Abstract
Background
Talazoparib (BMN673) is a new poly(ADP-ribose) polymerase inhibitor that has been FDA approved for patients suffering from metastatic breast cancer with germline BRCA mutations.
Method and Results
In the current study, an accurate and efficient liquid chromatography-tandem mass spectrometry (LC–MS/MS) analytical methodology was developed for TZB estimation in addition to its metabolic stability assessment. TZB and lapatinib (LAP) (which is chosen as an internal standard; IS) were separated using reversed phase elution system (Hypersil C18 column) with an isocratic mobile phase. The linearity range of the established method was 5–500 ng/mL (r2 ≥ 0.999) in the human liver microsomes (HLMs) matrix. Different parameters were calculated to confirm the method sensitivity (limit of quantification was 2.0 ng/mL), and reproducibility (intra- and inter-day precision and accuracy were below 3.1%) of our methodology. For evaluation of TZB metabolic stability in HLM matrix, intrinsic clearance (9.59 µL/min/mg) and in vitro half-life (72.7 mins) were calculated. TZB treatment discontinuations were reported due to adverse events and dose accumulation, so in silico metabolic vulnerability (experimental and in silico) and toxicity assessment (in silico) of TZB were performed utilizing P450 Metabolism and DEREK modules of StarDrop software.
Conclusion
TZB is slowly metabolized by the liver. TZB was reported to be minimally metabolized by the liver that approved our outcomes. We do recommend that plasma levels be monitored in cases when talazoparib is used for a long period of time, since it is possible for TZB to bioaccumulate after multiple doses to toxic levels. According to our knowledge, the current method is considered the first LC-MS/MS methodology for evaluating TZB metabolic stability. Further drug discovery studies can be done depending on this concept allowing the designing of new series of compounds with more safety profile through reducing side effects and improving metabolic behavior.
Keywords: talazoparib, metabolic stability evaluation, human liver microsomes, tandem mass spectrometry, DEREK, StarDrop software
Introduction
Breast cancer is considered the major reason of death in women, and as stated by the American Cancer Society, in 2018, there were around 41,400 deaths associated with breast cancer in the USA.1 The frequent germline mutations linked to breast cancer are in the ovarian and breast cancer susceptibility gene 1 (BRCA1) or BRCA.2,3 Around 10% of breast cancer patients were diagnosed with germline BRCA1/2 mutations, that make their disease more vulnerable to a poly(ADP-ribose) polymerase (PARP) inhibitors (e.g. talazoparib) that is considered as a new class of drugs.4
PARP inhibitors are ADP ribose molecular analogs that stop the interaction among the PARP enzyme and ADP ribose, and also perform as a PARP capturing that not only affects DNA repair, transcription and replication as it also has direct fatal breaks of double-strand DNA through collapsing stalled replication forks during S-phase. Presently, there are two PARP FDA-approved drugs for metastatic breast cancer: olaparib (Lynparza, AstraZeneca Inc.) and talazoparib (Talzenna, Pfizer).5
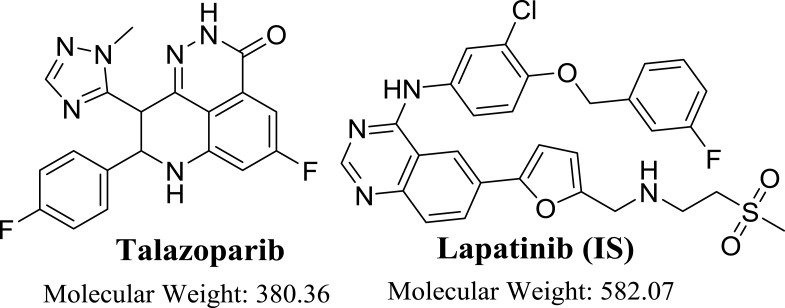
Talazoparib (BMN673, Figure 1) is utilized for the treatment of patients with metastatic breast cancer that were diagnosed with germline BRCA mutations. It has been FDA approved after a successful Phase III trial exhibited greater progression-free survival if compared to chemotherapy. TZB exhibited more potent antitumor responses at very lower concentrations if compared to other PARP inhibitors6,7 and also exhibited a higher degree of catalytic inhibition of PARP enzymes. TZB has also showed 100 fold potency at PARP–DNA complexes trapping in preclinical studies if compared to olaparib.6–8
Figure 1.
Chemical structure of talazoparib and lapatinib (IS).
Literature review revealed that no published work was found for TZB quantification in HLMs or TZB metabolic stability investigation. Consequently, these outcomes prompted us for establishing a reliable and accurate analytical method for estimation of TZB level. Accordingly, an isocratic LC-MS/MS analytical method was developed for the assessment of TZB concentration in HLMs matrix. The discussed procedure provided about 99.5 ± 2.43% recoveries.
For estimation of in vivo metabolic clearance rate using in vitro intrinsic clearance, three models were used including dispersion, parallel tube and venous equilibrium.9,10 In the current work, the metabolic stability of TZB including intrinsic clearance and in vitro half-life [t1/2] in HLMs were computed according to “in vitro half-life” approach, using the “well-stirred” model11,12 as it is considered the most widely used model in in vitro drug metabolism experiments. These parameters could be utilized for calculation of different physiological parameters (e.g, in vivo t1/2 and hepatic clearance). TZB treatment discontinuations were reported due to adverse events and dose accumulation.13 So, metabolic stability (in silico and experimental) and toxicological study (in silico) were done. The estimation of a drug bioavailability gives a picture of its metabolic fate. If it is a rapidly metabolized drug, it will show a low in vivo bioavailability value thus short duration of action.14 TZB is a very slow extraction ratio drug that is slowly excreted from the body compared to other previous studied TKIs in our laboratory.15–18 This reveals the possibility of dose accumulation inside the body, so TZB conc. should be monitored carefully.
Experimental
Materials
Pooled HLMs (Product Number: M 0567, male human liver) was purchased from Sigma-Aldrich (USA) and stored at −70°C. HLMs consists of a mixture of liver microsomes pooled from various individual human donors. Standard powders are of analytical grade (AR) and organic solvents are HPLC grade. HPLC grade water (H2O) was arranged by in situ Milli-Q plus filtration system (USA). Talazoparib (99.89%) and lapatinib (99.83%) were procured from Med Chem. Express Company (USA). Acetonitrile, HLMs, pooled (M0567), formic acid (HCOOH) and ammonium formate (NH4COOH) were procured from Sigma-Aldrich company (USA).
LC-MS/MS Methodology
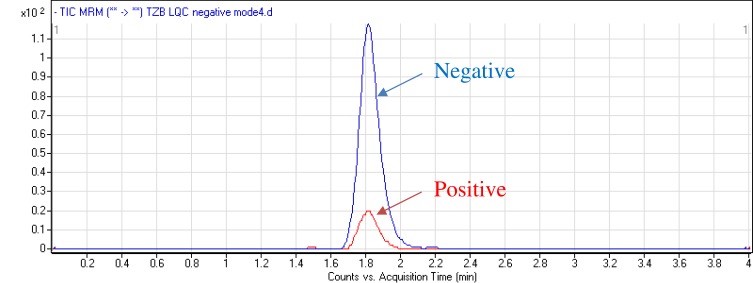
Chromatographic parameters were adjusted to attain the optimum separation of TZB and LAP (IS) with acceptable elution time of 4 mins. The total run time is divided into three segments: from 0.0 min to 1.0 min to waste, from 1.0 to 2.3 mins TZB MRM parameters and from 2.3 to 4.0 mins LAP MRM parameters. The aqueous part pH (10 mM ammonium formate) of the mobile phase was fixed at 3.8 as higher pH caused a peak tailing and retention time delay. The aqueous/organic part (acetonitrile) ratio was fixed at 32:75% as increase acetonitrile ratio resulted in bad resolution and overlapped peaks while acetonitrile decrease generated elution time increase. Various stationary phases were utilized as HILIC columns but TZB and LAP were not retained and optimum results were achieved using an Agilent Hypersil BDS-C18 column (125 mm length, 3.0 μm particle size and 2.0 mm internal diameter). MRM analyzer mode was utilized for TZB estimation to discard any interference from the HLM matrix components and increase the developed method sensitivity. The elution time for TZB and LAP was 4 mins with good separation. The chemical structure of TZB contains both acidic and basic groups, so it could be analyzed in both negative and positive mass analyzer modes. The intensity of the LQC peak of TZB at negative mode is high comparing to positive mode (Figure 2).
Figure 2.
MRM chromatograms of LQC of TZB (15 ng/mL) under positive and negative analyzer modes revealing high intensity at positive ESI mode.
A triple quadrupole (QqQ) mass analyzer with an electrospray ionization source (ESI) operated in the negative charge mode for TZB mass analysis and in the positive mode for LAP detection was used for analytes estimation. Nitrogen gas (11 L/min) was utilized for drying for nebulizer spray inside the ESI source and as a collision gas (55 psi) for product ion generation in the collision cell. Flow injection analysis was used for mass parameters optimization to attain the highest ion intensity so as to elevate sensitivity of the developed analytical methodology. ESI temperature and capillary voltage were fixed at 350°C and 4000 V, respectively. Data analysis was performed utilizing the Agilent Mass Hunter software.
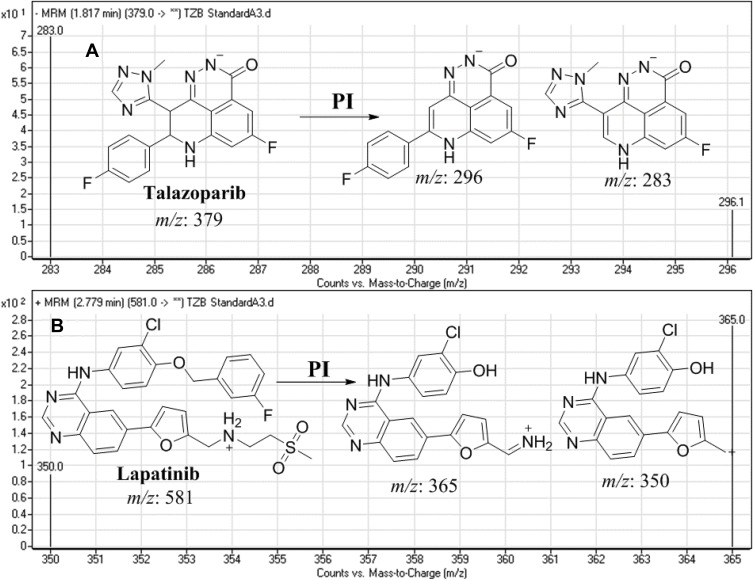
TZB was estimated using multiple reaction monitoring (MRM) mass analyzer mode for the mass transitions (parent to daughter fragments) from 379→296 and 379→283 for TZB and 581→365 and 581→350 for LAP). The fragmentor voltage (FV) was fixed at 140 and 145 V with collision energy (CE) of 30 eV and 32 eV for TZB, and FV of 140 V and 142 V with CE of 30 eV and 32 eV for LAP. MRM mode was utilized for TZB quantification to remove any interference from the HLM matrix constituents and increase the LC-MS/MS method sensitivity (Figure 3).
Figure 3.
MRM mass transitions of (A) talazoparib (TZB) and (B) lapatinib (IS) showing fragmentation pattern.
TZB and LAP Working Solutions
TZB and LAP are dimethyl sulfoxide soluble. TZB working solution 1 (WK1, 200 µg/mL) was prepared by dilution of the TZB (2 mg/mL) 10 times with mobile phase. TZB WK2 (20 µg/mL) was prepared by dilution of the (WK1, 200 µg/mL) 10 times with mobile phase. LAP sock solution was prepared in DMSO (1 mg/mL) then serially diluted with mobile phase to prepare LAP WK3 (2 µg/mL).
Establishing TZB Calibration Curve
TZB WK2 (20 µg/mL) was mixed with a 30 µL HLM matrix (1 mg protein) in phosphate buffer (total volume is 1 mL) to prepare 9 calibration levels: 5, 10, 30, 50, 80, 100, 200, 300 and 500 ng/mL that were used for calibration curve creation. Other three concentrations (15, 150, and 400 ng/mL) were chosen as the low-quality control (LQC), medium-quality control (MQC), and high-quality control (HQC), respectively. One hundred µL of LAP WK3 was then added to each calibration standards and quality controls. Protein precipitation method was utilized for analytes extraction (TZB and LAP) according to the following steps: 1 – Two mL of acetonitrile (precipitating agent) was added to prepared standards. 2 – Shaking for tubes for 5 mins in a shaker to ensure good mixing. 3 – Centrifugation for standards at 14,000 rpm for 12 mins was performed in a thermostated centrifuge (4°C) to discard precipitated proteins. 4 – One mL of each supernatant was filtered through a 0.22 µm syringe filter. 5 – Filtrates were transferred to 1.5 mL HPLC vials. 6 – One µL of each sample was injected into the LC-MS/MS system. Control samples were prepared following the same steps without utilizing the HLM matrix to confirm the absence of probable interference from the HLMs matrix. A linear calibration curve was established by plotting the peak area ratio of TZB to LAP (y-axis) and nominal values of TZB (x-axis). The linearity of the described methodology was confirmed by computing the linear regression equation.
Method Validation
Various parameters were computed for verifying validation of the established methodology. The analytical LC-MS/MS method was validated following FDA guidelines. The least squared statistical method (y = ax + b) was utilized for proving the linearity of the established calibration curve that was confirmed by the r2. LC-MS/MS methodology showed linearity in the range of 5 ng/mL to 500 ng/mL.
TZB Metabolic Stability
The TZB metabolic stability was analyzed by calculating the TZB remaining conc. after incubation with HLMs. Briefly, 1 µM of TZB was incubated with 30 µL HLMs (1 mg microsomal protein) in 1 mL phosphate buffer and the same experiment was repeated for three times to confirm results. Also, negative controls either in the absence of TZB or NADPH were utilized to validate the outcomes and to approve that the decrease in TZB conc. is metabolically mediated. The metabolic reaction was done at pH 7.4 (phosphate buffer) and 3.3 mM magnesium chloride. First, pre-incubation was done at 37°C for 10 mins. Second, the initiation of the metabolic reactions was done using NADPH (1 mM) for specific time. Third, stopping of the reaction was performed at time intervals: 0.0, 1.0, 2.5, 5.0, 7.5, 15, 30, 40 and 50 mins by adding 2 mL acetonitrile. From the generated data after sample analysis, TZB metabolic stability curve was constructed. The same experiments were repeated in the absence of NADPH or HLMs to confirm that the rate measured was metabolically mediated. The % TZB remaining is plotted versus incubation time. From this plot, time points in the linear range are selected to plot the natural logarithm (ln) of % TZB remaining versus time. The slope of the linear part indicates the rate constant for the disappearance of TZB that was utilized for in vitro t1/2 calculation following the next equation:
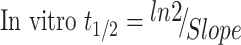 |
Then, TZB Clint was computed by applying the next equation:19
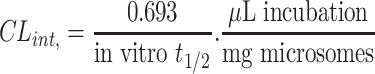 |
In silico prediction of TZB metabolic stability and toxicity using P450 Metabolism and DEREK modules of StarDrop software.
Identification of TZB vulnerability for metabolism was indicated by the composite site lability (CSL). The outcomes from the WhichP450™ module are presented by the pie chart and utilized for the identification of most likely cyp450 isoform that has the main role in TZB metabolism. Screening for the predicted toxicity of TZB was performed using DEREK software that was also utilized to screen for structural alerts for TZB.
Results and Discussions
LC–MS/MS Methodology
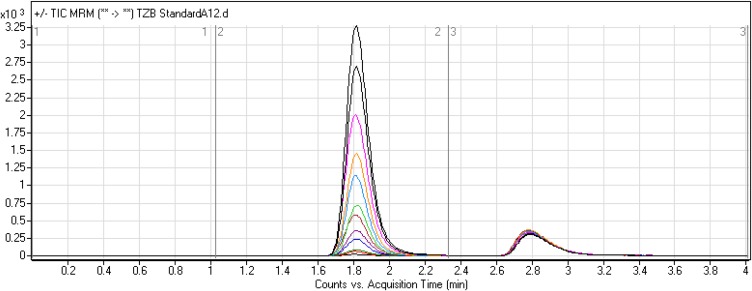
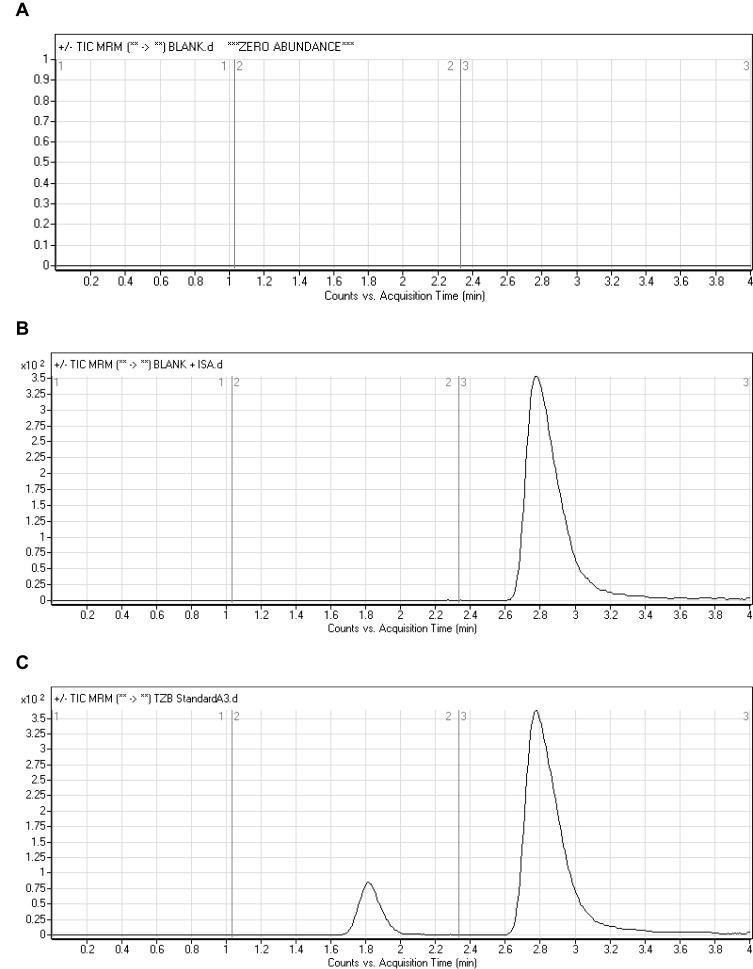
LAP was chosen as IS in TZB quantification as the same method of extraction could be applied for both TZB and LAP in the HLM matrix (TZB and LAP recoveries were 99.62 ± 2.3% and 97.2 ± 1.3%, respectively) and the elution time of LAP is near to that of TZB. The established procedure is fast with 4 mins run time (Figure 4). Both TZB and LAP are TKIs and were not concurrent given to the same patient, so the developed LC-MS/MS method could be used for pharmacokinetics or therapeutic drug monitoring (TDM) for patients under TZB treatment. Figure 5C shows the TZB LQC MRM chromatograms.
Figure 4.
Overlayed MRM chromatograms of TZB calibration levels.
Figure 5.
MRM chromatograms of (A) blank HLMs, (B) blank HLMs with IS, and (C) LQC of TZB (15 ng/mL).
Validation Parameters
Specificity
Figure 5 shows good separation of the TZB and LAP peaks and the absence of endogenous peaks in the blank HLM matrix at the same retention time of TZB and LAP that confirmed the current analytical method specificity. No carry over effect of TZB and LAP was observed in the MRM mass spectra chromatograms. TZB and LAP chromatographic peaks were eluted at 1.82 mins and 2.78 mins, respectively.
Sensitivity and Linearity
The linearity range and r2 for the proposed analytical method were 5–500 ng/mL and ≥0.9999, respectively. The line regression equation of TZB calibration curve was y = 1.4443x - 2.8203 R2 = 0.9995. LLQC peak exhibited high signal-to-noise (S/N) ratio and a perfect peak shape confirming the sensitivity of the LC-MS/MS method. RSD values of the six repetitions of each standard level were <3.81% (Table 1). The calculated LOD and LOQ were 0.61 ng/mL and 2.0 ng/mL, respectively. Back calculations for the 12 TZB standards (calibration standards and QC samples) in the HLM matrix firmly established the successfulness of the depicted analytical method.
Table 1.
Talazoparib (TZB) Back-Calculated Calibration Standards
| TZB Conc. (ng/mL) | Meana | SD | RSD (%) | Accuracy (%) |
|---|---|---|---|---|
| 5 (LLQC) | 4.94 | 0.13 | 2.60 | −1.18 |
| 10 | 9.47 | 0.26 | 2.79 | −5.32 |
| 15 (LQC) | 14.63 | 0.36 | 2.46 | −2.46 |
| 30 | 30.46 | 0.66 | 2.18 | 1.55 |
| 50 | 49.18 | 1.87 | 3.81 | −1.65 |
| 80 | 79.74 | 1.26 | 1.58 | −0.32 |
| 100 | 104.58 | 1.54 | 1.47 | 4.58 |
| 150 (MQC) | 151.60 | 1.90 | 1.26 | 1.07 |
| 200 | 199.02 | 1.56 | 0.78 | −0.49 |
| 300 | 294.03 | 2.90 | 0.99 | −1.99 |
| 400 (HQC) | 401.07 | 3.99 | 0.99 | 0.27 |
| 500 | 502.57 | 7.69 | 1.53 | 0.51 |
Note: aMean of six replicates.
Precision and Accuracy
Accuracies and precisions values are acceptable according to the FDA guidelines.20 The values of intra-day and inter-day precision and accuracy of the adopted methodology were 0.67 to 4.7 and −1.66 to 1.47, respectively (Table 2). The average TZB recoveries were 99.62 ± 2.3%.
Table 2.
Inter-Day and Intra-Day (Precision and Accuracy) of the Developed Methodology
| HLM Matrix | 5 ng/mL (LLQC) | 15 ng/mL (LQC) | 150 ng/mL (MQC) | 400 ng/mL (HQC) | ||||
|---|---|---|---|---|---|---|---|---|
| Inter-Day Assaya | Intra-Day Assayb | Inter-Day Assay | Intra-Day Assay | Inter-Day Assay | Intra-Day Assay | Inter-Day Assay | Intra-Day Assay | |
| Mean | 4.99 | 4.98 | 14.85 | 14.75 | 151.6 | 152.20 | 400.95 | 402.21 |
| SD | 0.23 | 0.15 | 0.37 | 0.27 | 1.90 | 2.02 | 6.12 | 2.71 |
| Precision (% RSD) | 4.70 | 3.04 | 2.49 | 1.82 | 1.26 | 1.33 | 1.53 | 0.67 |
| % Accuracy | −0.18 | −0.38 | −0.98 | −1.66 | 1.07 | 1.47 | 0.24 | 0.55 |
Notes: aAverage of six repeats for 3 days. bAverage of 12 repeats on the same day.
Extraction Recovery and Matrix Effects
Table 3 shows the recovery percentages of the QC samples for computing the TZB concentration in the HLM matrix. The recovery of TZB in the spiked HLM matrix was 99.62 ± 2.31% (relative standard deviation [RSD] <2.21%). LAP recovery was 97.2 ± 1.3. Matrix effect absence on TZB or LAP was validated by analyzing two HLMs batches (set 1), which were spiked with the TZB LQC (15 ng/mL) and LAP (100 ng/mL). Set 2 batches were prepared using the mobile phase instead of the HLM matrix. Matrix effect factor was computed by applying the next equation:
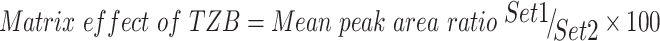 |
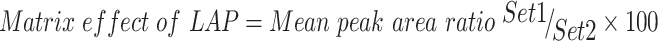 |
Table 3.
TZB Samples Recovery in HLMs
| Conc. of TZB (ng/mL) | HLMs Matrix | |||
|---|---|---|---|---|
| 5 | 15 | 150 | 400 | |
| Meana | 4.94 | 14.63 | 151.60 | 401.07 |
| SD | 0.13 | 0.36 | 1.90 | 3.99 |
| Precision (RSD %) | 2.60 | 2.46 | 1.26 | 0.99 |
| Recovery (%) | 98.82 | 97.54 | 101.07 | 100.27 |
Note: aMean of six repeats.
The tested HLMs containing TZB and LAP exhibited matrix effects of 99.62 ± 2.31% and 97.2 ± 1.3, respectively. IS normalized matrix effect (IS normalized MF) was calculated by applying the next equation:
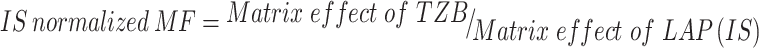 |
The IS normalized MF was 1.01 and it lies in the satisfactory range.21 Hence, these findings revealed that the HLM matrix had no obvious impact on the ionization of TZB and LAP.
Stability
We evaluated the stability of TZB in HLMs matrix (1 mg protein/1 mL phosphate buffer) under common laboratory storage conditions. TZB in HLM matrix (without NADPH) showed good stability after storage at −20°C for 30 days. Measured values were 96.8–102.12% for TZB. Stability data for TZB are described in Table 4. We did not observe the degradation of TZB under the mentioned conditions.
Table 4.
Stability of TZB in HLMs Matrix (1 mg/1 mL Phosphate Buffer) Under Different Laboratory Conditions
| TZB Conc. (ng/mL) | Mean (ng/mL) | Recovery % | Precision (RSD %) |
|---|---|---|---|
| Room Temperature for 8 hrs | |||
| 5 | 5.07±0.26 | 100.42 | 5.18 |
| 15 | 14.79±0.27 | 98.60 | 1.85 |
| 150 | 151.00±3.80 | 99.67 | 2.52 |
| 400 | 401.27±3.59 | 101.32 | 0.90 |
| Three Freeze-Thaw Cycles | |||
| 5 | 5.11±0.07 | 102.12 | 1.40 |
| 15 | 14.90±0.19 | 99.33 | 1.26 |
| 150 | 148.46±2.07 | 98.97 | 1.39 |
| 400 | 395.80±3.52 | 98.95 | 0.89 |
| Stored at 4°C for 24 hrs | |||
| 5 | 4.84±0.15 | 96.80 | 3.05 |
| 15 | 14.68±0.32 | 97.87 | 2.18 |
| 150 | 149.26±5.52 | 99.51 | 3.70 |
| 400 | 395.80±3.70 | 98.95 | 0.94 |
| Stored at −20°C for 30 Days | |||
| 5 | 4.87±0.13 | 97.34 | 2.73 |
| 15 | 14.57±0.34 | 97.14 | 2.34 |
| 150 | 144.58±2.32 | 96.39 | 1.60 |
| 400 | 395.80±4.74 | 98.95 | 1.20 |
Metabolic Stability
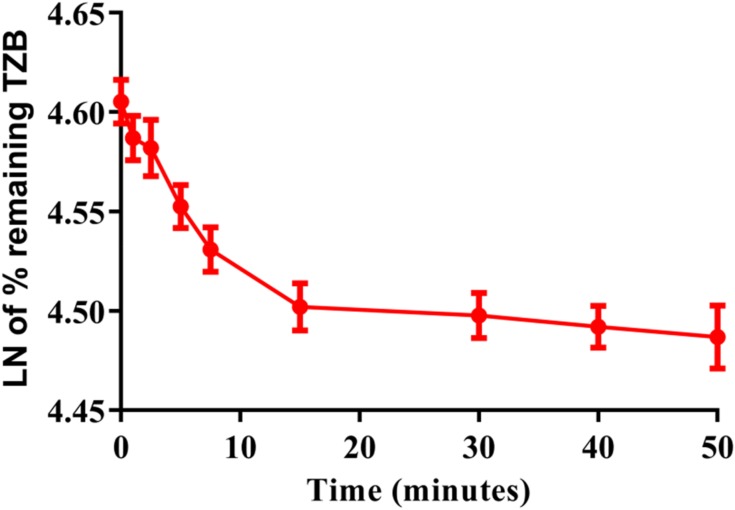
A conc. of TZB (1 µM) was incubated with HLMs (1mg/mL). The conc. of TZB at 1 1 µM were used to be sure that it is less than Michaelis-Menten constant so as to establish a linear relationship between incubation time and the metabolic rate. The conc. of HLMs at 1 mg/mL microsomal protein was used to be sure that no non-specific protein binding will exist. After incubation, extraction and purification of TZB, the conc. was calculated by displacing the peak area ratios in the calibration regression equation. The metabolic stability curve was constructed by plotting the ln percentage remaining of TZB (y-axis) versus the incubation time (x-axis) (Figure 6). The regression equation for the linear part of the curve was y = −0.0096x + 4.6022 with r2 = 0.9833 that was used for in vitro t1/2 calculation (Table 5).22–25
Figure 6.
TZB Metabolic stability curve in HLMs.
Table 5.
Parameters of TZB Metabolic Stability
| Time (min.) X Axis |
Conc. (ng/mL) | Y Axis Ln of TZB % Remaining | Linear Part of the Curve (0 to 15 mins) | Value |
|---|---|---|---|---|
| 0 | 473.00 | 4.61 | Regression equation | y = −0.0096x + 4.6022 |
| 1 | 471.12 | 4.59 | ||
| 2.5 | 470.61 | 4.58 | r2 | 0.9833 |
| 5 | 467.58 | 4.55 | ||
| 7.5 | 465.36 | 4.53 | Slope | 0.0096 |
| 15 | 462.40 | 4.50 | ||
| 30 | 461.95 | 4.50 | t1/2 | 72.7 mins |
| 40 | 461.37 | 4.49 | Clint | 9.59 µL/min/kg |
| 50 | 460.84 | 4.49 |
Using the next equations:
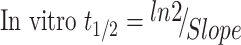 |
Slope was 0.0096.
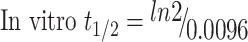 |
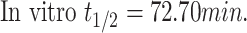 |
TZB intrinsic clearance was computed following the in vitro t1/2 method14 by using the next equation:19
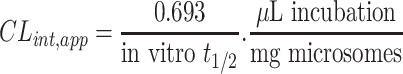 |
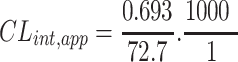 |
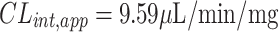 |
In vitro t1/2 and Clint were 72.7 min and 9.59 µL/min/mg, respectively.
TZB was reported to be minimally metabolized by the liver that approved our outcomes.26 Renal excretion of talazoparib as unchanged was considered a major route of elimination and in vivo t1/2 is expected to be long. We do recommend that plasma levels be monitored in cases where these drugs are used for long periods of time, since it is possible for TZB to bioaccumulate after multiple doses to toxic levels.
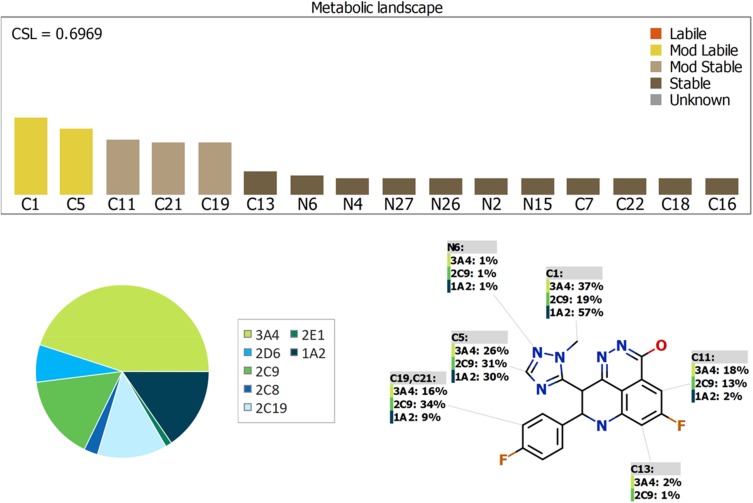
Results of in silico TZB Metabolic Vulnerability Prediction
The Metabolic Landscape for TZB indicates the lability of each site with respect to metabolism by CYP3A4 in absolute terms, to guide the prediction of TZB metabolites and also the optimization of chemical structure for improving metabolic stability. This indicates that C1 and C5 on the 1H-1,2,4-triazole ring are predicted to be the moderate labile sites of metabolism that matched with experimental work indicating the metabolic stability of the TZB. The other metabolic soft spots are stable. The CSL is shown in the top-right of the metabolic landscape. The result from the WhichP450™ module, shown by the pie chart used for indication of most likely cyp450 isoform that has a major role in TZB metabolism (Figure 7). Cyp3A4 was found to have a major role in TZB metabolism. In silico results were supported by the experimental work that indicated the metabolic stability of TZB.
Figure 7.
Proposed metabolic sites for TZB by StarDrop WhichP450™ module.
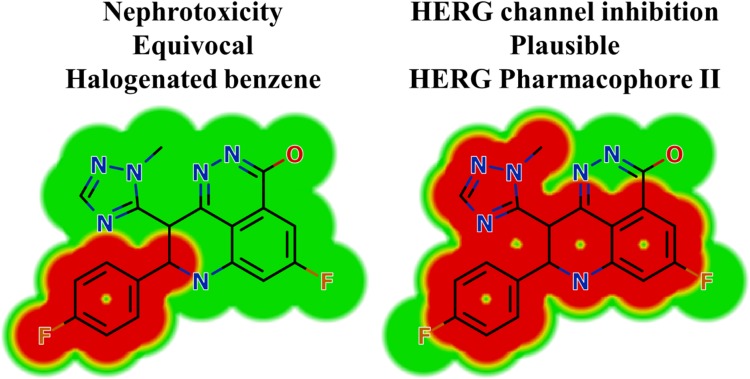
Results of in silico TZB Structural Alerts Sites and Toxicity Prediction
In silico toxicity assessment of TZB metabolites was carried out using DEREK software and structural modification was proposed to reduce their side effects using StarDrop software (Figure 3). TZB shows structural alerts as seen in Figure 3. That caused the proposed side effects that include nephrotoxicity and HERG channel inhibition due to halogenated benzene and HERG pharmacophore II (Figure 8).
Figure 8.
DEREK outcomes showing structural alerts with the proposed side effects of TZB. Red color indicates the structural alerts.
Conclusions
An analytical LC-MS/MS method was described and validated for determining TZB. The developed method showed good sensitivity, was ecofriendly (owing to using little volume of acetonitrile), fast, accurate, and exhibited high recovery. The LC-MS/MS methodology was applied for the evaluation of TZB metabolic stability in HLM matrix. Our findings demonstrated that the metabolic stability of TZB showed moderate Clint (9.59 µL·min−1·kg−1) and long in vitro t1/2 value (72.7 mins) that generated the TZB slow cleared from the blood by the liver. The experimental data were supported using in silico WhichP450™ and DEREK modules of StarDrop software. Further drug discovery studies can be done depending on this concept allowing the development of new series of drugs with increased safety profile without affecting its pharmacological action. In silico toxicological study for TZB was performed using DEREK software that revealed structural alerts and proposed side effects. From these results, we can predict that this drug when given to patients, liver will have a minor role in the drug excretion and may be accumulated after multiple doses.
Acknowledgments
The authors thank the Deanship of Scientific Research at King Saud University for funding this work through Research Group Project No. RG-1435-025.
Abbreviations
ACN, acetonitrile; TZB, talazoparib; LAP, lapatinib; CLint, intrinsic clearance; ESI, electrospray ionization; HLMs, human liver microsomes; TKIs, tyrosine kinase inhibitors; LC-MS/MS, liquid chromatography-tandem mass spectrometry; LOD, limit of detection; LOQ, limit of quantification; MRM, multiple reaction monitoring; PI, product ion; t1/2, in vitro half-life.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378(9801):1461–1484. doi: 10.1016/S0140-6736(11)61351-2 [DOI] [PubMed] [Google Scholar]
- 3.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402–2416. doi: 10.1001/jama.2017.7112 [DOI] [PubMed] [Google Scholar]
- 4.Bochar DA, Wang L, Beniya H, et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102(2):257–265. doi: 10.1016/S0092-8674(00)00030-1 [DOI] [PubMed] [Google Scholar]
- 5.Exman P, Barroso-Sousa R, Tolaney SM. Evidence to date: talazoparib in the treatment of breast cancer. Onco Targets Ther. 2019;12:5177–5187. doi: 10.2147/OTT.S184971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Y, Rehman FL, Feng Y, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19(18):5003–5015. doi: 10.1158/1078-0432.CCR-13-1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murai J, Huang SY, Renaud A, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13(2):433–443. doi: 10.1158/1535-7163.MCT-13-0803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bono J, Ramanathan RK, Mina L, et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 2017;7(6):620–629. doi: 10.1158/2159-8290.CD-16-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowland M, Benet LZ, Graham GG. Clearance concepts in pharmacokinetics. J Pharmacokinet Biopharm. 1973;1(2):123–136. doi: 10.1007/BF01059626 [DOI] [PubMed] [Google Scholar]
- 10.Wilkinson GR, Shand DG. Commentary: a physiological approach to hepatic drug clearance. Clin Pharmacol Ther. 1975;18(4):377–390. doi: 10.1002/cpt.1975.18.issue-4 [DOI] [PubMed] [Google Scholar]
- 11.Houston JB. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem Pharmacol. 1994;47(9):1469–1479. doi: 10.1016/0006-2952(94)90520-7 [DOI] [PubMed] [Google Scholar]
- 12.Obach RS, Baxter JG, Liston TE, et al. The prediction of human pharmacokinetic parameters from preclinical and in vitro metabolism data. J Pharmacol Exp Ther. 1997;283(1):46–58. [PubMed] [Google Scholar]
- 13.Hoffman J, Chakrabarti J, Plotka A, et al. Talazoparib has no clinically relevant effect on QTc interval in patients with advanced solid tumors. Anticancer Drugs. 2019;30(5):523–532. doi: 10.1097/CAD.0000000000000772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baranczewski P, Stanczak A, Sundberg K, et al. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol Rep. 2006;58(4):453–472. [PubMed] [Google Scholar]
- 15.Darwish HW, Attwa MW, Kadi AA. Rapid validated liquid chromatographic method coupled with Tandem mass spectrometry for quantification of nintedanib in human plasma. Trop J Pharm Res. 2016;15(11):2467–2473. doi: 10.4314/tjpr.v15i11.23 [DOI] [Google Scholar]
- 16.Darwish HW, Kadi AA, Attwa MW, Almutairi HS. Investigation of metabolic stability of the novel ALK inhibitor brigatinib by liquid chromatography-tandem mass spectrometry. Clin Chim Acta. 2018;480:180–185. doi: 10.1016/j.cca.2018.02.016 [DOI] [PubMed] [Google Scholar]
- 17.Attwa MW, Kadi AA, Darwish HW, Amer SM, Alrabiah H. A reliable and stable method for the determination of foretinib in human plasma by LC-MS/MS: application to metabolic stability investigation and excretion rate. Eur J Mass Spectrom. 2018;24(4):344–351. doi: 10.1177/1469066718768327 [DOI] [PubMed] [Google Scholar]
- 18.Amer SM, Kadi AA, Darwish HW, Attwa MW. Liquid chromatography-tandem mass spectrometry method for the quantification of vandetanib in human plasma and rat liver microsomes matrices: metabolic stability investigation. Chem Cent J. 2017;11(1):45. doi: 10.1186/s13065-017-0274-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manzo A, Montanino A, Costanzo R, et al. Chapter 33 - EGFR mutations: best results from second- and third-generation tyrosine kinase inhibitors In: Dammacco F, Silvestris F, editors. Oncogenomics. Academic Press; 2019. 477–486. [Google Scholar]
- 20.U.S. Food and Drugs Adminstration. Bioanalytical method validation, guidance for industry. 2018. Available from: https://www.fda.gov/media/70858/download. Accessed January 02, 2019.
- 21.Kadian N, Raju KSR, Rashid M, Malik MY, Taneja I, Wahajuddin M. Comparative assessment of bioanalytical method validation guidelines for pharmaceutical industry. J Pharm Biomed Anal. 2016;126:83–97. doi: 10.1016/j.jpba.2016.03.052 [DOI] [PubMed] [Google Scholar]
- 22.Scott WJ, Hentemann MF, Rowley RB, et al. Discovery and SAR of novel 2,3-dihydroimidazo[1,2-c]quinazoline PI3K Inhibitors: identification of copanlisib (BAY 80-6946). Chem Med Chem. 2016;11(14):1517–1530. doi: 10.1002/cmdc.201600148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attwa Mohamed W, Kadi AA, Darwish HW, Abdelhameed AS. Investigation of the metabolic stability of olmutinib by validated LC-MS/MS: quantification in human plasma. RSC Adv. 2018;8(70):40387–40394. doi: 10.1039/C8RA08161A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alrabiah H, Kadi AA, Attwa Mohamed W, Abdelhameed AS. A simple liquid chromatography-tandem mass spectrometry method to accurately determine the novel third-generation EGFR-TKI naquotinib with its applicability to metabolic stability assessment. RSC Adv. 2019;9(9):4862–4869. doi: 10.1039/C8RA09812C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alanazi MM, Alkahtani HM, Almehizia AA, Attwa MW, Bakheit AH, Darwish HW. Validated liquid chromatography-tandem mass spectrometry for simultaneous quantification of foretinib and lapatinib, and application to metabolic stability investigation. RSC Adv. 2019;9(34):19325–19332. doi: 10.1039/C9RA03251G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y, Chung C-H, Plotka A, et al. A Phase 1 mass balance study of 14C-labeled talazoparib in patients with advanced solid tumors. J Clin Pharmacol. 2019;59(9):1195–1203. doi: 10.1002/jcph.1415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- U.S. Food and Drugs Adminstration. Bioanalytical method validation, guidance for industry. 2018. Available from: https://www.fda.gov/media/70858/download. Accessed January 02, 2019.