Abstract
Cytokinin oxidase/dehydrogenase (CKX) is a key enzyme responsible for the degradation of endogenous cytokinins. However, the origins and roles of CKX genes in angiosperm evolution remain unclear. Based on comprehensive bioinformatic and transgenic plant analyses, we demonstrate that the CKXs of land plants most likely originated from an ancient chlamydial endosymbiont during primary endosymbiosis. We refer to the CKXs retaining evolutionarily ancient characteristics as “ancient CKXs” and those that have expanded and functionally diverged in angiosperms as “non-ancient CKXs”. We show that the expression of some non-ancient CKXs is rapidly inducible within 15 min upon the dehydration of Arabidopsis, while the ancient CKX (AtCKX7) is not drought responsive. Tobacco plants overexpressing a non-ancient CKX display improved oxidative and drought tolerance and root growth. Previous mutant studies have shown that non-ancient CKXs regulate organ development, particularly that of flowers. Furthermore, ancient CKXs preferentially degrade cis-zeatin (cZ)-type cytokinins, while non-ancient CKXs preferentially target N6-(Δ2-isopentenyl) adenines (iPs) and trans-zeatins (tZs). Based on the results of this work, an accompanying study (Wang et al. 10.1038/s41438-019-0211-x) and previous studies, we hypothesize that non-ancient CKXs and their preferred substrates of iP/tZ-type cytokinins regulate angiosperm organ development and environmental stress responses, while ancient CKXs and their preferred substrates of cZs play a housekeeping role, which echoes the conclusions and hypothesis described in the accompanying report (Wang, X. et al. Evolution and roles of cytokinin genes in angiosperms 1: Doancient IPTs play housekeeping while non-ancient IPTs play regulatory roles? Hortic Res 7, (2020). 10.1038/s41438-019-0211-x).
Subject terms: Cytokinin, Plant evolution
Introduction
Cytokinins are a major class of hormones that regulate many developmental processes in plants, including cell division, shoot and root growth, and vascular and gametophyte development1. Cytokinins also play important roles in the responses of plants to biotic and abiotic stresses. Natural cytokinins are N6-substituted adenine derivatives, and their most common forms in plants are N6-(Δ2-isopentenyl) adenines (iPs), trans-zeatins (tZs), and cis-zeatins (cZs). These cytokinin forms exhibit different levels of physiological activity and are differentially distributed among land plant lineages and in various tissues or stages of angiosperms2.
Angiosperms exhibit an efficient system for controlling the homeostasis of endogenous cytokinins in different organs and tissues. The major enzymes involved in the regulation of cytokinin content are isopentenyltransferases (IPTs) and “LONELY GUY” (LOGs, named by Kurakawa et al.3) cytokinin-specific phosphoribohydrolases for biosynthesis, cytokinin glucosyltransferases for conjugation, and cytokinin oxidases/dehydrogenases (CKXs) for irreversible degradation4. The ATP/ADP- and tRNA-IPT genes encode the main enzymes responsible for the biosynthesis of tZ- and cZ-type cytokinins in angiosperms, respectively5. IPT activity and active cytokinins have been detected in various algae6,7, suggesting that cytokinin biosynthesis already existed before plants were established on land. However, only tRNA-IPT genes have been identified in seedless plants8, and ATP/ADP-IPT genes are proposed to have arisen during the evolution of angiosperms, probably due to the need for higher levels of cytokinins4.
CKXs play an essential role in decreasing endogenous cytokinin content. Cytokinin glucosyltransferases can also reduce active cytokinins by conjugation, but their effects are mostly reversible, and changes in their gene expression do not always lead to variations in active cytokinin levels or phenotypes1,9. In contrast, CKXs are the only known proteins that specifically degrade cytokinins, including their ribosides and some glucosides10. The manipulation of CKX gene expression can substantially alter the levels of active cytokinins, resulting in physiological or developmental changes11–13. CKX isoforms show differences in their subcellular localization, substrate preference and other properties10. The overexpression of individual Arabidopsis CKX (AtCKX) genes results in various root phenotypes11,13, suggesting that CKX isoforms play different roles in plant organ development.
CKX genes have also been shown to be involved in plant tolerance to abiotic stresses12,14–16. Carabelli et al.17 demonstrated that AtCKX6 is critical for the growth arrest of the leaf primordium, which contributes to extension growth under shaded conditions. The constitutive or root-specific overexpression of AtCKX1-4 in Arabidopsis and other species results in improved drought, salt and heat tolerance compared with wild-type plants12,14,18.
Despite the importance of CKX genes in angiosperms, their origin and evolution are poorly studied. In addition to land plants, CKX activity and homologous CKX sequences have been detected in bacteria such as Rhodococcus fascians19 but not in any algae. The plant CKX genes were once suggested to have been derived from cyanobacteria20, along with thousands of other nuclear genes transferred from plastids21. Furthermore, among the angiosperm CKX genes, using Arabidopsis as an example, AtCKX7 has been proposed to be an evolutionarily ancient isoform based on the comparison of protein characteristics and the changes observed in AtCKX-overexpressing plants between different AtCKX members13. We refer to AtCKX7 and its orthologs in other angiosperms as ancient CKXs. Our systematic survey and phylogenetic analysis of CKX genes from bacteria, archaea, and eukaryotes provide evidence supporting the hypothesis that all land plant CKX genes are derived from a single CKX with a chlamydial origin. We therefore refer to the genes of angiosperms that have expanded and functionally diverged from the ancient genes as non-ancient CKXs. We show that the non-ancient CKX genes are expressed in a tissue/organ-specific manner and exhibit rapid positive responses to dehydration and that their overexpression increases plant stress tolerance and root growth and development. On the other hand, transcriptome and qPCR analyses in several angiosperm species show that the expression of ancient CKXs is generally constitutive in tissues/organs and is non-stress responsive. Our findings provide important insights into the origin, evolution, and possible roles of CKX genes and give rise to the hypothesis that non-ancient CKXs and their preferred substrates, iPs and tZs, play regulatory roles in organ development and stress tolerance in angiosperms, while ancient CKXs and their preferred substrates, cZs, likely play a housekeeping role.
Results
CKX genes are largely restricted to land plants and bacteria
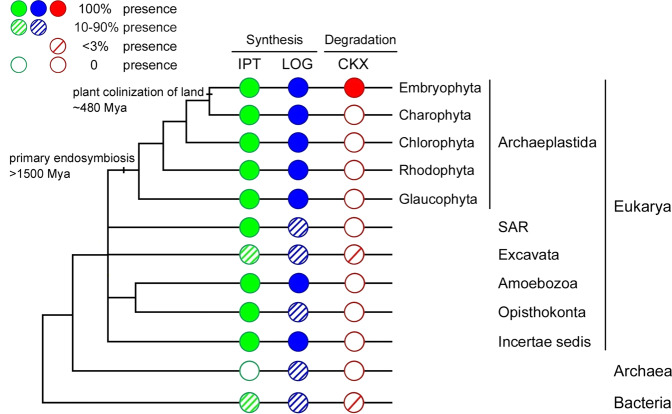
CKX genes have been previously reported in land plants and bacteria but seldom in other organisms4. To systematically investigate the taxonomic distribution of CKX genes, we first conducted exhaustive HMMER, BLASTP, or TBLASTN searches for the Cytokin-bind domain (Pfam: PF09265), which is characteristic of CKX proteins, in organisms outside of land plants and bacteria. Only a single homolog, NgrCKX1, in the excavate species Naegleria gruberi (Fig. 1; Table S1), could be identified among these organisms. We could not detect any other CKX homologs in over 50 other excavate species whose complete genomes are publicly available.
Fig. 1. Distribution of cytokinin biosynthesis (IPT and LOG) and degradation (CKX) genes in living organisms, showing that CKX genes are uniquely restricted to land plants, Excavata and a small number of bacteria.
Cytokinin biosynthesis and degradation genes and their homologs (IPTs, green; LOGs, blue; and CKXs, red circles) are present in all (100%, solid circles), a high percentage (10–90%, multiple-slashes filled circles), a low percentage (<3%, one-slash filled circles), or none (open circles) of the lineages within each taxon. Fifty complete genomes from representative lineages of bacteria, archaea, and eukaryotes (see Table S2) were sampled to estimate the percentages of the presence of IPT and LOG genes. Branch length is not proportional to evolutionary time.
We then searched the sequences of land plants and bacteria. CKX genes were identified in all available complete proteomes of land plants (Table S3). To compensate for the deficiency of complete proteomes from seedless land plants, we also searched the 1KP database, which contains transcriptomic sequences of many nonvascular plants (liverworts, mosses, and hornworts) and seedless vascular plants (lycophytes and monilophytes). At least one CKX gene was detected in 158 (94.6%) species across major lineages of 167 nonvascular and seedless vascular plants; no CKX homologs could be found in the remaining species, most likely because of their incomplete transcriptomic sequences (Table S4). As such, it is highly likely that CKX genes are ubiquitously present in all major lineages of land plants. On the other hand, homologous CKX sequences were identified in less than 2.5% of bacterial proteomes/genomes (240 out of more than 9,700) downloaded from the GenBank and JGI databases. The bacteria containing putative CKX genes predominantly come from Actinobacteria, Proteobacteria (alpha-, beta-, delta-, and gamma-) and Cyanobacteria, plus a few from Chlamydiae, Chloroflexi, and unclassified bacteria. No CKX homolog could be identified in other bacterial phyla. Collectively, the results of our survey demonstrate that CKX genes are ubiquitously present in land plants, one excavate species and a small percentage of bacteria, providing evidence that the CKX genes of land plants and bacteria are closely related, which is likely attributable to gene transfer between the two groups.
We also investigated the taxonomic distribution of cytokinin biosynthesis genes (IPTs and LOGs). Homologs of both IPT and LOG genes could be detected in a majority of the major lineages of bacteria and eukaryotes and several groups of archaea (Fig. 1; Table S2). Such a wide distribution strongly suggests that IPTs and LOGs have very ancient origins and are inherited among major taxa by conventional vertical descent, in sharp contrast to the CKX genes responsible for cytokinin degradation.
Land plant CKX genes are likely derived from Chlamydiae
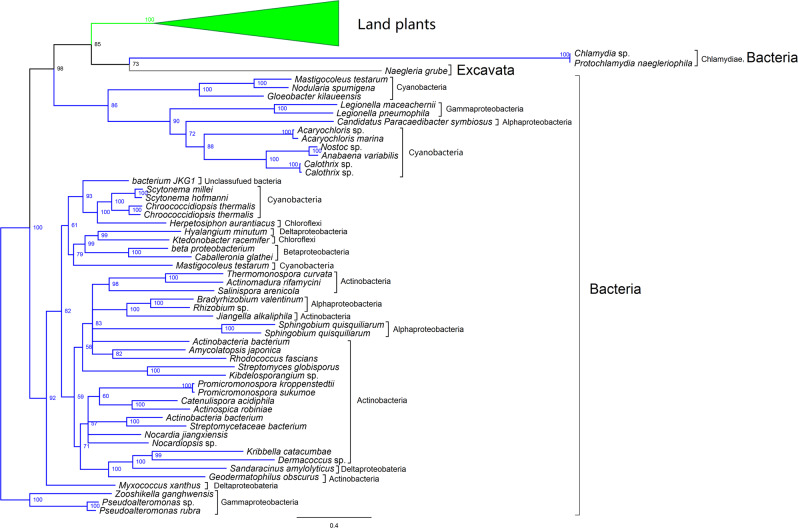
We further performed phylogenetic analyses to understand the evolutionary relationships of the CKX genes. The phylogenies constructed from ML and Bayesian analyses provided similar relationships among land plant, bacterial and excavate CKXs (Figs. 2, S1 and S2). All land plant CKXs form a well-supported clade, which is a sister clade to a monophyletic group comprising chlamydial homologs and NgrCKX1. This large group of plant, chlamydial and Naeglaria sequences is in turn affiliated with cyanobacterial and proteobacterial homologs. Given the intimate physical association of Chlamydiae and Naegleria22, it is likely that NgrCKX1 was acquired from Chlamydiae. The close relationship between land plant and chlamydial CKXs is reminiscent of other genes of chlamydial origin frequently reported in photosynthetic eukaryotes23–25. Therefore, land plant CKX genes may also be derived from Chlamydiae.
Fig. 2. Phylogenetic analyses demonstrate a close relationship between land plant and chlamydial CKXs.
The reduced MrBayes phylogeny (see Fig. S1 for the full tree topology) constructed from the two domain regions of the CKX proteins of six representative land plants, one excavate, and 49 selected bacteria (see Table S1) is shown. Support values (Bayesian posterior probabilities×100) greater than 50 are indicated at the nodes. Color coding: Green, land plants; blue, bacteria; gray, excavate.
We compared the CKX sequence divergence between land plants, Chlamydiae, and other bacteria to assess whether the observed relationship of land plant and chlamydial CKX genes might be due to an artifact. The Ks value (synonymous nucleotide substitutions per synonymous site), which has been used as a proxy for divergence time between duplicates26, was calculated between each putative CKX gene pair sampled for the phylogenetic analysis. The results indicated that the average Ks value of the CKX pairs between land plants and Chlamydiae is lower than those between land plants and other bacterial groups (Table S5). In contrast, the highest Ka (nonsynonymous substitutions per nonsynonymous site) and Ka/Ks values are observed between land plants and Chlamydiae. These results suggest that the divergence of land plant and chlamydial CKXs occurred relatively late and that the chlamydial CKX protein sequences evolved faster than those of other bacteria. Due to these high Ka values, the amino acid similarities between the chlamydial and land plant CKXs are not high (approximately 25%). However, chlamydial CKXs contain the two key domains: the Cytokin-bind and FAD_binding_4 domains, and share core motifs with land plant CKXs (Fig. S3). Although Ks values may be affected by different evolutionary rates or become saturated over a long timescale, our Ka/Ks and sequence structure analyses suggest the closest relationship between land plant and chlamydial CKXs, thus indicating a chlamydial origin of land plant CKXs.
Evolutionary patterns of CKX genes in angiosperms
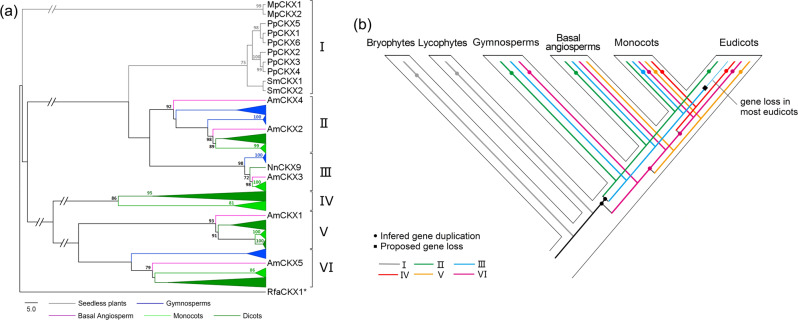
We next investigated the evolution of the CKX genes in land plants after their transfer from Chlamydiae. Twenty-one species with available whole-genome sequences (Table S6) were sampled from all major lineages of land plants. A total of 184 CKX genes were identified. The number of CKX genes varied from two in the liverwort Marchantia polymorpha and the lycophyte Selaginella moellendorffii to 17 in the core eudicot Glycine max (Table S6), indicating the expansion of the CKX genes during land plant evolution.
Phylogenetic analysis of land plant CKXs was performed using the actinobacterial R. fascians homolog as an outgroup (Fig. S4). In the phylogeny, the CKX sequences of seedless plants (including M. polymorpha, Physcomitrella patens and S. moellendorffii) clustered with their paralogs from the same species (Group I in Figs. 3a, S4 and Table S6), suggesting that the CKX gene may have been retained in a single copy before the divergence of seedless plants and seed plants (Fig. 3b). Seed plant CKXs can be divided into five major groups (Groups II-VI in Figs. 3a, S4 and Table S6). Among these groups, Groups II and III appear to be more closely related to Group I, consisting of the seedless plant CKX homologs, than to the other groups (Figs. 3, S4), although the bootstrap support for this relationship was modest. Motif composition analysis showed a closer relationship between Group I and Group II, rather than between Group I and Group III27. Furthermore, the ancient AtCKX7 isoform of Arabidopsis13 belongs to Group II. Biochemical and other characterizations of ZmCKXs in maize suggest that the Group II protein ZmCKX10 shows similar characteristics to AtCKX7 and that the ZmCKXs in Groups III-VI show distinct features28,29. Therefore, Group II is expected to be the only group containing ancient CKXs in angiosperms. Hereafter, we refer to this group as the ancient CKX group and to the other groups that contain expanded and functionally diverged angiosperm CKXs as non-ancient CKX groups.
Fig. 3. Phylogenetic analysis and evolutionary history of CKX genes in land plants.
a Reduced PhyML phylogeny (see Fig. S4 for the full tree topology) of the CKX proteins from 21 representative land plants constructed based on the Cytokin-bind domain region. Bootstrap values greater than 70 are indicated at the nodes. The color coding for the plant lineages is indicated at the bottom. The abbreviations for the species names in front of the gene names are indicated in Table S6. b Inferred expansion history of the gene family in land plants. Inferred duplications and proposed losses are indicated with solid circles and black diamonds, respectively. Colored lines represent the respective groups in a, Fig. S4 and Table S6.
Next, we focused on the comparison of ancient and non-ancient CKX genes in angiosperms in their expansion and expression patterns. The ancient CKX genes are generally conservatively retained in a single copy per species, with the exception of a few of these genes in angiosperms that have undergone recent polyploidization events, as observed in G. max. In contrast, most non-ancient CKX genes have undergone significant expansions. Our synteny analysis using MCScanX30 showed that 54 of 128 non-ancient CKX genes in angiosperms likely resulted from WGD/segmental duplication and 20 from tandem or proximal duplication (Table S7). Based on our analysis, most of the WGD/segmental CKX duplicates may have more likely been derived from WGD rather than segmental duplication because (1) all the WGD/segmental duplicates except for ClCKX2/7 come from angiosperms that have experienced lineage-specific polyploidization events; (2) the pairwise Ks values of the WGD/segmental duplicates are in agreement with the ages of these polyploidization events (Table S7); and (3) the vast majority of the randomly selected collinear regions containing WGD/segmental duplicates are involved in the WGD remnants reported in previous studies (data not shown). Therefore, WGD duplication is likely a major mechanism of the expansion of non-ancient CKX genes in angiosperms.
To investigate the expression patterns of the CKX genes, we summarized the data reported previously in Arabidopsis31, maize32, and woodland strawberry33. The results demonstrate that in tissues/organs, ancient CKX genes (i.e., ZmCKX10) are nearly constitutively expressed, whereas non-ancient CKX genes exhibit divergent expression patterns (Fig. S5a, c, e). The non-ancient CKX genes have also diversified in response to abiotic stresses (Fig. S5b, d, f). At least one member of the duplicate CKXs in each non-ancient CKX group responded positively to abiotic stress treatment. For instance, within the duplicate pair AtCKX1/6 in the angiosperm-derived Group VI, the expression level of AtCKX6 was significantly increased under drought, cold or salinity stress conditions (Fig. S5b). In contrast, the ancient CKXs, such as AtCKX7 and ZmCKX10, showed no significant response to abiotic stresses (Fig. S5b, d). These results suggest that in angiosperms, the ancient CKXs likely maintain constitutive and non-abiotic stress-responsive expression patterns throughout the plant, while the non-ancient CKXs showing functional divergence after duplication confer tissue/organ-specific and abiotic stimulus-responsive expression patterns.
Expression patterns of CKX genes upon dehydration
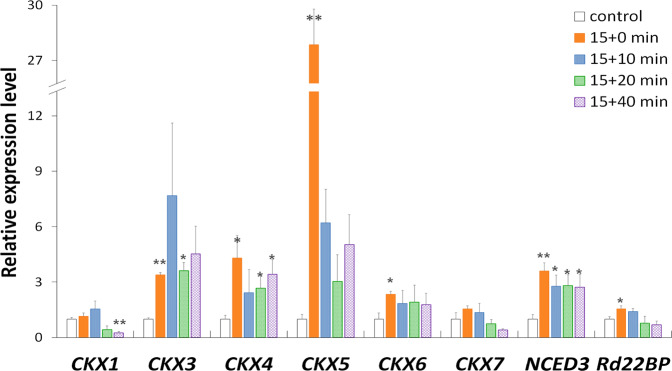
To verify the differences in abiotic stress responses between the ancient and non-ancient CKX genes, we examined the expression profile of AtCKX genes in the very early stages of dehydration treatment. Arabidopsis leaves were air-dried for 15 min, resulting in approximately 10% water loss. At the end of the 15 min dehydration period, significant increases in the expression of two drought-induced marker genes, NCED3 and Rd22BP1, were observed (Fig. 4), indicating that the dehydration treatment was effective. Among the six expressed AtCKX genes, four of the five non-ancient CKXs, AtCKX3-6, showed significantly increased expression levels (Fig. 4). This increase in expression declined 10–20 min after the initiation of dehydration, but the expression levels of AtCKX3-6 throughout the duration of the stress treatment were consistently higher than those under the control condition. The rapid upregulation of most non-ancient AtCKX genes at very early stages of dehydration strongly suggests that non-ancient CKXs play an important role in the plant response to water stress. On the other hand, the ancient CKX AtCKX7 did not present significant increases in expression under dehydration, indicating that the ancient CKX genes are less involved in the regulation of abiotic stress responses in angiosperms.
Fig. 4. Rapid responses of representative Arabidopsis CKX genes in leaves under dehydration.
Leaves from three-week-old Arabidopsis seedlings were exposed to fanning for accelerated dehydration for approximately 15 min to achieve 10% water loss, followed with 0 (15+0), 10 (15+10), 20 (15+20) or 40 (15+40) min of incubation in sealed plastic bags. Detached leaves placed under 100% relative humidity without treatment were used as controls, in which expression levels were set to 1. The Tub8 gene was used as an endogenous reference gene. The error bar indicates the standard error of the mean (SEM) (n = 3 biological replicates), and an asterisk indicates a significant difference between treated leaves and the control (* for p < 0.05, ** for p < 0.01).
Non-ancient CKX genes play a role in the oxidative and drought stress tolerance of angiosperms
We further investigated the role of the non-ancient CKX genes in plant tolerance to abiotic stresses. We generated transgenic tobacco plants overexpressing the non-ancient CKX gene AtCKX2 under the control of the 35S promoter. The CKX-overexpressing plants displayed typical cytokinin-deficient phenotypes, including an expanded root system with increased root branching and a narrower leaf shape than the wild-type plants (Figs. 5a, S6). However, the height of the transgenic plants was similar to that of the wild-type plants when either cultured on MS medium or grown in the soil (Figs. 5a, S7).
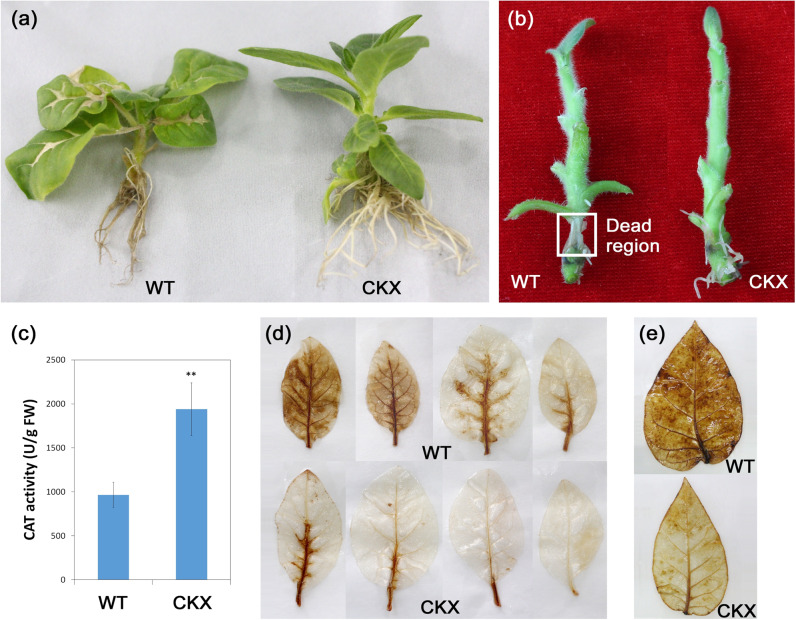
Fig. 5. Improved oxidative stress tolerance in transgenic tobacco plants overexpressing a non-ancient CKX gene.
a,b More dead tissues were observed in WT (left) than 35S:AtCKX2-overexpressing (CKX2-10, right) tobacco plants under oxidative stress for 5 days. c Lower catalase (CAT) enzyme activities were observed in the leaves of WT plants than in the leaves of CKX2-10 plants under oxidative stress for 36 h. The error bars indicate SEM values (n = 3 biological replicates), and asterisks indicate significant differences between WT and transgenic plants (** for p < 0.01). d, e More H2O2 accumulation was observed in WT leaves (upper) than in CKX2-10 leaves (lower) under oxidative stress for 36 h (d) or under drought treatment (e; see Fig. S7) after 14 days.
Oxidative stress tolerance is a basis of tolerance to many abiotic stresses, such as drought and extreme temperatures. We examined the oxidative stress tolerance of the wild-type (WT) and CKX transgenic plants through the application of methyl viologen (MV), a chemical that can generate reactive oxygen species (ROS) in plant cells. Tissue death starting in the petioles followed by the veins of WT leaves and necrosis in WT stem tissues were observed after 2–3 days of culture in MV-containing media (Fig. 5a, b). In contrast, the transgenic plants remained healthy with no obvious symptoms under the same conditions. Significantly higher levels of H2O2 were detected in WT plant leaves than in their transgenic counterparts after 36 h of exposure to MV (Fig. 5d). We also observed an approximately 2-fold increase in ROS-scavenging activity (catalase enzymes) in the transgenic leaves (Fig. 5c). These results show that the overexpression of a non-ancient CKX gene can lead to increased ROS-scavenging activity in plant cells and increased plant tolerance to oxidative stress.
Increased tolerance was also observed in the CKX-overexpressing plants compared with the WT plants under drought conditions. Leaves were harvested from WT and CKX transgenic plants at two weeks after watering was ceased. Much less H2O2 accumulation was detected in transgenic leaves than in the WT controls (Fig. 5e), indicating that the CKX-overexpressing plants show higher tolerance to oxidative stress induced by drought. Moreover, although the wild-type and CKX transgenic plants displayed similar degrees of wilting and stem shrinking after one month of drought treatment (Fig. S7b, d), the transgenic plants showed significantly faster and better recovery than the WT plants (Fig. S7c, e). More than 90% of the 60 transgenic plants recovered from drought stress, as indicated by fully turgid leaves and stems, within 4 days after rewatering. In contrast, 75% of the 60 wild-type plants failed to recover, and the rest took 5 days or longer to reach full turgor. Therefore, the overexpression of a non-ancient CKX resulted in improved plant tolerance to drought, in addition to oxidative stress (Table 1).
Table 1.
Comparison of stress-related functions between ancient and non-ancient AtCKX genes.
| Phenotype | Ancient AtCKX7 | Non-ancient AtCKXs | Reference | |
|---|---|---|---|---|
| Rapid drought responsea | No | Yes | This study | |
| Stress toleranceb | Oxidative stress | Not reported | Increased | This study |
| Drought | Not reported | Increased | This study,12,18 | |
| High temperature | Not reported | Increased | 14,18 | |
| Salinity | Not reported | Increased | 12 | |
| Root growthb | Primary roots | Reduced | Increased | This study,11,13 |
| Lateral roots | Reduced | Increased | This study,11,13 | |
| Protein property | Subcellular localizationc | Cytosol | ER, vacuole or apoplast | 11,13,40 |
| Substrate preferenced | cZs | iPs and/or tZs | 2,28 |
aRapid drought response refers to the response to drought treatment within 15 min
bResults of CKX overexpression under the control of the 35S promoter are shown
cThe reduction of cytokinins by cytosolic AtCKX7 resulted in the early termination of primary root growth13, whereas reduction by other AtCKXs resulted in increased primary and lateral root growth and development (Fig. S6)11,38
dUnder stress conditions, the levels of tZs were reduced, but the levels of cZs were increased12,41, suggesting a negative role of tZs, but not cZs, in the stress response
Discussion
Origin of land plant CKX genes
Our analyses show that, with the sole exception of the excavate N. gruberi, CKX homologs are restricted to land plants and bacteria. Although the existence of putative CKX homologs has been suggested in several other eukaryotes, such as a few microalgal species34, no CKX homologs could be identified in the complete genomes of these species. Hence, the ability to degrade cytokinins in these other eukaryotes may not involve CKX proteins. The distribution of CKX homologs mainly in land plants and bacteria suggests that HGT has likely occurred during the evolution of this gene, though other possible scenarios (particularly differential gene losses) cannot be completely excluded. Furthermore, the identification of CKXs in multiple bacterial phyla indicates that CKX genes may have evolved in ancient bacteria and subsequently spread to land plants.
A well-known route of gene transfer from bacteria to plants is endosymbiotic gene transfer (EGT) from mitochondria and plastids21. Schmülling et al.20 once hypothesized a plastid origin of plant CKX genes due to the presence of CKX homologs in cyanobacteria. Nevertheless, the phylogenies constructed here and previously by others4 do not support that hypothesis. Instead, our analyses indicate that land plant CKXs are most closely related to chlamydial sequences, suggesting a chlamydial origin of these genes. Chlamydiae are a group of obligate intracellular bacteria living in animals, free-living amoebae and the environment25. Chlamydial genes in photosynthetic eukaryotes have been reported from several genome analyses22–25. Because of the nature of Chlamydiae as obligate intracellular endosymbionts, it has been suggested that an ancient chlamydial endosymbiont was present in the most recent common ancestor of Plantae. Thus, genes were transferred from this chlamydial endosymbiont to the nucleus of the host cell, which in turn facilitated the establishment of cyanobacterial endosymbionts (i.e., plastids). We conclude that the original/ancient CKX in land plants was most likely derived from a gene in the ancient chlamydial endosymbiont and that its expansion and diversification produced the non-ancient CKXs, which might contribute more to the regulation of organ development and abiotic stress responses in angiosperms.
Evolution of CKX genes in angiosperms
It is unclear from the available data when the CKX gene was transferred from the chlamydial endosymbiont to the nuclear genome of plants and coopted into their cytokinin metabolism. Nevertheless, neither a CKX homolog nor CKX activity has been detected in any algal lineage, suggesting that cytokinin degradation by CKXs in Plantae is most likely restricted to land plants. Our analyses indicate that the single ancient CKX gene underwent duplications in the common ancestor of seed plants, after which further duplications and divergence occurred in angiosperms, giving rise to non-ancient CKXs with different features from the ancient genes (Fig. 3b). The percentage of CKX genes among the total genes in most sampled angiosperms was nearly stable (Table S6). More notably, in the vast majority of sampled angiosperms, the ratio of CKX to IPT genes (the IPT genes encode key enzymes responsible for the biosynthesis of cytokinins) is restricted to a value of approximately 1 (Table S8). This result indicates that the numbers of CKX and IPT genes in angiosperm genomes may be kept in balance and vary concomitantly. As such, the retention of CKX and IPT duplicates in angiosperms likely follows the gene balance hypothesis, which postulates that complex subunits or proteins in a regulatory cascade need to be maintained in dosage balance to avoid negative consequences35. CKX proteins exhibit opposite roles to IPTs in the control of endogenous cytokinin content. Thus, the dosage balance between CKX and IPT genes may be critical for the regulation of endogenous cytokinins in angiosperms.
Functional divergence of CKX genes in angiosperms
There are two types of IPT genes, tRNA-IPTs (ancient cytokinin biosynthesis genes) and ATP/ADP-IPTs (non-ancient cytokinin biosynthesis genes), in angiosperms. In an accompanying article on this topic (Wang et al.36), we show that in angiosperms, the tRNA-IPT (ancient) genes are conservatively retained, constitutively expressed throughout the plant, and nonresponsive or only slightly responsive to environmental stresses. On the other hand, the expression of ATP/ADP-IPTs (non-ancient), which emerged and diverged in angiosperms, is tissue specific and is rapidly downregulated under various stress conditions. It is particularly interesting that we found similar characteristics between the ancient/non-ancient CKX genes and the ancient/non-ancient IPT (tRNA-type and ATP/ADP-type) genes in terms of their evolution, tissue/organ expression and environmental stress responses in angiosperms (Table 2). Based on several lines of evidence discussed below, we hypothesize that, similar to the ATP/ADP-IPT (non-ancient) genes (Wang et al.36), the non-ancient CKXs play regulatory roles in organ development and the adaption of angiosperms to environmental stresses, whereas the ancient CKXs more likely exhibit housekeeping functions, similar to the tRNA-IPT (ancient) genes.
Table 2.
Evolutionary history, protein characteristics and proposed roles of cytokinin metabolism genes in angiosperms.
| tRNA-IPTs | ATP/ADP-IPTs | Ancient CKXs | Non-ancient CKXs | |
|---|---|---|---|---|
| Products or substratesa | cZs | iPs/tZs | Preferentially cZs | Preferentially iPs/tZs |
| Origin | Ancient IPT in the last common ancestor of eukaryotes | Class II tRNA-IPT in the common ancestor of angiosperms | Chlamydial CKX during the primary endosymbiosis | Ancient CKX in the last common ancestor of land plants |
| Expansion & functional divergence in angiosperms | No | Yes | No | Yes |
| Organ development of angiosperms | Mutations lead to small and often chlorotic plants5 | Altered expression leads to plants with abnormal organ development5 | Overexpression leads to no significant changes after root growth is restored two weeks after germination13,b | Altered expression leads to plants with abnormal organ development37,61 |
| Responses to abiotic stresses | Slightly responsive or nonresponsive | Downregulated | Nonresponsive | Rapidly induced |
| Abiotic stress tolerance | Not reported | Knockout mutations lead to increased drought or salt tolerance12; stress-, maturation-, or senescence-induced overexpression of the Agrobacterium AMP-IPT leads to increased iPs/tZs and improved tolerance to drought, salt and heat stresses12,62 | Not reported | Overexpression leads to improved tolerance to drought, salt62, heat14, or oxidative stresses, and knockout mutations of an inflorescence meristem-specific CKX lead to improved salt tolerance63 |
| Proposed roles in angiosperms | Housekeeping | Regulatory | Housekeeping | Regulatory |
aProducts of IPTs or substrates of CKXs
bPlants overexpressing the ancient AtCKX7 gene showed small shoots and early-terminating primary roots within two weeks after germination. Thereafter, root growth was restored, and no significant changes were observed in plant growth or developmental patterns at later stages13
First, the differences in copy numbers and expression patterns of the ancient and non-ancient CKX genes support the notion that these two types of CKX genes (ancient and non-ancient) functionally diversified in angiosperms. Our results show that the ancient CKX genes are conservatively retained in 1 or 2 copies in most sampled angiosperms, whereas the non-ancient CKX genes have undergone significant expansions and display large variations in gene numbers (from 3 to 14) among different species. More importantly, based on the analyses of the data reported previously in Arabidopsis31, maize32, and woodland strawberry33, the ancient CKX genes are nearly constitutively expressed across all tissues/stages, while the non-ancient CKXs exhibit highly diversified expression patterns (Fig. S5). In particular, the expression levels of most non-ancient CKXs vary significantly among different tissues/organs or developmental stages. The relatively constitutive expression of the ancient CKK genes suggests that these genes likely play a housekeeping role in maintaining basic cellular functions, while the highly diversified expression patterns of the non-ancient CKXs suggest regulatory roles in plant growth and development.
Second, plants with altered expression of ancient and non-ancient CKXs exhibit distinct phenotypic changes. With respect to organ development, the knockout of non-ancient CKXs leads to increased floral and ovule primordium formation and more flowers, siliques, and seeds but abnormal floral organ development, based on the observation of Arabidopsis ckx3 ckx5 double mutants37. Constitutive overexpression of single non-ancient CKXs improves primary root growth and lateral root formation while substantially reducing shoot development, decreasing the activity of vegetative and floral shoot apical meristems and leaf primordia, and causing lower fertility11. In contrast, young seedlings constitutively overexpressing the ancient AtCKX7 gene produce normal primary roots immediately after germination, but their growth ceases three days later13. The root growth of the AtCKX7-overexpressing seedlings is restored after one week. The aerial parts of the AtCKX7-overexpressing plants are relatively small before their root growth is restored. However, there are no significant changes observed in plant growth and developmental patterns at later stages after root growth is restored13. Furthermore, under tissue culture conditions, no developmental changes except for reduced root sizes have been observed in the isolated roots of transgenic tomato plants that overexpress an ancient CKX gene, ZmCKX1029. These results also support the housekeeping and regulatory roles of ancient and non-ancient CKXs, respectively, in the organ development of angiosperms.
Third, the ancient and non-ancient CKX genes exhibit differential response patterns upon environmental stress treatments (Table 1). According to our analyses of CKX expression data in this and previous studies31–33, the ancient copies appear to be constitutively expressed in tissues/organs and to show no responses to abiotic stresses (Figs. 4, S5). In contrast, the non-ancient CKXs display rapid and diversified responses to stresses. Among the species containing lineage-specific duplicates in the ancient CKX group (e.g., woodland strawberry), one member (FveCKX6) displays nearly the same pattern as the singleton ancient CKX genes AtCKX7 and ZmCKX10, while the other member (FveCKX7) shows diversified expression in tissues/organs and positive responses to abiotic stresses, similar to the members of the non-ancient CKX groups. These findings suggest that the ancient CKX genes tend to be retained with a conserved copy number and play a housekeeping role, while the non-ancient CKXs are functionally diversified and more involved in the adaptation of angiosperms to environmental stresses.
Fourth, the manipulation of the expression of ancient and non-ancient CKXs likely leads to different changes in stress tolerance (Table 1). This and previous studies have demonstrated that the overexpression of a non-ancient CKX gene improves plant tolerance to drought, osmotic, or heat stress (Fig. S7)12,18,38. Moreover, our experiments revealed that transgenic plants overexpressing a non-ancient CKX gene exhibit increased oxidative stress tolerance (Fig. 5). Lower H2O2 accumulation under drought (Fig. 5e) and increased antioxidant defense against abiotic stresses have also been detected. Environmental stresses usually lead to an increase in ROS generation, which triggers oxidative stress in plants39. Increased oxidative stress tolerance provides a basis for the improvement of tolerance to other stresses. Furthermore, the improved root growth and biomass (Fig. S6)11,38 of the plants overexpressing a non-ancient CKX play a role in the improvement of overall drought tolerance. On the other hand, the suppression of root growth following the overexpression of an ancient CKX suggests that ancient CKXs may not play a positive role in the environmental stress tolerance of angiosperms.
In addition, ancient and non-ancient CKXs present differential subcellular localizations (Table 1). Studies on AtCKXs have revealed that the ancient AtCKX7 uniquely localizes to the cytosol, whereas the non-ancient AtCKXs predominantly localize to the endoplasmic reticulum (ER), vacuole or apoplast11,13,40. This discrepancy in subcellular localizations between ancient and non-ancient CKXs is likely related to their respective housekeeping and regulatory roles in angiosperms. Taken together, these above lines of evidence support our hypothesis that the non-ancient CKX genes are more involved in the regulation of organ development and the adaptation of angiosperms to environmental stresses, whereas the ancient CKXs likely contribute more to maintaining basic cellular functions.
The ancient and non-ancient CKXs also exhibit different substrate specificities as well. Non-ancient CKXs, such as AtCKX4 and ZmCKX1, present a greater substrate preference for tZ- or iP-type cytokinins, while ancient CKXs prefer cZs11,28,29. Experiments in CKX-overexpressing plants also show that the non-ancient AtCKXs degrade tZs much more efficiently than the ancient AtCKX7, whereas AtCKX7 preferentially degrades cZs13. We have proposed the hypothesis in the accompanying article in this issue (Wang et al.36) that iPs/tZs play more regulatory roles in organ development and stress responses, while cZs play a housekeeping role in maintaining basic cellular functions, based on the analyses of the corresponding genes responsible for their biosynthesis (ATP/ADP-IPTs and tRNA-IPTs). In this study, we provide further supporting evidence for this hypothesis from comparative analyses of non-ancient and ancient CKXs (Table 2). We have demonstrated above that the non-ancient CKXs, preferentially degrading iPs/tZs, play a regulatory role, while the ancient CKXs, preferentially degrading cZs, play a housekeeping role in the organ development of angiosperms. We have also shown that non-ancient CKXs are rapidly induced, whereas ancient CKXs are not responsive under environmental stress conditions. These results are consistent with the observations that the levels of iPs/tZs are drastically reduced, while those of cZs are either not changed or increased in response to environmental stresses41. Moreover, plants overexpressing non-ancient AtCKXs show substantially reduced levels of iP/tZ-cytokinins and exhibited strong drought- or salt stress-tolerant phenotypes, regardless of their cZ levels12. These findings support the notion that iPs/tZs, rather than cZs, play a regulatory role in abiotic stress response/tolerance in angiosperms. In addition, due to increases in cZ concentrations under stress conditions, it has been suggested that cZs may play a role in maintaining basic cellular functions under growth-limiting conditions2,41. However, because cZs have been shown to present much less cytokinin activity than iPs and tZs42,43, the role of cZs under stress conditions needs to be experimentally verified.
In conclusion, based on the results from the accompanying (Wang et al.36) and the current studies, we propose that in angiosperms, the ancient tRNA-IPTs and CKXs and their products or preferred substrates, the cZ-type cytokinins, play a housekeeping role to maintain basic cellular functions. On the other hand, the non-ancient ATP/ADP-IPTs and CKXs and their products or preferred substrates, the iP- and tZ-type cytokinins, contribute more to the regulation of organ development and abiotic stress responses. Our results and hypotheses shed light on the differential roles of the ancient and non-ancient IPT and CKX genes and associated cZ-, iP-/tZ-type cytokinins in plant growth and development and the responses to abiotic stresses. We hope that our hypothesis may bring about more interest regarding the elucidation of the functions of both types of cytokinins as well as the genes involved in their biosynthesis and metabolism in angiosperms.
Material and methods
Data retrieval
An exhaustive search for CKX genes was performed in online GenBank databases (http://blast.ncbi.nlm.nih.gov/) and our customized local databases. All available complete proteomes and corresponding genomic sequences from land plants, bacteria, and other organisms were downloaded from the NCBI (ftp://ftp.ncbi.nlm.nih.gov/) and JGI databases (http://genome.jgi.doe.gov/). Additional searches of CKX genes in green and red algae, glaucophytes and seedless land plants were performed online in the database of the One Thousand Plants Consortium (1KP; https://www.bioinfodata.org/Blast4OneKP/). To estimate the percentages of presence of IPT and LOG genes, 44 complete genomes from representative lineages of bacteria, archaea, and eukaryotes as well as another six species from the major lineages of land plants, were sampled according to recently proposed classification systems (Table S2)44,45. The transcriptomic data used for the expression analyses of FveCKX genes in the tissues and organs of woodland strawberry (Fragaria vesca L.) were downloaded from the SGR database (http://bioinformatics.towson.edu/strawberry/)33.
Homolog identification
The full-alignment Hidden Markov Model (HMM) profiles of the Cytokin-bind domains (Pfam: PF09265), IPPT domains (Pfam: PF01715), and Lysine_decarbox domains (Pfam: PF03641) of the CKX, IPT, and LOG proteins, respectively, were downloaded from the Pfam database v28.046. These HMM profiles were used as queries to search for homologs in all available proteome sequences of bacteria, archaea, and eukaryotes using hmmsearch47 (E-value cut-off 10−4). Additional searches of the CKXs were performed in the online GenBank protein databases of archaea and eukaryotes other than land plants using BLASTP. Next, the sequences of all matching proteins were verified using the Pfam database (http://pfam.xfam.org/)46 and the Simple Modular Architecture Research Tool database (SMART; http://smart.embl-heidelberg.de/)48 with an E-value cut-off of 10−10. The identified protein sequences that contained the core domains of all known CKXs [both Cytokin-bind (Pfam: PF09265) and FAD_binding_4 (Pfam: PF01565) domains], IPTs [single IPPT (Pfam: PF01715) domain], and LOGs [single Lysine_decarbox (Pfam: PF03641) domain] were regarded as putative homologs in the study.
For homolog searches of CKX proteins against genomic or other nucleotide sequences, the full-alignment HMM profile of the Cytokin-bind domain was used as a query in TBLASTN searches with an E-value cut-off of 10−4. To include all possible functional homologs, hit sequences longer than 10% of the domain (30 amino acids) were selected for further verification. The nucleotide hit sequences together with their 5′ and 3′ flanking regions (each 5000 bp) were used for gene annotation by FGENESH (http://linux1.softberry.com/berry.phtml). The genes containing the hit sequences or genes that were reannotated using FGENESH were considered as putative candidates. The domain structure of their protein sequences was assessed using the Pfam and SMART databases. The proteins containing both Cytokin-bind and FAD_binding_4 domains were regarded as CKX homologs.
Sequence alignment and phylogenetic analysis
For phylogenetic analyses of CKX genes in eukaryotes and bacteria, six representative land plants, one excavate, and 49 bacteria from all major bacterial groups that contained CKX homologs were sampled (Table S1). The FAD_binding_4 and Cytokin-bind domain regions of the CKX proteins were aligned using ClustalX 2.149, followed by manual inspection and refinement. Gaps and ambiguously aligned sites were removed manually. The ClustalX parameters included a gap opening penalty of 5 and a gap extension penalty of 3. The WAG+G+F substitution model was identified as the optimal model of amino acid sequence evolution using the program MODELGENERATOR50 with four gamma categories. The phylogenetic trees were then constructed via Bayesian51 and maximum-likelihood (ML)52 analyses. The Bayesian analysis was performed with MrBayes 3.251, where WAG was selected as the evolutionary model, and the number of discrete categories used to approximate the gamma distribution was set to 4 (rates = gamma, ngammacat = 4). The ML analyses were conducted with PhyML node-by-node SH test53 and RaxML54 bootstrap replicates using the WAG + F + G model, and 100 bootstrap replicates were selected to calculate the bootstrap support for the ML trees.
To investigate the evolution of land plant CKXs, 21 species with available whole-genome sequences were sampled among all major lineages of land plants (Table S6). CKX genes were identified using the above methods, and the phylogeny was constructed using PhyML based on the multiple sequence alignment of Cytokin-bind domain sequences. A functional bacterial CKX (RfaCKX1) was used as an outgroup. The ModelGenerator program was also used to identify the best-fitting model.
Ka/Ks evaluation and synteny analysis
Ks and Ka values were calculated using the program yn00 from the PAML package55. The CKX alignments between the six land plants and 49 bacteria sampled in the MrBayes phylogeny were used. Nucleotide sequences were forced to fit the amino acid CKX alignments using PAL2NAL49. To determine the mechanisms responsible for the non-ancient CKX genes in angiosperms, all against all BLASTP searches (E-value cut-off of 10−10, top five matches) were first performed in each proteome of the 16 angiosperms sampled in the phylogeny of land plant CKXs (Table S6). Syntenic blocks were detected, and the origins of all non-ancient CKX genes in angiosperms obtained from BLASTP searches were classified using McScanX30. The mechanisms of the non-ancient CKX duplications were then retrieved. The pairwise Ks values of proximal, tandem and WGD/segmental CKX duplicates were calculated using the CKX alignments of each of the 16 angiosperms with the yn00 program55.
Plant materials, tobacco transformation and growth conditions
The Arabidopsis Col-0 plants used for the dehydration treatment were grown in soil at 20 °C (dark)–24 °C (light) under 120 µmol m−2 s−1 irradiance, 65% relative humidity and a 16-h light/8-h dark photoperiod in a growth chamber.
An Arabidopsis non-ancient CKX gene, AtCKX2, was used to examine CKX overexpression in tobacco plants (Nicotiana tabacum L. var. xanthi). For plasmid construction, the full-length open reading frame of the AtCKX2 cDNA was cloned into the Apa1 and BamH1 sites of the binary vector pCambia2301 (Clontech) under the control of the cauliflower mosaic virus (CaMV) 35S promoter to yield an overexpression plasmid for non-ancient CKXs. The correct orientations were confirmed by restriction digestion analysis and sequencing. The tobacco leaf-disc explants were used for transformation by Agrobacterium tumefaciens strain LBA4404 carrying the binary vector pCambia2301.
After rooting in Murashige and Skoog (MS) medium containing 50 mg L−1 kanamycin, the transgenic plants were verified and vegetatively propagated in MS medium. Twenty-six independent lines were obtained, displaying different levels of cytokinin-deficient phenotypes56. Two lines (CKX2-10 and CKX2-21) showing obvious typical cytokinin-deficient phenotypes were selected and propagated for further oxidative and drought stress experiments. Free-hand sections of the shoots of wild-type (WT), 35S:AtCKX2 and Agrobacterium IPT-overexpressing tobacco plants were cultured on MS medium57 in Magenta boxes at 22 °C (dark)–26 °C (light) under 120 µmol m−2 s−1 light and a 16-h light/8-h dark photoperiod in a growth chamber. Photographs of the roots of these plants were taken after four weeks of growth.
Leaf dehydration treatment and qRT-PCR analysis
The rosette leaves of 3-week-old Arabidopsis Col-0 plants were harvested and placed between a piece of gauze and a piece of filter paper and air-dried using a fan for accelerated dehydration for approximately 15 min until 10% water loss was achieved. The leaves were then divided into four samples, which were immediately packed in foil and plunged into liquid nitrogen after 0, 10, 20, or 40 min of incubation. Detached leaves that were placed on wet filter paper under 100% relative humidity were used as controls.
Total RNA was extracted from the samples using the SV Total RNA Isolation System (Takara). cDNA was synthesized using the PrimeScript RT reagent Kit with a DNA Eraser kit (Perfect Real-Time) (Takara) according to the manufacturer’s instructions. qRT-PCR was performed in a Bio-Rad IQ5 Real-Time PCR System (Bio-Rad) using SYBR Premix Ex Taq (Takara). Three biological replicates were conducted, and the results for each sample were normalized using Tub8 as an internal reference gene. Transcription levels are presented as 2-ΔCt values, where ΔCt represents the difference between the cycle threshold values of the target and the reference genes58. The statistical significance of the differences between the treated leaves and the control was estimated using the one-tailed t-test. The primers used for the qRT-PCR analyses are provided in Table S9.
Oxidative treatment using methyl viologen (MV)
Free-hand sections of the shoots of WT or transgenic plants were cultured for three weeks. Thereafter, one WT and one 35S:AtCKX2 tobacco plant were transferred to Murashige and Skoog medium57 containing 20 μM MV in each box. Fifteen CKX2-10, fifteen CKX2-21, and thirty WT plants were used for each experiment. The experiments were repeated three times, and all the experiments showed similar results. Leaves were harvested after 36 h for the detection of H2O2 accumulation and antioxidant enzyme activities with three biological replicates. Photographs of representative oxidatively stressed plants (WT and CKX2-10) were taken after 5 days of treatment.
Drought stress treatment
After one month of growth in MS media, one WT and one 35S:AtCKX2 tobacco plant generated from the free-hand sections of the shoots were transferred to each pot. The plants were grown at 22 °C (dark)–26 °C (light) under 160 µmol m−2 s−1 irradiance, 65% relative humidity and a 16-h light/8-h dark photoperiod in a growth chamber and fertilized once a week with 300 mL of 1% solid fertilizer (20% N, 20% P, 20% K, 0.05% Mg and 0.025% Mn). After one month, watering was stopped, followed by one month of drought treatment. Ten CKX2-10, ten CKX2-21, and twenty WT plants were used for each experiment. The experiments were repeated three times, and all the experiments showed similar results. On Day 14, leaves with no sign of wilting were harvested for the detection of H2O2 accumulation with three biological replicates. Photographs of the drought-stressed plants (WT and CKX2-10) were taken on Day 31 before rewatering. Then, the plants were rewatered (the soil was saturated with water), and photographs were taken on the 3rd and 14th days after rewatering.
DAB staining and enzyme assays
H2O2 production by the oxidatively or drought-stressed leaves was detected by 3,3′-diaminobenzidine (DAB) polymerization59. For catalase enzyme analyses, frozen oxidatively stressed leaf samples (0.2 g) were homogenized in 1.6 mL of sodium phosphate buffer (50 mM, pH 7.8). The homogenate was then centrifuged for 20 min at 12,000 × g, and the supernatant was used for the detection of enzyme activity according to Zhou et al.60.
Supplementary information
Acknowledgements
The authors thank Dr. Qun Sun of Sichuan University and members of the Y.L. laboratory for discussions and comments on the manuscript. This work was supported by the National Natural Science Foundation of China [grant number 31471860 to J.D. and number 31401842 to H.W.]; and the Open Project of the State Key Laboratory of Crop Genetics and Germplasm Enhancement [grant number ZW2014008 to H.W.].
Author contributions
Y.L. and J.D. designed the experiments; X.W. and S.L. performed the experiments and data analyses; D.L. participated in data analyses; J.D., X.W. and Y.L. wrote the manuscript; T.G., H.W., R.T., J.H., Y.L. and R.M. revised the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Yi Li is a no pay visiting professor at Nanjing Agricultural University.
These authors contributed equally: Xiaojing Wang, Jing Ding
Contributor Information
Jing Ding, Email: jding@njau.edu.cn.
Jinling Huang, Email: huangj@ecu.edu.
Yi Li, Email: yi.li@uconn.edu.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-0246-z).
References
- 1.Kieber JJ, Schaller GE. Cytokinins. Arabidopsis Book. 2014;11:e0168. doi: 10.1199/tab.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gajdosova S, et al. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 2011;62:2827–2840. doi: 10.1093/jxb/erq457. [DOI] [PubMed] [Google Scholar]
- 3.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 4.Frebort I, Kowalska M, Hluska T, Frebortova J, Galuszka P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011;62:2431–2452. doi: 10.1093/jxb/err004. [DOI] [PubMed] [Google Scholar]
- 5.Miyawaki K, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl Acad. Sci. USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stirk W, Van Staden J. Flow of cytokinins through the environment. Plant Growth Regul. 2010;62:101–116. doi: 10.1007/s10725-010-9481-x. [DOI] [Google Scholar]
- 7.Lu Y, Xu J. Phytohormones in microalgae: a new opportunity for microalgal biotechnology? Trends Plant Sci. 2015;20:273–282. doi: 10.1016/j.tplants.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Lindner AC, et al. Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 2014;65:2533–2543. doi: 10.1093/jxb/eru142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Ma XM, Kojima M, Sakakibara H, Hou BK. Glucosyltransferase UGT76C1 finely modulates cytokinin responses via cytokinin N-glucosylation in Arabidopsis thaliana. Plant Physiol. Biochem. 2013;65:9–16. doi: 10.1016/j.plaphy.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Avalbaev AM, Somov KA, Yuldashev RA, Shakirova FM. Cytokinin oxidase is key enzyme of cytokinin degradation. Biochemistry (Moscow.) 2012;77:1354–1361. doi: 10.1134/S0006297912120024. [DOI] [PubMed] [Google Scholar]
- 11.Werner T, et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiyama R, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23:2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köllmer I, Novák O, Strnad M, Schmülling T, Werner T. Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 2014;78:359–371. doi: 10.1111/tpj.12477. [DOI] [PubMed] [Google Scholar]
- 14.Macková H, et al. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 2013;64:2805–2815. doi: 10.1093/jxb/ert131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mytinova Z, et al. Effect of abiotic stresses on the activity of antioxidative enzymes and contents of phytohormones in wild type and AtCKX2 transgenic tobacco plants. Biol. Plant. 2010;54:461–470. doi: 10.1007/s10535-010-0082-3. [DOI] [Google Scholar]
- 16.Prerostova S, et al. Cytokinins: Their impact on molecular and growth responses to drought stress and recovery in. Arabidopsis. Front. Plant Sci. 2018;9:e655. doi: 10.3389/fpls.2018.00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carabelli M, et al. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Gene Dev. 2007;21:1863–1868. doi: 10.1101/gad.432607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lubovská Z, Dobrá J, Štorchová H, Wilhelmová N, Vanková R. Cytokinin oxidase/dehydrogenase overexpression modifies antioxidant defense against heat, drought and their combination in Nicotiana tabacum plants. J. Plant Physiol. 2014;171:1625–1633. doi: 10.1016/j.jplph.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 19.Pertry I, et al. Rhodococcus fascians impacts plant development through the dynamic fas-mediated production of a cytokinin mix. Mol. Plant Microbe. 2010;23:1164–1174. doi: 10.1094/MPMI-23-9-1164. [DOI] [PubMed] [Google Scholar]
- 20.Schmülling T, Werner T, Riefler M, Krupková E, Bartrina y Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003;116:241–252. doi: 10.1007/s10265-003-0096-4. [DOI] [PubMed] [Google Scholar]
- 21.Bock R. The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Casson N, Michel R, Müller KD, Aubert JD, Greub G. Protochlamydia naegleriophila as etiologic agent of pneumonia. Emerg. Infect. Dis. 2008;14:168–172. doi: 10.3201/eid1401.070980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Gogarten JP. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 2007;8:e99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker B, Hoef-Emden K, Melkonian M. Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol. Biol. 2008;8:e203. doi: 10.1186/1471-2148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collingro A, et al. Unity in variety—the pan-genome of the Chlamydiae. Mol. Biol. Evol. 2011;28:3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc. Natl Acad. Sci. USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu R, et al. Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase (CKX) gene family. J. Plant Growth Regul. 2010;29:428–440. doi: 10.1007/s00344-010-9155-y. [DOI] [Google Scholar]
- 28.Galuszka P, et al. Biochemical characterization of cytokinin oxidases/ dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J. Plant Growth Regul. 2007;26:255–267. doi: 10.1007/s00344-007-9008-5. [DOI] [Google Scholar]
- 29.Šmehilová M, et al. Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J. Exp. Bot. 2009;60:2701–2712. doi: 10.1093/jxb/erp126. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsui A, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 32.Vyroubalová Š, et al. Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol. 2009;151:433–447. doi: 10.1104/pp.109.142489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darwish O, et al. SGR: an online genomic resource for the woodland strawberry. BMC Plant Biol. 2013;13:e223. doi: 10.1186/1471-2229-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, et al. Antagonistic roles of abscisic acid and cytokinin during response to nitrogen depletion in oleaginous microalga Nannochloropsis oceanica expand the evolutionary breadth of phytohormone function. Plant J. 2009;80:52–68. doi: 10.1111/tpj.12615. [DOI] [PubMed] [Google Scholar]
- 35.Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X. et al. Evolution and roles of cytokinin genes in angiosperms 1: Do ancient IPTs play housekeeping while non-ancient IPTs play regulatory roles? Hortic Res7, (2020). 10.1038/s41438-019-0211-x. [DOI] [PMC free article] [PubMed]
- 37.Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23:69–80. doi: 10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner T, et al. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell. 2010;22:3905–3920. doi: 10.1105/tpc.109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 40.Niemann MC, et al. The cytokinin oxidase/dehydrogenase CKX1 is a membrane-bound protein requiring homooligomerization in the endoplasmic reticulum for its cellular activity. Plant Physiol. 2018;176:2024–2039. doi: 10.1104/pp.17.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer M, et al. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015;66:4873–4884. doi: 10.1093/jxb/erv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz RY, Skoog F, Playtis AJ, Leonard NJ. Cytokinins: synthesis and biological activity of geometric and position isomers of zeatin. Plant Physiol. 1972;50:702–705. doi: 10.1104/pp.50.6.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mok MC, Mok DW, Armstrong DJ. Differential cytokinin structure-activity relationships in. Phaseolus. Plant Physiol. 1978;61:72–75. doi: 10.1104/pp.61.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adl SM, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–514. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hug LA, et al. A new view of the tree of life. Nat. Microbiol. 2016;1:e16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 46.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 48.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:302–305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006;6:e29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 52.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 53.Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- 54.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 55.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 56.Werner T, Motyka V, Strnad M, Schmülling T. Regulation of plant growth by cytokinin. Proc. Natl Acad. Sci. USA. 2001;98:10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 58.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 59.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J. 1997;11:1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- 60.Zhou YH, Yu JQ, Huang LF, Nogues S. The relationship between CO2 assimilation, photosynthetic electron transport and water-water cycle in chill-exposed cucumber leaves under low light and subsequent recovery. Plant Cell Environ. 2004;27:1503–1514. doi: 10.1111/j.1365-3040.2004.01255.x. [DOI] [Google Scholar]
- 61.Nieminen K, et al. Cytokinin signaling regulates cambial development in poplar. Proc. Natl Acad. Sci. USA. 2008;105:20032–20037. doi: 10.1073/pnas.0805617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17:172–179. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 63.Joshi R, et al. Knockdown of an inflorescence meristem-specific cytokinin oxidase–OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2018;41:936–946. doi: 10.1111/pce.12947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.