Abstract
Background
Lung adenocarcinoma (LUAD) is the most common subtype of lung malignancy and is the leading cause of cancer-related mortalities worldwide. N6-methyladenosine (m6A), the most prevalent internal modification of mRNAs, plays crucial roles in regulating mRNA splicing, exportation, localization, translation, and stability. This study assessed the expression patterns and prognostic value of m6A-related genes in LUAD.
Material/Methods
The expression data of 509 LUAD samples and 20 normal samples were obtained from the Cancer Genome Atlas (TCGA) to determine the mRNA expression levels of m6A-related genomic targets. mRNA expression of 6 LUAD datasets was obtained from the Gene Expression Omnibus (GEO) repository. Subsequently, the Human Protein Atlas (HPA) and tissue microarray (TMA) cohort were used to verify the expression pattern of m6A-related genes at mRNA and protein level. The t test was used to analyze correlations between m6A-related genes and clinical features. Finally, survival analysis was performed to assess the prognostic value of m6A-related genes in LUAD patients.
Results
We found that KIAA1429, RBM15, METTL3, HNRNPC, HNRNPA2B1, YTHDF1, and YTHDF2 were upregulated in TCGA-LUAD databases. The analysis of 7 GEO databases was consistent with the TCGA. YTHDF1 was overexpressed in LUAD patients and YTHDF2 was overexpressed in the great majority of cases. METTL3, YTHDF1, and YTHDF2 were associated with better OS and RFS.
Conclusions
m6A-related genes were differentially expressed in LUAD compared to matched normal patients. The m6A-related genes METTL3, YTHDF1, and YTHDF2 could serve as novel biomarkers for the prognosis of LUAD.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; Prognosis; Puromycin Aminonucleoside
Background
Lung cancer (LC) ranks as the first cause of cancer-related mortalities worldwide, with an approximate 5-year survival rate of 16.6%. Lung adenocarcinoma (LUAD) is the most frequent histological manifestation of lung cancer [1,2]. Multiple therapies, such as surgical resection, chemotherapy, radiotherapy, and molecular targeted therapy, have been developed in clinical treatment of LUAD in the past decades, but the overall survival time of LUAD patients has not significantly improved, primarily due to the lack of useful molecular biomarkers [3]. Thus, a comprehensive understanding of the molecular mechanism underlying LUAD occurrence and progression is vital to improve diagnosis and prognosis at the outset. N6-methyladenosine (m6A) is a universal modification of RNA molecules in eukaryotes. However, the biological importance of m6A remains mostly unknown. Alteration of m6A plays a crucial role in mRNA splicing, export, translation, and stability [4]. Related research has revealed that m6A exerts regulatory roles in RNA modulation by acting as “writers”, “erasers”, and “readers” [5]. Many enzymes take part in the m6A system, such as methylases methyltransferase-like 3 (METTL3) and Heterogeneous Nuclear Ribonucleoprotein A2/B1 (HNRNPA2B1). Moreover, there is a significant association between m6A and diseases. Recent studies reported m6A modification is involved in the progression of various human diseases, such as obesity [6] and cancer [7], and in embryonic development [8]. However, the expression and prognostic value of m6A-related genes in LUAD have not been reported.
In this study, we profiled the mRNA expression patterns of m6A-related genes in lung adenocarcinoma. The data used in this study were obtained from the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) repository. Further transcription-mediated amplification (TMA) and hybridization protection assay analysis demonstrated that these molecules consistently depict similar expression patterns at the protein level. We also evaluated the correlations between m6A-related proteins and clinical features, including race, age, sex, and pathological stage, in LUAD patients from the TCGA database. Survival analysis showed the robustness of m6A dysregulation in LUAD patients.
Material and Methods
Acquisition and processing of LUAD datasets
The TCGA-LUAD and corresponding clinical data of 509 LUAD samples and 20 normal samples were downloaded from the Cancer Genome Atlas (TCGA) repository (https://gdc-portal.nci.nih.gov/). mRNA expression of 6 LUAD datasets (GSE27262, GSE10072, GSE31210, GSE33532, GSE40791, and GSE43767) were obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). Translational-level validation of m6A-related genes was done using the Human Protein Atlas (HPA) database (http://www.proteinatlas.org/).
Patients and specimens
The LUAD tissue microarray (TMA) cohort was composed of 21 LUAD samples and their matched normal tissues. All samples were obtained from the ZZU TMA cohort (First Affiliated Hospital of Zhengzhou University, from April 2016 to December 2016) [9]. None of the patients had ever received chemotherapy, radiotherapy, or immunotherapy before surgery. Our study was permitted by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University and written informed consents were signed before operations for all patients.
Immunohistochemical (IHC) analysis
IHC staining was performed using a SignalStain® Boost IHC Detection kit following the manufacturer’s instructions [10]. Briefly, the TMA sections were deparaffinized in xylene and rehydrated through graded alcohol. The antigen was then retrieved in Target Retrieval Solution (Dako, CA, USA) and 10% normal goat serum was used to block nonspecific binding. Subsequently, the sections were incubated with corresponding primary antibodies (Proteintech Group, China) overnight at 4°C. The secondary antibody (Proteintech Group, China) was added for incubation for 1 h and then the nucleus was stained with the SignalStain® DAB Substrate Kit (CST, USA). The whole sections were evaluated by 2 pathologists who were unaware of the clinical data of all patients. The samples were scored based on the proportion and intensity of cell staining. Scores greater than 1 were regarded as high expression, while scores of 0 were regarded as low expression. The Supplementary Table 1 shows the specific information of antibodies used in our study.
Statistical analysis
All statistical analyses were done using SPSS 23.0 software (IBM Corp., Armonk, NY, USA) and GraphPad Prism 7 (San Diego, CA, USA). Correlation between the expression of m6A-related genes and clinical variables of LUAD patients were estimated by t test. The Pearson’s χ2 test or Fisher’s exact test were used to evaluate TMA results. Kaplan-Meier and log-rank test were conducted for survival analysis. R studio was used to obtain each gene’s best cutoff value and survival curves. In addition, univariate and multivariate Cox regression analyses were conducted to select independent prognostic factors for OS and RFS. P values (two-sided) less than 0.05 were considered as statistically significant.
Results
The mRNA expression pattern of m6A-related genes in LUAD
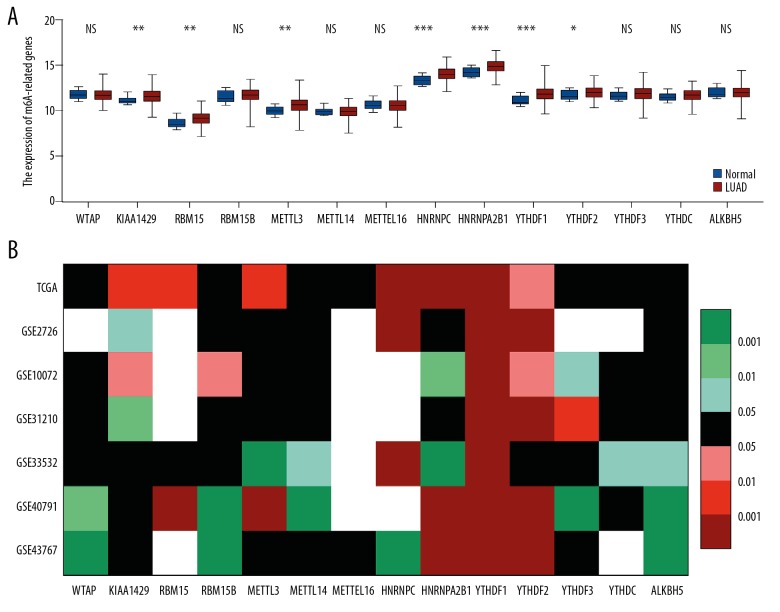
The expression pattern of all known m6A-related genes was analyzed in lung adenocarcinoma (LUAD) based on TCGA datasets. The analysis results revealed that several m6A-related genes were dysregulated in lung adenocarcinoma tissues compared with normal tissues. In LUAD, m6A “writers”, including KIAA1429, RBM15, and METTL3, were upregulated. However, the expression of m6A “readers” was remarkably increased in tumor tissues compared with corresponding normal tissues, except for YTHDF3 and YTHDC, whose expression was not significantly different between the 2 tissue types. In m6A “erasers”, the difference in expression of ALKBH5 between tumor tissues and normal tissues was not significant (Figure 1A). Subsequently, 6 GEO datasets were applied to further verify the expression pattern of m6A-related genes in LUAD tissues. Consistent with TCGA analysis, YTHDF1 was overexpressed in all 6 GEO datasets, while YTHDF2 was upregulated in 5 GEO datasets. In contrast, the expression of WTAP, RBM15B, METTL14, METTL16, YTHDF3, YTHDC, and ALKBH5 was not significantly different between LUAD and non-tumor tissues. The details are shown in Figure 1B. Generally, these results suggested that m6A-related genes are frequently dysregulated in various LUAD clinical cohorts.
Figure 1.
Fourteen m6A-related genes were differentially expressed in LUAD patients. The t test (unpaired, two-tailed) was used for statistical analysis. (A) mRNA expression of m6A-related genes in TCGA data analysis in LUAD tissues and paired normal tissues. (B) The heatmap indicates the expression of m6A-related genes in TCGA and 6 GEO databases. TCGA – The Cancer Genome Atlas; LUAD – lung adenocarcinoma; NS – not significant; * P<0.05; ** P<0.01; *** P<0.001.
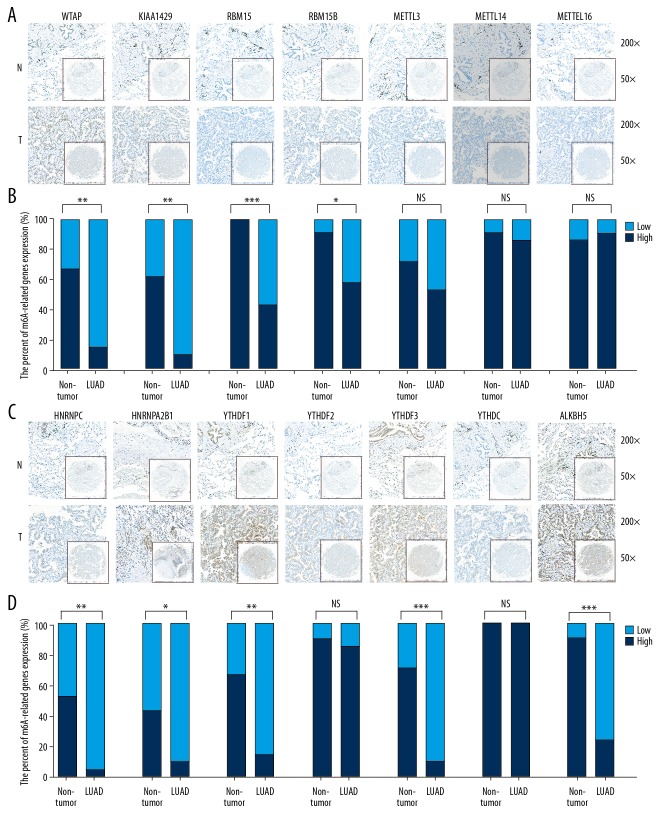
The protein expression profile of m6A-related genes in LUAD
Considering the complexity of post-transcriptional regulation, we further used IHC to analyze the expression differences of m6A-related genes at the protein level. A TMA cohort containing 21 LUAD tissues and their corresponding normal tissues was used in the IHC analysis. The results indicated that the protein expression of 4 m6A “writers” – WTAP, KIAA1429, RBM15, and RBM15B – was upregulated in LUAD tissues compared to normal tissues (Figure 2A, 2B). mRNA expression analysis indicated that m6A “readers” were upregulated and similar protein expression results were obtained except for YTHDF2 and YTHDC1. The protein expression of the other 4 m6A “readers” – HNRNPC, HNRNPA2B1, YTHDF1, and YTHDF3 – was upregulated in LUAD tissues as compared to non-tumor tissues (Figure 2C, 2D). In m6A “erasers”, the expression of ALKBH5 was upregulated in LUAD tissues at the protein level (Figure 2C, 2D). Upregulated genes depicted a similar expression pattern at both the mRNA level and at the protein levels. For instance, the protein expression levels of the 7 highly-expressed mRNAs were higher in LUAD tissues than in normal tissues except for METTL3 and YTHDF2 (Figure 2). However, other molecules like METTL14, METTL16, and YTHDC, which demonstrate similar mRNA expression levels between LUAD tissues and non-tumor tissues, also did not show significant differences at the protein level. The protein expression of m6A-related genes obtained in the HPA database (Figure 3) was similar to the one reflected in our TMA cohort.
Figure 2.
The protein expression of m6A-related genes in LUAD tissues and paired non-tumor samples. (A–C) The protein expression in LUAD tissues and paired non-tumor samples. (B–D) Some proteins, including WTAP, KIAA1429, RBM15, RBM15B, HNRNPC, HNRNPA2B1, YTHDF1, YTHDF3, and ALKBH5, were overexpressed in LUAD tissues. The t test was conducted for statistically analysis. LUAD – lung adenocarcinoma; NS – not significant; N – normal; T – tumor; * P<0.05; ** P<0.01; *** P<0.001.
Figure 3.
The protein expression of m6A-related genes in HPA cohort, which were largely similarly to our TMA cohort.
The clinicopathological data of m6A-associated genes in LUAD patients
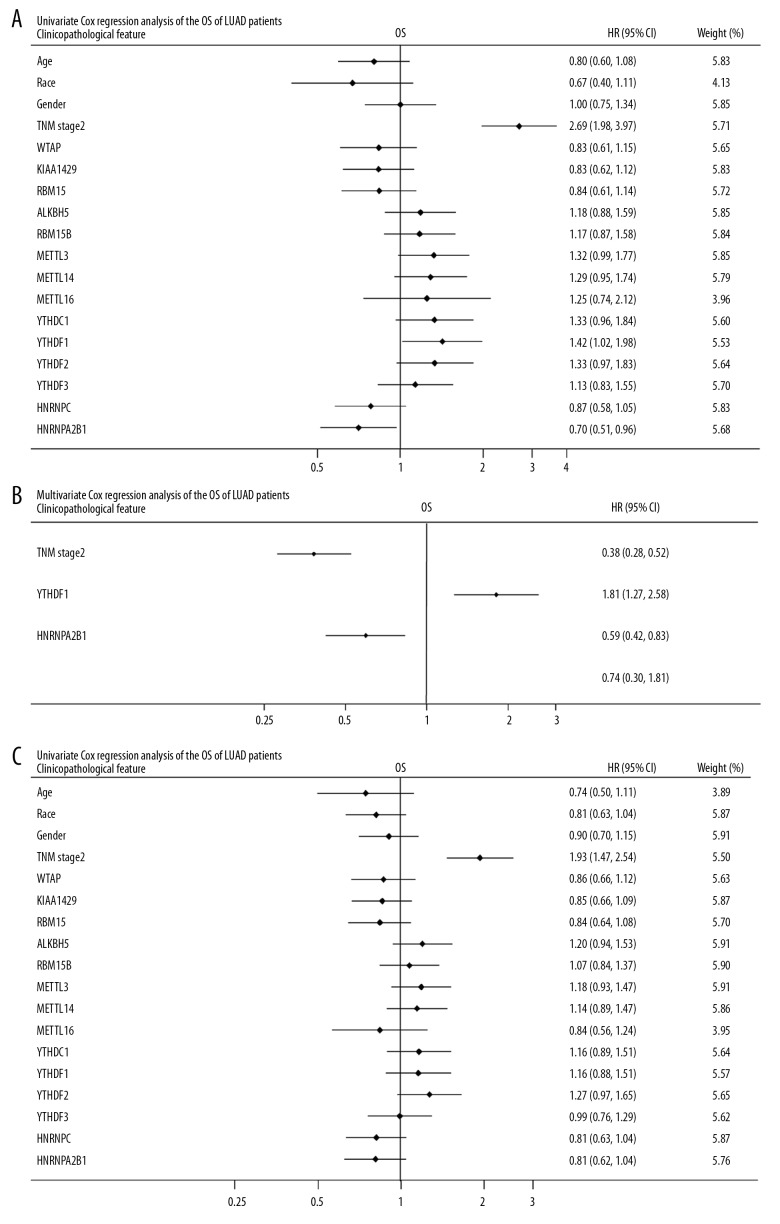
The clinicopathological data of LUAD obtained from the TCGA data portal is detailed in Figure 4. We observed that METTL3 was differentially expressed in LUAD patients among different races and ages. KIAA1429 was upregulated in male LUAD patients compared to female patients. METTL14 and YTHDF2 were differentially expressed in different TNM stages. The results of univariate and multivariate Cox analyses of TCGA-LUAD databases, as displayed in forest plots, are shown in Figure 5. These results confirm that m6A-related genes were differentially expressed in the LUAD cohort.
Figure 4.
The clinicopathological data of LUAD TCGA data are shown in detail. The t test was used for comparison between 2 groups. P<0.05 was considered to indicate statistical significance. (A) METTL3 was differentially expressed in LUAD patients of different races. (B) METTL3 was differentially expressed at different ages. (C) KIAA1429 was upregulated in male LUAD patients compared to female patients. (D) METTL14 and YTHDF2 were differentially expressed in different TNM stages. LUAD – lung adenocarcinoma; NS – not significant; * P<0.05; ** P<0.01; *** P<0.001.
Figure 5.
Forest plot depicting correlations between the indicated factors and the OS and RFS of TCGA-LUAD cohort. R studio was used to determine the best cutoff value. The correlations between m6A-related genes and OS, as well as RFS, were performed by univariate and multivariate Cox regression analyses. (A) The univariate analysis of the overall survival rate. (B) The multivariate analysis of the overall survival rate revealed that TNM stage 2, YTHDF1, and HNRNPA2B1 were independent predictors of OS rate. (C) The univariate analysis of recurrence-free survival rate. TCGA – The Cancer Genome Atlas; LUAD – lung adenocarcinoma; OS – overall survival; RFS – recurrence-free survival.
M6A dysregulation generates predictive signatures in LUAD patients
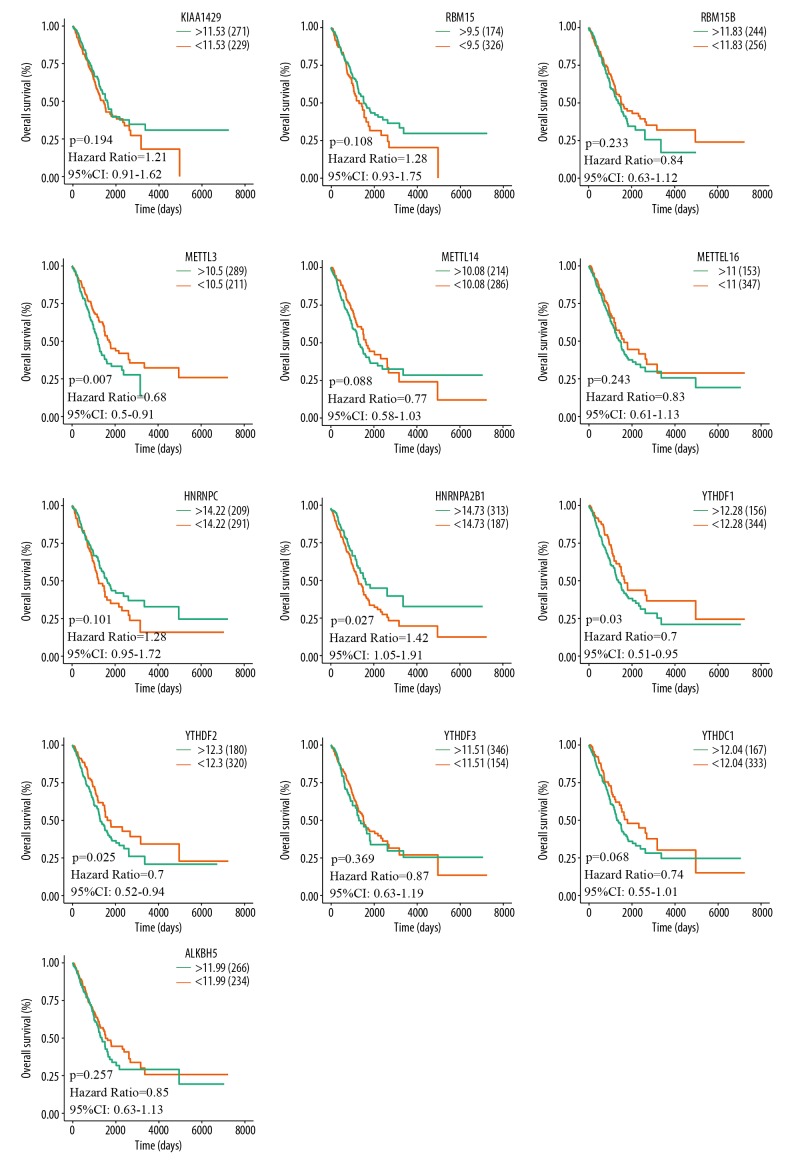
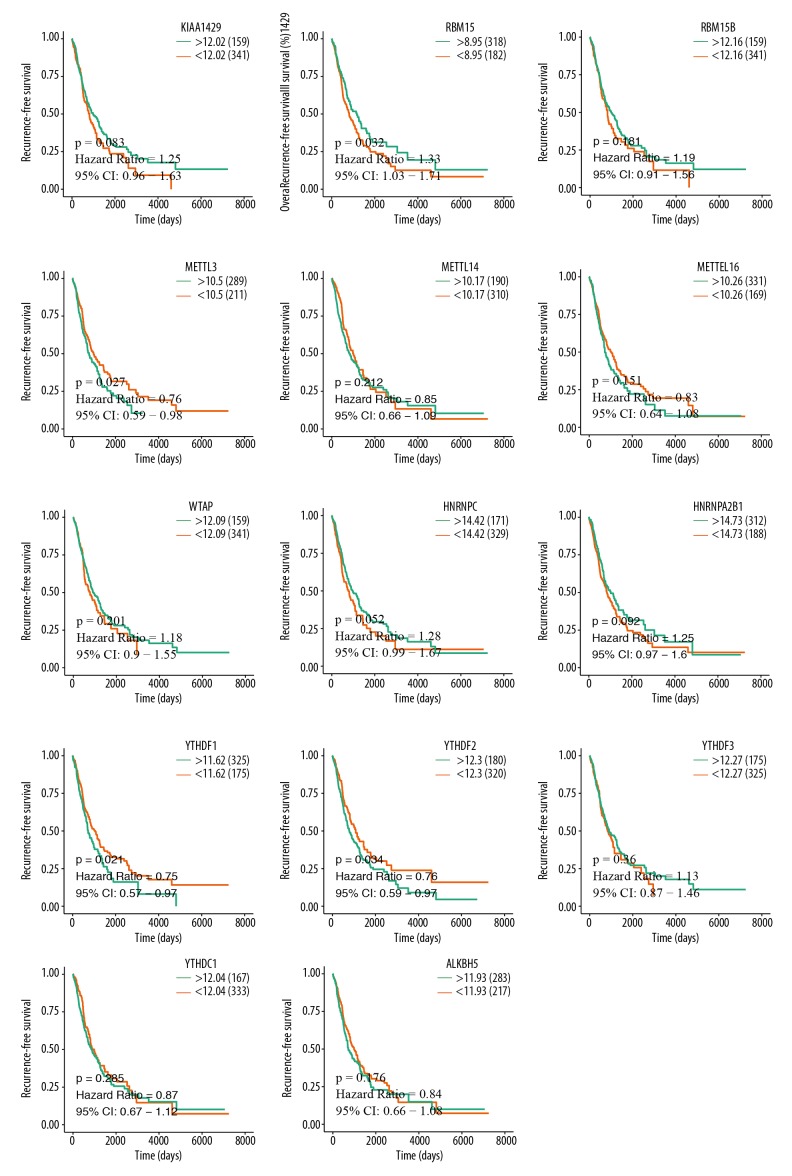
An assessment of whether the abnormal expression of m6A-related genes was correlated with the clinical progression and outcomes of LUAD patients was done by analyzing the overall survival and recurrence-free survival data of the TCGA-LUAD cohort. The analysis results revealed that METTL3 (P=0.007), HNRNPA2B1 (P=0.027), YTHDF1 (P=0.03), and YTHDF2 (P=0.025) were significantly associated with overall survival (Figure 6). Also, RBM15 (P=0.032), METTL3 (P=0.027), YTHDF1 (P=0.021), YTHDF2 (P=0.034) were obviously associated with recurrence-free survival (Figure 7). These results indicated that METTL3, YTHDF1, and YTHDF2 expression are associated with the prognosis of LUAD patients.
Figure 6.
Analysis of the relationships between m6A-related genes expression and OS in LUAD TCGA samples was performed using R studio. Patients with upregulation of METTL3, YTHDF1, and YTHDF2 were correlated with better OS rate and patients with overexpression of HNRNPA2B1 were associated with worse OS rate. TCGA – The Cancer Genome Atlas; LUAD – lung adenocarcinoma; OS, overall survival.
Figure 7.
Analysis of the relationships between m6A-related genes expression and RFS in LUAD TCGA samples was performed using R studio. Patients with increased expression of RBM15 were significantly associated with poor RFS, and better RFS was related to elevated METTL3, YTHDF1, and YTHDF2 expression. TCGA – The Cancer Genome Atlas; LUAD – lung adenocarcinoma; RFS – recurrence-free survival.
Discussion
Lung adenocarcinoma (LUAD) is one of the most common causes of global cancer-related mortalities worldwide [11]. Numerous studies have revealed that the occurrence and progression of tumors can be facilitated by genomic and epigenetic alterations [12], such as DNA methylation [13]. For instance, modifications in m6A, the most common form of mRNA modification, have been implicated in tumor proliferation, invasion, and metastasis. The findings of Lin et al. revealed that METTL3, a modified form of m6A, promotes the proliferation and mobility of gastric cancer cells [14]. Also, Zhou et al. verified that the alternation of m6A is associated with a pathologic stage in clear cell renal carcinoma [15].
In the present study, we assessed whether m6A-related genes could serve as novel biomarkers for LUAD. From the TCGA databases, we found that some m6A-associated genes, including KIAA1429, RBM15, METTL3, HNRNPC, HNRNPA2B1, YTHDF1, and YTHDF2, are upregulated in LUAD compared to matched normal patients. Consistent with these findings, analysis of data from 6 GEO databases revealed that YTHDF1 and YTHDF2 were also overexpressed in LUAD patients. Previous studies have shown that expression of m6A-related genes is dysregulated in several cancers, including breast cancer [16], lung cancer [17], and glioma [18]. For example, the expression of METTL3 is significantly upregulated in non-small-cell lung carcinoma (NSCLC) [19]. A previous study reported that m6A is associated with the differentiation pathways that determine the fate of stem cells. Notably, the pathways involved in the maintenance and differentiation of embryonic stem cells have been directly linked to the acquisition of stem cell properties in both solid and hematological malignancies. Subsequently, m6A alterations could have a role in cancer development [20].
Moreover, the results of this study indicate that METTL3, YTHDF1, YTHDF2, and HNRNPA2B1 are correlated with the overall survival rate. We also found that METTL3, YTHDF1, YTHDF2, and RBM15 are linked to the rate of recurrence-free survival. Overexpression of ALKBH5 has been associated with a lower survival time of glioblastoma [21]. Moreover, Chen et al. found that the RNA methyltransferase METTL3 promotes the progression of liver cancer through YTHDF2-dependent post-transcriptional silencing of suppressors of cytokine signaling 2 (SOCS2) [22]. In the present study, patients with high expression of METTL3, YTHDF1, and YTHDF2 were associated with better OS while patients with overexpression of HNRNPA2B1 were linked with poor OS. Whereas patients with high expression of RBM15 were associated with poor RFS, patients with elevated expression of METTL3, YTHDF1, and YTHDF2 were associated with better RFS. These findings suggest that METTL3, YTHDF1, and YTHDF2 are promising LUAD prognostic biomarkers. Notably, METTL3 has been found to promote protein translation of oncogenes in lung cancer cells through methyltransferase-independent activity [23]. Recent studies have found that t METTL3 knockdown triggers apoptosis and modulation of p53 signaling in cancer cells, indicating that METTL3 is essential in the survival of cancer cells [24–26]. The YTH domain-containing proteins, such as YTHDF1 and YTHDF2, are recognized as m6A “reader” proteins [27]. YTHDF1 enhances the efficiency of mRNA translation, while YTHDF2 influences mRNA stability [28,29]. Recent studies have highlighted that RNA methylation affects cancer self-renewal and cell fate [30–32]. Also, methylation of RNA is involved in cancer stem cell (CSC) pluripotency, extensive cell proliferation, metastasis, and potentially in tumor immunity. Subsequently, RNA methylation could provide a novel therapeutic avenue, and should, therefore, be explored further [33–35]. The identification of consistently altered gene expression patterns is an essential tool in basic and clinical cancer research. In this study, we demonstrated for the first time that m6A-related genes are partially upregulated in lung adenocarcinoma and are potential diagnostic biomarkers and prognostic indicators in lung adenocarcinoma.
Modification in m6A form is the most ubiquitous regulation mechanism in RNAs processing, which is mediated by 3 categories of RNA-modification enzymes. The enzyme categories include M6A “writers,” M6A “erasers,” and M6A “readers.” M6A “writers” such as METTL3 modify RNA by methylation, but the methylation status of RNAs can be reversed by m6A “erasers” like FTO and ALKBH5 [7,36]. On the other hand, m6A “reader” proteins recognize m6A modification by binding specific domains, followed by RNA splicing, mRNA decay, and translation regulation [37,38]. These mechanisms facilitate m6A-related molecules to manipulate carcinoma progression and deterioration. However, the current body of knowledge on the mechanistic link between m6A and human carcinogenesis is limited. Therefore, elucidation of the function of m6A in tumor progression is of significance and could shed light on future clinical cancer therapy and drug developments.
Conclusions
The expression patterns of m6A-related genes differ substantially between LUAD and matched normal tissues. The functions of m6A-related genes METTL3, YTHDF1, and YTHDF2 appear to be associated with the prognosis of LUAD.
Supplementary Data
Supplementary Table 1.
Information on antibodies used in this study.
| Group | Antibody | IHC | Specificity | Company |
|---|---|---|---|---|
| Writers | WTAP | 1: 500 | Mouse monoclonal | Proteintech Group, China |
| KIAA1429 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| RBM15 | 1: 500 | Rabbit polyclonal | Proteintech Group, China | |
| RBM15B | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| METTL3 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| METTL14 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| METTL16 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| Erasers | ALKBH5 | 1: 500 | Rabbit polyclonal | Proteintech Group, China |
| Readers | HNRNPA2B1 | 1: 500 | Rabbit polyclonal | Proteintech Group, China |
| HNRNPC | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDF1 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDF2 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDF3 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDC1 | 1: 200 | Rabbit polyclonal | Proteintech Group, China |
IHC – immunohistochemistry.
Footnotes
Source of support: This work was supported by funds from the National Natural Science Foundation of China (81702757 and 81702346)
Conflicts of interest
None.
References
- 1.Lee JJ, Park S, Park H, et al. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell. 2019;177(7):1842–57. doi: 10.1016/j.cell.2019.05.013. e21. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Cao L, Wang X, et al. Neuron-specific enolase is an independent prognostic factor in resected lung adenocarcinoma patients with anaplastic lymphoma kinase gene rearrangements. Med Sci Monit. 2019;25:675–90. doi: 10.12659/MSM.913054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Wang R, Yu C, et al. FOX-A1 contributes to acquisition of chemoresistance in human lung adenocarcinoma via transactivation of SOX5. EBioMedicine. 2019;44:150–61. doi: 10.1016/j.ebiom.2019.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwok CT, Marshall AD, Rasko JE, Wong JJ. Genetic alterations of m(6)A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol. 2017;10:39. doi: 10.1186/s13045-017-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Zhang YC, Huang C, et al. m(6)A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics. 2019;17(2):154–68. doi: 10.1016/j.gpb.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rong ZX, Li Z, He JJ, et al. Downregulation of fat mass and obesity associated (FTO) promotes the progression of intrahepatic cholangiocarcinoma. Front Oncol. 2019;9:369. doi: 10.3389/fonc.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Li Y, Yue M, et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Liu L, Dong Z, et al. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am J Transl Res. 2019;11:3972–91. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu W, Li H, Yu Y, et al. Enolase-1 serves as a biomarker of diagnosis and prognosis in hepatocellular carcinoma patients. Cancer Manag Res. 2018;10:5735–45. doi: 10.2147/CMAR.S182183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Yuan Y, Li Y, et al. An inverse interaction between HOXA11 and HOXA11-AS is associated with cisplatin resistance in lung adenocarcinoma. Epigenetics. 2019;14:949–60. doi: 10.1080/15592294.2019.1625673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10584–97. doi: 10.3748/wjg.v21.i37.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, Liu G, Zhou F, et al. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18:1–14. doi: 10.1007/s10238-017-0467-0. [DOI] [PubMed] [Google Scholar]
- 14.Lin S, Liu J, Jiang W, et al. METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Wars) 2019;14:25–31. doi: 10.1515/med-2019-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Wang J, Hong B, et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma – a retrospective study using TCGA database. Aging (Albany NY) 2019;11:1633–47. doi: 10.18632/aging.101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L, Wu D, Ning J, et al. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19:326. doi: 10.1186/s12885-019-5538-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Ren D, Du Z, et al. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–64. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 18.Chai RC, Wu F, Wang QX, et al. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY) 2019;11:1204–25. doi: 10.18632/aging.101829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du M, Zhang Y, Mao Y, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–89. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 20.Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–6. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Zhao BS, Zhou A, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen M, Wei L, Law CT, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 23.Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177–87. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou R, Gao Y, Lv D, et al. METTL3 mediated m(6)A modification plays an oncogenic role in cutaneous squamous cell carcinoma by regulating DeltaNp63. Biochem Biophys Res Commun. 2019;515(2):310–17. doi: 10.1016/j.bbrc.2019.05.155. [DOI] [PubMed] [Google Scholar]
- 25.Cheng M, Sheng L, Gao Q, et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-kappaB/MYC signaling network. Oncogene. 2019;38:3667–80. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 26.Hua W, Zhao Y, Jin X, et al. METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecol Oncol. 2018;151:356–65. doi: 10.1016/j.ygyno.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Du K, Zhang L, Lee T, Sun T. m(6)A RNA methylation controls neural development and is involved in human diseases. Mol Neurobiol. 2019;56:1596–606. doi: 10.1007/s12035-018-1138-1. [DOI] [PubMed] [Google Scholar]
- 28.Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. 2018;90:6380–84. doi: 10.1021/acs.analchem.8b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao S, Sun H, Xu C. YTH domain: A family of N(6)-methyladenosine (m(6)A) readers. Genomics Proteomics Bioinformatics. 2018;16:99–107. doi: 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang G, Leu JS, Ma L, et al. Methylation of RNA N(6)-methyladenosine in modulation of cytokine responses and tumorigenesis. Cytokine. 2019;118:35–41. doi: 10.1016/j.cyto.2018.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Dixit D, Xie Q, Rich JN, Zhao JC. Messenger RNA methylation regulates glioblastoma tumorigenesis. Cancer Cell. 2017;31:474–75. doi: 10.1016/j.ccell.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen WC, Lai YC, Li LH, et al. Methylation and PTEN activation in dental pulp mesenchymal stem cells promotes osteogenesis and reduces oncogenesis. Nat Commun. 2019;10:2226. doi: 10.1038/s41467-019-10197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai Y, Yang C, Wu R, et al. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vu LP, Cheng Y, Kharas MG. The biology of m(6)A RNA methylation in normal and malignant hematopoiesis. Cancer Discov. 2019;9:25–33. doi: 10.1158/2159-8290.CD-18-0959. [DOI] [PubMed] [Google Scholar]
- 36.Song H, Feng X, Zhang H, et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15(8):1419–37. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan Q, Liu PY, Haase J, et al. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–92. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 38.Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 2018;28:507–17. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Information on antibodies used in this study.
| Group | Antibody | IHC | Specificity | Company |
|---|---|---|---|---|
| Writers | WTAP | 1: 500 | Mouse monoclonal | Proteintech Group, China |
| KIAA1429 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| RBM15 | 1: 500 | Rabbit polyclonal | Proteintech Group, China | |
| RBM15B | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| METTL3 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| METTL14 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| METTL16 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| Erasers | ALKBH5 | 1: 500 | Rabbit polyclonal | Proteintech Group, China |
| Readers | HNRNPA2B1 | 1: 500 | Rabbit polyclonal | Proteintech Group, China |
| HNRNPC | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDF1 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDF2 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDF3 | 1: 200 | Rabbit polyclonal | Proteintech Group, China | |
| YTHDC1 | 1: 200 | Rabbit polyclonal | Proteintech Group, China |
IHC – immunohistochemistry.