Abstract
SOX11 is a valuable marker to identify biologically and clinically relevant groups of mantle cell lymphoma such as cyclin D1 negative and leukemic non-nodal mantle cell lymphoma (MCL). We aimed to establish a sensitive in situ hybridization analysis of SOX11 mRNA allowing its quantification within the histopathological context and compare it with immunohistochemistry and real-time quantitative reverse transcription-PCR (RT-qPCR). Furthermore, TP53 status was correlated with SOX11 mRNA levels. Sixty-six cases were investigated; 58 conventional mantle cell lymphomas (cMCL), including six cyclin D1 negative (46 classic, 12 blas-toid) and eight leukemic non-nodal mantle cell lymphomas (nnMCL). RNAscope was used for the in situ hybridization and the results scored as 0 to 4. MCL cases with SOX11 positivity by immunohistochemistry (IHC) were positive by RNA in situ hybridization (RNAscope) but with different scores. RT-qPCR showed a good correlation with the median of the grouped scores but had a wide variation in individual cases. The SOX11 negative leukemic non-nodal mantle cell lymphomas were also negative by RNAscope. TP53 was mutated in 13/63 (21%) cases, including 5/7 (71%) leukemic non-nodal and 8/56 (14%) cMCL. Interestingly, of the TP53 mutated cases, nine were in the RNAscope negative/low SOX11 group (9/15; 60%) and four in the high SOX11 group (4/36; 11%) (P=0.0007). In conclusion, RNAscope is a reliable method to evaluate SOX11 mRNA levels. This study demonstrates the broad range of SOX11 mRNA levels in MCL. An important finding is the significant correlation of TP53 mutations with negative/low SOX11 mRNA level both in leukemic nnMCL and cMCL.
Introduction
MCL is a mature B-cell neoplasm, which accounts for 3-10% of all non-Hodgkińs lymphomas. The genetic hallmark of MCL is the t(11;14)(q13;q32) chromosomal translocation that juxtaposes the immunoglobulin heavy chain gene (IGH) on 14q32 to the CCND1 gene on 11q13 leading to a deregulated overexpression of CCND1 mRNA and cyclin D1 protein, in 95% of the cases.1,2 The existence of cyclin D1 negative MCL (D1-MCL) was demonstrated by gene expression profile3 (GEP) and further studies based on real-time quantitative RT-qPCR and fluorescence in situ hybridization (FISH) analysis demonstrated that the majority of these cases carry a CCDN2 translocation4–7 and few cases have a CCND3/IGH or IGK/IGL rearrangement.8,9 Importantly, cyclin D1 positive and negative MCL have the same morpho-logic, pathologic, clinical and molecular features and are characterized by the expression of the transcription factor Sex determining region Y-box 11 (SOX11).10 Due to the positivity of SOX11 in D1-MCL6,11,12 and the negativity in other mature B-cell neoplasms, the immunohistochemical analysis of SOX11 has become a valuable marker for the identification of the different subsets of MCL.13 Although MCL has been traditionally considered an aggressive and incurable disease, an indolent variant has been recognized that presents with leukemic nnMCL, and treatment is usually deferred for months or years.14,15 Interestingly, nnMCL, in contrast to cMCL, is characterized by a lack of SOX11 expression, the presence of IGHV mutations and usually low genomic complexity, indicating that these two variants follow different pathways of lymphomagenesis.15–17 The lack of SOX11 expression in nnMCL is believed to play an important role for the indolent behavior of the disease.11,14,16,18–20 SOX11 is a transcription factor that belongs to the SOX gene family and is widely expressed in embryogenesis, but not in normal hematopoiesis.21 In MCL, it is controversial whether SOX11 acts as an oncogene or as a tumor suppressor gene. SOX11 has been proposed to act as an oncogene inducing cell proliferation, enforcing PAX5 expression and inhibiting terminal B-cell differentiation into plasma cells via PRDM1 and BCL6.16,22 Additionally, it has been shown to promote tumor angiogenesis through the PDGFA axis.23 More recently, in a murine model, it was demonstrated that SOX11 overexpression in B-cells promotes B-cell receptor signaling and results in a disease phenotype similar to MCL.24 In contrast, it has been shown that SOX11-binding targets could repress proliferation, and therefore, it has been suggested that SOX11 in MCL may act as a tumor suppressor gene.25 The prognostic relevance of SOX11 in MCL is equally controversially discussed. Although originally it was thought that SOX11 negative cases carried an indolent clinical course, other studies have shown that high SOX11 expression is associated with improved survival in a subset of MCL patients, and low or negative SOX11 expression with poor overall survival.25–28 Further work identified a group of MCL lacking SOX11 expression with a dismal prognosis.11 Interestingly, many of these cases were associated with TP53 mutations. TP53 mutations are preferentially associated with blastoid morphology (up to 30%),29 but also occur in both cMCL with classic morphology and nnMCL, and associated with a poor prognosis.30–32
SOX11 expression is usually investigated by immunohis-tochemistry (IHC) in tissue specimens; however, IHC is not a quantitative technique. RNAscope is a relatively new in situ hybridization (ISH) technique that allows a highly sensitive visualization of molecular markers in the morphological context by target-specific amplification of signals with suppression of the background.33 In recent years, some studies have analyzed this technology in different tissues and found it to be a method comparable to immunohistochem-istry and RT-qPCR.34–36
By integrating TP53 status and SOX11 expression in the diagnostic workup of MCL the risk stratification could be improved. Given the lack of reliable quantification of IHC and the conflicting results concerning the prognostic role of SOX11, we aimed to establish a sensitive ISH analysis of SOX11 mRNA allowing its quantification within the histopathological context and to compare it with IHC and RT-qPCR analyses. Furthermore, the TP53 status was investigated by IHC and next-generation sequencing (NGS) and correlated with SOX11 mRNA expression levels.
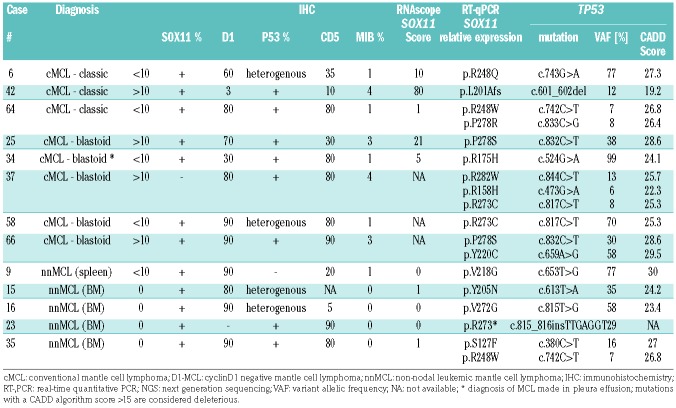
Methods
Patient selection
Sixty-six cases with the diagnosis of MCL were selected from the files of the Institute of Pathology, University of Tübingen. In addition, 12 cases with the diagnosis of small cell B-cell lymphoma (chronic lymphocytic leukemia (CLL), follicular lymphoma (FL) and marginal zone B-cell lymphoma (MZL), four cases each) were used as controls. The study was approved by the local ethics committee (TÜ-730/2018BO2).
Histology and immunohistochemistry
Hematoxylin and Eosin (H&E) sections and all immunohistochemical stains were reviewed. BM trephines were formalin-fixed, decalcified in EDTA, and paraffin-embedded (FFPE). Immunohistochemical stains were performed using an automated immunostainer (Ventana Medical Systems, Tuscon, USA), according to the manufacturer’s protocol. The following antibodies were used: SOX11 (MRQ-58) (Medac Diagnostika, Wedel, Germany) and p53 (DO-7) (Novocastra Liquid, Leica Biosystems, Newcastle, UK). SOX11 stain was scored as negative (0%), low (1-10% positive cells) and positive (>10% positive cells). p53 was considered positive when >20% cells were strongly positive.
RNAscope
The mRNA ISH was performed using the RNAscope 2.5 VS reagent kit-RED with custom designed SOX11-RNA, as target (Advanced Cell Diagnostics, ACD, Hayward, CA), according to the manufacturer’s protocol, as previously described33 (Online Supplementary Material and Methods). The RNAscope procedure was performed in the Ventana Discovery XT autostainer for open procedures. Tissue mRNA preservation was assessed by performing RNAscope analysis of mRNA of the housekeeping gene peptidylpropyl isomerase B (PPIB) (Online Supplementary Figure S1).
RNAscope scores
The results were scored according to the guidelines described in the manufactureŕs protocol. Briefly, staining score 0 was defined as: no staining or less than 1 dot to every 10 cells (40× magnification), score 1: 1-3 dots/cell (visible at 20-40× magnification), score 2: 4-9 dots/cell, and very few dot clusters (visible at 20-40× magnification), score 3: 10-15 dots/cell, less than 10% positive cells have dot clusters (visible at 20× magnification), and score 4: >15 dots/cell, more than 10% positive cells have dot clusters (visible at 20× magnification). Score 0-1 was considered negative/low, score 2 intermediate, and score 3-4 high SOX11 mRNA expression.
RNA isolation and Real-time quantitative PCR (RT-qPCR)
RNA was isolated using the Maxwell® 16 LEV RNA FFPE Purification Kit and the Maxwell® 16 Instrument (Promega, Madison, WI, USA). RT-qPCR was performed to quantify SOX11 mRNA levels (see Online Supplementary Material and Methods). Data were analyzed using the 2−ΔΔCp method and the mean of the SOX11 negative cases was defined as calibrator.
Next generation sequencing of TP53 gene
NGS was performed using the Ion AmpliSeq™ TP53 Panel (Life Technologies) on the Ion GeneStudio™ S5 Prime System according to the manufacturer’s protocol (see Online Supplementary Material and Methods).
Statistical analysis
Statistical analysis was performed with JMP software version 10 (SAS Institute GmbH) using Fisher exact or χ2 test for comparison of nominal data. Statistical significance was concluded for values of P<0.05.
Results
Clinical and Morphological features
Table 1 summarizes the clinical and morphological features. A total of 66 patients with the diagnosis of MCL were included in the study of which 27 were female and 39 males with a median age of 72 years (range 47-91 years). Main biopsy sites were lymph nodes (LN) in 31 cases, bone marrow (BM) in 14 cases, and 21 extranodal sites including one spleen, four tonsils and two intestinal biopsies.
Table 1.
Clinical and histological data of the 66 cases.
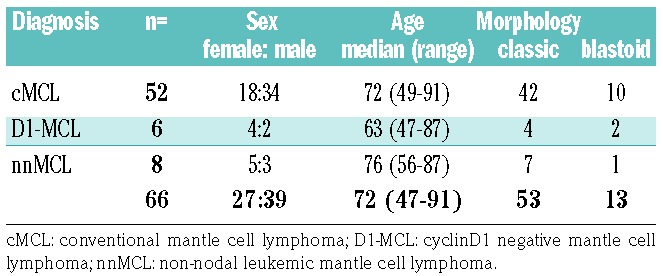
Of the 66 cases, 58 (88%) were classified as cMCL with predominantly nodal involvement, but also BM infiltration. Forty-six of these cases showed a classic morphology (46/58, 79%), and 12 cases (12/58, 21%) were classified as blastoid. Fifty-two cases (52/58; 90%) were cyclin D1 and SOX11 positive, whereas six cases (6/58; 10%) were cyclin D1 negative (four classic and two blastoid) but positive for SOX11. In eight of 66 cases (12%), the diagnosis of nnMCL was made based on peripheral blood and BM involvement without or minimal nodal disease. One case classified as nnMCL presented mainly with splenic involvement. The nnMCL cases were SOX11 negative and cyclin D1 positive. In 63 cases immunohistochemical analysis for p53 was performed. In 11 cases (17%) p53 was strongly expressed in >20% of tumor cells suggesting a TP53 mutation. Forty-seven cases (75%) were considered P53 negative (≤20% of tumor cells). Five cases (8%) were equivocal with weak, heterogeneous staining in the majority of the tumor cells. The proliferation rate assessed with MIB1 showed a low proliferation (<10%) in 10 cases, intermediate proliferation (10-29%) in 34 cases, and high proliferation (>30%) in 19 cases. The 12 cases with blas-toid morphology showed a median proliferation rate of 80%. CD5 was analyzed in 65 cases, 54 cases were CD5 positive whereas 11 cases (17%) remained negative.
RNAscope analysis for SOX11 mRNA
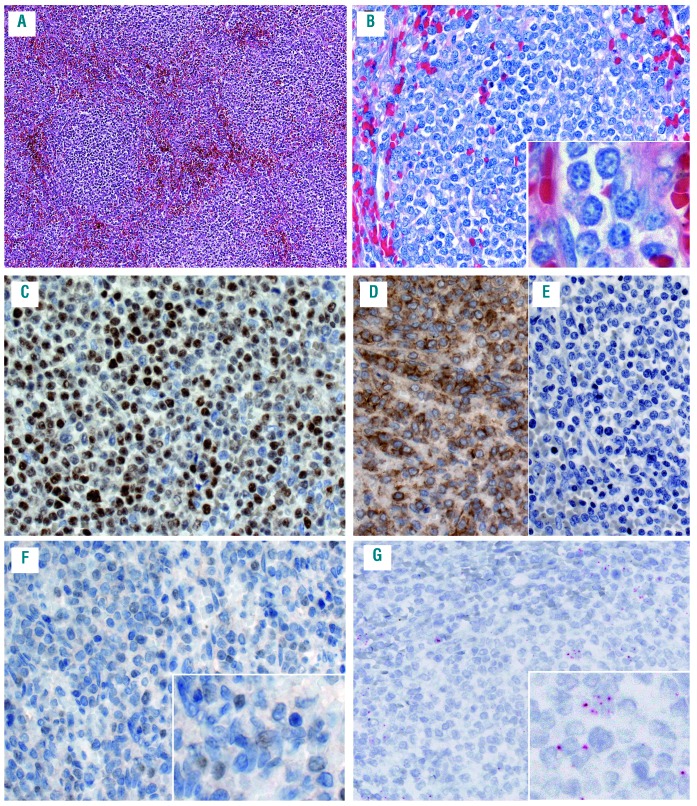
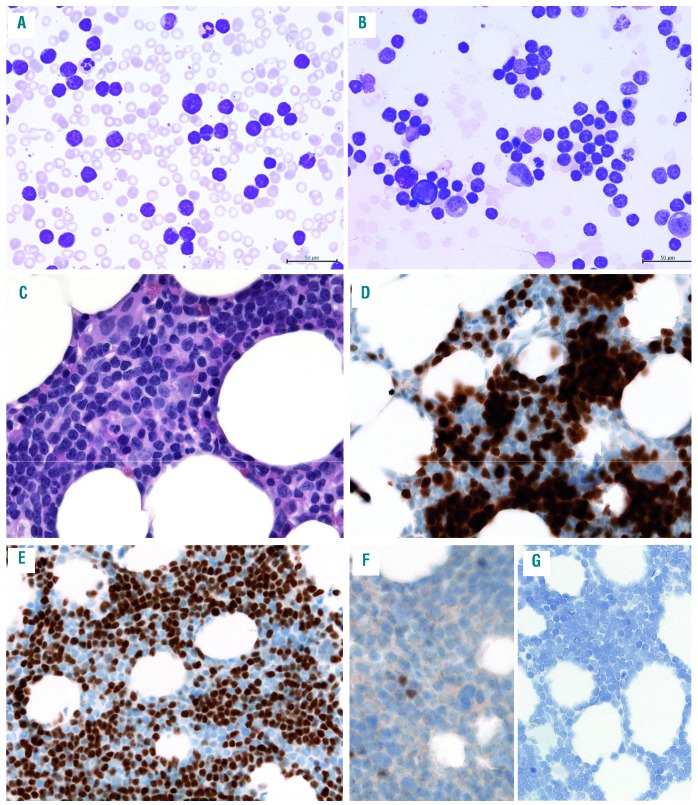
Of the 66 cases analyzed with RNAscope for SOX11 mRNA, 63 cases were informative, whereas two BM biopsies and one lymph node core biopsy were considered not evaluable due to the lack or low expression of the housekeeping gene PPIB used as positive control. The poor mRNA preservation in the two BM most probably is secondary to the decalcification process. The interpretation and quantification of RNAscope was easy to perform. The cases were scored according to the quantity of punctate dots, as described above (Figure 1A-E; Table 2). Twenty-two cases were classified as score 4 (cyc D1+ n=18; cyc D1- n=4), 14 cases as score 3 (cyc D1+ n=13; cyc D1- n=1) 12 cases as score 2 (cyc D1+ n=11; cyc D1-, n=1), eight cases as score 1 (cMCL cyc D1+ n=7; nnMCL n=1) and seven cases as score 0 (nnMCL n=7). All small B-cell lymphomas used as controls were negative. Eight of 12 (67%) MCL cases with blastoid morphology and 26 of 44 (59%) MCL with classic morphology showed high SOX11 mRNA expression (score 3 or 4). Interestingly, 5/6 (83%) cases with D1-MCL were in the high SOX11 mRNA expression group. In contrast, 15 (24%) cases had low/negative SOX11 mRNA expression including the 8/8 (100%) nnMCL cases and 7/55 (13%) cMCL cases. The overall correlation between IHC and RNAscope was very good. Two cases that were considered as SOX11 negative by IHC were scored as 1 with RNAscope. One of these cases was a case with primary splenic presentation (Figure 2A) with plasmacytic differentiation (Figure 2B) and kappa immunoglobulin light chain restriction (Figure 2D-E). The SOX11 IHC was equivocal whereas the SOX11 RNAscope demonstrated clear signals corresponding to score 1 (Figure 2F-G). This case was classified as nnMCL with low SOX11 mRNA expression. The remaining seven cases classified as nnMCL in BM were SOX11 negative by IHC and had a score 0 with RNAscope.
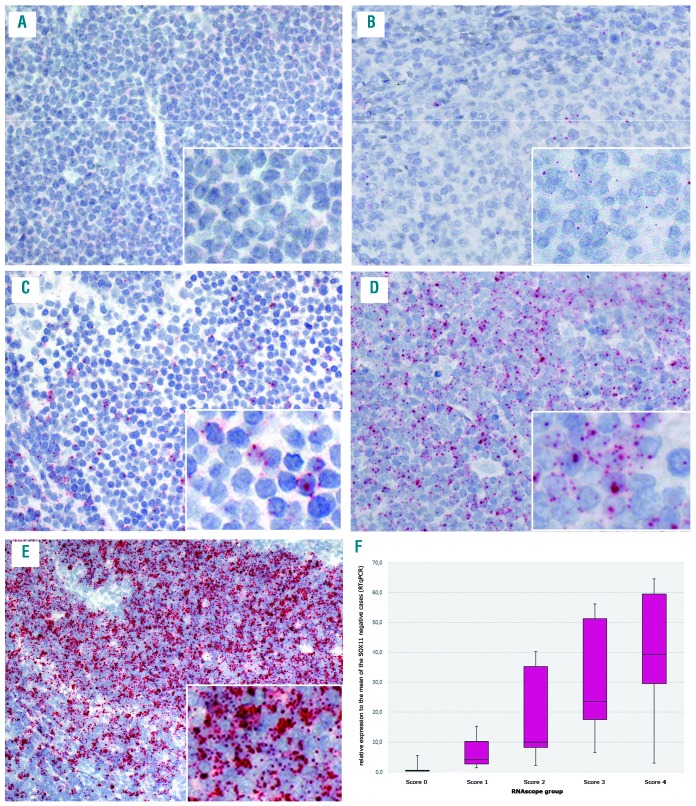
Figure 1.
Illustration of the staining pattern in the different RNAscope scores and correlation to the real-time quantitative PCR (RT-qPCR) results, which shows a decrease of SOX11-mRNA levels with decreasing staining scores. (A) score 0, no staining (case #16); (B) score 1, 1-3 dots/cell (case #9); (C) score 2, 4-9 dots/cell, very few clusters (case #5); (D) score 3, 10-15 dots/cell, less than 10% positive cells with dot clusters (case #2); (E) score 4, 15 dots/cell, more than 10% positive cells with dot clusters (case #11); (F) box plot of SOX11 mRNA levels measured by RT-qPCR according to the score groups showing the third quartile and first quartile range of the data and the median for the particular score (P=0.0002). A-E original magnification 200× and insert with 400×.
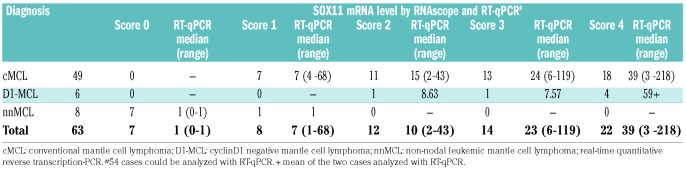
Table 2.
SOX11 RNAscope and RT-qPCR analyses in the three MCL subgroups.
Figure 2.
Mantle cell lymphoma (MCL) with splenic involvement and plasmacytoid differentiation (case #9). (A) the spleen is characterized by a nodular infiltration of the white pulp (hematoxylin and eosin); (B) Giemsa stain highlights the cytologic features. The infiltrate is composed of medium-sized cells with a rim of basophilic cytoplasm, dispersed nuclear chromatin and occasional small nucleoli; (C) the cells are positive for cyclin D1; (D) the tumor cells are positive for lambda-stain (D) and negative for kappa (E); (F) the SOX11 immunohistochemistry (IHC) was interpreted as negative at diagnosis, but upon review occasional weakly positive tumor cells could be detected; (G) the SOX11 RNA in situ hybridization (RNAscope) shows a score 1 with 1-3 dots/cell. A original magnification 50×, B-G original magnification 200×, insert original magnification 630×.
SOX11 mRNA analysis with RT-qPCR
In 54 cases, for which material was available, RT-qPCR was performed. The SOX11 mRNA levels are shown in Figure 1F. The analysis showed a wide range of SOX11 mRNA levels among the cases. cMCL cases cyclin D1 and SOX11 positive by IHC showed a median level of 30-fold SOX11 mRNA level compared to the mean of the SOX11 negative cases (range 2-218), whereas the four analyzed D1-MCL cases showed a median SOX11 mRNA level of 12 (range 8-102). nnMCL negative for SOX11 by IHC showed the lowest SOX11 mRNA levels (median 1; range 0-1).
Correlation of SOX11 mRNA levels between RNAscope and RT-qPCR
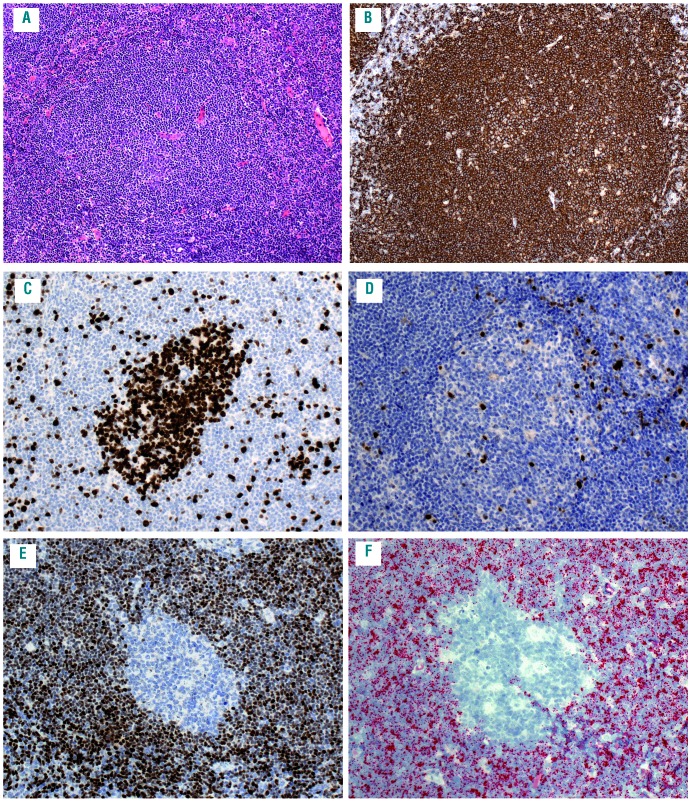
The correlation of SOX11 mRNA levels between RNAscope and RT-qPCR was further analyzed (Table 2). A median SOX11 mRNA level of 39-fold (range 3-218) was found for score 4, 23-fold (range 6-119) for score 3, 10-fold (range 2-43) for score 2, seven-fold (range 1-68) for score 1, and 1-fold (range 0-1) for score 0. The correlation between RNAscope and mRNA levels obtained by RT-qPCR analysis showed a significant association between the median of the single groups (P=0.0002) (Figure 1F). However, there was a broad range of SOX11 mRNA levels, as measured by RT-qPCR in each of the groups. In part, this could be explained by a dilution effect due to the high content of reactive cells in some cases with partial infiltration. This is well demonstrated in a case of D1-MCL with mantle zone growth pattern and partial involvement of the LN (Figure 3A-D). IHC and RNAscope revealed similar patterns and intensities (Figure 3E-F); however, RT-qPCR showed relatively low levels of SOX11 mRNA.16 In most cases; however, there was no clear explanation for the variability between RNAscope and RT-qPCR. In the two BM cases with a lack of signals in the RNAscope assay, RT-qPCR detected SOX11 mRNA and SOX11 was also positive by IHC. This indicates that pre-analytical parameters may differ in their influence on the two mRNA quantification methods. Taken together, although both methods showed an overall good correlation, RNAscope was considered more reliable for the quantification of SOX11 mRNA in individual cases.
Figure 3.
Classical cyclin D1 negative mantle cell lymphoma (D1-MCL) with mantle zone pattern (case #10). (A) the architecture is characterized by the expansion of the follicle mantle zone with tumor cells and a central “naked” germinal center (hematoxylin and eosin), (B) CD20 highlights the expanded mantle area; (C) MIB1 shows a high proliferation in the residual germinal center whereas the proliferation of the tumor cells is 5%; (D) the tumor cells are negative for cyclin D1; (E) the SOX11 stain demonstrates a strong positivity in the tumor cells, while the germinal center cells are negative; (F) the SOX11 RNA in situ hybridization (RNAscope) shows a score 4 with 15 dots/cell and >10% cells with dot clusters. All original magnification 200×.
Correlation between SOX11 and TP53 status
Table 3 and Online Supplementary Table S1 summarize the data of the TP53 mutated group. In thirty-seven cases, all exons of TP53 were sequenced by NGS including all p53 positive cases (11 cases), the five equivocal cases and 20 p53 negative cases including all remaining blastoid cases. TP53 mutations were demonstrated in 13 cases; 11 considered p53 positive, and two considered p53 negative; one of these a nnMCL case revealed complete negativity by IHC. The five equivocal and all remaining negative cases were TP53 wild-type. Although the cut-off for p53 positivity was set on >20%, 10 cases showed strong p53 expression in more than 60% of the tumor cells. TP53 mutations were identified in eight of 56 cMCL cases (14%), five of them with blastoid morphology (Figure 4). Four of these eight cases were in the group of negative/low SOX11 expression. In contrast, five of seven evaluable cases (71%) of nnMCL had TP53 mutations (14% vs. 71%; P=0.003). Four of these cases were diagnosed in BM and presented with striking lymphocytosis in the peripheral blood (Figure 5). One case had a blastoid morphology. Altogether, nine of the TP53 mutated cases were in the negative/low SOX11 group (RNAscope 0-1) (9/15; 60%) and four in the high SOX11 group (RNAscope 3-4) (4/36; 11%). This difference was statistically significant (P=0.0007). Within the group of cMCL four TP53 mutated cases where in the low SOX11 group (4/7; 57%) and four in the high SOX11 group (4/36; 11%), and the difference was statistically significant (P=0.01) Of the cMCL, the TP53 mutated cases showed the lowest SOX11 mRNA scores including two cases with blastoid morphology (Figure 4).
Table 3.
Histological, immunohistochemical and molecular data of 13 cases with TP53 mutation.
Figure 4.
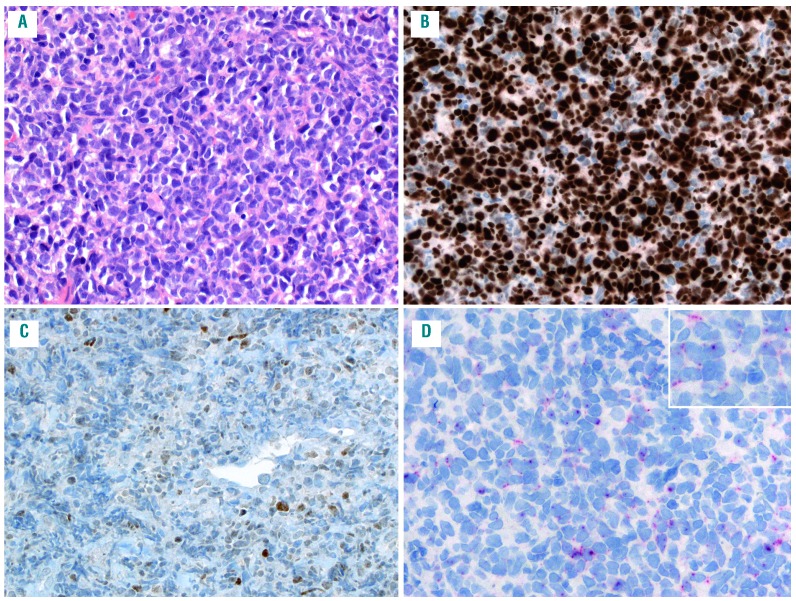
Mantle cell lymphoma (MCL) with blastoid morphology, low SOX11 expression and TP53 mutation (case # 58). (A) the infiltrate is characterized by a diffuse, monotonous population of medium-sized to large cells with irregular nuclei, fine chromatin and narrow cytoplasm (hematoxylin and eosin); (B) p53 is strongly positive in the majority of the tumor cells; (C) the SOX11 stain shows a rather weak heterogeneous positivity in the tumor cells (<10%); (D) SOX11 RNAscope shows a score 1 with 1-3 dots/cell and no dot clusters. All original magnification 200×.
Figure 5.
Leukemic non-nodal disease (nnMCL) mantle cell lymphoma (MCL) (case #15). (A-B) The cytology of the peripheral blood (A) and the bone marrow (B) shows a striking increase of small lymphocytes with rather round nuclei, condensed chromatin and scant cytoplasm; (C) Bone marrow biopsy shows a lymphoid infiltrate composed of small lymphocytes with round nuclei and scant cytoplasm (hematoxylin and eosin); (D) the tumor cells are positive for cyclin D1; (E) p53 is positive in 80% of the tumor cells; (F) the SOX11 stain is negative with only isolated positive cells; (G) the SOX11 RNA in situ hybridization (RNAscope) shows a score 0 with no staining. All original magnification 200×.
Discussion
In this study, we investigated 66 cases of MCL for the expression levels of SOX11 mRNA with RNAscope ISH assay and compared it with IHC and RT-qPCR. Additionally, the TP53 status was investigated and correlated with SOX11 mRNA levels. We show that RNAscope ISH assay is a reliable method to quantitate SOX11 mRNA levels within the histopathological context. Importantly, and in overall agreement with the RT-qPCR results, we demonstrated that SOX11 mRNA levels show a very broad range in cMCL and confirmed the negative or low mRNA levels in nnMCL cases. An interesting finding of this study was the correlation of TP53 mutations with low/negative SOX11 mRNA scores in all investigated subtypes of MCL (P=0.0007).
RNAscope is a relatively new ISH technology with a probe design strategy that allows the visualization of RNA expression in the context of preserved tissue morphology.33,39 In recent years, several studies confirmed the suitability of this method for the quantification of mRNA levels in FFPE tissues; however, SOX11 has not been investigated.34,40,41 The diagnostic importance of SOX11 expression for identifying specific subsets of MCL has been confirmed in many studies; however, its relevance as a prognostic marker is controversial. One possible explanation is that IHC, as a predominantly used technique, is not a quantitative method and does not provide reliable information about SOX11 expression levels. The current gold standard for quantitative gene expression analysis is RT-qPCR.42 However, this method lacks the morphological context, and in our study, a wide range of SOX11 mRNA level was found that not always correlated with the RNAscope assay. Similar findings were described by Lord et al., who also found a wide range of SOX11 mRNA lev els by RT-qPCR and were not able to define a natural cutoff that could stratify cases with low protein expression by IHC.43 The highly variable SOX11 expression in MCL could indicate that not only the presence or absence of the protein, but also its level may be important for the behavior of the malignant cells. Nevertheless, a similar number of cases, with high expression of SOX11 mRNA, was found in the group with blastoid morphology when compared with classic morphology (62% vs. 59%).
The oncogenic potential of SOX11 has been extensively studied in the last years.16,22–24,27,44,45 SOX11 has been shown to promote tumor growth via the induction of angiogenesis by regulating PDGFA.23 Recently, it has also been described that SOX11 binds and transcriptionally regulates two genes important for the modulation of microen-vironment-tumor interactions, CXCR4 (C-X-C motif chemokine receptor 4) and PTK2, encoding for focal adhesion kinase (FAK).44 These genes are essential for tumor cell migration and adhesion to stromal cells in the bone marrow.46 Furthermore, SOX11 has been described to correlate also with high proliferation activity and aggressive behaviour of the disease.22,47,48 The accurate quantification of SOX11 expression opens the possibility to investigate the potential influence of SOX11 levels, and thus, might help to refine both the biological functions in MCL and to determine whether it can be used as a prognostic marker. The correlation between RNAscope, IHC and RT-qPCR was in general good; however, the automated RNAscope technology, direct visualization in tissue specimens and easy quantification of the signals makes this technology very attractive for diagnosis and research purposes. The caveat of any RNA-based technology including RNAscope is that good mRNA preservation is needed, and this depends on good fixation procedures. However, the low dropout rate in our study, with only two decalcified BM specimens not evaluable based on the positive control, documents the robust performance of RNAscope in routine archival tissue specimens.
An intriguing finding in this study was the higher frequency of TP53 mutations in cases with low/negative SOX11 mRNA levels both in cMCL as well as nnMCL. TP53 mutations are rarely observed in MCL with classic morphology,11 but are frequently found in highly proliferative MCL with blastoid morphology (up to 30%), and associated with aggressive clinical course.29,30,49 A recent study comparing the gene signature of cMCL and nnMCL demonstrated that the incidence of TP53 mutations is similar in both subgroups (36% and 38%, respectively).50 Nevertheless, an interesting observation was the report of a group of cMCL cases that lacks SOX11 expression, carries TP53 mutations and has a dismal prognosis.11 This finding was recently confirmed in a study from the European MCL network that demonstrated that p53 overexpression is preferentially found in SOX11 negative cMCL cases (50% vs. 13%).26 A caveat of the latter study is that molecular analysis to demonstrate the presence of TP53 mutations was not performed. Although, p53 stain is a very good surrogate marker of gene mutation when strong, homogeneous staining is considered as positive, a group of cases with negative or low p53 staining due to deletions or frameshift mutations are missed as demonstrated in this study (2/13; 15%). Accordingly, of the TP53 mutated cases nine were in the negative/low SOX11 mRNA level group and four in the high mRNA level group (60% vs. 11%, P=0.0007). Our results further confirm previous reports suggesting that TP53 overexpression/mutations frequently occur in SOX11 negative/low cases.11,26 It is worth mentioning that in the two previous studies only nodal cMCL cases with no previous history of leukemic manifestation were included. Nevertheless, it has been suggested that these cases might correspond to a selected subset of progressed nnMCL tumors.17,48 However, contrary to the cases reported by Nygren et al. and Aukema et al., in this study eight nnMCL cases with typical presentation diagnosed in the BM and one in spleen were included. Although the TP53 mutated nnMCL cases showed striking lymphocytosis in the peripheral blood, they lacked significant lymphadenopathy and rarely blastoid morphology was observed. The high incidence of nnMCL with TP53 mutation in this study (5/7; 71%) most probably represents a natural bias since patients with nnMCL will usually seek medical attention only when the disease progresses and becomes symptomatic. Nevertheless, we also confirmed the presence of a group of nodal cMCL with both classic and blastoid morphology with TP53 mutations and low SOX11 mRNA level. The reason why TP53 mutations are found preferentially in the negative/low SOX11 group is not clear. The possible interaction between SOX11 and p53, if any, and the influence of loss of SOX11 in p53 function/mutation have not been explored and warrant further investigation.
In conclusion our data demonstrate that RNAscope ISH assay is a reliable method for the detection and quantification of target RNA and can be used to evaluate SOX11 mRNA expression accurately. This study also revealed a broad range of SOX11 expression in MCL, indicating that SOX11 expression levels may be relevant for the behaviour of the disease. The association of TP53 mutations with negative/low SOX11 mRNA expression in different MCL subtypes is an important finding, which needs to be explored in further studies.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/105/3/754
References
- 1.Bosch F, Jares P, Campo E, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood.1994;84(8):2726–2732. [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Seto M, Müller-Hermelink H-K. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. eds. Mantle Cell Lymphoma. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues, Lyon: IARC Press. 2017(Lyon IARC Press; ):285–290. [Google Scholar]
- 3.Rosenwald A, Wright G, Wiestner A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell.2003;3(2):185–197. [DOI] [PubMed] [Google Scholar]
- 4.Fu K, Weisenburger DD, Greiner TC, et al. Cyclin D1-negative mantle cell lymphoma: a clinicopathologic study based on gene expression profiling. Blood.2005; 106(13):4315–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quintanilla-Martinez L, Slotta-Huspenina J, Koch I, et al. Differential diagnosis of cyclin D2+ mantle cell lymphoma based on fluorescence in situ hybridization and quantitative real-time-PCR. Haematologica.2009; 94(11):1595–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salaverria I, Royo C, Carvajal-Cuenca A, et al. CCND2 rearrangements are the most frequent genetic events in cyclin D1(-) mantle cell lymphoma. Blood.2013;121(8):1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gesk S, Klapper W, Martin-Subero JI, et al. A chromosomal translocation in cyclin D1-negative/cyclin D2-positive mantle cell lymphoma fuses the CCND2 gene to the IGK locus. Blood.2006;108(3):1109–1110. [DOI] [PubMed] [Google Scholar]
- 8.Wlodarska I, Dierickx D, Vanhentenrijk V, et al. Translocations targeting CCND2, CCND3, and MYCN do occur in t(11;14)-negative mantle cell lymphomas. Blood.2008;111(12):5683–5690. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Garcia D, Navarro A, Valdes-Mas R, et al. CCND2 and CCND3 hijack immunoglobulin light chain enhancers in cyclin D1-negative mantle cell lymphoma. Blood.2019;133(9):940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ek S, Dictor M, Jerkeman M, et al. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood.2008;111(2):800–805. [DOI] [PubMed] [Google Scholar]
- 11.Nygren L, Baumgartner Wennerholm S, et al. Prognostic role of SOX11 in a population-based cohort of mantle cell lymphoma. Blood.2012;119(18):4215–4223. [DOI] [PubMed] [Google Scholar]
- 12.Mozos A, Royo C, Hartmann E, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica.2009;94(11):1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander B, Quintanilla-Martinez L, Ott G, et al. Mantle cell lymphoma--a spectrum from indolent to aggressive disease. Virchows Arch.2016;468(3):245–257. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez V, Salamero O, Espinet B, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res.2010;70(4):1408–1418. [DOI] [PubMed] [Google Scholar]
- 15.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer.2007;7(10):750–762. [DOI] [PubMed] [Google Scholar]
- 16.Palomero J, Vegliante MC, Eguileor A, et al. SOX11 defines two different subtypes of mantle cell lymphoma through transcriptional regulation of BCL6. Leukemia.2016;30(7):1596–1599. [DOI] [PubMed] [Google Scholar]
- 17.Bea S, Amador V. Role of SOX11 and genetic events cooperating with cyclin D1 in mantle cell lymphoma. Curr Oncol Rep.2017;19(6):43. [DOI] [PubMed] [Google Scholar]
- 18.Ribera-Cortada I, Martinez D, Amador V, et al. Plasma cell and terminal B-cell differentiation in mantle cell lymphoma mainly occur in the SOX11-negative subtype. Mod Pathol.2015;28(11):1435–1447. [DOI] [PubMed] [Google Scholar]
- 19.Wasik AM, Lord M, Wang X, et al. SOXC transcription factors in mantle cell lymphoma: the role of promoter methylation in SOX11 expression. Sci Rep. 2013;3:1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res.2012;72(20):5307–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegner M. All purpose Sox: The many roles of Sox proteins in gene expression. Int J Biochem Cell Biol.2010;42(3):381–390. [DOI] [PubMed] [Google Scholar]
- 22.Vegliante MC, Palomero J, Perez-Galan P, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood.2013;121(12):2175–2185. [DOI] [PubMed] [Google Scholar]
- 23.Palomero J, Vegliante MC, Rodriguez ML, et al. SOX11 promotes tumor angiogenesis through transcriptional regulation of PDGFA in mantle cell lymphoma. Blood.2014;124(14):2235–2247. [DOI] [PubMed] [Google Scholar]
- 24.Kuo PY, Jatiani SS, Rahman AH, et al. SOX11 augments BCR signaling to drive MCL-like tumor development. Blood.2018;131(20):2247–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo PY, Leshchenko VV, Fazzari MJ, et al. High-resolution chromatin immunoprecipi-tation (ChIP) sequencing reveals novel binding targets and prognostic role for SOX11 in mantle cell lymphoma. Oncogene. 2015;34(10):1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aukema SM, Hoster E, Rosenwald A, et al. Expression of TP53 is associated with the outcome of MCL independent of MIPI and Ki-67 in trials of the European MCL Network. Blood.2018;131(4):417–420. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Wang Y, Yu Z, et al. SOX11 regulates the pro-apoptosis signal pathway and predicts a favorable prognosis of mantle cell lymphoma. Int J Hematol. 2017;106(2):212–220. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Asplund AC, Porwit A, et al. The subcellular Sox11 distribution pattern iden tifies subsets of mantle cell lymphoma: correlation to overall survival. Br J Haematol.2008;143(2):248–252. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez L, Fest T, Cazorla M, et al. p53 gene mutations and protein overexpression are associated with aggressive variants of mantle cell lymphomas. Blood.1996;87(8):3351–3359. [PubMed] [Google Scholar]
- 30.Slotta-Huspenina J, Koch I, de Leval L, et al. The impact of cyclin D1 mRNA isoforms, morphology and p53 in mantle cell lymphoma: p53 alterations and blastoid morphology are strong predictors of a high proliferation index. Haematologica.2012;97(9):1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Royo C, Navarro A, Clot G, et al. Non-nodal type of mantle cell lymphoma is a specific biological and clinical subgroup of the disease. Leukemia.2012;26(8):1895–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bea S, Valdes-Mas R, Navarro A, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci U S A.2013;110(45):18250–18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn.2012;14(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu X, Guo S, Song W, et al. Estrogen receptor alpha (ERalpha) status evaluation using RNAscope in situ hybridization: a reliable and complementary method for IHC in breast cancer tissues. Hum Pathol.2017;61: 121–129. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Wang MX, Su N, et al. RNAscope for in situ detection of transcriptionally active human papillomavirus in head and neck squamous cell carcinoma. J Vis Exp. 2014(85). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roe CJ, Siddiqui MT, Lawson D, et al. RNA In Situ Hybridization for Epstein-Barr virus and cytomegalovirus: comparison with in situ hybridization and immunohistochem-istry. Appl Immunohistochem Mol Morphol.2019;27(2):155–159. [DOI] [PubMed] [Google Scholar]
- 37.Quintanilla-Martinez L, Pittaluga S, Miething C, et al. NPM-ALK-dependent expression of the transcription factor CCAAT/enhancer binding protein beta in ALK-positive anaplastic large cell lymphoma. Blood.2006;108(6):2029–2036. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt J, Gong S, Marafioti T, et al. Genome-wide analysis of pediatric-type follicular lymphoma reveals low genetic complexity and recurrent alterations of TNFRSF14 gene. Blood.2016;128(8):1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson CM, Zhang B, Miller M, et al. Fully automated RNAscope in situ hybridization assays for formalin-fixed paraffin-embedded cells and tissues. J Cell Biochem.2016;117(10):2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmid E, Klotz M, Steiner-Hahn K, et al. Detection of MET mRNA in gastric cancer in situ. Comparison with immunohisto-chemistry and sandwich immunoassays. Biotech Histochem.2017;92(6):425–435. [DOI] [PubMed] [Google Scholar]
- 41.Bingham V, McIlreavey L, Greene C, et al. RNAscope in situ hybridization confirms mRNA integrity in formalin-fixed, paraffin-embedded cancer tissue samples. Oncotarget.2017;8(55):93392–93403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. BioTechniques.2005;39(1):75–85. [DOI] [PubMed] [Google Scholar]
- 43.Lord M, Wasik AM, Christensson B, et al. The utility of mRNA analysis in defining SOX11 expression levels in mantle cell lymphoma and reactive lymph nodes. Haematologica.2015;100(9):e369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balsas P, Palomero J, Eguileor A, et al. SOX11 promotes tumor protective microenvironment interactions through CXCR4 and FAK regulation in mantle cell lymphoma. Blood.2017;130(4):501–513. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Bjorklund S, Wasik AM, et al. Gene expression profiling and chromatin immunoprecipitation identify DBN1, SET-MAR and HIG2 as direct targets of SOX11 in mantle cell lymphoma. PloS One.2010;5(11):e14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudelius M, Rosenfeldt MT, Leich E, et al. Inhibition of focal adhesion kinase overcomes resistance of mantle cell lymphoma to ibrutinib in the bone marrow microenvironment. Haematologica.2018;103(1):116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrando AA. SOX11 is a mantle cell lymphoma oncogene. Blood.2013;121(12): 2169–2170. [DOI] [PubMed] [Google Scholar]
- 48.Beekman R, Amador V, Campo E. SOX11, a key oncogenic factor in mantle cell lymphoma. Curr Opin Hematol.2018;25(4): 299–306. [DOI] [PubMed] [Google Scholar]
- 49.Greiner TC, Moynihan MJ, Chan WC, et al. p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood.1996;87(10):4302–4310. [PubMed] [Google Scholar]
- 50.Clot G, Jares P, Giné E, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood.2018;132(4): 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]