Abstract
The pathogenesis of acute myeloid leukemia (AML) involves serial acquisition of mutations controlling several cellular processes, requiring combination therapies affecting key downstream survival nodes in order to treat the disease effectively. The BCL2 selective inhibitor venetoclax has potent anti-leukemia efficacy; however, resistance can occur due to its inability to inhibit MCL1, which is stabilized by the MAPK pathway. In this study, we aimed to determine the anti-leukemia efficacy of concomitant targeting of the BCL2 and MAPK pathways by venetoclax and the MEK1/2 inhibitor cobimetinib, respectively. The combination demonstrated synergy in seven of 11 AML cell lines, including those resistant to single agents, and showed growth-inhibitory activity in over 60% of primary samples from patients with diverse genetic alterations. The combination markedly impaired leukemia progenitor functions, while maintaining normal progenitors. Mass cytometry data revealed that BCL2 protein is enriched in leukemia stem/progenitor cells, primarily in venetoclax-sensitive samples, and that cobimetinib suppressed cytokine-induced pERK and pS6 signaling pathways. Through proteomic profiling studies, we identified several pathways inhibited downstream of MAPK that contribute to the synergy of the combination. In OCI-AML3 cells, the combination downregulated MCL1 protein levels and disrupted both BCL2:BIM and MCL1:BIM complexes, releasing BIM to induce cell death. RNA sequencing identified several enriched pathways, including MYC, mTORC1, and p53 in cells sensitive to the drug combination. In vivo, the venetoclax-cobimetinib combination reduced leukemia burden in xenograft models using genetically engineered OCI-AML3 and MOLM13 cells. Our data thus provide a rationale for combinatorial blockade of MEK and BCL2 pathways in AML.
Introduction
Acute myeloid leukemia (AML) is characterized by the uncontrolled proliferation and arrested differentiation of abnormal stem and progenitor cells. Standard induction chemotherapy induces a high rate of complete remission but fails to improve overall survival especially in elderly patients with AML.1,2 Genes significantly mutated in AML can be organized into several functional categories that are associated with enhanced proliferation, impaired differentiation, deregulated chromatin modification, and DNA methylation.1,3 Therefore, co-targeting downstream pathways that contribute to leukemogenesis may deliver the greatest clinical efficacy.
The anti-apoptotic protein BCL2 has been studied extensively for its role in leukemic transformation and chemoresistance. BCL2 is highly expressed in AML leukemia stem cells (LSC) containing low levels of reactive oxygen species which are resistant to chemotherapy.4,5 BCL2 inhibitors have been shown to eradicate AML LSC and sensitize chronic myeloid leukemia LSC to tyrosine kinase inhibitors. Oncogenic dependency on BCL2 was found in AML patients carrying mutations in IDH1 and IDH2.6,7 A recent study used BH3 profiling to discover that co-inhibition of BCL2 with tyrosine kinase inhibitors facilitated eradication of genetically diverse AML in patient-derived xenograft (PDX) models.8 We have reported the anti-leukemia potency of venetoclax (ABT-199/GDC-0199), an orally bioavailable BH3 mimetic that selectively binds with high affinity to BCL2, but lacks affinity for BCL-XL and MCL1, in AML models.9 In a phase II clinical trial, venetoclax monotherapy had clinical activity in patients with relapsed or refractory AML with a tolerable safety profile.10 However, the inability of venetoclax to inhibit MCL1 causes resistance in leukemia cells that require MCL1 for survival.11,12
The RAF/MEK/ERK (MAPK) cascade, a major effector pathway activated in 70%-80% of patients with AML, is activated by upstream mutant proteins such as FLT3, KIT, and RAS.13–15 The MAPK pathway regulates BCL2 family proteins by stabilizing anti-apoptotic MCL111,16 and inactivating pro-apoptotic BIM (BCL2L11).17 Monotherapy with MEK inhibitors has had limited clinical efficacy.15 Recently it was shown that MAPK signaling activation contributed to primary resistance to an IDH2 inhibitor18 and acquired resistance to venetoclax,19 suggesting that combination regimens that include MEK inhibitors could be efficacious in these patients. Cobimetinib (GDC-0973) is an allosteric MEK inhibitor with antitumor activity in BRAF- and KRAS-mutant tumor cells,20 and was recently approved to treat patients with metastatic melanoma. Its anti-AML efficacy, particularly in combination with venetoclax, is unknown.
Furthermore, biomarkers predictive of response to this combination in AML are unknown. Suppression of downstream pERK does not predict sensitivity to MEK inhibition.21 In melanoma, it has been demonstrated that mTORC1/2 and pS6 activities are associated with acquired resistance to MEK inhibitors and suppression of pS6 may serve as a biomarker to predict clinical response to MEK inhibitors.22,23 The role of pS6 in response to MEK and BCL2 inhibition has not been addressed.
In this study, we evaluated the anti-leukemia effects of concomitant BCL2 and MAPK blockade by venetoclax and cobimetinib in AML cell lines, primary patients’ samples, and xenograft murine models. Through the use of reverse-phase protein arrays (RPPA) and RNA sequencing, we identified pharmacodynamic markers that correlated with the efficacy of the combination treatment, in particular activated pS6 (Ser235/236), which discriminated combination-sensitive from -insensitive AML cells. Our data support the rationale for dual inhibition of the BCL2 and MEK pathways.
Methods
Patients’ samples, acute myeloid leukemia cell lines, and reagents
Bone marrow and peripheral blood samples were collected from patients with AML or healthy donors after informed con sent had been obtained in accordance with the Institutional Review Board of The University of Texas MD Anderson Cancer Center. The cell line culture methodology is described in the Online Supplementary Methods.
Assays and other methods
Details of the CellTiter-Glo proliferation assay, colony-forming cell assay, electrochemiluminescent enzyme-linked immunosorbent assay, RPPA, and RNA sequencing are provided in the Online Supplementary Methods. Antibody conjugation for mass cytometry staining and the spanning-tree progression analysis of density-normalized events (SPADE) analysis are also explained in detail in the Online Supplementary Methods.
Apoptosis in primary acute myeloid leukemia samples
As previously reported,24 18 primary AML peripheral blood mononuclear cells or AML PDX samples were cultured in LSC medium. Viable AML CD45dim blast cells were enumerated by using CountBright counting beads (Cat. C36950; Invitrogen, Carlsbad, CA, USA) with concurrent annexin V and DAPI detection on a Gallios Flow Cytometer (Beckman Coulter, Indianapolis, IN, USA). Data analysis and additional details are included in the Online Supplementary Methods.
In vivo study of cobimetinib and venetoclax in acute myeloid leukemia xenograft mouse models
The animal studies were performed in accordance with guidelines approved by the Institutional Animal Care and Use Committee at MD Anderson. Nonobese diabetic/severe combined immunodeficient gamma IL3-GM-SF (NSG-SGM3 or NSGS) mice (female, 8-10 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME, USA). The mice were injected intravenously with OCI-AML3-Luci-GFP (1.0×106) cells, which were lentivirally transduced with firefly luciferase. Leukemia engraftment was confirmed 1 week after injection through a noninvasive in vivo bioluminescence imaging (BLI) system (Xenogen, Alameda, CA, USA) after injection of a D-luciferin (4 mg/mouse) substrate. Mice were distributed into four groups (11 mice/group) with comparable tumor burden and dosed daily for 4 weeks with one of the following oral preparations: vehicle, cobimetinib (10 mg/kg), venetoclax (100 mg/kg), or cobimetinib plus venetoclax. BLI was performed weekly to determine the extent of engraftment. Survival was monitored as an endpoint. A similar MOLM13 model is described in the Online Supplementary Methods.
Statistical analyses
The Student t-test was used to analyze the statistical significance of differences between groups, both in vitro and in vivo. All statistical tests were two-sided, and the results are expressed as the mean ± standard deviation. A P value ≤0.05 was considered statistically significant. The RPPA and RNA-sequencing data analysis are described in the Online Supplementary Methods.
Results
Cobimetinib and venetoclax demonstrate synergistic anti-leukemia efficacy in acute myeloid leukemia cell lines in vitro
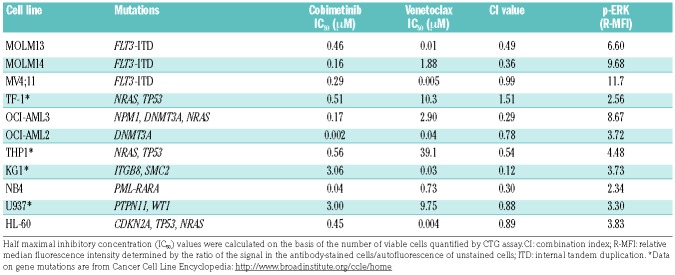
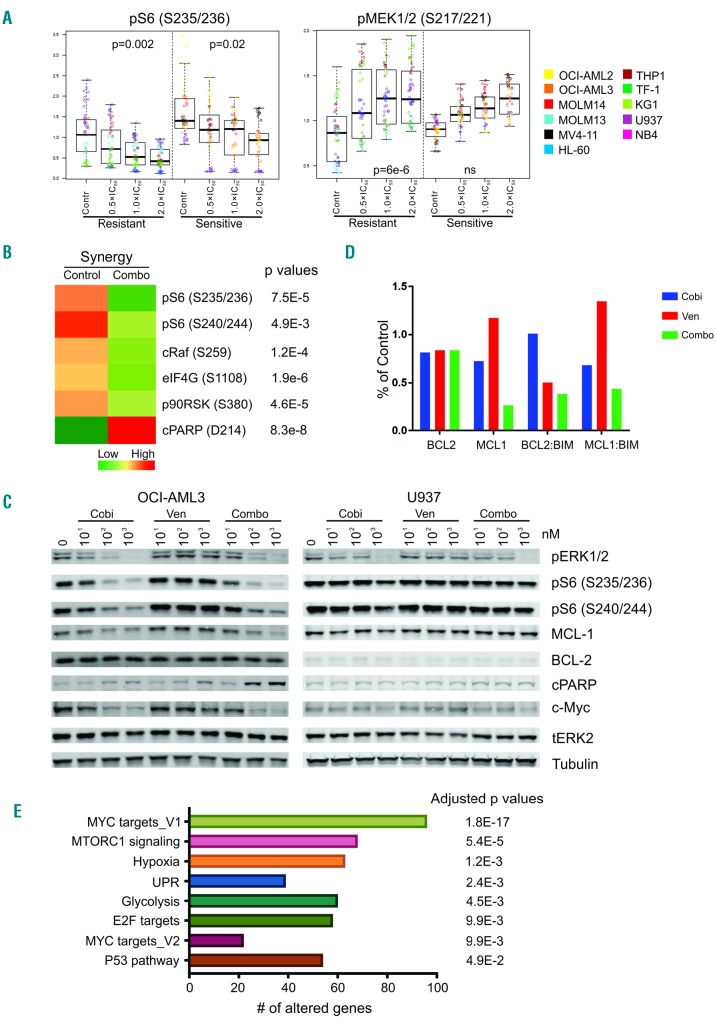
To assess the anti-leukemia activity of cobimetinib and venetoclax as single agents or in combination, we studied their effects on cell proliferation of 11 AML cell lines (Table 1). The median inhibitory concentration (IC50) values of single agents were determined in a dose-response manner using CellTiter-Glo (CTG) assays after 72 h of drug treatment. The IC50 values of cobimetinib (range, 0.002 μM - 3.0 μM) did not correlate with either the status of RAS mutations or the basal levels of p-ERK1/2 determined by flow cytometry (Table 1). To assess pharmacological interactions between cobimetinib and venetoclax, incremental doses were applied based on the IC50 value of each drug. In seven of the 11 cell lines, combination of the agents elicited synergistic growth inhibition based on the Chou-Talalay method of analysis [combination index (CI) <0.8].25 Cell lines with IC50 values below the selected cutoff values (0.3 μM for cobimetinib20 and 0.1 μM for venetoclax9) were defined as sensitive to the single agent. Patterns of response to single agents and the combination were distinct. Notably, while synergy was observed in both venetoclax-resistant (MOLM14, OCI-AML3, NB4) and cobimetinib-resistant cell lines (KG1, MOLM13), the lowest CI value (0.12) was seen in venetoclaxsensitive/cobimetinib-resistant AML cells (KG1) (Figure 1).
Table 1.
Cytotoxicity of cobimetinib and venetoclax in acute myeloid leukemia cell lines.
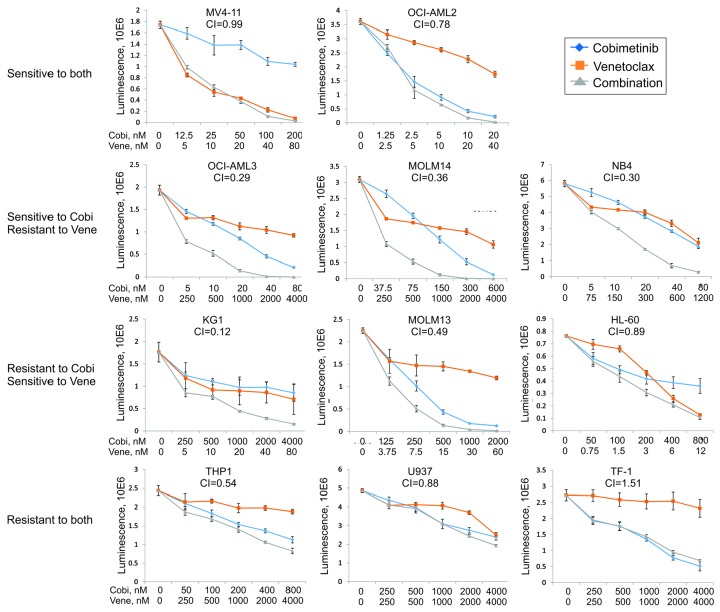
Figure 1.
Anti-leukemia efficacy of cobimetinib and venetoclax in acute myeloid leukemia cell lines. Eleven acute myeloid leukemia cell lines were left untreated or treated with cobimetinib or venetoclax as single agents at 0.001, 0.01, 0.1, or 1.0 μM for 72 h. Calcusyn software was used to calculate the median inhibitory concentration (IC50) values. Combinations of the two drugs were then tested on the same cell lines at dose ranges of 0.25, 0.5, 1, 2, and 4 times the IC50 value of each compound. The combination index of each combination in each cell line was calculated on the basis on the luminescent intensity that correlated with number of viable cells determined by the CellTiter-Glo assay. Responses to treatment were categorized into four patterns: (i) sensitive to both drugs; (ii) sensitive only to cobimetinib and showing synergy for the combination; (iii) sensitive only to venetoclax and showing synergy for the combination; (iv) resistant to both drugs. AML: acute myeloid leukemia; Cobi: cobimetinib; Ven: venetoclax; CI: combination index
Cobimetinib and venetoclax demonstrate on-target suppression of cell proliferation and clonogenic potential of leukemia progenitors in a subset of primary acute myeloid leukemia cells ex vivo
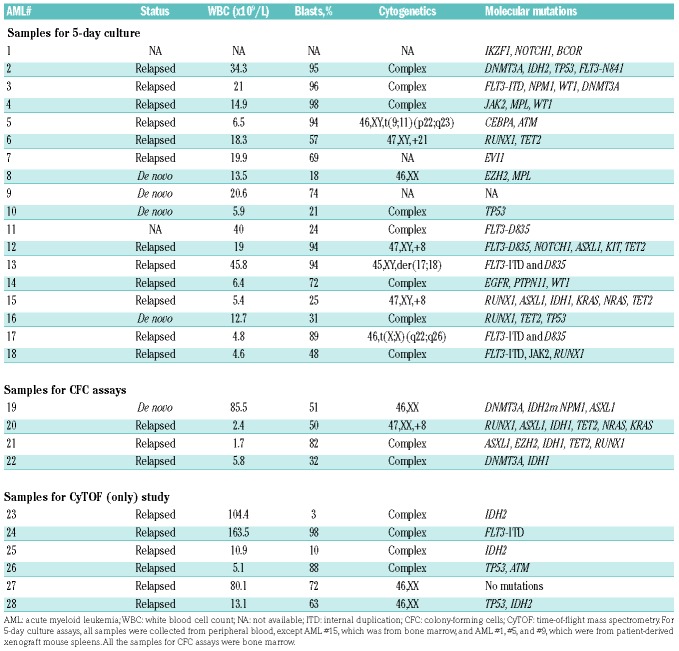
The anti-leukemia activities of cobimetinib and venetoclax were examined in 18 primary samples with diverse genetic alterations, collected from patients with AML or spleen from PDX models (Table 2). Primary AML blasts were treated with cobimetinib and venetoclax alone or in combination at 0.1 μM for 5 days in LSC medium to maintain the immature state of the leukemia cells.24 Cobimetinib alone induced minimal cell death (specific apoptosis, 6.7 ± 5.9%), which was significantly enhanced when the drug was given in combination with venetoclax (27.7 ± 20.2%, P=0.001) (Figure 2A, left). Cobimetinib inhibited cell proliferation in the majority of cases (34.2 ± 23.7%), and this suppression was more pronounced when the drug was combined with venetoclax (60.2 ± 28.8%, P<0.001) (Figure 2A, right). Venetoclax as a single agent reduced viable cell numbers by more than 50% in six cases (33.3%). Three of the four AML samples demonstrating over 50% growth inhibition by the cobimetinib treatment carried the FLT3-ITD and/or D835 point mutation (AML 12, 13, and 17). As previously reported, IDH-mutant AML samples were highly sensitive to venetoclax as a single agent (AML 2 and 15). Over 60% (11 of 18) of the patients’ samples responded to the combination treatment, including those insensitive to either compound alone (AML 1, 8 and 11). Importantly, induction of apoptosis in AML stem/progenitor CD34+CD38−CD123+ population following the combination treatment was observed in two out of four AML samples tested (Online Supplementary Figure S1). The clonogenic potential of myeloid progenitors was significantly suppressed by the combination (82.5 ± 20.0%), as compared to cobimetinib (38.3 ± 14.6%, P=0.01) or venetoclax (41.9 ± 18.6%, P<0.05) alone. Normal progenitor function was minimally affected (Figure 2B and Online Supplementary Figure S2).
Table 2.
Clinical information for primary acute myeloid leukemia patients’ samples.
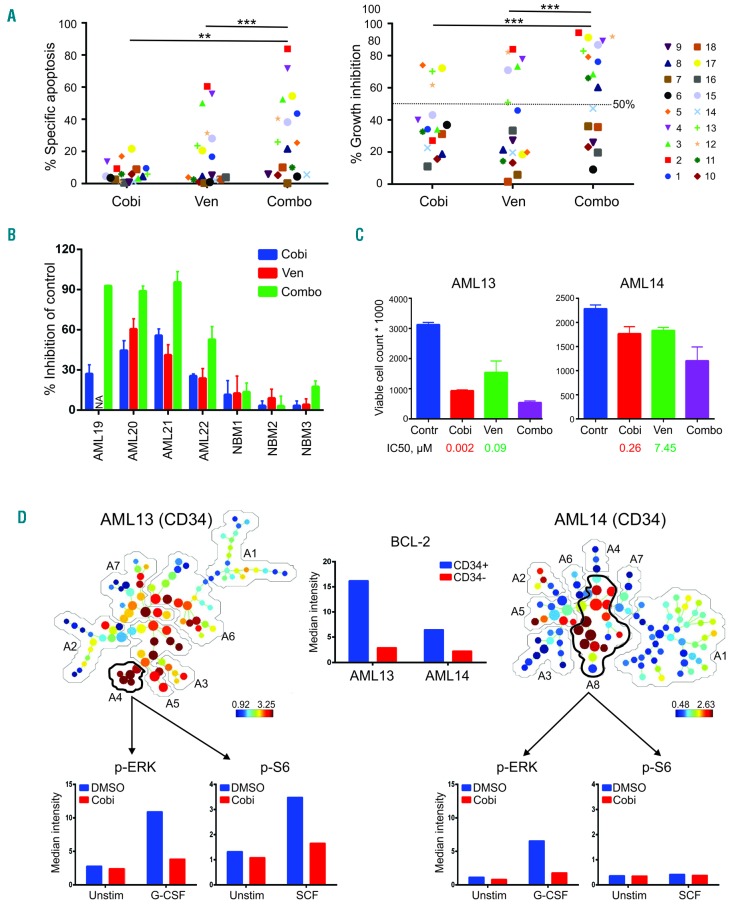
Figure 2.
Treatment with cobimetinib and venetoclax causes on-target suppression of cell proliferation and impairs leukemia progenitor function in a subset of primary acute myeloid leukemia cases. (A) Primary acute myeloid leukemia (AML) peripheral blood mononuclear or bone marrow cells from AML cases were cultured in serum-free expansion medium supplemented with BIT 9500 Serum Substitute and cytokines, including stem cell factor (SCF; 100 ng/mL), Flt3 ligand (50 ng/mL), interleukin 3 (IL3; 20 ng/mL), and granulocyte colony-stimulating factor (G-CSF; 20 ng/mL) as well as StemRegenin 1 (SR1; 1 μM). Cells were left untreated or treated with cobimetinib (Cobi) or venetoclax (Van), both at 0.1 μM, as single agents or in combination (Combo). After culture for 5 days, cells were stained with CD45-PE, Annexin-V-APC, and DAPI. Apoptotic leukemia blasts (CD45dimAnnexin-V+) were isolated by flow cytometry. Results are expressed as percentage of specific apoptosis calculated by the formula: 100 × (% apoptosis of treated cells – % apoptosis of control cells)/(100 – % apoptosis of control cells). Percentage of growth inhibition was calculated on the basis of the number of control viable cells (Annexin-V−/DAPI−). **P<0.01; ***P<0.001. (B) Mononuclear cells collected from AML patients (100,000 cells) or healthy donors (50,000 cells; NBM) were plated in methylcellulose, then treated with venetoclax or cobimetinib (both at 0.1 μM) as single agents or in combination. Colonies were scored on day 14. Data are presented as percentage inhibition compared to control groups. (C) The absolute cell counts of AML13 and AML14 samples as determined in (A) are shown in comparison with those of untreated controls (Contr), with median inhibitory concentration (IC50) values indicated for each sample. (D) AML13 and AML14 samples were treated with cobimetinib 1.0 μM overnight followed by 10 min with or without (Unstim) stimulation with SCF or G-CSF (100 ng/mL). Cells were fixed, permeabilized, and processed for time-of-lfight mass spectrometry. Spanning-tree progression analysis of density-nor-malized events (SPADE) trees were generated by using markers shown in Online Supplementary Figure S2. The leukemia stem/progenitor populations were manually annotated and highlighted by analysis of all surface markers. The median intensities of pERK and pS6 in gated populations are shown. BCL2 expression in CD34+ and CD34− fractions in both samples is shown. DMSO: dimethylsulfoxide.
To test the on-target efficacy of both agents, we developed a 28-parameter mass cytometry [time-of-flight mass spectrometry (CyTOF)] panel comprising antibodies against surface antigens to define AML stem/progenitor fractions and intracellular proteins of the BCL2 family and various signaling pathways26 (Online Supplementary Table S1). The CyTOF study was performed in AML13 (sensitive to the combination) and AML14 (resistant to the combination) samples (Figure 2C). SPADE trees were built and annotated using all cell surface markers (Online Supplementary Table S1); the positive markers were included in the heat maps (Online Supplementary Figure S3). BCL2 protein levels were significantly enriched in CD34+ stem/progenitor cells compared to CD34− cells and BCL2 was expressed at a higher level in the venetoclax-sensitive sample (AML13) than in the venetoclax-resistant sample (AML14), consistent with our published data9 (Figure 2D). These results support the notion that venetoclax preferentially target LSC in AML. As previously reported, the cancer signaling network relies on the manner in which cancer cells respond to external stimuli rather than their basal phosphorylation state.27 Therefore, following cobimetinib treatment, we stimulated primary AML cells with granulocyte colony-stimulating factor (G-CSF) or stem cell factor (SCF) to study MEK downstream signaling pathways under conditions mimicking a cytokine-rich bone marrow microenvironment. The SPADE trees were colored based on expression levels of CD34. In AML13, annotation 4 (6.5% of total viable cells) represented the leukemia stem/progenitor cell population by phenotypically positive expression of CD34, CD123, CD25, CD135, and CD64. Annotation 8 accounted for 53.4% of total viable cells in AML14, and was highly positive for expression of CD34, CD123, CD117, CD135, and CD64 (Online Supplementary Figure S3). In both samples, we observed low basal levels of pERK, which increased following G-CSF stimulation (3.9-fold in AML13 and 5.7-fold increase in AML14). G-CSF-stimulated pERK in both patients’ samples was largely inhibited by cobimetinib despite differential responses in proliferation assays (Figure 2C, D), indicating that suppression of pERK does not predict sensitivity to MEK inhibition and is consistent with previous reports.21 Several studies have shown that suppression of mTORC1 and downstream pathways (especially S6) predicted sensitivity to MEK inhibition.21,22 We found that pS6 was highly activated by SCF and effectively suppressed by cobimetinib in the cobimetinib-sensitive AML sample, whereas the cobimetinib-resistant AML sample did not demonstrate a response to SCF. As in this study, we treated cells overnight and transiently stimulated then for 10 min to look into activation of signal transduction pathways (Figure 2). Due to limited exposure to the inhibitors (2 h), we were unable to detect changes in frequencies of AML stem/progenitor cells (Online Supplementary Figure S4).
Transcriptomic and proteomic profiles identify pharmacodynamic markers underlying responses to targeted agents
To identify the pharmacodynamic markers underlying the observed drug responses, we treated the 11 AML cell lines (Table 1) with cobimetinib and venetoclax as single agents or in combination for 24 h at doses that were 0.5, 1 or 2 times their IC50 values, followed by RPPA and RNA sequencing analysis. As already described, cells with IC50 values <0.3 μM for cobimetinib or <0.1 μM for venetoclax were categorized as sensitive and those with IC50 values above these cutoffs were considered resistant. For the combination groups, CI values <0.8 were considered synergistic.
Quantification of 90 proteins by RPPA identified several biomarkers that correlated with in vitro drug responses. For example, S6 phosphorylation at Ser235/236 was significantly reduced in both cobimetinib-sensitive and -resistant cell lines compared to untreated cells, with sensitive cells displaying higher basal phosphorylation at Ser235/236. Significant pMEK induction was observed in cobimetinib-resistant cell lines (Figure 3A). Several signaling pathways were highly activated under basal conditions in cobimetinib-sensitive cells compared to resistant cells, including pS6 (Ser235/236), pRSK, pERK, p38MAPK and pPTEN (Online Supplementary Figure S5A). Proteins indicating responses to venetoclax treatment were largely limited to the caspase-dependent apoptotic cascade (data not shown). Higher levels of BAX and BCL2 and lower levels of BIM and pS6 (Ser240/244) correlated with sensitivity to venetoclax (Online Supplementary Figure S5B). In cell lines in which cobimetinib and venetoclax had a synergistic effect, several MEK downstream pathways were significantly downregulated and cleaved poly (ADP-ribose) polymerase (PARP) was detected, indicating induction of apoptosis (Figure 3B). These changes were not identified in cell lines in which a synergistic effect did not occur. The heat maps of the complete RPPA datasets are shown in Online Supplementary Figure S6.
Figure 3.
Pharmacodynamic markers of drug response identified through reverse-phase protein arrays and RNA sequencing. Acute myeloid leukemia (AML) cell lines were left untreated or treated with cobimetinib or venetoclax as single agents or in combination at 0.5, 1, or 2 times the median inhibitory concentration (IC50) value of each compound in each cell line for 24 h. Cell pellets were harvested after treatment and subjected to reverse-phase protein array (RPPA) analysis as previously reported. (A) Plots depict proteins differentially expressed between cobimetinib-sensitive and cobimetinib-resistant cells. (B) The mean values of corresponding proteins in cell lines showing synergy to the combination treatment (CI<0.8 as presented in Table 1) are shown in the heatmap. Only the proteins showing significant differences (P<0.05) between control and treated groups are shown. C: untreated control; T: treated; S: sensitive; R: resistant; Syn: synergy. (C) Cells were treated with cobimetinib (Cobi), venetoclax (Ven), or a combination (Combo) at 10, 100 and 1000 nM for 4 h and subjected to lysis; proteins were separated and probed with the antibodies indicated. (D) AML cell lines were left untreated (control) or treated with cobimetinib or venetoclax as single agents or in combination at 10 times the median inhibitory concentration (IC50) value of each compound in each cell line for 4 h. Cell pellets were harvested after treatment and subjected to electrochemiluminescent enzyme-linked immunosorbent assay. The levels of BCL2, MCL1, BCL2:BIM and MCL1:BIM complexes were plotted based on percentages of the levels in the control group. (E) AML cells were treated and processed as described above for the RPPA assay. RNA was isolated using the RNeasy kit and sent for mRNA sequencing. The enriched pathways in cell types showing synergy in response to the combination are shown.
Western blotting was performed to validate the RPPA data in four cell lines representing different response patterns (Table 1, Figure 3C and Online Supplementary Figure S5C). Suppression of pERK by cobimetinib was observed in both sensitive (OCI-AML3 and MV4;11) and resistant (MOLM13 and U937) cells at 0.01 μM, irrespective of response patterns. pS6 (Ser235/236 or Ser240/244) was inhibited by a low dose (0.1 μM) of cobimetinib in sensitive OCI-AML3 cells but not in resistant U937 cells (Figure 3C), consistent with our RPPA findings noted above. These data also indicate direct suppression of mTORC1 signaling by cobimetinib as the Ser240/244 site is regulated exclusively by mTORC1. MYC was downregulated by cobimetinib alone or in combination with venetoclax in OCI-AML3, but not in U937 cells. Cell death characterized by elevated levels of cleaved PARP was observed in the combination group in OCI-AML3 cells, consistent with pro-apoptotic synergy (Figure 3C). To capture the dynamic interactions of pro- and anti-apoptotic BCL2 family members, a Meso Scale Discovery assay was performed in OCI-AML3 cells (Figure 3D). High basal levels of BCL2 and BCL2:BIM complexes were identified. After venetoclax treatment, BCL2:BIM complexes were disrupted and MCL1 protein levels were upregulated, resulting in increased MCL1:BIM complexes. The combination of cobimetinib with venetoclax suppressed both BCL2:BIM and MCL1:BIM complexes, enabling release of free BIM to induce cell death (Figure 3D). In addition, cobimetinib treatment induced total BIM protein levels in MV4;11 cells, thereby priming the cells for death (Online Supplementary Figure S7).
To refine our search for potential biomarkers correlating with response to the venetoclax-cobimetinib combination, we performed RNA sequencing and evaluated differential gene expression after exposure to the drugs. The aim was to identify hallmark cancer pathways significantly altered specifically in cells sensitive to the drug combination (Figure 3E and Online Supplementary Table S2). Our analysis demonstrated that several downstream pathways, including MYC, E2F and their target genes, were significantly altered after treatment in cells that responded synergistically to the combination. Consistent with western blot data, mTORC1 signaling was also altered in cells showing synergistic responses. Hypoxia and unfolded protein response (UPR) pathways were also significantly enriched, possibly downstream of mTOR/4EBP1/eIF4E signaling, which directs protein synthesis of HIF-1α,28 and can trigger the UPR.29 Glycolysis, another enriched pathway, is regulated by the ERK signaling pathway through RNK126-mediated ubiquitination of pyruvate dehydrogenase kinase, which may account for resistance to apopto-sis.30
The combination of cobimetinib and venetoclax reduces leukemia burden in acute myeloid leukemia models in vivo
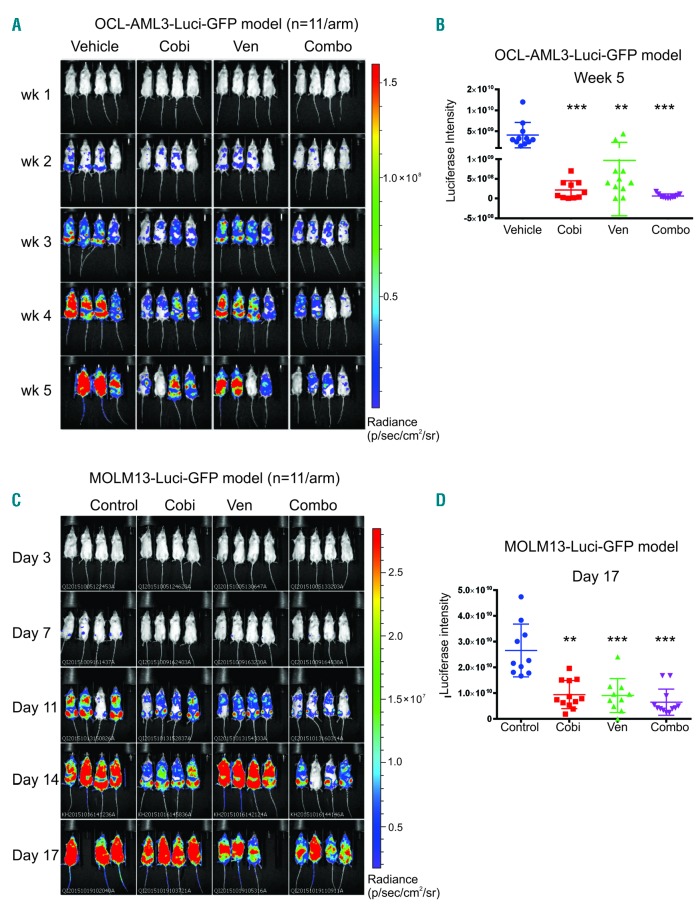
To test the efficacy of cobimetinib and venetoclax in vivo, we induced leukemia in NSGS mice by injecting the animals with genetically engineered OCI-AML3/Luc/GFP cells. Leukemia engraftment was confirmed 1 week after injection using BLI. Mice were randomly distributed into four arms and dosed orally with vehicle, cobimetinib (10 mg/kg), venetoclax (100 mg/kg), or cobimetnib plus venetoclax daily for 28 days. BLI demonstrated that the leukemia burden was significantly reduced in treated groups compared to controls over time (Figure 4A). At week 5, the tumor reduction was significantly greater in the groups that received single-agent cobimetinib (P<0.001) or cobimetnib plus venetoclax (P<0.001) than in the control group (Figure 4B). The tumor reduction was greater following combination treatment than following venetoclax (P<0.05) or cobimetinib (P<0.05) alone. All drug treatments, including combinations, were tolerated in vivo based on minimal changes in body weights (data not shown).
Figure 4.
In vivo administration of cobimetinib in combination with venetoclax demonstrated anti-leukemia efficacy in acute myeloid leukemia xenograft mouse models. (A) NSGS mice were injected intravenously with OCI-AML3-Luci-GFP cells (1.0×106). Leukemia engraftment was confirmed 1 week later through a noninvasive in vivo bioluminescence imaging (BLI) system following injection with a D-luciferin (4 mg/mouse) substrate. Mice were dosed daily with oral vehicle or an orally active form of cobimetinib (Cobi; 10 mg/kg) or venetoclax (Ven; 100 mg/kg) or their combination (Combo) for 4 weeks. BLI data over time are shown. (B) Luciferase intensity [mean ± standard deviation(SD)] at week 5. Human CD45 engraftment in bone marrow and spleen was determined by time-of-flight mass spectrometry (C) BLI data over time from the leukemia model established with MOLM13-Luc-GFP cells (1×106 per animal) in NSGS mice. Mice received treatment as for the OCI-AML3/Luc/GFP model for 14 days. (D) Quantification of BLI signals (mean ± SD) on day 17 in the MOLM13 model. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
As a second AML cell line-derived xenograft model, we introduced genetically engineered MOLM3/Luc/GFP cells into NSGS mice and initiated treatment as for the OCI-AML3/Luc/GFP model. Again, BLI demonstrated significantly reduced leukemia burden in the treated groups compared to controls and the reduction was more pronounced in the groups treated with single-agent venetoclax or cobimetinib plus venetoclax (Figure 4C, D). Additionally, human CD45 engraftment and cell counts in both bone marrow and spleen demonstrated a trend toward decreased tumor burden in mice treated with the drug combination compared to that in mice treated with either agent alone (Online Supplementary Figure S8A, B). As in the OCI-AML3/Luc xenograft model, all drug treatments were tolerated based on minimal changes in body weights. We performed additional PDX studies in NSG mice using an AML PDX generated from primary sample AML11 (Table 2). The mice were treated with the same doses of drugs as those used in the cell line models. As shown in Online Supplementary Figure S9, the combination therapy extended survival in the AML11 PDX model. These data demonstrate that the combination of cobimetinib plus venetoclax potently suppresses leukemia burden in tumor-bearing mice in vivo at tolerable doses.
Discussion
Although gain-of-function mutations often represent secondary events in the pathogenesis of AML,31,32 they are required for AML maintenance and are therefore attractive therapeutic targets.33 While MEK inhibitors have demonstrated limited activity in AML as single agents,15,34 preclinical studies with first generation MEK and BCL2 inhibitors demonstrated synergistic induction of apoptosis by suppression of MCL1 following MEK inhibition.11,35
In this study, five of the 11 AML cell lines tested were sensitive to cobimetinib, including two that harbored a FLT3-ITD mutation (MOLM14 and MV4;11) and one with an NRAS mutation (OCI-AML3). Consistent with previous reports, the baseline levels of ERK phosphorylation did not correlate with response to cobimetinib.21,36 Venetoclax as a single agent had activity in five of the cell lines tested while the combination with cobimetinib was synergistic in seven of the cell lines, including those that were resistant to each agent alone.
To extend our preliminary findings in cell lines, we studied a selection of genetically diverse primary AML patients’ samples. Venetoclax induced pronounced apoptosis (>50%) in only three of the samples (16.7%), a rate similar to that of clinical responses to venetoclax monotherapy (19%),10 possibly reflecting protective properties of the tumor microenvironment, as our culture conditions were cytokine-rich. Cobimetinib induced very limited cell death in all AML samples, consistent with previous reports that MEK inhibitors preferentially suppress proliferation and promote differentiation, rather than induce death.33,37,38 Remarkably, over 60% of patients’ samples responded to the combination therapy, notably including samples that were insensitive to both agents on their own. Moreover, these responders carried diverse genetic alterations that affect leukemia cell proliferation (FLT3, RAS), differentiation (RUNX1), genomic stability (NPM1), and epigenetic modifications (TET2, IDH1 and IDH2). Clonogenic assays demonstrated that the combination markedly impaired the colony-forming functions of AML progenitors, while normal progenitors were only minimally affected.
CyTOF has proven to be a powerful approach for identifying functional proteins in diverse cell populations at single-cell levels.39 Several groups, including ours, have studied the feasibility of CyTOF in AML.26,40,41 In this study, we utilized CyTOF combined with SPADE software42 to investigate the efficacy of cobimetinib and venetoclax in two primary patients’ samples: one responder and one non-responder. BCL2 was highly expressed in CD34+ stem/progenitor cells compared to the CD34− cells, underlying the critical need for BCL2 inhibition to eliminate LSC. The venetoclax-sensitive sample displayed a higher level of BCL2 protein than the resistant sample. Cobimetinib inhibited G-CSF-induced pERK irrespective of response status. In line with several studies reporting that suppression of mTORC1 and its downstream pathways (specifically S6) predicted sensitivity to MEK inhibition,21,22 our data also demonstrated that the pS6 signaling pathway was suppressed in the cobimetinib-responding sample, suggesting that S6 phosphorylation may be a more predictive pharmacodynamic marker for MEK inhibition. However, the latter requires validation in a larger cohort of samples from patients.
The distinct response patterns in AML cell lines and patients’ samples led us to search for additional pharmaco-dynamic markers correlating with drug responses using proteomic and transcriptomic profiling. In line with the findings of an extensive study of MEK inhibition,20 we observed bypass induction of pMEK signaling upon MEK inhibition, which was more pronounced in cobimetinib-resistant cell lines. Several signaling pathways were highly activated in cobimetinib-sensitive cell lines, including pS6, pERK, p38MAPK, and pPTEN. Lauchle and colleagues demonstrated that leukemia clones with pre-existing resistance to MEK inhibition displayed reduced p38 kinase activity and increased RasGRP1 levels.13 It was also previously reported that the RSK signaling pathway, which is downstream of MAPK, regulates an mTOR-independent pathway to induce S6 phosphorylation.43 Western blotting analysis performed to validate the RPPA data showed that S6 phosphorylation at both Ser235/236 and Ser240/244 sites was markedly suppressed in cobimetinib-sensitive OCI-AML3 and MV4-11 cells. In OCI-AML3 cells, the combination treatment resulted in significant cell death characterized by elevated levels of cleaved PARP, which could be attributed to disruption of BCL2:BIM complexes, releasing BIM to trigger apoptosis. We also observed BIM induction in MV4-11 cells, underscoring its critical role in the efficacy of the combination of BCL2 and MEK inhibitors.37,38,44 Although RPPA data showed no modulation of MCL1 after cobimetinib treatment, both western blot and Meso Scale Discovery assays showed downregulation of MCL1 in OCL-AML3 cells and upregulation of MCL1 after venetoclax treatment. These data suggest that increased MCL1 levels induced by venetoclax favor the formation of MCL1:BIM complexes were disrupted, freeing BIM to initiate apoptosis. Consistent with these findings, we recently showed that MCL-1 degradation associated with MDM2 inhibition occurs through MEK/ERK suppression and GSK3 activation.45 The downregulation of MYC levels by cobimetinib also suggests a MEK/ERK-GSK3β link, as ubiquitination and degradation of MYC requires phosphorylation at T58 by GSK3β.46 Furthermore, RNA sequencing analyses revealed enhanced expression of MYC and E2F target genes in cells demonstrating a synergistic response to the cobimetinib-venetoclax combination. This finding is consistent with a previous report that MEK inhibition sensitized cells to ABT-263-induced apoptosis by promoting a G1 cell cycle arrest.37 Glycolysis and oxidative phosphorylation (OXPHOS) are known to be regulated by ERK signaling through RNK126-mediated ubiquitination of pyruvate dehydrogenase kinase.30 Potent anti-tumor efficacy has been demonstrated in melanoma cells through combined inhibition of BCL2, OXPHOS and MAPK signaling.47 Alterations in p53 and UPR pathways identified by transcriptome analysis may also account for synergy between MEK and BCL2 inhibition.48 These proposed mechanisms of actions are summarized in Online Supplementary Figure S10. However these models require further validation in controlled mechanistic studies.
The potency of the cobimetinib and venetoclax combination was further demonstrated in vivo using models established with OCI-AML3 (resistant to venetoclax) and MOLM13 (resistant to cobimetinib) leukemia cells. Although we observed strong synergistic effects in both cell lines in vitro, the combination did not confer significant survival benefits in the in vivo models. This may be due to protection against cell death provided by the microenvironment, as we have observed in patients’ samples cultured in cytokine-rich medium. Similar to our in vitro observations, the OCI-AML3 xenograft model is hypersensitive to cobimetinib, and we found no significant survival differences between animals that received single-agent cobimetinib and those that received the combination. In the very aggressive MOLM13 model, in which untreated mice die 3 weeks after cell injection, the combination reduced but did not eliminate leukemia burden markedly on day 17.
In summary, combinatorial blockade of the MAPK and BCL2 pathways promotes cell death and suppresses proliferation in the majority of primary AML cells. This anti-leukemia efficacy is associated with the simultaneous inhibition of BCL2 by venetoclax and the downregulation of MCL1 mediated by cobimetinib, which together enable the release of the pro-death protein BIM. These preclinical data provided a strong mechanistic rationale for evaluating the combination of cobimetinib with venetoclax in a phase I trial now enrolling elderly patients with relapsed/refractory AML (NCT02670044), and initial data have included objective clinical responses.49
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/3/697
References
- 1.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
- 2.Bose P, Vachhani P, Cortes JE. Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat Options Oncol. 2017;18(3):17. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N, Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagadinou ED, Sach A, Callahan K, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter BZ, Mak PY, Mu H, et al. Combined targeting of BCL-2 and BCR-ABL tyrosine kinase eradicates chronic myeloid leukemia stem cells. Sci Transl Med. 2016;8(355): 355ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan SM, Thomas D, Corces-Zimmerman MR, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med. 2015;21(2):178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goff DJ, Court Recart A, Sadarangani A, et al. A pan-BCL2 inhibitor renders bone-marrow-resident human leukemia stem cells sensitive to tyrosine kinase inhibition. Cell Stem Cell. 2013;12(3):316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito Y, Mochizuki Y, Ogahara I, et al. Overcoming mutational complexity in acute myeloid leukemia by inhibition of critical pathways. Sci Transl Med. 2017;9(413). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan R, Hogdal LJ, Benito JM, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4(3):362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konopleva M, Pollyea DA, Potluri J, et al. Efficacy and biological correlates of response in a phase II study of venetoclax monotherapy in patients with acute myelogenous leukemia. Cancer Discov. 2016;6(10):1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konopleva M, Milella M, Ruvolo P, et al. MEK inhibition enhances ABT-737-induced leukemia cell apoptosis via prevention of ERK-activated MCL-1 induction and modulation of MCL-1/BIM complex. Leukemia. 2012;26(4):778–787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Lin KH, Winter PS, Xie A, et al. Targeting MCL-1/BCL-XL forestalls the acquisition of resistance to ABT-199 in acute myeloid leukemia. Sci Rep. 2016;6:27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauchle JO, Kim D, Le DT, et al. Response and resistance to MEK inhibition in leukaemias initiated by hyperactive Ras. Nature. 2009;461(7262):411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CC, Shah NP. The role of kinase inhibitors in the treatment of patients with acute myeloid leukemia. Am Soc Clin Oncol Educ Book. 2013;313–318. [DOI] [PubMed] [Google Scholar]
- 15.Jain N, Curran E, Iyengar NM, et al. Phase II study of the oral MEK inhibitor selumetinib in advanced acute myelogenous leukemia: a University of Chicago phase II consortium trial. Clin Cancer Res. 2014;20(2):490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh YY, Chen R, Hessler J, et al. Up-regulation of CDK9 kinase activity and Mcl-1 stability contributes to the acquired resistance to cyclin-dependent kinase inhibitors in leukemia. Oncotarget. 2015;6(5):2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei XY, Dai Y, Tenorio S, et al. MEK1/2 inhibitors potentiate UCN-01 lethality in human multiple myeloma cells through a Bim-dependent mechanism. Blood. 2007; 110(6):2092–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amatangelo MD, Quek L, Shih A, et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 2017;130(6):732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Han L, Shi C, et al. Upregulation of MAPK/MCL-1 maintaining mitochondrial oxidative phosphorylation confers acquired resistance to BCL-2 inhibitor venetoclax in AML. Blood. 2016; 128(22):101–101. [Google Scholar]
- 20.Hatzivassiliou G, Haling JR, Chen H, et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013;501(7466):232–236. [DOI] [PubMed] [Google Scholar]
- 21.Hirashita Y, Tsukamoto Y, Yanagihara K, et al. Reduced phosphorylation of ribosomal protein S6 is associated with sensitivity to MEK inhibition in gastric cancer cells. Cancer Sci. 2016;107(12):1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corcoran RB, Rothenberg SM, Hata AN, et al. TORC1 suppression predicts responsiveness to RAF and MEK inhibition in BRAF-mutant melanoma. Sci Transl Med. 2013;5(196):196ra198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teh JLF, Cheng PF, Purwin TJ, et al. In vivo E2F reporting reveals efficacious schedules of MEK1/2-CDK4/6 targeting and mTOR-S6 resistance mechanisms. Cancer Discov. 2018;8(5):568–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pabst C, Krosl J, Fares I, et al. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11(4):436–442. [DOI] [PubMed] [Google Scholar]
- 25.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2): 440–446. [DOI] [PubMed] [Google Scholar]
- 26.Han L, Qiu P, Zeng Z, et al. Single-cell mass cytometry reveals intracellular survival/pro-liferative signaling in FLT3-ITD-mutated AML stem/progenitor cells. Cytometry A. 2015;87(4):346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irish JM, Hovland R, Krutzik PO, et al. Single cell profiling of potentiated phosphoprotein networks in cancer cells. Cell. 2004;118(2): 217–228. [DOI] [PubMed] [Google Scholar]
- 28.Dodd KM, Yang J, Shen MH, Sampson JR, Tee AR. mTORC1 drives HIF-1alpha and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene. 2015;34(17):2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croft A, Tay KH, Boyd SC, et al. Oncogenic activation of MEK/ERK primes melanoma cells for adaptation to endoplasmic reticulum stress. J Invest Dermatol. 2014;134(2): 488–497. [DOI] [PubMed] [Google Scholar]
- 30.Yoshino S, Hara T, Nakaoka HJ, et al. The ERK signaling target RNF126 regulates anoikis resistance in cancer cells by changing the mitochondrial metabolic flux. Cell Discov. 2016;2:16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess MR, Hwang E, Firestone AJ, et al. Preclinical efficacy of MEK inhibition in Nrasmutant AML. Blood. 2014;124(26): 3947–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borthakur G, Popplewell L, Boyiadzis M, et al. Activity of the oral mitogen-activated protein kinase kinase inhibitor trametinib in RAS-mutant relapsed or refractory myeloid malignancies. Cancer. 2016;122(12):1871–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konopleva M, Contractor R, Tsao T, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10(5):375–388. [DOI] [PubMed] [Google Scholar]
- 36.Ricciardi MR, Scerpa MC, Bergamo P, et al. Therapeutic potential of MEK inhibition in acute myelogenous leukemia: rationale for "vertical" and "lateral" combination strategies. J Mol Med (Berl). 2012;90(10):1133–1144. [DOI] [PubMed] [Google Scholar]
- 37.Airiau K, Prouzet-Mauleon V, Rousseau B, et al. Synergistic cooperation between ABT-263 and MEK1/2 inhibitor: effect on apoptosis and proliferation of acute myeloid leukemia cells. Oncotarget. 2016;7(1):845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corcoran RB, Cheng KA, Hata AN, et al. Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell. 2013;23(1):121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011; 332(6030):687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisher DAC, Malkova O, Engle EK, et al. Mass cytometry analysis reveals hyperactive NF kappa B signaling in myelofibrosis and secondary acute myeloid leukemia. Leukemia. 2017;31(9):1962–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine JH, Simonds EF, Bendall SC, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roux PP, Shahbazian D, Vu H, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282(19):14056–14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sale MJ, Cook SJ. The BH3 mimetic ABT-263 synergizes with the MEK1/2 inhibitor selumetinib/AZD6244 to promote BIM-dependent tumour cell death and inhibit acquired resistance. Biochem J. 2013;450(2): 285–294. [DOI] [PubMed] [Google Scholar]
- 45.Pan R, Ruvolo V, Mu H, et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell. 2017;32(6):748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai WB, Aiba I, Long Y, et al. Activation of Ras/PI3K/ERK pathway induces c-Myc sta-bilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res. 2012;72(10):2622–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serasinghe MN, Gelles JD, Li K, et al. Dual suppression of inner and outer mitochondrial membrane functions augments apoptotic responses to oncogenic MAPK inhibition. Cell Death Dis. 2018;9(2):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer 2004;4(12):966–977. [DOI] [PubMed] [Google Scholar]
- 49.Daver N, Pollyea DA, Yee KWL, et al. Preliminary results from a phase Ib study evaluating BCL-2 inhibitor venetoclax in combination with MEK inhibitor cobimetinib or MDM2 inhibitor idasanutlin in patients with relapsed or refractory (R/R) AML. Blood. 2017;130 (Suppl 1):813. [Google Scholar]