 was distributed as a singleton. The strains isolated at the Hospital Clínic were distributed in the four clonal complexes, while the strains at the Hospital del Mar and at the Hospital of Milan were distributed in clonal complexes CC5 and CC22. The most frequent clonal complexes were CC22 and CC5, which accounted for 55 and 30% of local MRSA strains respectively.
Fig. 2.
Phylogenetic tree of the different sequence types and their corresponding clonal complexes. a Phylogenetic tree of all sequence types found in the isolated MRSA strains. b Sequence types, alleles for the different housekeeping genes (per sequence type), and clonal complexes where included. The included genes are as follows: arcC (carbamate kinase), aroE (shikimate dehydrogenase), glpF (glycerol kinase), gmK (guanylate kinase), pta (phosphate acetyltransferase), tpi (triosephosphate isomerase), yqil (acetyl coenzyme A acetyltransferase)
Fig. 3.
Clonal complexes where the strains are located. a CC5, where the founder is ST5. Within this complex, we find ST8, (Strains 3, 5, and 9), ST83 (Strain 15), ST403 (Strain 17), and ST1221 (Strain 18). b CC22, where the founder is ST22 (Strains 7, 8, 10, 11, 12, 14, and 19). Within this complex, we also found ST217 (Strains 4, 16, and 20) and ST954 (Strain 1). c CC45, where ST45 (Strain 6) is its founder. d CC59 was predicted from ST87 (Strain 2) and ST59. Finally, ST1535 (Strain 13) did not belong to any clonal complex and was recorded as a singleton. Abbreviations: CC, clonal complex; MRSA, methicillin resistant Staphylococcus aureus; ST, sequence types
Discussion
The present study reports several important findings regarding the antimicrobial susceptibility, resistance mechanisms, sequence type distributions, and clonality of MRSA strains obtained from ICU respiratory infections in Spain and Italy. At the participating ICUs, S. aureus was not found to be resistant to first-line antibiotics such as linezolid and vancomycin. Although the prevalence of MRSA in the participating centres was low, the mechanisms of resistance described may also be representative for sites with high MRSA prevalence because the MRSA collected during this study corresponded to highly disseminated clonal complexes (CC22 and CC5). In addition, these strains harbored a wide range of antimicrobial mechanisms to second-line antibiotics, including ciprofloxacin, erythromycin, gentamicin, tetracycline, clindamycin, and fusidic acid.
Although our strains did not show resistance to linezolid, the detection of subpopulations resistant to this antimicrobial is a finding that merits comment. Hetero-resistance (HR) is an unstable phenomenon with a high incidence in several bacterial strains, according to recent reports. It is considered unstable because subpopulations defined as hetero-resistant in one susceptibility test may no longer appear as such if the test is repeated. The recent finding of plasmid-associated HR mechanisms emphasizes the problem, since these HR mechanisms may spread horizontally between pathogens. The lack of routine determination by many laboratories and the decrease in antimicrobial activity may have clinical implications. Another important feature is that the decrease in antimicrobial activity caused by this phenomenon is not reflected in the MIC value. Previous research also suggests that HR can indeed be responsible for treatment failure in S. aureus infections. However, this phenomenon is not detected by established procedures and new methods are needed for rapid identification of HR in pathogenic bacteria [13].
Few reports on MRSA resistance mechanisms contain an exhaustive evaluation of antibiotics. In our study we often identified a combination of several resistance mechanisms for MRSA strains, such as spontaneous mutations that decrease bacterial replication, transferable and chromosomal efflux pumps, and antimicrobial or target-modifying enzymes. In terms of antimicrobial susceptibility, similar results have been reported in other studies. For instance, in a series of MRSA strains isolated from different samples, Kitti et al. [14] found high levels of resistance to ciprofloxacin (72.1%), erythromycin (86.9%), gentamicin (72.1%), and clindamycin (86.9%).
In agreement with Sierra et al. [15], we found mutations in the quinolone resistance-determining regions (gyrA, grlA, gyrB, grlB) of ciprofloxacin-resistant S. aureus strains. Several reports have indicated that topoisomerase IV is the primary target for quinolone resistance in Gram-positive microorganisms, including S. aureus with DNA-gyrase acting as secondary target, with specific point mutations at GrlA (subunit A of the topoisomerase IV) and GyrA (subunit A of the DNA-gyrase) as the most relevant [16]. However, our results differ because the mutation S144P in grlA gene may be a polymorphism, given that it is found in both susceptible and resistant strains. This means that the primary target in our strains is the DNA-gyrase. Studies in Japan have not shown mutations in the gyrB and grlB genes [17], which in any case tend to be infrequent in MRSA strains. Nevertheless, some of our strains showed more than three mutation points in each gene. Therefore, further studies are needed to confirm whether these mutations determine resistance or genetic polymorphisms. In addition, in gyrA and grlA, some rare mutations were described (Table 2). Because antibiotic combinations are used during nosocomial pneumonia treatment, these strains are exposed to strong antibiotic selection pressure which may contribute to the high number of mutations found here compared with prior studies. For instance, it has been demonstrated that hospital-acquired MRSA harbors higher levels of antimicrobial resistance than community-acquired MRSA [18].
Analyzing 206 strains of S. aureus from different centers in Canada, China, and France, Martineau et al. attributed erythromycin resistance to ermA (98%), ermB (21%), ermC (2.4%), and msrA (1%) genes. In our study, erythromycin resistance in S. aureus was mediated by the ermA (15%), ermC (62%) and msrA (23%) genes. In an Algerian study, erythromycin resistance was much lower (37.8%) than in our study (65%). However, the authors of the Algerian study included S. aureus from food, nosocomial, and community-acquired infections and identified only the ermC gene [6, 19]; the fact that we only isolated strains from nosocomial pneumonia, whereas the other studies used different sources, could explain the differences observed.
Similarly, Yilmaz et al. also included S. aureus from different clinical samples and found the ermC and ermA genes in strains with resistance to clindamycin [20]. They reported a lower prevalence (6%) of resistance to clindamycin compared to ours (20%), and detected the lnuA gene instead of the erm genes [20, 21]. Some S. aureus strains have shown inducible resistance to clindamycin after exposure to subinhibitory concentrations of erythromycin [22]. In our study, the D-test [10] revealed two MRSA isolates with inducible resistance to clindamycin. This finding is important because clindamycin is used in the treatment not only of pneumonia but also of muscle, bone, skin, and soft tissue infection.
In our study, gentamicin resistance was related to the aac(6′)/aph(2′′). Choi et al. detected higher proportions of the aac(6′)/aph(2′′) gene in MRSA isolates from blood, sputum, urine, and pus samples (65%) than we did in ETT specimens (35%), but they also found prevalences of ant(4′)-Ia and aph(3′)-IIIa of 41 and 9% respectively [23]. Yilmaz et al. found that four of six MRSA isolates carried the same aac(6′)/aph(2′′) gene. Nevertheless, our findings are consistent with those of Martineau et al., who observed a higher number of S. aureus isolates, among which all those with gentamicin resistance had the aac(6′)/aph(2′′) gene [6].
Tetracycline resistance in S. aureus at our ICU was mediated only by the tetK gene. By contrast, other studies have found different proportions of involvement of the tetK or tetM genes alone or in combination; for instance, Strommenger et al. identified ten strains of S. aureus carrying tetK, tetM, or both genes [7]. Yilmaz et al. identified nine strains of S. aureus with the tetM gene and ten with the tetK gene [20]. Finally, Achek et al. detected both the tetK and tetM genes in ten S. aureus isolates from clinical samples [19].
Although fusB was initially thought to be the only gene to encode a protein capable of protecting EF-G, a whole family of related fusB-like proteins has since been described. Thus, mutations in two more genes (fusC and fusD) can lead to staphylococcal resistance to fusidic acid [24]. Several studies have also reported an increase in resistance to fusidic acid. We suggest a chromosomal location of the fusB gene because the primers we used were developed in previous studies by O’Neil et al. in which the fusB gene was detected in total DNA preparations but not in plasmid DNA preparations, indicating a chromosomal location for this resistance determinant (different fusB genes have been discovered on plasmid pUB101 and plasmid pUB102). Some previous data suggest that chromosomal fusB was associated with epidemic strains of S. aureus [25]. Interestingly, in another study fusB-type resistance (fusB and fusC) was found in 87% of MRSA isolates [24], with an association between fusB and clonal complexes CC45 and CC97. By contrast, we found only one strain with fusB, and this was the singleton ST1535. Fusidic acid is a topical drug that is used for the treatment of staphylococcal skin infections, but its increased use appears to have led to the emergence and dissemination of resistant staphylococci [26].
The molecular epidemiology of MRSA in bloodstream infections has been described previously, but less frequently in respiratory infections contracted in the ICU [27, 28]. CC22 is one of the largest circulating clonal complexes associated with hospital-acquired MRSA in Europe (UK) and Asia (Kuala Lumpur, China) [11, 29], while studies of nosocomial pneumonia indicate that CC5 is associated with MRSA strains originating mostly from the US, Europe (Portugal), Asia (China), Africa (Algeria) and Latin America (Argentina and Chile) [18, 19, 30, 31]. Despite the marked heterogeneity of the sequence types in this study, CC22 and CC5 were the main clonal complexes detected. Specific resistance mechanisms can be associated with clonality, since a higher number of these mechanisms were found in the widely expanded CC5 and CC22 clones than in the CC45 and CC59 clones [32]. Consistent with our results, previous studies (in the US, Portugal and Japan) have found CC5 and CC59 to be associated with extensive multi-drug resistance, but not CC45 [32]. Another important point to stress is that these CCs had previously been associated with virulent S. aureus strains.
The heterogeneity of MRSA sequence types at each hospital suggests that ICU cross-transmission has decreased, probably due to the introduction of VAP prevention bundles, isolation measures, and hospital hygiene measures over the last 10 years. Thus, our study indicates that other sources of MRSA transmission such as nasal carriage constitutes risk factors for ICU and nosocomial pneumonia. Although we did not assess nasal MRSA carriage in our study, it has been shown to be an independent risk factor for ICU pneumonia in previous work [33].
This study has some limitations. First, the number of strains is relatively low because S. aureus and MRSA are infrequent causes of nosocomial pneumonia in Spain. However, we also included strains from Italy and found a high heterogeneity of sequence types, which may be representative of the current clones circulating as causes of hospital-acquired MRSA in Europe. Second, although we did not assess the virulence of our MRSA strains, some of the clonal complexes identified, such as the CC59 and CC45, have been shown to be closely related to virulent strains. Immune evasion cluster (IEC) genes have been associated with CC59 (IEC-hemolysin genes) and CC45 (IEC-enterotoxin-hemolysin genes) [32].
Despite the limitations mentioned, we think that this study is important for establishing the epidemiology of S. aureus. Little recent information is available on the resistance mechanisms of action of S. aureus strains obtained from mechanically ventilated patients, and it is unclear whether or not these mechanisms are associated with particular circulating S. aureus clones. We also observed the presence of linezolid hetero-resistance and high resistance to second-line antibiotics in MRSA strains isolated from endotracheal tubes in humans mechanically ventilated for long periods in the ICU. These findings show that MRSA infection is still relevant in southern Europe, with a high capacity of resistance to different antimicrobials, an extensive battery of resistance mechanisms, and a wide clonal variability.
Conclusions
The high level of second-line antimicrobial resistance represents a major problem for the treatment of nosocomial respiratory infections due to MRSA, which display decreased susceptibility to linezolid and vancomycin. Nevertheless, the mechanisms of resistance reported may be useful for the design of new strategies for preventing MRSA. The wide genotypic diversity found reinforces the central role of infection control measures for preventing nosocomial MRSA transmission in the ICU.
Acknowledgments
We thank Ceccato A., Viña L., Li Bassi G., Israel T., Nicolas J.M., Zavala E., Fernández J., Rovira I. Ferrer M. for providing ETT from ICU in mechanical ventilated patients with MRSA respiratory infections, and Dr. Joaquim Ruiz for professional advice.
Authors’ contributions
AT, LFB, RCO, AM, RLA and JV participated in the protocol development, study design and study management. RCO, LBF and RLA participated in data interpretation and writing of the manuscript. LFB, RCO, AM and NV, participated in the study of antimicrobial susceptibility. RCO assessed the mechanisms of resistance. LFB, RLA, YL, AM and RCO participated in the MLST including the analysis of gene sequences. RLA performed the phylogenetic analysis. LM participated in the identification of strains. MP, FAL, PC, AT and JV obtained the respiratory specimens and critically reviewed the manuscript. All authors participated in data collection and reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by CIBER de enfermedades respiratorias-Ciberes (CB 06/06/0028), Ciberes is an initiative of ISCIII, unrestricted grant from Pfizer (WI173058), EUROPE ASPIRE award 2011, SGR, IDIBAPS and ICREA Academy Award to Prof. Antoni Torres.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study. Hospital Clinic ethical committee reference number: 2012/7927.
Consent for publication
Not applicable.
Competing interests
A. Torres has received grants from MedImmune, Cubist, Bayer, Theravance, and Polyphor and personal fees as Advisory Board member from Bayer, Roche, The Medicines CO, and Curetis. He has received personal speaker’s bureau fees from GSK, Pfizer, Astra Zeneca, and Biotest Advisory Board, unconnected to the study submitted here.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Roberto Cabrera and Laia Fernández-Barat contributed equally to this work.
Contributor Information
Laia Fernández-Barat, Email: lfernan1@clinic.cat.
Antoni Torres, Email: atorres@clinic.cat.
References
- 1.Torres A, Niederman MS, Chastre J, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: Guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociacion Latinoamericana del Torax (ALAT). Eur Respir J. 2017;50.
- 2.European Centre for Disease Prevention and Control . ECDC. Annual epidemiological report for 2016. Stockholm: ECDC; 2018. Healthcare-associated infections acquired in intensive care units. [Google Scholar]
- 3.Azhar A, Rasool S, Haque A, et al. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J Med Microbiol. 2017;66:1328–31. [DOI] [PubMed]
- 4.Kochan K, Nethercott C, Perez-Guaita D, et al. Detection of antimicrobial resistance-related changes in biochemical composition of Staphylococcus aureus by means of Atomic Force Microscopy - Infrared Spectroscopy. Anal Chem. 2019;91:15397–403. [DOI] [PubMed]
- 5.Liapikou A, Torres A. Emerging drugs for nosocomial pneumonia. Expert Opin Emerg Drugs. 2016;21:331–341. doi: 10.1080/14728214.2016.1206077. [DOI] [PubMed] [Google Scholar]
- 6.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–238. doi: 10.1128/AAC.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strommenger B, Kettlitz C, Werner G, et al. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J Clin Microbiol. 2003;41:4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Barat L, Motos A, Panigada M, et al. Comparative efficacy of linezolid and vancomycin for endotracheal tube MRSA biofilms from ICU patients. Crit Care. 2019;23:251. doi: 10.1186/s13054-019-2523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 10.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint tables for interpretation of MICs and zone diameters. 2015. [Google Scholar]
- 11.Enright MC, Day NP, DAVIES CE, et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil EJ, Li BC, Aanensen DM, et al. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicoloff H, Hjort K, Levin BR, et al. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol. 2019;4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 14.Kitti T, Seng R, Saiprom N, et al. Molecular characteristics of methicillin-resistant staphylococci clinical isolates from a tertiary Hospital in Northern Thailand. Can J Infect Dis Med Microbiol. 2018;2018:8457012. doi: 10.1155/2018/8457012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sierra JM, Marco F, Ruiz J, et al. Correlation between the activity of different fluoroquinolones and the presence of mechanisms of quinolone resistance in epidemiologically related and unrelated strains of methicillin-susceptible and -resistant Staphylococcus aureus. Clin Microbiol Infect. 2002;8:781–790. doi: 10.1046/j.1469-0691.2002.00400.x. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 17.Nakaminami H, Sato-Nakaminami K, Noguchi N. A novel GyrB mutation in meticillin-resistant Staphylococcus aureus (MRSA) confers a high level of resistance to third-generation quinolones. Int J Antimicrob Agents. 2014;43:478–479. doi: 10.1016/j.ijantimicag.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Peng H, Liu D, Ma Y, et al. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012-2017. Sci Rep. 2018;8:17916. [DOI] [PMC free article] [PubMed]
- 19.Achek R, Hotzel H, Cantekin Z, et al. Emerging of antimicrobial resistance in staphylococci isolated from clinical and food samples in Algeria. BMC Res Notes. 2018;11:663. doi: 10.1186/s13104-018-3762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yilmaz ES, Aslantas O. Antimicrobial resistance and underlying mechanisms in Staphylococcus aureus isolates. Asian Pac J Trop Med. 2017;10:1059–1064. doi: 10.1016/j.apjtm.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Lina G, Quaglia A, Reverdy ME, et al. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/AAC.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller MA, Sader HS, Rhomberg PR, et al. In vitro activity of Tedizolid in comparison with other oral and intravenous agents against a collection of community-acquired methicillin-resistant Staphylococcus aureus (2014-2015) in the United States. Microb Drug Resist. 2019;25:938–43. [DOI] [PubMed]
- 23.Choi SM, Kim SH, Kim HJ, et al. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J Korean Med Sci. 2003;18:631–636. doi: 10.3346/jkms.2003.18.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaws FB, Larsen AR, Skov RL, et al. Distribution of fusidic acid resistance determinants in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:1173–1176. doi: 10.1128/AAC.00817-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill AJ, Larsen AR, Henriksen AS, et al. A fusidic acid-resistant epidemic strain of Staphylococcus aureus carries the fusB determinant, whereas fusA mutations are prevalent in other resistant isolates. Antimicrob Agents Chemother. 2004;48:3594–3597. doi: 10.1128/AAC.48.9.3594-3597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JT, Huang IW, Chang SC, et al. Increasing resistance to fusidic acid among clinical isolates of MRSA. J Antimicrob Chemother. 2017;72:616–618. doi: 10.1093/jac/dkw430. [DOI] [PubMed] [Google Scholar]
- 27.Perez-Montarelo D, Viedma E, Larrosa N, et al. Molecular epidemiology of Staphylococcus aureus bacteremia: Association of Molecular Factors with the source of infection. Front Microbiol. 2018;9:2210. doi: 10.3389/fmicb.2018.02210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hetem DJ, Derde LP, Empel J, et al. Molecular epidemiology of MRSA in 13 ICUs from eight European countries. J Antimicrob Chemother. 2016;71:45–52. doi: 10.1093/jac/dkv298. [DOI] [PubMed] [Google Scholar]
- 29.Niek WK, Teh CSJ, Idris N, et al. Predominance of ST22-MRSA-IV clone and emergence of clones for methicillin-resistant Staphylococcus aureus clinical isolates collected from a tertiary teaching hospital over a two-year period. Jpn J Infect Dis. 2019;72:228–36. [DOI] [PubMed]
- 30.Mendes RE, Deshpande LM, Smyth DS, et al. Characterization of methicillin-resistant Staphylococcus aureus strains recovered from a phase IV clinical trial for linezolid versus vancomycin for treatment of nosocomial pneumonia. J Clin Microbiol. 2012;50:3694–3702. doi: 10.1128/JCM.02024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Liu Q, Zhang H, et al. Molecular characteristics of community-associated Staphylococcus aureus isolates from pediatric patients with bloodstream infections between 2012 and 2017 in Shanghai, China. Front Microbiol. 2018;9:1211. doi: 10.3389/fmicb.2018.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Han C, Huang X, et al. A molecular epidemiological study of methicillin-resistant and methicillin-susceptible Staphylococcus aureus contamination in the airport environment. Infect Drug Resist. 2018;11:2363–2375. doi: 10.2147/IDR.S178584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma XX, Sun DD, Wang S, et al. Nasal carriage of methicillin-resistant Staphylococcus aureus among preclinical medical students: epidemiologic and molecular characteristics of methicillin-resistant S. aureus clones. Diagn Microbiol Infect Dis. 2011;70:22–30. doi: 10.1016/j.diagmicrobio.2010.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.