Abstract
PURPOSE
To estimate age-specific relative and absolute cancer risks of breast cancer and to estimate risks of ovarian, pancreatic, male breast, prostate, and colorectal cancers associated with germline PALB2 pathogenic variants (PVs) because these risks have not been extensively characterized.
METHODS
We analyzed data from 524 families with PALB2 PVs from 21 countries. Complex segregation analysis was used to estimate relative risks (RRs; relative to country-specific population incidences) and absolute risks of cancers. The models allowed for residual familial aggregation of breast and ovarian cancer and were adjusted for the family-specific ascertainment schemes.
RESULTS
We found associations between PALB2 PVs and risk of female breast cancer (RR, 7.18; 95% CI, 5.82 to 8.85; P = 6.5 × 10−76), ovarian cancer (RR, 2.91; 95% CI, 1.40 to 6.04; P = 4.1 × 10−3), pancreatic cancer (RR, 2.37; 95% CI, 1.24 to 4.50; P = 8.7 × 10−3), and male breast cancer (RR, 7.34; 95% CI, 1.28 to 42.18; P = 2.6 × 10−2). There was no evidence for increased risks of prostate or colorectal cancer. The breast cancer RRs declined with age (P for trend = 2.0 × 10−3). After adjusting for family ascertainment, breast cancer risk estimates on the basis of multiple case families were similar to the estimates from families ascertained through population-based studies (P for difference = .41). On the basis of the combined data, the estimated risks to age 80 years were 53% (95% CI, 44% to 63%) for female breast cancer, 5% (95% CI, 2% to 10%) for ovarian cancer, 2%-3% (95% CI females, 1% to 4%; 95% CI males, 2% to 5%) for pancreatic cancer, and 1% (95% CI, 0.2% to 5%) for male breast cancer.
CONCLUSION
These results confirm PALB2 as a major breast cancer susceptibility gene and establish substantial associations between germline PALB2 PVs and ovarian, pancreatic, and male breast cancers. These findings will facilitate incorporation of PALB2 into risk prediction models and optimize the clinical cancer risk management of PALB2 PV carriers.
INTRODUCTION
Germline pathogenic variants (PVs) in PALB21 were first associated with an increased risk of breast cancer (BC) more than a decade ago.2-4 This was confirmed by multiple studies that culminated into a large international study by the PALB2 Interest Group (PALB2-IG), which estimated the absolute risk of BC to be 14% by 50 years of age and 44% by 80 years of age on the basis of data from 154 families.5 PALB2 is now included on BC gene panels,6 and clinical testing for germline PALB2 PVs in the context of female BC is standard of care,7 although gaps in our understanding of risk for other cancers remain.
Beyond BC, germline PVs in PALB2 have been associated with pancreatic cancer (PaC)8,9 and gastric cancer.10-12 Possible associations with ovarian (OC)13 and colorectal cancer (CRC)14 have been suggested, but the statistical evidence is weak. Guidelines for the management of PALB2-associated BC risk exist,7,15 but risk estimates for other cancers are based on small numbers and have large imprecision. Here, we use cancer family history data from 524 families comprising 17,906 individuals to refine age-specific cancer risks for BC and, for the first time to our knowledge, to estimate risks of OC, PaC, male breast cancer (MBC), prostate cancer (PrC), and CRC.
METHODS
Families
Data on 764 families were obtained through study groups that participated in PALB2-IG. Families included at least 1 member with a PALB2 PV, and those with a known PV in BRCA1/BRCA2 were excluded. Variants were considered pathogenic only if they were predicted to lead to a truncated protein, and PALB2 missense variants were excluded. Studies were grouped using two types of ascertainment schemes: through cancer family clinics or families participating in research studies on the basis of having multiple affected members and through BC or OC series unselected for cancer family history. Participants provided informed consent in accordance with institutional review board policies and local practices at each participating center. The Data Supplement lists families by study group and details of study-specific ascertainment criteria.
Statistical Analysis
Complex segregation analysis was used to estimate cancer-specific relative risks (RRs) by fitting genetic models to the cancer inheritance patterns and observed genotypes in families. We estimated RRs for BC, OC, MBC, PaC, PrC, CRC, and all other cancers combined. Pedigree likelihoods were constructed and maximized using the pedigree analysis software Mendel version 3.3.16
For the main analysis, family members were followed from birth until age at diagnosis of first cancer (excluding nonmelanoma skin cancer) because cancer incidence can change after first cancer diagnosis. Otherwise, they were followed until age at death, age at last follow-up, age at risk-reducing mastectomy (RRM) in the BC analyses, risk-reducing salpingo-oophorectomy (RRBSO) in the OC analyses (if RRM/RRBSO occurred at least 1 year before cancer diagnosis), or age 80 years, whichever occurred first. Individuals diagnosed with BC, OC, MBC, PaC, PrC, or CRC were assumed to be affected by that cancer type at the age of diagnosis. Individuals with another subsequent cancer diagnosis were censored at the cancer diagnosis at their youngest age and for the purpose of the analysis, were considered to be affected with other cancer (Data Supplement). Noninformative families, in which no additional information beyond the data relevant to the ascertainment was available, were excluded from the analysis.
Two types of genetic susceptibility models were fitted: a single gene model that assumed that all familial aggregation of cancer is due to PALB2 and a mixed single-gene/polygenic model that also allowed for a residual familial component because of other unobserved genetic effects in addition to PALB2. We fitted these models using country- and cohort-specific population age-specific incidences and constrained the overall cancer age-specific incidences over all assumed genetic effects in the model to agree with the population age-specific incidences17 (Data Supplement).
Because family ascertainment criteria varied across studies, we adjusted for ascertainment for each family separately using an ascertainment-free approach in which likelihoods are computed conditional on any data that may be relevant to the ascertainment, which ensures consistent estimates18-20 (Data Supplement). Nested models were compared using the likelihood ratio test (LRT), and non-nested models were compared using the Akaike information criterion (AIC). Equivalence of RR estimates between multiple-case and population-based families was assessed using the LRT. All statistical tests were two sided. To adjust for the testing of associations with 7 cancer types, we calculated the Benjamini-Hochberg (BH)–adjusted P value for a false discovery rate of .05.21 We also derived the posterior distribution for the effect estimate (relative risks) for nominally significant associations to estimate the probability that the true effect is greater than an RR of 1.5.
RESULTS
Families
A total of 764 families with at least one member with a PALB2 PV were identified through the PALB2-IG (Data Supplement). After adjustment for ascertainment and excluding the noninformative families, 524 families from 44 study centers in 21 countries were included in the analysis. Of these, 363 were multiple-case families, and 161 were from population-based studies of individuals with BC or OC. The eligible families included 8,830 females (852 with PALB2 PVs) and 9,076 males (124 with PALB2 PVs; Data Supplement). One hundred sixty-one different PVs were identified, the most frequent being c.3113G>A (61 families). Twenty-three deletions or duplications of whole exons were observed, all of which were clustered in the PALB2 WD40 domain (Data Supplement).
Risk Models
The genetic models that included a residual (polygenic) familial component for BC or OC provided a better fit to the data than the single gene (AIC for single gene model, 10,687.50 v 10,662.08 for the BC polygenic model and 10,681.93 for the OC polygenic model). Therefore, the results presented herein are based on the models that assumed a single gene plus residual familial component for BC or OC.
BC Risk
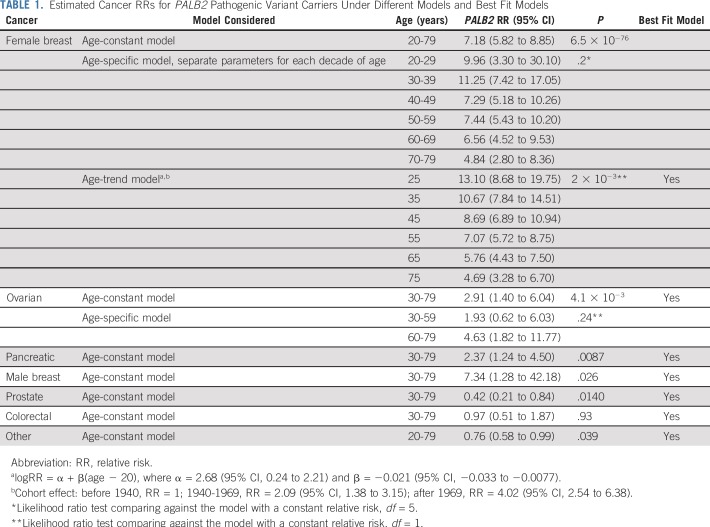
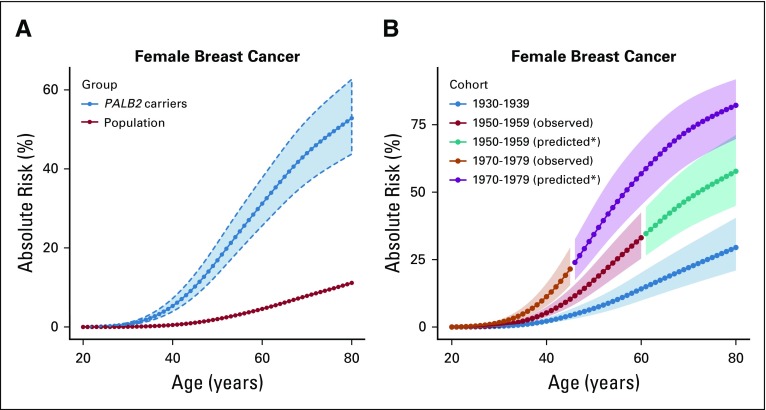
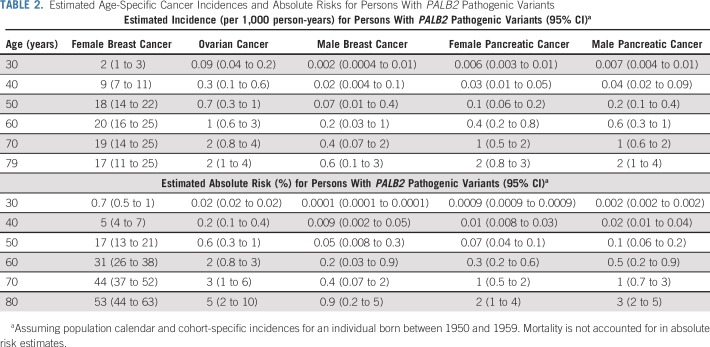
The estimated BC RR was 7.18 (95% CI, 5.82 to 8.85; P = 6.5 × 10−76; BH-adjusted P = 4.6 × 10−75) when it was assumed to be constant with age (Table 1). When separate RRs were estimated for each decade of age, there was a suggestion that the RRs decreased with age; however, this model did not fit significantly better than the model with a constant RR (LRT, df = 5; P = .20; Table 1). We also fitted a model where the logRR was assumed to be a linear function of age from 20 to 79 years (AIC, 10,654.54; Table 1). This model gave a better fit than the model with a constant logRR (P = 2.0 × 10−3) or the model where logRR was assumed to be a linear function up to age 50 years and constant thereafter, which allowed for a threshold effect (AIC, 10,656.38). Under the linear trend model, the BC logRR estimate decreased with age (P = 2.0 × 10−3) from 13.10 at age 25 years to 4.69 at age 75 years. The absolute risk of developing BC was 16.9% (95% CI, 13.3% to 21.3%) to age 50 years and 52.8% (95% CI, 43.7% to 62.7%) to age 80 years, assuming that all women had the calendar period incidences experienced by a woman born during 1950-1959 (Fig 1A; Table 2).
TABLE 1.
Estimated Cancer RRs for PALB2 Pathogenic Variant Carriers Under Different Models and Best Fit Models
FIG 1.
Estimated absolute risk of developing breast cancer for women with germline PALB2 pathogenic variants (PVs) by age under (A) a model that assumes no cohort effect (blue, the risk for women with PALB2 PVs; red, the risk in the United Kingdom general population, assuming that population incidences are applicable to individuals born between 1950 and 1959) and (B) a model that allows for cohort-specific relative risk parameters. The dotted curves and shaded area show the 95% CI. (*) Assuming that the relative risk estimates apply to the unobserved age ranges for women born in these cohorts.
TABLE 2.
Estimated Age-Specific Cancer Incidences and Absolute Risks for Persons With PALB2 Pathogenic Variants
We investigated whether BC risks varied by birth cohort. Compared with women born before 1940, the estimated RR was 2.09 (95% CI, 1.38 to 3.15) for women born during 1940-1969 and 4.02 (95% CI, 2.54 to 6.38) for women born after 1969. Under this model, the absolute risk of developing BC was estimated to be 6.9% (95% CI, 4.6% to 10.2%) to age 50 years and 29.5% (95% CI, 21.0% to 40.4%) to age 80 years for those born in 1930-1939 and 17.4% (95% CI, 12.9% to 23.1%) to age 50 years and 57.7% (95% CI, 45.0% to 71.2%) to age 80 years for those born in 1950-1959. The risk to age 50 years was 34.3% (95% CI, 25.7% to 44.9%) for those born after 1969 (Fig 1B).
OC Risk
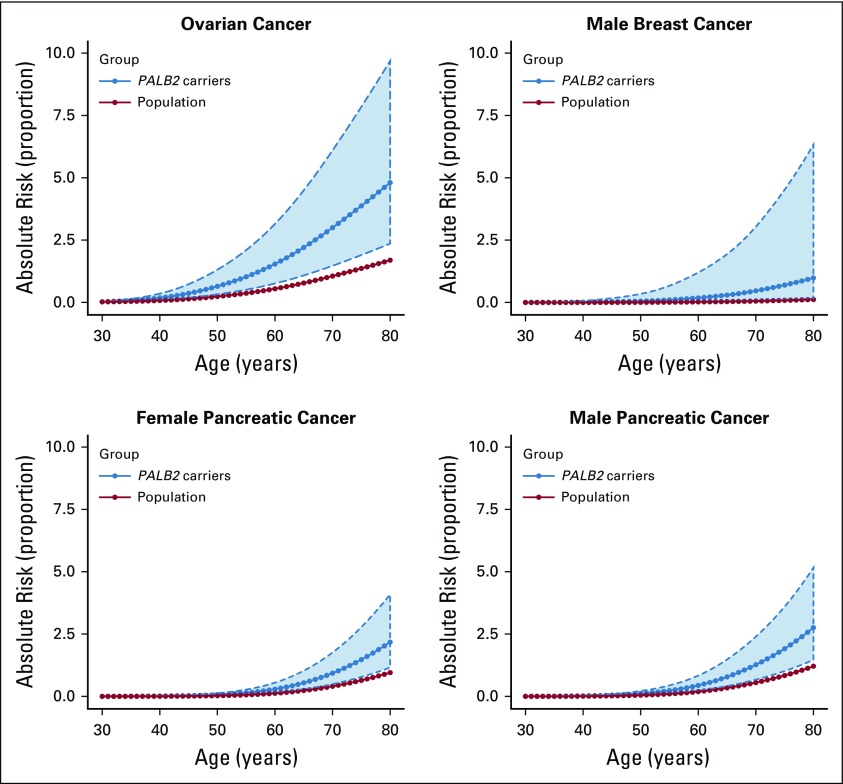
The estimated OC RR was 2.91 (95% CI, 1.40 to 6.04; P = 4.1 × 10−3; BH-adjusted P = .014) when the RR was assumed to be constant with age (Table 1). There was a suggestion of a higher OC RR in ages 60-79 years (RR, 4.63; 95% CI, 1.82 to 11.77) compared with ages 30-59 years (RR, 1.93; 95% CI, 0.62 to 6.03), but this model did not fit significantly better than the model with a constant RR (LRT, df = 1; P = 0.24). The absolute risk of developing OC for women born during 1950-1959 was 0.6% (95% CI, 0.3% to 1.3%) to age 50 years and 4.8% (95% CI, 2.4% to 9.7%) to age 80 years (Fig 2; Table 2).
FIG 2.
Estimated absolute risk of developing ovarian, pancreatic, and male breast cancer for individuals with PALB2 pathogenic variants PVs and in the general population by age (assuming that population incidences are applicable to individuals born between 1950 and 1959). The dotted curves and shaded area show the 95% CI.
PaC Risk
The RR of PaC was estimated to be 2.37 (95% CI, 1.24 to 4.50; P = .0087; BH-adjusted P = .020; Table 1). The number of individuals with PaC was too small to obtain age-specific RR estimates with any precision. Under this model, the absolute risk of developing PaC to age 80 years for a person born during 1950-1959 was 2.2% (95% CI, 1.2% to 4.2%) for females and 2.8% (95% CI, 1.5% to 5.3%) for males (Fig 2; Table 2).
MBC Risk
The estimated MBC RR was 7.34 (95% CI, 1.28 to 42.18; P = .026; BH-adjusted P = .036; Table 1), and the corresponding absolute risk of developing MBC to age 80 years for men born during 1950-1959 was 0.9% (95% CI, 0.2% to 4.9%; Fig 2; Table 2).
PrC, CRC, and Other Cancer Risk
The PrC RR was estimated to be 0.42 (95% CI, 0.21 to 0.84; P = .014; BH-adjusted P = .025). There was no significant association with CRC (RR, 0.97; 95% CI, 0.51 to 1.87; P = .93; BH-adjusted P = .93; Table 1). The results remained similar when separate CRC RRs were estimated for males and females (LRT, P = .74). The estimated RR of all other cancers was 0.76 (95% CI, 0.58 to 0.99; P = .039; BH-adjusted P = .046).
Predicted Risks by Family History
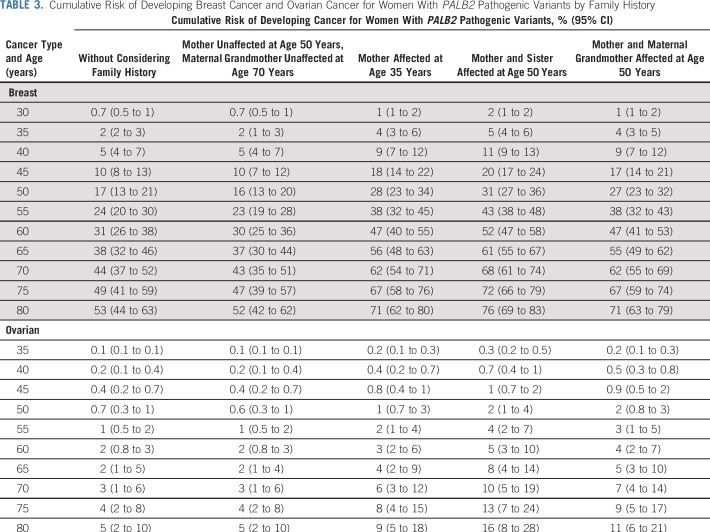
The most parsimonious models included a residual familial component for BC or OC. As a result, the predicted absolute risks of developing BC or OC differed by cancer family history. For example, the predicted absolute risk of developing BC by age 80 years varies from 52% (95% CI, 42% to 62%) for a female with an unaffected mother at age 50 years and unaffected maternal grandmother at age 70 years to 76% (95% CI, 69% to 83%) for a female with two affected first-degree relatives (Table 3). Similarly, the predicted risk of developing OC by age 80 years varies from 5% (95% CI, 2% to 10%) for a female with no family history of OC in first- and second-degree relatives to 16% (95% CI, 8% to 28%) for a female whose mother and sister developed OC at age 50 years (Table 3).
TABLE 3.
Cumulative Risk of Developing Breast Cancer and Ovarian Cancer for Women With PALB2 Pathogenic Variants by Family History
DISCUSSION
Robust quantification of cancer risks is critical for the optimum clinical management of persons with germline PVs in PALB2. Using the largest worldwide collection of people with PALB2 PVs (976 from 524 families) to our knowledge, we have firmly established the place of PALB2 as an important nonsyndromic BC gene after BRCA1 and BRCA2. We also found significantly increased risks of OC, PaC, and MBC, and for the first time to our knowledge, we provide risk estimates for these. The posterior probabilities for the RR parameter estimates being > 1.5 were 0.96 for MBC, 0.89 for OC, and 0.87 for PaC (Data Supplement). No increased risks for PrC, CRC, or other cancers were identified.
Previously published studies provided BC odds ratio (OR) or hazard ratio estimates for women with PALB2 PVs that ranged from 3.40 to 12.67 (Data Supplement). This variation is likely due to differences in study designs and chance caused by small sample sizes. Here, by using a modified segregation analysis approach that adjusts appropriately for ascertainment, the estimated BC RR was found to vary from 13.1 at young ages to 4.69 for older ages, all in the range of other reported estimates. The absolute risk of developing BC to age 80 years was 53% (95% CI, 44% to 63%; Fig 1A; Table 1). Both the RR and the present absolute risk estimates were somewhat higher than those reported in the previous PALB2-IG study in 154 families,5 which shared 77 families with the current study. When risks were estimated separately for multiple-case families and population-based families, the BC risk estimates were slightly higher for multiple-case families but not significantly different after adjusting for ascertainment (P = .41; Data Supplement).
There has been conflicting evidence for the role of PALB2 in OC predisposition; 2 observational studies that implicated an association with PALB2 lacked unaffected or matched controls.22,23 Other studies reported RRs of 0.96-5.53, but none were significant.5,13,24,25 Here, we show that PALB2 PVs are associated with a moderate risk of OC (RR, 2.91; P = 4.1 × 10−3) and that the estimated absolute risk of developing OC to age 80 years was approximately 5%.
Models that allow for a residual familial component in addition to the PALB2-attributable risk provided a better fit to the data for both BC and OC. This is consistent with previous analyses of BC and OC risks for both PALB2 and BRCA1/BRCA2 and strongly suggests other genetic or environmental factors shared in families that modify these risks for PALB2.5,26-29 The combined effects of common genetic variants identified through genome-wide association studies, summarized as a polygenic risk score (PRS), have been shown to modify BC and OC risks women with BRCA1/BRCA2 PVs,30 which explains part of this residual familial component. It is likely that a PRS will also modify the risk associated with PALB2 PVs, thus further improving risk prediction.
We included cohort- and country-specific cancer population incidences in our models to reflect the baseline cancer incidence changes over time and across countries. Despite this, the BC RR estimates varied by both birth cohort and age, with higher RRs observed for more recent birth cohorts and younger ages, consistent with previous findings.5,31,32 The higher RR of BC for women born more recently might reflect under-reporting of cancers in earlier decades; changes in lifestyle, reproductive, or other environmental factors; or more intensive cancer surveillance in recent decades. No evidence for variation in OC risks by age or birth cohort was observed, but the number of individuals with OC (n = 104) limited statistical power.
The absolute risks presented here were obtained by applying estimated RRs to United Kingdom population cancer incidences, so they would be applicable to women from populations with similar age-specific cancer incidences. If the RRs are assumed to be constant across populations, then the estimated absolute risk will be lower for populations with lower cancer incidences.
Previous observational studies of PALB2 in familial PaC reported conflicting results.8,9,33-36 The current analysis confirms the association with PaC and is the first in our knowledge to quantify it, with an RR estimate of 2.30 (albeit with wide confidence limits), which translates to an absolute risk of 2%-3% by age 80 years (Fig 2; Table 2). Previous studies observed a higher prevalence of PALB2 PVs in MBC,5,37-39 and the results presented here confirm an increased MBC risk (RR, 7.34; 95% CI, 1.28 to 42.18).
No previous study that we know of has demonstrated statistically significant associations of PALB2 with PrC risk,25,40-42 and our analysis points to a weak association with decreased risk. Because families were primarily ascertained through female individuals with BC and OC, this result might reflect under-reporting of PrC in these families, and the same phenomenon could explain the slightly decreased risk for all other cancers. Studies have observed germline PALB2 PVs in patients with CRC who underwent gene panel testing,14,43 and while a case-control analysis found a higher frequency of PALB2 PVs in cases with CRC (OR estimate, 3.4), the evidence of association was weak (P = .034), and the results were not replicated in cases with early-onset CRC.44 Here, we did not find evidence of an association with CRC.
The current study has several limitations. Retrospective kin-cohort studies are susceptible to possible biases related to self-reported family histories of cancer. Under-reporting of cancer in families is a common problem,45 which might partly explain the results for cancers beyond breast, ovary, and pancreas. Of the individuals with cancer in the data set, age at diagnosis was missing for 5.5% and could not be inferred by other available information. We assumed that these individuals developed the cancer at the average age at diagnosis of the corresponding cancer in the data set. To examine the effect of this assumption, we performed a sensitivity analysis that censored those individuals at age 0 (ie, effectively ignoring these diagnoses from the analysis). The results remained similar for all cancers except PaC, where the estimated RR was attenuated to 1.84 (95% CI, 0.87 to 3.91) as a result of excluding 10 of the 99 individuals with PaC (Data Supplement). The risk of a second primary BC in women previously diagnosed with PALB2-associated BC could not be determined from the available data, although it remains an important issue to assess in future studies.
PALB2 interacts closely with BRCA1 and BRCA2 in the homologous recombination (HR) DNA repair pathway where the sequence of recruitment to DNA is BRCA1, PALB2, and then BRCA2.46 This suggests that PALB2 and BRCA2 might have similar cancer risks because BRCA2 needs PALB2 to be recruited in HR repair. Our results show a similar BC birth cohort effect to that previously observed in women with BRCA1/BRCA2 PVs,32 and the BC-specific age incidences follow a similar pattern to that seen in BRCA247 (Table 2), where incidences increase with age and reach a constant level from age 50 years onward. The observed associations with MBC and PaC and the moderate risk of OC are also reminiscent of the pattern seen in BRCA2, which presumably reflects tissue-specific differences in DNA repair mechanisms and highlights the importance of conducting such studies for each susceptibility gene.
The cumulative risk estimates for BC in women with PALB2 PVs overlap with BRCA1/BRCA2, for whom RRM is typically offered as an option, and here we provide critical data that allow refinement of RRM guidelines for PALB2. Risk estimates for OC are somewhat lower than for BRCA1/BRCA2, and here the family history of OC would be an important factor when considering RRBSO. Given the similarity in the cancer spectrum and underlying biology, we expect that cancer drugs effective in persons with BRCA1 or BRCA2 PVs may also be effective for those with PALB2 PVs,48,49 and clinical trials currently are addressing this (eg, ClinicalTrials.gov identifier: NCT03344965).
To our knowledge, this is the largest study of PALB2-associated cancer risks to date, and has allowed us to refine BC risk estimates and, for the first time, to provide estimates for OC, PaC, and MBC risk. This advance in knowledge warrants the inclusion of PALB2 in cancer gene panels and will facilitate better cancer risk management of women and men with germline PVs in this gene.
ACKNOWLEDGMENT
We thank the members of the PALB2-IG for their very helpful comments and suggestions. We are grateful to all clinicians, genetics counselors, and other health care professionals who have contributed families with PALB2 to PROMPT. The City of Hope Clinical Cancer Genomics Community Research Network was supported by award number RC4A153828 (principal investigator, J.N.W.) from the National Cancer Institute (NCI) and the Office of the Director, National Institutes of Health (NIH). J.N.W. was also supported by the Breast Cancer Research Foundation and the Dr Norman & Melinda Payson Professorship in Medical Oncology. T.S. was supported by the NCI grant K08CA234394. A.M.D. is supported by Cancer Research UK grant C8197/A16565. The HEBCS study was supported by a Helsinki University Hospital research grant, the Sigrid Jusélius Foundation, and the Finnish Cancer Society. K.B.M.C. is supported by grant G.A044.10 from the Fund for Scientific Research–Flanders. SWE-BRCA (The Swedish BRCA1 & BRCA2 Study Collaborators): Gothenburg, Sahlgrenska University Hospital: Zakaria Einbeigi; Linköping University Hospital: Marie Stenmark-Askmalm and Ekaterina Kuchinskaya; Lund University Hospital: Hans Ehrencrona, Therese Törngren, Anders Kvist, and Åke Borg; Stockholm, Karolinska University Hospital: Brita Arver, Annika Lindblom, and Emma Tham; Umeå University Hospital: Beatrice Melin; and Uppsala University Hospital: Ylva Paulsson-Karlsson. Z.K., P.K., J.S., and M.J. were supported by grants from the Ministry of Health of the Czech Republic (NV16-29959A) and Charles University projects PROGRES Q28/LF1 and SVV2019/260367. We thank clinical geneticists Kamila Vesela, Ales Panczak, and Jaroslav Kotlas from the Institute of Biology and Medical Genetics and Michal Vocka from the Department of Oncology, First Faculty of Medicine, Charles University, and General University Hospital in Prague for their valuable contribution to the study. This study was supported by Research Council of Lithuania grant SEN18/2015. A.S.G.L. was supported by grants from the National Medical Research Council (NMRC) of Singapore (NMRC/0763/2003, NMRC/1194/2008, NMRC/CBRG/0034/2013). S.N. is partially supported by the Morris and Horowitz Families Endowed Professorship. P.C. and J.L.B. represent the WECARE Study Collaborative Group, which is supported by NCI grants CA083178, CA097397, CA114236, and CA129639. S.D. is funded by Susan G Komen. F.C. was supported by NIH grants CA128978 and CA116167, an NIH Specialized Program of Research Excellence in Breast Cancer grant (CA116201), and the Breast Cancer Research Foundation. This study was partially supported by grants from Associazione Italiana per la Ricerca sul Cancro to P.Pe. (AIRC-IG #4017) and P.R. (AIRC-IG #15547) and from Ricerca Finalizzata–Bando 2010 from Ministero della Salute, Italy (C.T. and P.Pe.), and by funds from the Italian citizens who allocated a 5/1,000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori according to Italian laws (INT-Institutional Strategic Projects “5x1000”; S.M.). L.O. was supported by Associazione Italiana per la Ricerca sul Cancro grant AIRC-IG 2018-ID. 21389 and Italian Ministry of Education, Universities and Research–Dipartimenti di Eccellenza-L. 232/2016. D.G.E. is supported by the NIHR Manchester Biomedical Research Centre (IS-BRC-1215–20007). We thank all the collaborating cancer clinics of the French National Study GENESIS (GENE SISters); the GENESIS principal investigators D. Stoppa-Lyonnet and N. Andrieu; the Investigation Platform (PIGE), S. Eon-Marchais, M.G. Dondon, D. Le Gal, J. Beauvallet, N. Mebirouk, and E. Cavaciuti; as well as L. Barjhoux (Biological Resource Centre). The GENESIS study was supported by the Ligue Nationale Contre le Cancer (grants PRE05/DSL and PRE07/DSL), the Institut National du Cancer (INCa grant No. b2008-029/LL-LC), and the Site de Recherche Intégrée sur le Cancer (grant INCa-DGOS-4654). E.R.M. acknowledges funding from the European Research Council (Advanced Researcher Award), NIHR (Senior Investigator Award and Cambridge NIHR Biomedical Research Centre), and Cancer Research UK Cambridge Cancer Centre. The views expressed are those of the authors and not necessarily those of the NHS or Department of Health. The University of Cambridge has received salary support for E.R.M. from the NHS in the East of England through the Clinical Academic Reserve. We thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from National Health and Medical Research Council (NHMRC), the National Breast Cancer Foundation, Cancer Australia, and NIH) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by a grant from the National Breast Cancer Foundation and previously by NHMRC; the Queensland Cancer Fund; the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia; and the Cancer Foundation of Western Australia. We thank the Breast Cancer Family Registry (BCFR), which is funded by the NCI, NIH. T.N.-D. is supported by a Career Development Fellowship from the National Breast Cancer Foundation (Australia, ECF-17-001). We thank Eric Rosenthal and the team at Myriad Genetics for their help in recruiting patients. This work was supported by NCI grant UM1 CA164920. The content of this article does not necessarily reflect the views or policies of the NCI or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government or the BCFR. I.L.A. holds the Anne and Max Tanenbaum Chair in Molecular Medicine at Mount Sinai Hospital and the University of Toronto. The Australian site of the BCFR was also supported by the NHMRC of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation, and the Victorian Breast Cancer Research Consortium. This work was also supported by the NHMRC (project grant APP1029974) and the Victorian Breast Cancer Research Consortium. M.So. is an NHMRC Senior Research Fellow, and J.L.H. is an NHMRC Senior Principal Research Fellow. T.P. is supported in part by the Ingram Professorship and the Kleeberg Foundation. R.W. is supported by the Academy of Finland (project grant 318337 and Center of Excellence grant 251314), the Finnish Cancer Foundation, the Sigrid Jusélius Foundation, the University of Oulu, and the special Government Funding of Oulu University Hospital grants. K.Py. is supported by Academy of Finland Research Fellow grant 314183 and the Finnish Cancer Foundation. A.Ma. is supported by special Government Funding of Kuopio University Hospital grants, the Cancer Fund of North Savo, the Finnish Cancer Foundation, and the strategic funding of the University of Eastern Finland. The MyBrCa study was funded by research grants from the Wellcome trust (203477/Z/16/Z), Ministry of Higher Education to University Malaya (UM.c/Hir/MOHe/06), Estée Lauder group of companies, Sime Darby Foundation, PETRONAS Foundation, Cancer Research UK (c1287/a16563, c8197/a16565, and c12292/a20861), and the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement 634935 (BriDgeS), and the PerSPectiVe project was funded by the government of Canada through Genome Canada and the Canadian Institutes of Health Research, the Ministère de l’Économie, de la Science et de l’Innovation du Québec through Genome Québec, and the Quebec Breast Cancer Foundation. The University of Miami Caribbean Women’s Cancer Study was funded by the Susan G Komen Foundation. P.P. receives salary support from the National Health Service (NHS) in the East of England through the Clinical Academic Reserve. S.H.L.G. is funded by the CDMRP Ovarian Cancer program (W81XWH-18-1-0072). J.B. was supported by the Carlos III National Health Institute funded by FEDER funds–a way to build Europe (PI16/11363). W.D.F. is funded by Susan G Komen and CIHR Foundation Grant FDN 148390. The analysis and data management for this project was support by Cancer Research UK grant C12292/A20861. M.T. was funded by the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement n.310018.
Footnotes
Support by the European Research Council and Cancer Research UK.
DATA SHARING
All mutation data will be deposited in the Leiden Open Variation Database https://www.lovd.nl (open access)
AUTHOR CONTRIBUTIONS
Conception and design: Arto Mannermaa, John L. Hopper, Paul Pharoah, Zaki El-Haffaf, Tuya Pal, Melissa Southey, Robert Winqvist, Douglas F. Easton, William D. Foulkes, Antonis C. Antoniou, Marc Tischkowitz
Financial support: Ellen L. Goode, George Zogopoulos, John L. Hopper, Melissa Southey, Antonis C. Antoniou, Marc Tischkowitz
Administrative support: Alicja Doroszuk, D. Gareth Evans, Arto Mannermaa, John L. Hopper, Jeffrey N. Weitzel, Melissa Southey, Marc Tischkowitz
Provision of study material or patients: Alison M. Dunning, Mitul Shah, Muriel A. Adank, Julian Adlard, Peter Ang, Jonine L. Bernstein, Kristie Bobolis, Åke Borg, Kathleen B.M. Claes, Patrick Concannon, Adeline Cuggia, Francesca Damiola, Antoine de Pauw, Orland Diez, D. Gareth Evans, Florentia Fostira, Judy Garber, Stephen B. Gruber, Eric Hahnen, Zdenek Kleibl, Petra Kleiblova, Irene Konstantopoulou, Anders Kvist, Holly Laduca, Ann S.G. Lee, Fabienne Lesueur, Eamonn R. Maher, Arto Mannermaa, Siranoush Manoukian, Wendy McKinnon, Nur Aishah Mohd Taib, Katherine L. Nathanson, Susan Neuhausen, Pei Sze Ng, Florian Obermair, Kenneth Offit, Olufunmilayo I. Olopade, Judith Penkert, Katri Pylkäs, Paolo Radice, Susan J. Ramus, Anne-Bine Skytte, Thomas Slavin, Jana Soukupova, Gary Unzeitig, Lydia Usha, James Whitworth, Marie Wood, Cheng Har Yip, Sook-Yee Yoon, Amal Yussuf, George Zogopoulos, John L. Hopper, Georgia Chenevix-Trench, Paul Pharoah, Sophia H.L. George, Paul James, Marketa Janatova, Heli Nevanlinna, Rita Schmutzler, Soo-Hwang Teo, Mark Robson, Fergus Couch, Jeffrey N. Weitzel, Melissa Southey, Robert Winqvist, Douglas F. Easton, William D. Foulkes, Antonis C. Antoniou, Marc Tischkowitz
Collection and assembly of data: Goska Leslie, Alicja Doroszuk, Sandra Schneider, Jamie Allen, Brennan Decker, Alison M. Dunning, James Redman, James Scarth, Inga Plaskocinska, Craig Luccarini, Mitul Shah, Karen Pooley, Muriel A. Adank, Julian Adlard, Irene L. Andrulis, Peter Ang, Julian Barwell, Jonine L. Bernstein, Kristie Bobolis, Åke Borg, Carl Blomqvist, Kathleen B.M. Claes, Patrick Concannon, Adeline Cuggia, Julie O. Culver, Francesca Damiola, Antoine de Pauw, Orland Diez, Jill S. Dolinsky, Susan M. Domchek, Christoph Engel, D. Gareth Evans, Judy Garber, Lisa Golmard, Ellen L. Goode, Stephen B. Gruber, Eric Hahnen, Christopher Hake, Tuomas Heikkinen, Judith E. Hurley, Ramunas Janavicius, Zdenek Kleibl, Petra Kleiblova, Irene Konstantopoulou, Anders Kvist, Holly Laduca, Ann S.G. Lee, Fabienne Lesueur, Eamonn R. Maher, Arto Mannermaa, Siranoush Manoukian, Rachel McFarland, Wendy McKinnon, Alfons Meindl, Kelly Metcalfe, Nur Aishah Mohd Taib, Jukka Moilanen, Katherine L. Nathanson, Susan Neuhausen, Pei Sze Ng, Tu Nguyen-Dumont, Sarah M. Nielsen, Florian Obermair, Kenneth Offit, Olufunmilayo I. Olopade, Laura Ottini, Judith Penkert, Katri Pylkäs, Paolo Radice, Susan J. Ramus, Vilius Rudaitis, Lucy Side, Rachel Silva-Smith, Valentina Silvestri, Anne-Bine Skytte, Thomas Slavin, Jana Soukupova, Carlo Tondini, Alison H. Trainer, Gary Unzeitig, Lydia Usha, James Whitworth, Marie Wood, Cheng Har Yip, Sook-Yee Yoon, Amal Yussuf, George Zogopoulos, David Goldgar, John L. Hopper, Georgia Chenevix-Trench, Paul Pharoah, Sophia H.L. George, Judith Balmaña, Claude Houdayer, Paul James, Zaki El-Haffaf, Hans Ehrencrona, Marketa Janatova, Paolo Peterlongo, Heli Nevanlinna, Rita Schmutzler, Soo-Hwang Teo, Mark Robson, Tuya Pal, Fergus Couch, Jeffrey N. Weitzel, Aaron Elliott, Melissa Southey, Robert Winqvist, William D. Foulkes, Antonis C. Antoniou, Marc Tischkowitz
Data analysis and interpretation: Xin Yang, Jamie Allen, Brennan Decker, Leila Dorling, Andrew Lee, Jonine L. Bernstein, Carl Blomqvist, Susan M. Domchek, D. Gareth Evans, Florentia Fostira, Eric Hahnen, Irene Konstantopoulou, Arto Mannermaa, Tu Nguyen-Dumont, Thomas Slavin, George Zogopoulos, Georgia Chenevix-Trench, Zaki El-Haffaf, Tuya Pal, Jeffrey N. Weitzel, Melissa Southey, Robert Winqvist, Antonis C. Antoniou, Marc Tischkowitz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Cancer Risks Associated With Germline PALB2 Pathogenic Variants: An International Study of 524 Families
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/journal/jco/site/ifc.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Alicja Doroszuk
Employment: AstraZeneca
Research Funding: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Brennan Decker
Stock and Other Ownership Interests: Avidea Technologies
Consulting or Advisory Role: Foundation Medicine, Avidea Technologies
Inga Plaskocinska
Employment: IQVIA
Honoraria: IQVIA
Andrew Lee
Patents, Royalties, Other Intellectual Property: Inventor of the BOADICEA model, which is commercialized by Cambridge Enterprise (part of the University of Cambridge). Currently, the model is licensed to the company FamHis. I have received royalties from this commercialization.
Peter Ang
Consulting or Advisory Role: Roche Pharma AG, Eli Lilly, Novartis
Julian Barwell
Honoraria: AstraZeneca, Merck
Consulting or Advisory Role: Deerfield Management
Travel, Accommodations, Expenses: AstraZeneca
Åke Borg
Honoraria: Roche
Travel, Accommodations, Expenses: Roche, AstraZeneca
Kathleen B.M. Claes
Consulting or Advisory Role: AstraZeneca (Ins)
Patrick Concannon
Stock and Other Ownership Interests: Amgen
Patents, Royalties, Other Intellectual Property: 6,458,534 Gene associated with Nijmegen breakage syndrome, a gene product and methods for their use filed April 27, 1999; 5,955,279 Ataxia-telangiectasia: mutations in the ATM gene filed June 13, 1997; 5,770,372 Detection of mutations in the human ATM gene filed November 20, 1996
Other Relationship: 10X Genomics (Inst), CELLINK (Inst), Canon USA (Inst), Illumina (Inst)
Adeline Cuggia
Consulting or Advisory Role: AstraZeneca
Jill S. Dolinsky
Employment: Ambry Genetics
Susan M. Domchek
Honoraria: AstraZeneca, Clovis Oncology, Bristol-Myers Squibb
Research Funding: AstraZeneca (Inst), Clovis Oncology (Inst), PharmaMar (Inst)
D. Gareth Evans
Honoraria: AstraZeneca
Judy Garber
Consulting or Advisory Role: Novartis (I), GTx (I), Helix BioPharma, Konica Minolta, Aleta BioTherapeutics (I), H3 Biomedicine (I), Kronos Bio (I)
Research Funding: Novartis (I), Ambry Genetics, Invitae Genetics, Myriad Genetics
Other Relationship: Susan G. Komen for the Cure (I), American Association for Cancer Research, Diane Helis Henry Medical Foundation (I), James P. Wilmot Foundation (I), Adrienne Helis Malvin Medical Research Foundation (I), Breast Cancer Research Foundation, Facing our Risk of Cancer Empowered
Stephen B. Gruber
Employment: Brogent
Leadership: Brogent
Stock and Other Ownership Interests: Brogent, Fulgent Therapeutics
Consulting or Advisory Role: Myriad Genetics, Fulgent Therapeutics
Research Funding: Myriad Genetics (Inst)
Tuomas Heikkinen
Employment: Bayer AG
Irene Konstantopoulou
Speakers’ Bureau: AstraZeneca
Research Funding: AstraZeneca
Holly Laduca
Employment: Ambry Genetics
Stock and Other Ownership Interests: Ambry Genetics
Eamonn R. Maher
Consulting or Advisory Role: Illumina
Travel, Accommodations, Expenses: Illumina
Sarah M. Nielsen
Employment: Invitae
Stock and Other Ownership Interests: Invitae
Consulting or Advisory Role: AstraZeneca, Merck, Myriad Genetics
Speakers’ Bureau: AstraZeneca
Travel, Accommodations, Expenses: Myriad Genetics, AstraZeneca
Olufunmilayo I. Olopade
Employment: CancerIQ (I)
Leadership: CancerIQ
Stock and Other Ownership Interests: CancerIQ, Tempus
Research Funding: Novartis (Inst), Roche (Inst), Genentech (Inst)
Other Relationship: Tempus, Color Genomics, Roche, Genentech, Bio Ventures for Global Health
Open Payments Link: https://openpaymentsdata.cms.gov/physician/olopade
Vilius Rudaitis
Travel, Accommodations, Expenses: MSD Oncology, Roche
Lucy Side
Research Funding: Myriad Genetics (Inst)
Carlo Tondini
Consulting or Advisory Role: Myriad Genetics
Speakers’ Bureau: Amgen
Travel, Accommodations, Expenses: Roche, Genentech, Novartis, Celgene
Lydia Usha
Consulting or Advisory Role: Agendia, Myriad Genetics
Patents, Royalties, Other Intellectual Property: Patent in relationship to an adverse effect of a pharmaceutical agent. The patent has not generated any royalties or other reimbursements (Inst).
Travel, Accommodations, Expenses: Myriad Genetics
Thomas van Overeem Hansen
Honoraria: Pfizer
James Whitworth
Consulting or Advisory Role: SellmerDiers Sperm Bank
Marie Wood
Consulting or Advisory Role: Heron Therapeutics (Inst), AstraZeneca (Inst)
Sook-Yee Yoon
Research Funding: AstraZeneca (Inst)
Amal Yussuf
Employment: Ambry Genetics
George Zogopoulos
Consulting or Advisory Role: Ipsen, AstraZeneca
Research Funding: Diazon Pharmaceuticals
Patents, Royalties, Other Intellectual Property: TC-PTP inhibitors as APC activators for immunotherapy
Travel, Accommodations, Expenses: Baxalta
Paul Pharoah
Patents, Royalties, Other Intellectual Property: The PREDICT breast cancer prognostic model is licensed to OncoAssist by the University of Cambridge. I receive a share of the fees. I receive a share of the fees for a patent held by the University of Cambridge of a 7-single nucleotide polymorphism polygenic risk assay.
Judith Balmaña
Consulting or Advisory Role: AstraZeneca, Pfizer
Research Funding: AstraZeneca (Inst), PharmaMar (Inst)
Patents, Royalties, Other Intellectual Property: European patent request submitted (EP17382884.9) not related to this work
Travel, Accommodations, Expenses: AstraZeneca, PharmaMar
Hans Ehrencrona
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Rita Schmutzler
Honoraria: AstraZeneca, Clovis Oncology
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Soo-Hwang Teo
Speakers’ Bureau: AstraZeneca
Mark Robson
Honoraria: AstraZeneca
Consulting or Advisory Role: McKesson, AstraZeneca
Research Funding: AstraZeneca (Inst), Myriad Genetics (Inst), Invitae (Inst), AbbVie (Inst), Tesaro (Inst), Medivation (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Pfizer
Other Relationship: Research to Practice, Clinical Care Options, Physician Education Resource
Uncompensated Relationships: Merck, Pfizer, Daiichi Sankyo
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669/summary
Fergus Couch
Consulting or Advisory Role: AstraZeneca
Speakers’ Bureau: Ambry Genetics, QIAGEN, GRAIL
Travel, Accommodations, Expenses: GRAIL, QIAGEN
Other Relationship: Ambry Genetics
Jeffrey N. Weitzel
Speakers’ Bureau: AstraZeneca
Aaron Elliott
Employment: Ambry Genetics
Leadership: Ambry Genetics
Douglas F. Easton
Patents, Royalties, Other Intellectual Property: Royalties from BOADICEA risk prediction tool (Inst)
William D. Foulkes
Research Funding: AstraZeneca (Inst)
Antonis C. Antoniou
Patents, Royalties, Other Intellectual Property: Inventor of the BOADICEA model, which has been licensed to Cambridge Enterprise for commercialization. May receive royalties if commercialization is realized.
No other potential conflicts of interest were reported.
REFERENCES
- 1.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 2.Erkko H, Xia B, Nikkilä J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 3.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor A, Brady AF, Frayling IM, et al. Consensus for genes to be included on cancer panel tests offered by UK genetics services: Guidelines of the UK Cancer Genetics Group. J Med Genet. 2018;55:372–377. doi: 10.1136/jmedgenet-2017-105188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic. Version 1.2020, December 4, 2019.
- 8.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fewings E, Larionov A, Redman J, et al. Germline pathogenic variants in PALB2 and other cancer-predisposing genes in families with hereditary diffuse gastric cancer without CDH1 mutation: A whole-exome sequencing study. Lancet Gastroenterol Hepatol. 2018;3:489–498. doi: 10.1016/S2468-1253(18)30079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang KL, Mashl RJ, Wu Y, et al: Pathogenic germline variants in 10,389 adult cancers. Cell 173:355-370.e14, 2018. [DOI] [PMC free article] [PubMed]
- 12. Sahasrabudhe R, Lott P, Bohorquez M, et al: Germline mutations in PALB2, BRCA1, and RAD51C, which regulate DNA recombination repair, in patients with gastric cancer. Gastroenterology 152:983-986.e6, 2017. [DOI] [PMC free article] [PubMed]
- 13.Ramus SJ, Song H, Dicks E, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107:djv214. doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearlman R, Frankel WL, Swanson B, et al. Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2017;3:464–471. doi: 10.1001/jamaoncol.2016.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tung N, Domchek SM, Stadler Z, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13:581–588. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange K, Weeks D, Boehnke M. Programs for pedigree analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–472. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 17.Antoniou AC, Pharoah PD, McMullan G, et al. Evidence for further breast cancer susceptibility genes in addition to BRCA1 and BRCA2 in a population-based study. Genet Epidemiol. 2001;21:1–18. doi: 10.1002/gepi.1014. [DOI] [PubMed] [Google Scholar]
- 18.Cannings C, Thompson EA. Ascertainment in the sequential sampling of pedigrees. Clin Genet. 1977;12:208–212. doi: 10.1111/j.1399-0004.1977.tb00928.x. [DOI] [PubMed] [Google Scholar]
- 19.Ewens WJ, Shute NC. A resolution of the ascertainment sampling problem. I. Theory. Theor Popul Biol. 1986;30:388–412. doi: 10.1016/0040-5809(86)90042-0. [DOI] [PubMed] [Google Scholar]
- 20.Shute NC, Ewens WJ. A resolution of the ascertainment sampling problem. III. Pedigrees. Am J Hum Genet. 1988;43:387–395. [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 22.Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. doi: 10.1001/jamaoncol.2018.2956. Lu HM, Li S, Black MH, et al: Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing. JAMA Oncol, 5:51-57, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southey MC, Goldgar DE, Winqvist R, et al. PALB2, CHEK2 and ATM rare variants and cancer risk: Data from COGS. J Med Genet. 2016;53:800–811. doi: 10.1136/jmedgenet-2016-103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. doi: 10.1038/sj.bjc.6604305. Antoniou AC, Cunningham AP, Peto J, et al: The BOADICEA model of genetic susceptibility to breast and ovarian cancers: Updates and extensions. Br J Cancer 98:1457-1466, 2008 [Erratum: Br J Cancer 98:2015, 2008] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Begg CB. On the use of familial aggregation in population-based case probands for calculating penetrance. J Natl Cancer Inst. 2002;94:1221–1226. doi: 10.1093/jnci/94.16.1221. [DOI] [PubMed] [Google Scholar]
- 28.Begg CB, Haile RW, Borg A, et al. Variation of breast cancer risk among BRCA1/2 carriers. JAMA. 2008;299:194–201. doi: 10.1001/jama.2007.55-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy-Lahad E, Lahad A, Eisenberg S, et al. A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc Natl Acad Sci U S A. 2001;98:3232–3236. doi: 10.1073/pnas.051624098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuchenbaecker KB, McGuffog L, Barrowdale D, et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2017;109:djw302. doi: 10.1093/jnci/djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: A combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foulkes WD, Brunet JS, Wong N, et al. Change in the penetrance of founder BRCA1/2 mutations? A retrospective cohort study. J Med Genet. 2002;39:407–409. doi: 10.1136/jmg.39.6.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010;78:490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhen DB, Rabe KG, Gallinger S, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: A PACGENE study. Genet Med. 2015;17:569–577. doi: 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant RC, Al-Sukhni W, Borgida AE, et al. Exome sequencing identifies nonsegregating nonsense ATM and PALB2 variants in familial pancreatic cancer. Hum Genomics. 2013;7:11. doi: 10.1186/1479-7364-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harinck F, Kluijt I, van Mil SE, et al. Routine testing for PALB2 mutations in familial pancreatic cancer families and breast cancer families with pancreatic cancer is not indicated. Eur J Hum Genet. 2012;20:577–579. doi: 10.1038/ejhg.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanco A, de la Hoya M, Balmaña J, et al. Detection of a large rearrangement in PALB2 in Spanish breast cancer families with male breast cancer. Breast Cancer Res Treat. 2012;132:307–315. doi: 10.1007/s10549-011-1842-2. [DOI] [PubMed] [Google Scholar]
- 38.Ding YC, Steele L, Kuan CJ, et al. Mutations in BRCA2 and PALB2 in male breast cancer cases from the United States. Breast Cancer Res Treat. 2011;126:771–778. doi: 10.1007/s10549-010-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizzolo P, Zelli V, Silvestri V, et al. Insight into genetic susceptibility to male breast cancer by multigene panel testing: Results from a multicenter study in Italy. Int J Cancer. 2019;145:390–400. doi: 10.1002/ijc.32106. [DOI] [PubMed] [Google Scholar]
- 40. Eeles RA, Olama AA, Benlloch S, et al: Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45:385-391, 2013. [DOI] [PMC free article] [PubMed]
- 41.Pakkanen S, Wahlfors T, Siltanen S, et al. PALB2 variants in hereditary and unselected Finnish prostate cancer cases. J Negat Results Biomed. 2009;8:12. doi: 10.1186/1477-5751-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tischkowitz M, Sabbaghian N, Ray AM, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in hereditary prostate cancer. Prostate. 2008;68:675–678. doi: 10.1002/pros.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurgelun MB, Kulke MH, Fuchs CS, et al. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35:1086–1095. doi: 10.1200/JCO.2016.71.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.AlDubayan SH, Giannakis M, Moore ND, et al. Inherited DNA-repair defects in colorectal cancer. Am J Hum Genet. 2018;102:401–414. doi: 10.1016/j.ajhg.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziogas A, Anton-Culver H. Validation of family history data in cancer family registries. Am J Prev Med. 2003;24:190–198. doi: 10.1016/s0749-3797(02)00593-7. [DOI] [PubMed] [Google Scholar]
- 46.Ducy M, Sesma-Sanz L, Guitton-Sert L, et al. The tumor suppressor PALB2: Inside out. Trends Biochem Sci. 2019;44:226–240. doi: 10.1016/j.tibs.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 48.Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10:e9172. doi: 10.15252/emmm.201809172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foo TK, Tischkowitz M, Simhadri S, et al. Compromised BRCA1-PALB2 interaction is associated with breast cancer risk. Oncogene. 2017;36:4161–4170. doi: 10.1038/onc.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]