Abstract
Neoplastic transformation of germinal center B (GCB) cells may give rise to a variety of different B cell lymphoma subtypes, most of which show substantial heterogeneity in terms of genetic alterations and clinical features. The mutations observed in cancer-related genes in GCB cells are related to abnormalities in the immunogenetic mechanisms associated with germinal center reaction. Recent studies have rapidly identified genomic alterations in B cell lymphomas that may be useful for better subclassification, noninvasive diagnosis, and prediction of response to therapy. The WHO recognizes different lymphoma subsets classified within 2 major categories of B cell lymphoma: Hodgkin’s lymphoma (HL) and B cell non-Hodgkin’s lymphoma (NHL), each with distinct genetic aberrations, including chromosomal translocations, copy number abnormalities, or point mutations. Next-generation sequencing-based technologies have allowed cancer researchers to identify somatic mutations and gene expression signatures at a rapid pace so that novel diagnostic or prognostic biomarkers, as well as therapeutic targets, can be discovered much faster than before. Indeed, deep sequencing studies have recently revealed that lymphoma-specific somatic mutations may be detected in cell-free circulating DNA obtained from the peripheral blood of B cell lymphoma patients, suggesting the possibility of minimally invasive diagnosis, monitoring, and predicting response to therapy of B cell lymphoma patients. In this study, the current status of the recurrent genetic aberrations observed during diagnosis and/or relapse in HL and the major subtypes of B cell NHL (i.e. diffuse large B cell lymphoma, follicular lymphoma, mantle cell lymphoma, and Burkitt lymphoma) are discussed to shed light on their potential use as noninvasive diagnostic or prognostic biomarkers and to reveal their role in lymphomagenesis as a target in therapy for newly diagnosed and chemotherapy-resistant cases.
Keywords: Hodgkin’s lymphoma, B cell non-Hodgkin’s lymphoma, genetic alterations
1. Introduction
The human immune system uses complex defense mechanisms made up of cellular and humoral components that defend our body against a variety of pathogens. Our body is constantly protected against various microbial organisms with the help of a functional immune system. Lymphocytes are a group of white blood cells and critical cellular components of the immune system which protect us from pathogenic microorganisms and tumor formation. Lymphocytes fall into three main categories: B, T, and NK cells. B lymphocytes differentiate into plasma cells which secrete antibodies against specific antigens displayed on pathogens (Slifka et al., 1998). On the other hand, in addition to their immunoregulatory roles, T and NK cells are cytotoxic cells with the capacity to kill virus-infected or neoplastic cells by stimulating apoptosis (Bevan, 2004; Vivier et al., 2008). Proper functioning of the immune system depends on a fine balance between activation and termination of their activation in a timely manner; otherwise, dysregulated immune cell homeostasis may lead to uncontrolled growth of immunocytes. Somatic mutations of genes regulating cancer-associated biological processes including, but not limited to, cell cycle or apoptosis in lymphocytes may lead to uncontrolled cell growth, thereby resulting in the development of various types of lymphoma.
The majority of lymphomas (90%–95%) originate from B cells undergoing neoplastic transformation (Küppers, 2005); on the other hand, the cells of origin of the remaining lymphomas are either T or NK cells. B cell lymphomas are usually associated with defects in immunogenetic mechanisms operating as a part of the germinal center reaction, which plays a role in T cell dependent B cell activation and immunity (Küppers, 2005). Lymphoma is classified into two main groups: Hodgkin’s lymphoma (HL) and non-Hodgkin’s lymphoma (NHL). The first type of lymphoma described in history was HL, which was discovered by Thomas Hodgkin (van Gijn and Gijselhart, 2012). HL constitutes approximately 10% of all lymphomas (Küppers, 2005). The different types of lymphoma that were discovered after HL include other B cell lymphoma subtypes (e.g., diffuse large B cell lymphoma and follicular lymphoma), all of which are collectively much more frequent than subtypes of T or NK cell lymphomas. Subtypes of B cell lymphoma show substantial heterogeneity with respect to genetic and clinical characteristics. In the following sections, we discuss the somatic mutations involved in the formation of various subtypes of B cell lymphoma.
2. Genetic alterations identified in Hodgkin’s lymphoma
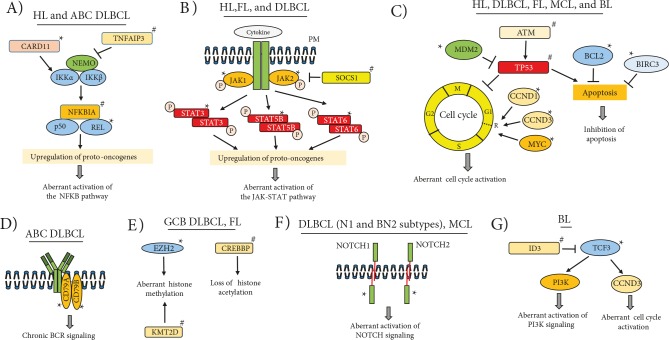
HL, first described by Thomas Hodgkin in 1832, is also known as Hodgkin’s disease (van Gijn and Gijselhart, 2012). The World Health Organization classifies HLs into two main categories: nodular lymphocyte predominant HL and classical Hodgkin’s lymphoma (cHL) (Swerdlow et al., 2016). cHL represents 90%–95% of HLs and has poorer prognosis compared to the other category (Shanbhag and Ambinder, 2018). It has four subtypes: 1) nodular sclerosis (NSCHL); 2) lymphocyte-rich (LRCHL); 3) mixed-cellularity (MCCHL), and 4) lymphocyte-depleted (LDCHL). HL develops with clonal proliferation of multinucleated, special type germinal center B cells called Reed–Sternberg cells (Kanzler et al., 1996). Hodgkin Reed–Sternberg (HRS) cells are tumor cells with no B cell characteristics and do not express the B cell receptor (BCR) on their surface (Farrell and Jarrett, 2011). Under normal conditions, these GCB cells are expected to undergo apoptosis in the absence of BCR signaling. This suggests that survival signals normally originating from active BCR signaling need to be sustained through alternative pathway(s) in HRS cells to compensate for the lack of BCR expression. In this context, HRS cells keep surviving due to deregulation of various signal transduction pathways and transcription factors, especially nuclear factor NF-κB (Figure 1A). REL locus amplifications are responsible for activation in roughly 50% of HL cases with constitutively active NF-κB signaling (Barth et al., 2003). In a significant proportion of cases, irregular activation of the NF-κB signaling pathway is believed to be related to loss-of-function mutations in tumor suppressor genes (e.g., NFKBIA, TNFAIP3) that inhibit the NF-κB signaling pathway (Weniger and Küppers, 2016).
Figure 1.
Mutated genes and affected pathways or biological processes in Hodgkin’s lymphoma and B cell non-Hodgkin’s lymphoma subtypes. HL: Hodgkin’s lymphoma; DLBCL: diffuse large B cell lymphoma; ABC: activated B cell type; GCB: germinal center B cell type; FL: follicular lymphoma; MCL: mantle cell lymphoma; BL: Burkitt lymphoma; *: gain of function mutations; #: loss-of-function mutations; P: phosphate; R: restriction point; PM: plasma membrane.
Genetic analysis of cHL tumors is often difficult as tumor cellularity is less than 5% in biopsies. A recent study involving the microdissection method revealed mutated genes in the JAK-STAT signaling pathway (i.e. STAT3, STAT5B, JAK1, JAK2, and PTPN1) in 87% of cHL cases (Tiacci et al., 2018) (Figure 1B). Moreover, in a significant fraction of cHL cases, it has been reported that the JAK-STAT5 signaling pathway is activated due to mutations in SOCS1, an inhibitor of this pathway (Weniger et al., 2006). The LMP2A gene encoded by the Epstein-Barr virus (EBV) was shown to contribute to the development of EBV+ HLs through inducing transcriptional changes similar to those in HRS cells (Portis et al., 2003). Chromosomal translocations involving the BCL6 oncogene were reported in approximately half of nodular lymphocyte predominant HL cases (Wlodarska et al., 2003). In the majority of HLs, gains of MDM2 or loss-of-function mutations of TP53 have been reported, which suggests that the P53 signaling pathway is probably not functional in cases with these genetic aberrations (Küppers, 2009) (Figure 1C). Table 1 shows the recurrent genetic alterations and the dysregulated biological pathways in HL.
Table 1.
The major genetic aberrations identified in Hodgkin’s lymphoma.
| Lymphomatype | Gene | Genetic aberration | Frequency of mutated cases (%) | Dysregulated biological process or pathway | References |
| Hodgkin’s lymphoma | REL | Amplification | ~50 | NF-κB pathway | Barth et al., 2003 |
| NFKBIA, NFKBIE, TNFAIP3 | Point mutations, deletions | 50–60 | Weniger et al., 2016 | ||
| MDM2 | Gains | 60 | P53-dependent biological processes (e.g., cell cycle arrest and apoptosis) | Küppers, 2009 | |
| TP53 | Point mutations, deletions | 10 | |||
| Classical Hodgkin’s lymphoma | JAK1, STAT3, STAT5B | Missense mutations | 15 | JAK-STAT pathway | Tiacci et al., 2018 |
| JAK2 | Gains | 32 | |||
| PTPN1 | Splice-acceptor, missense mutations | 6 | |||
| SOCS1 | Frameshift mutations, disruptive in-frame deletions, splice donor, missense | 47 | JAK-STAT5pathway | Weniger et al., 2006;Tiacci et al., 2018 | |
| EBV+ Hodgkin’s lymphoma | LMP2A | EBV-encoded LMP2A mediated transcriptional changes | ~50 | Transcriptional signature similar to that of HRS* cells | Portis et al., 2003 |
*: Reed–Sternberg cells of Hodgkin’s lymphoma.
It has been known for a long time that HL cases respond to anti-CD30 antibody-drug conjugates, even in the presence of relapse (Falini et al., 1992; Schnell et al., 2002). However, until recently, the cell of origin of HRS cells in HL was not clear. A recent study by Weniger et al. showed that the transcriptomic profile of HRS cells is highly similar to that of CD30+ B cells, a rare B cell subset inside germinal centers (Weniger et al., 2018). In the same study, the transcriptional differences between the CD30+ B cell subset and HRS cells of HL were compared, and this showed significant downregulation of genomic stability regulators and cytokinesis.
3. Genetic alterations in B cell non-Hodgkin’s lymphoma
A great majority of NHL subtypes are B cell NHLs associated with abnormalities in immunogenetic mechanisms which operate in germinal center B cell reaction (Küppers, 2005). In this section, we address recurrent genetic alterations observed to date in different B cell NHL subtypes and oncogenic signaling pathway dysregulations associated with these alterations.
3.1. Diffuse large B cell lymphoma
Diffuse large B cell lymphoma (DLBCL) is the most common type of NHL, accounting for 30%–40% of all cases (Dubois and Jardin, 2018). Based on gene expression profiling, DLBCL consists of two distinct subtypes, namely germinal center B cell (GCB) type and activated B cell (ABC) type (Alizadeh et al., 2000). This study is very critical in terms of the value of DNA microarray-based gene expression profiling for identification of new disease subtypes with prognostic differences, which cannot be identified with routine histological evaluations. Another study confirmed that DLBCL cases can be subclassified into two different molecular groups as GCB and non-GCB type with the tissue microarray (TMA) method and immunohistochemical analysis of CD10, BCL6, and MUM1 protein expression (Hans et al., 2004). Subsequent studies showed that these two DLBCL subtypes are genetically and biologically distinct. Uncontrolled, constitutive activation of the prosurvival NF-κB signaling pathway was observed in the ABC DLBCL subtype but not in GCB DLBCL cases (Davis et al., 2001). Indeed, CARD11 mutations associated with activation of NF-kB were identified in approximately 10% of ABC DLBCL cases (Lenz et al., 2008) (Figure 1A). In ABC DLBCL cases, the NF-κB signaling pathway can continuously be activated by ‘chronic’ stimulation of the B cell receptor (BCR), and somatic mutations of two BCR subunits (i.e. CD79A and CD79B) may contribute to this oncogenic activation (Davis et al., 2010) (Figure 1D). In more than half of ABC DLBCL cases, deletions or inactivating mutations in the TNFAIP3 (A20) tumor suppressor gene and genetic defects in some other protooncogenes and tumor suppressor genes that regulate the NF-κB signaling pathway were found to have activated this signaling pathway irregularly (Compagno et al., 2009). Moreover, oncogenic MYD88 mutations leading to constitutive activation of the JAK-STAT signaling pathway were observed very frequently in ABC DLBCL cases (Ngo et al., 2011). A very recent study by Jain et al. identified a novel protooncogene called TCF4 (E2.2), which is located in the recurrently gained 18q21.2 locus in ABC DLBCL cases. Overexpression of TCF4 by Jain et al. showed direct transcriptional activation of MYC and IGHM genes in ABC DLBCL cell lines and cytotoxicity when it was knocked down (Jain et al., 2019).
t(14;18)(q32;q21) translocation that increases BCL2 expression was detected in 20% of DLBCL cases (Willis and Dyer, 2000). Importantly, point mutations and indels of BCL2 were reported in GCB type DLBCL cases (Schuetz et al., 2012). The BCL6 protooncogene was observed to be deregulated due to t(3;14)(q27;q32) translocation in 5%–10% of DLBCL cases (Willis and Dyer, 2000). Additionally, somatic mutations in BCL6 disrupting the negative autoregulation of its expression were reported in DLBCL cases (Pasqualucci et al., 2003). It is important to note that some of the most critical genetic alterations specifically detected in GCB DLBCL cases to date were the point mutations affecting the oncogenic histone methyltransferase EZH2 on the Tyr641 residue in 21.7% of GCB DLBCL cases (Morin et al., 2010) (Figure 1E). EZH2 Tyr641 gain-of-function mutations were shown to increase H3K27 trimethylation, and this chromatin modification may be responsible for the silencing of tumor suppressor genes in mutated DLBCL cases (Yap et al., 2011). The major genetic aberrations observed in DLBCL cases are shown in Table 2.
Table 2.
The major genetic aberrations observed in diffuse large B cell lymphoma.
| Lymphoma type | Gene | Genetic aberration | Frequency of mutated cases (%) | Dysregulated biological process or pathway | References |
| DLBCL | BCL2 | t(14;18)(q32;q21) | 20 | Intrinsic pathway of apoptosis | Willis et al., 2000 |
| GCB DLBCL | BCL2 | Point mutations, indels | 43 | Intrinsic pathway of apoptosis | Schuetz et al., 2012 |
| DLBCL | BCL6 | t(3;14)(q27;q32) | 5–10 | Germinal center B cell reaction | Willis et al., 2000 |
| DLBCL | BCL6 | Somatic point mutations | 16 | Disruption of negative autoregulation of BCL6 expression | Pascualucci et al., 2003 |
| ABC DLBCL | CARD11 | Missense mutations in coiled-coil domain | 9.6 | NF-κB pathway | Lenz et al., 2008 |
| CD79B | Y196 ITAM mutation, ITAM deletion | 21 | Chronic BCR signaling, NF-κB pathway | Davis et al., 2010;Schmitz et al., 2018 | |
| CD79A | ITAM deletion, splice site mutation | 2.9 | |||
| GCB DLBCL | EZH2 | Point mutations on Tyr641 | 21.7 | Trimethylation of Lys27of histone H3 (H3K27) | Morin et al., 2010 |
| DLBCL | MYC | t(8;14)(q24;q32) | 10 | G1 phase of the cell cycle | Willis et al., 2000 |
| ABC DLBCL | MYD88 | Missense mutations | 37 | JAK-STAT pathway | Ngo et al., 2011;Schmitz et al., 2018 |
| DLBCL (N1 andBN2 subtypes) | NOTCH1, NOTCH2 | Frameshift truncating mutations | 11.7 | Notch signaling pathway | Arcaini et al., 2015 |
| ABC DLBCL | TNFAIP3 (A20) | Nonsense mutations, frameshift indels, splice site mutations, deletions | ~55 | NF-κB pathway | Compagno et al., 2009 |
| ABC DLBCL | TCF4 | Gain/amplification | 40.7 | Transcriptional activatorof IGHM and MYC | Jain et al., 2019 |
ABC: Activated B cell type; GCB: germinal center B cell type; ITAM: immunoreceptor tyrosine-based activation motif; BCR: B cell receptor.
As a result of the above-mentioned studies that reveal differences in genomic, transcriptomic, biological, and clinical characteristics in ABC and GCB type DLBCL cases, the WHO provisionally defined the ABC and GCB types as 2 distinct subclasses of DLBCL. However, some other DLBCL subtypes were also recognized by WHO based on other characteristics such as presence of chronic inflammation or viral infection in primary tumors (Swerdlow et al., 2016). Although the molecular classification dividing DLBCL into the ABC and GCB subgroups is generally accepted, 10%–20% of cases cannot be classified using this method (Dubois and Jardin, 2018). While discussions on DLBCL subtypes persist, a quaternary molecular classification system for DLBCL has recently been proposed based on results of extensive genomic and transcriptomic analyses. Subtype nomenclature proposed for this classification is as follows: MCD for MYD88, L265P, and CD79B mutated cases; BN2 for BCL6 fusion genes or NOTCH2 mutated cases; N1 for NOTCH1 mutated cases; and EZB for EZH2 mutated or BCL2 translocated cases (Schmitz et al., 2018) (Figure 1F). This newly proposed classification method stemming from various oncogenic mutations frequently observed in DLBCL cases aims to ensure selection of more appropriate treatment approaches for genetically and clinically heterogeneous DLBCL cases, thereby improving ABC and GCB classification instead of replacing these two subtypes.
3.2. Follicular lymphoma
Follicular lymphoma (FL) is the most common type of indolent lymphoma, and it is the second most frequent lymphoma among all NHLs after DLBCL (Shankland et al., 2012). Like other B cell NHL subtypes, development of FL is thought to be associated with irregularities in somatic hypermutation and class switch recombination mechanisms functional during GCB reaction. The most characteristic genetic alteration of FL is the t(14; 18) (q32; q21) IGH/BCL2 translocation that brings the active enhancer of the immunoglobulin heavy chain gene (IGH) closer to the BCL2 gene (Tsujimoto et al., 1984). This translocation contributes to FL development through inhibition of apoptosis due to irregular, high expression of the antiapoptotic BCL2 gene product (Capaccioli et al., 1996). t(14; 18) (q32; q21) translocation is observed in 70% to 95% of FL cases, and is used as a diagnostic biomarker of FL that can be detected by routine cytogenetic analyses (Einerson et al., 2005). Recent genomic analyses of FL cases have revealed mutations related to lymphomagenesis in genes of the JAK-STAT and NF-κB signaling pathways as well as in genes involved in posttranslational modification of histones such as CREBBP, EZH2, and KMT2D (MLL2) (Okosun et al., 2014) (Figure 1E). More than 50% of FL cases may show histological transformation to more aggressive cancers such as DLBCL and Burkitt lymphoma (Kridel et al., 2012). The life expectancy in cases of transformed FL (tFL) is as short as 1.7 years (Al-Tourah et al., 2008); therefore, ongoing investigations focus on identification of genetic alterations associated with histological transformation. Somatic point mutations of BCL2 (Matolcsy et al., 1996; Correia et al., 2015), TP53 (Lo Coco et al., 1993), miR-142, and MEF2B (Bouska et al., 2017) genes were reported to be associated with this transformation. Table 3 lists the recurrent genetic alterations observed in FL and/or tFL cases.
Table 3.
Recurrent genetic alterations identified in follicular lymphoma, Burkitt lymphoma, and mantle cell lymphoma.
| Lymphomatype | Gene | Genetic aberration | Frequency of mutated cases (%) | Dysregulated biological process or pathway | References |
| Follicular and/or transformed follicular lymphoma | BCL2 | t(14;18)(q32;q21) | ~80 | Intrinsic pathway of apoptosis | Tsujimoto et al., 1984; Willis et al., 2000 |
| CREBBP | Somatic nonsynonymous mutations | 64 | Histone acetylation patterns | Okosun et al.,2014 | |
| EZH2 | 20 | H3K27me3 repressive marks | |||
| KMT2D (MLL2) | 81 | Histone lysine methylation patterns | |||
| SOCS1 | 8 | JAK-STAT signaling | |||
| STAT6 | 12 | ||||
| MEF2B | 20 | B cell transcription factor | Bouska et al., 2017 | ||
| PAPOLG | Gain/amplification | 24 in FL, 30 in tFL | Polyadenylation of transcripts | Kurşun et al., 2019 | |
| REL | 24 in FL, 30 in tFL | DNA damage inducedNFκB pathway | Hu et al., 2017 | ||
| MYC | 15.7 in FL, 29.1 in tFL | Cell cycle and other oncogenic pathways | Bouska et al.,2014 | ||
| TP53 | Deletion | 12 in FL, 22 in tFL | P53 dependent cell cycle arrest and apoptosis | ||
| TNFAIP3 | NF-κB pathway | ||||
| Burkittlymphoma | CCND3 | Missense, nonsense,indel mutations | 14.6 | G1-S cell cycle transition | Schmitz et al.,2012 |
| ID3 | Missense, nonsense, cyrptic splice site mutations | 58.5 | Pro-survival PI3 pathway, cell cycle | ||
| TCF3(E2A) | Missense mutations | 29.2 | Pro-survival PI3 pathway, cell cycle | ||
| MYC | t(8;14)(q24;q32) | 100 | c-MYC target genes involved in cell cycle regulation, metabolism etc. | Willis et al., 2000 | |
| Mantle cell lymphoma | ATM | Deletion, deleteriouspoint mutations | 75 | DNA damage response | Schaffner et al.,2000 |
| CCND1 | t(11;14)(q13; q32) | 95 | G1-S cell cycle transition | Willis et al., 2000 | |
| P53 | Missense mutations | 15 | P53 dependent cell cycle arrest and apoptosis | Greiner et al.,1996 | |
| NOTCH1 | Truncating mutations, frameshifting indels | 12 | NOTCH signaling pathway | Kridel et al., 2011 | |
| BIRC3 | Deletion, splice site mutations | 10 | Apoptosis | Beà et al., 2013 | |
| MEF2B | Missense mutations (p.K23R and p.N49S) | 7 | Unknown | ||
| NOTCH2 | Truncating mutations | 5 | NOTCH signaling pathway | ||
| WHSC1 | Missense mutations (p.E1099K and p.T1150A) | 14 | Altered methylation of H3K36 |
A recent article published by Andor et al. analyzed the transcriptome of 34,188 cells from 6 primary FL tumors to better understand the complexity of FL tumor biology for identification of novel, more effective targeted therapeutic approaches (Andor et al., 2019). In malignant B cell clones, the authors observed overexpression of BCL2 and downregulation of CD52, FCER2, and MHC class II genes compared with the expression in normal B cells from the same patients. In the same single-cell RNA-Seq study, the authors also focused on expression profiles in tumor-infiltrating T cells, which showed genes simultaneously expressed with immune checkpoint molecules in regulatory T cells. This single cell-based study showed different subclones, implying intratumoral genetic heterogeneity of primary FL tumor tissues.
3.3. Burkitt lymphoma
Burkitt lymphoma (BL) is an aggressive type of pediatric and adult B cell lymphoma originating from GCB cells (Deffenbacher et al., 2012). Pediatric BL is the most common type of childhood B cell NHL, accounting for 80% of all childhood B cell NHLs (Minard-Colin et al., 2015). BL is clinically classified into three subtypes: sporadic, endemic, and immunodeficiency-related (Piccaluga et al., 2011). The most characteristic genetic aberration associated with BL cases is the juxtaposition of the IgH enhancer and MYC protooncogene due to t(8; 14) translocation, which leads to high and constitutive expression of MYC (Joos et al., 1992). MYC-induced apoptosis in BL cells is not functional in one-third of BL cases due to deletions or point mutations of P53 (Gaidano et al., 1991). For P53 WT BL cases, MDM4, which is located in the recurrently gained 1q locus, has recently been found to be overexpressed and to inhibit MYC-induced apoptosis through inhibition of P53 (Hüllein et al., 2019).
Oncogenic CCND3 mutations stabilizing CCND3 protein isoforms and thereby disrupting cell cycle progression were identified in 38% of sporadic BL (sBL) cases (Schmitz et al., 2012). In the same study, mutations in the TCF3 (E2A) transcription factor or its negative regulator, ID3, were reported in 70% of sBL cases (Figure 1G). It was proposed that mutated TCF3 contributes to BL formation by stimulating survival through irregular activation of the phosphatidyl inositol-3-OH kinase (PI3K) signaling pathway (Schmitz et al., 2012). Another whole-genome and whole-exome sequencing-based study reported mutations in SWI/SNF family genes involved in chromatin remodeling such as ARID1A and SMARCA4, as well as those in CCT6B, SALL3, FTCD, and PC genes (Love et al., 2012). The major genetic aberrations and the biological processes affected by these alterations in BL cases are shown in Table 3.
One of the most critical characteristic features of BL is the presence of EBV in the genome of most BL tumors, especially in the endemic subtype (Shannon-Lowe et al., 2017). Integrative analyses of whole-genome and transcriptomic profiles have recently shown that EBV-positive tumors show a robust increase in abnormal somatic hypermutation in pediatric endemic and sporadic BL cases (Grande et al., 2019). This study has also shown genetic alterations in genes affecting BCR/PI3K signaling, MYC regulation, apoptosis, the SWI/SNF complex, and G-protein coupled receptor (GPCR) signaling. Another study also showed inactivating mutations in components of the GPCR signaling pathway (i.e. GNA13 and RHOA) genes, suggesting that this pathway may be a target of therapy for GNA13 or RHOA-mutated BL cases (O’Hayre et al., 2016).
3.4. Mantle cell lymphoma
Mantle cell lymphoma (MCL) is a type of lymphoid cancer originating from neoplastic transformation of naive B cells that do not encounter antigens (Cortelazzo et al., 2012). MCLs are highly heterogeneous with respect to their morphological, clinical, and pathological characteristics. One of the most characteristic genetic alterations of MCL is the t(11; 14)(q13; q32) chromosomal translocation, which increases CCND1 expression that stimulates the cell cycle (Cortelazzo et al., 2012) (Figure 1C). Interestingly, highly proliferative MCL tumors express shorter but more stable versions of CCND1, generated through genomic deletions or point mutations ending up with shorter 3’ untranslated regions, which in turn results in poor survival of MCL patients (Wiestner et al., 2007). A recent study by Mohanty et al. investigated the consequence of CCND1 coding sequence point mutations (i.e. E36K, Y44D, or C47S) and they observed improved stability of CCND1 protein in these mutated MCL cells (Mohanty et al., 2016).
MCL can be clinically indolent or aggressive; the 5-year overall survival rate is approximately 60% in the low-risk group, whereas it decreases significantly in moderate or high-risk groups (Hoster et al., 2008). MCL was proposed to develop in 2 distinct ways. Classical MCL usually consists of SOX11+ cases with no IGHV gene mutations. On the other hand, other MCLs derive from cells with IGHV mutation but not SOX11 expression (Swerdlow et al., 2016).
Previous studies have shown that deletions and inactivating mutations in the tumor suppressor ATM (Schaffner et al., 2000) or TP53 (Greiner et al., 1996) may play a role in MCL development (Figure 1C). The impact of inactivating mutations of P53 may not be limited with MCL lymphomagenesis. Eskelund et al. reported that P53 mutation status is an important factor for increasingly poor prognosis of younger MCL patients, for which P53 mutations confer resistance to rituximab or autologous stem cell transplantation (Eskelund et al., 2017). Whole-transcriptome analyses of MCL tumor samples and cell lines showed NOTCH1 mutations in 12% of MCL cases (Kridel et al., 2012). In another NGS-based study, cancer-associated mutations were observed in antiapoptotic BIRC3, Toll-like receptor 2 (TLR2), chromatin modification genes (WHSC1, MLL2, and MEF2B), and the NOTCH2 gene (Beà et al., 2013). Table 3 shows the genetic alterations associated with the pathogenesis of MCL.
4. Diagnostic, prognostic, and therapeutic implications of genetic alterations
Several studies revealed a clinical relevance for the identified genetic alterations in different subtypes of B cell NHL. These clinically relevant genetic alterations may be potentially useful in routine diagnosis as well as in risk stratification during diagnosis. The oncogenic chromosomal translocations originating from aberrantly operating class-switch recombination mechanisms are useful for genetic diagnosis of B cell NHL subtypes (Lenz et al., 2007). The presence of these chromosomal translocations that are specific for certain subtypes of B cell NHL can be used to evaluate clonality and to facilitate the diagnosis of the specific subtype of lymphoma. Interphase FISH is routinely used for detection of t(14;18) translocation (i.e. IGH-BCL2) in FL (Vaandrager et al., 2000), t(11;14) translocation (i.e. IGH-CCND1) in MCL (Monteil et al., 1996), or t(8;14) translocation (i.e. IgH-MYC) in BL (Siebert et al., 1998) for diagnostic purposes.
Some of the identified genetic alterations may be useful to predict patient survival. It has been known for some time that P53 mutations are associated with poor patient survival in aggressive B cell lymphomas (Ichikawa et al., 1997). Several studies have established a relationship between P53 mutations and poor prognosis for different B cell lymphoma subtypes. As an example, Zainuddin et al. reported an inferior survival for GCB DLBCL cases with P53 mutations (Zainuddin et al., 2009). Another report showed that P53-mutated MCL cases have a worse survival rate than that of unmutated ones (Halldórsdóttir et al., 2011). In support of these previous reports, a recent study based on metaanalyses showed a general prognostic significance for P53 mutations in B cell NHLs (Xu et al., 2017). MYC overexpression due to genetic aberrancies (e.g., translocation and amplification) can be used as a prognostic biomarker for certain B cell NHL subtypes. Based on a recent report, MYC amplifications or rearrangements can independently predict overall survival in patients of DLBCL (Quesada et al., 2017). Application of next-generation sequencing (NGS)-based technologies such as whole exome sequencing revealed the genomic mutational landscape of different types of B cell lymphomas. Detection of these mutations may provide useful information regarding prognosis or prediction of response to immunochemotherapy. In one of these NGS-based studies, FFPE tumor samples of DLBCL patients were screened for the presence of recurrently identified mutations using high throughput sequencing (Juskevicius et al., 2017). After that, a relationship was established between the identified mutations and overall progression-free and event-free survival in R-CHOP-treated DLBCL patients, which showed poorer prognosis for patients with CREBBP and EP300 mutations and a better prognosis for patients with SOCS1 mutations.
Somatic mutation status may add value to the traditionally used prognostic indexes in terms of better risk stratification of B cell NHLs. To address this possibility, Pastore et al. retrospectively analyzed the mutational status of tumor samples of 151 follicular lymphoma cases, which were obtained before patients received the first-line immunochemotherapy (Pastore et al., 2015). This report showed that evaluation of the mutation status of seven cancer-associated genes (i.e. EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP, and CARD11) with deep sequencing improved prognostication of follicular lymphoma patients receiving first-line immunochemotherapy when integrated into FILIP1 and ECOG prognostic indexes, which in turn outperformed prognostications performed with FILIPI alone or FILIPI combined with ECOG performance status.
Genetic alterations and transcriptional signatures may be used together or separately as biomarkers for improved diagnosis or prognosis of B cell lymphoma patients. Whether mutations or transcriptional signatures should be chosen as biomarkers depends on the potential of these genetic or transcriptional changes to separate B cell lymphoma subtypes into more distinct and refined diagnostic and/or prognostic subgroups. Subclassification of DLBCL cases into ABC and GCB subgroups using gene expression profiling may be one of the best examples not only in terms of improved cancer diagnosis and prognosis but also for identification of genetic alterations specific for each subtype. Lenz et al. showed that there are 30 genetic alterations observed in a DLBCL subtype specific manner (Lenz et al., 2008). For instance, INK4a/ARF tumor suppressor loci deletions were identified mainly in the ABC DLBCL subtype. On the other hand, PTEN deletions were observed in the GCB but not in the ABC subtype of DLBCL (Lenz et al., 2008). Overall, these observations suggest that using transcriptomic signatures for DLBCL subclassification during diagnosis may be superior to other classifications including genetic alterations. However, MYC/BCL2 coexpression, which is observed more commonly in the ABC subtype, was proposed as an independent prognostic factor regardless of the ABC versus GCB subtype classification (Hu et al., 2013). DLBCL cases with MYC/BCL2 coexpression showed remarkably poorer survival rates when compared to the rest of the cases. It should be noted that a distinct oncogenic gene expression signature was identified for the MYC/BCL2 coexpression group in this study. A transcriptional signature may be more useful than mutations for subdividing MCL cases into prognostically distinct diagnostic subgroups. A thirteen-gene signature including SOX11 was identified to clearly distinguish indolent and conventional subtypes of MCL (i.e. iMCL and cMCL, respectively). SOX11 protein expression status in MCL cases may be useful to distinguish indolent and conventional subtypes of MCL, which differ in clinicopathological characteristics, including patient survival (Fernàndez et al., 2010).
Identification of the aberrant signaling pathways in the subtypes of B cell lymphoma provides opportunities for development of targeted therapeutics with fewer side effects in comparison to traditional chemotherapeutic approaches. Recent clinical studies have focused on inhibition of oncogenic pathways for better treatment of B cell lymphoma patients. JAK-STAT pathway activation is frequently observed in cHL cases. Consequently, JAK1/2 inhibitors are already in clinical trials for treatment of advanced relapsed/refractory HL (Den Neste et al., 2018). A selective BCL2 inhibitor (venetoclax) was evaluated in 106 patients with relapsed or refractory B cell NHL (i.e. FL, DLBCL, and MCL) as a single agent for its activity and safety profile; however, future studies are needed maximize the therapeutic benefits for these patients (Davids et al., 2017). Genetic alterations in epigenetic regulatory genes offer significant therapeutic opportunities for treatment of FL and DLBCL patients with relapsed or refractory disease. To address this possibility, Italiano et al. recently investigated an EZH2 inhibitor (i.e. tazemetostat) in relapsed and refractory B cell NHL patients (Italiano et al., 2018). Based on these results, it was concluded that tazemetostat has a favorable safety profile and antitumor activity in patients with refractory B cell NHL. One of the most intriguing results regarding targeted therapeutics against a constitutively activated oncogenic pathway is selective inhibition of the chronic BCR signaling with ibrutinib in DLBCL. Consistent with the chronic activation of BCR signaling selectively in the ABC DLBCL subtype, ABC DLBCL patients responded significantly better to this BCR signaling inhibitor in comparison to patients with the GCB DLBCL subtype (Wilson et al., 2015). These and other similar studies show that genetic alterations that activate oncogenic signaling pathways may be pharmacologically targeted to improve currently available therapeutic options such that B cell lymphoma patients in advanced stages may benefit from more effective and safer treatment alternatives.
5. Chemotherapy resistance-associated genetic aberrations in B cell NHL
One of the most important challenges in the treatment of B cell lymphoma patients is the resistance that develops against chemotherapy or immunochemotherapy treatment. After inclusion of rituximab in the traditional CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) chemotherapy, CHOP therapy resistance was eliminated in many B cell lymphoma cases. Mounier et al. reported that R-CHOP therapy overcomes therapy resistance associated with BCL2 overexpression in elderly DLBCL patients (Mounier et al., 2003). A report by Liu et al. showed that PRDM1β is expressed in a subset of non-GCB-like DLBCL cases, and its expression is associated with poor survival in CHOP-treated but not R-CHOP-treated DLBCL cases. They further showed that rituximab downregulates PRDM1β expression in vitro (Liu et al., 2007). However, DLBCL cases may develop resistance to R-CHOP therapy. Yuan et al. showed that high microRNA-125b and microRNA-130a levels in the serum of DLBCL cases are associated with R-CHOP resistance in DLBCL cases (Yuan et al., 2016). Laursen et al. showed that high expression of CXCR4 on the tumor cell surface may be responsible for R-CHOP resistance in a subset of DLBCL cases (Laursen et al., 2019). Another study by Wilson et al. asked whether ABC DLBCL patients who have chemotherapy refractory disease can be treated through inhibition of BCR signaling. They performed a clinical trial with ibrutinib and observed a high response in those patients, especially in the presence of MYD88 mutations (Wilson et al., 2015).
Follicular lymphoma cases may transform into high-grade lymphomas called transformed follicular lymphomas (tFL) such as DLBCL or BL, which are often chemotherapy-resistant (Lossos and Gascoyne, 2011). Many genetic alterations in FL tumors such as deletions of P53, P16/CDK2NA, or deregulations of MYC have been associated with FL to tFL transformation (Lossos and Gascoyne, 2011). Given the increased chemoresistance after histological transformation, these genetic alterations may also be responsible for chemotherapy resistance. Based on genome-wide copy number analyses of 198 FL and 79 tFL cases using SNP arrays, Bouska et al. identified recurrent chromosomal gains or losses in FL cases that can be potentially associated with tFL transformation. These chromosomal gains and losses may restrict immune surveillance and deregulate P53 or NFκB pathways (Bouska et al., 2014). REL is one of the protooncogenes identified to reside on the frequently amplified 2p16.1-p15 locus in FL and tFL cases based on this study. Recently, Hu et al. showed that FL cases with high REL expression levels may be associated with chemotherapy resistance due to activation of DNA damage-induced repair and cell cycle arrest pathways; however, future studies are needed to investigate whether REL is a predictive biomarker of chemotherapy in FL and/or tFL cases (Hu et al., 2017). Another recent study systematically analyzed genes in the frequently amplified 2p16.1-p15 locus by integrating an SNP array and gene expression profiling data of previously published reports on FL and tFL cases to address whether there are genes other than REL that may act as protooncogenes during transformation to tFL (Kurşun and Küçük, 2019). This study showed that PAPOLG is a candidate protooncogene for FL development, and it may have a role during histological transformation to tFL.
Current efforts in MCL genomics focus on identification of mutations associated with chemotherapy or immunochemotherapy resistance. To identify genetic alterations associated with therapy resistance in MCL cases, Wu et al. performed genome-wide analyses of MCL tumors which revealed cancer-related mutations at the time of diagnosis, as well as relapse (Wu et al., 2016). They identified recurrent CARD11 mutations in 5.5% of MCL cases. More importantly, overexpression of mutated CARD11 constructs in MCL cell lines conferred resistance to ibrutinib and lenalidomide, which are inhibitors of BCR and NF-κB signaling, respectively. Similarly, CCND1 mutations described in MCL cases by Mohanty et al. conferred therapeutic resistance to ibrutinib, a bruton tyrosine kinase inhibitor used for MCL treatment (Mohanty et al., 2016). Agarwal et al. focused on the genomic profiles of MCL patients who respond or did not respond to ibrutinib and venetoclax combinatory therapy and identified a distinct genomic profile separating responders from nonresponders. Based on this very recent study, chemotherapy-resistant patients harbor 9p21.1–p24.3 locus deletion and/or mutations of the genes forming the SWI–SNF chromatin-remodeling complex (Agarwal et al., 2019). They observed that defective SWI–SNF complexes promote transcriptional upregulation of BCL-XL, which may confer resistance to the ibrutinib and venetoclax combination. This study is a good example of precision medicine and shows that individual patients’ tumor genotypes can be used to decide on the most appropriate therapy.
6. Noninvasive clinical applications of genomic alterations of B cell lymphoma for diagnosis, prognosis, or disease monitoring
Traditional diagnosis of B cell lymphoma subtypes requires immunohistochemical and histopathological analyses of tumor biopsy sections. Tumor biopsies are obtained from lymphoma patients using invasive methodologies, which may have the following specific disadvantages: 1) procedures are generally time-consuming; 2) tissue sampling is localized; therefore, the obtained tissue may not reflect the genetic characteristics and heterogeneity of the entire tumor tissue present in the patient; 3) tumor biopsies may not always be easy to obtain; 4) patients may have some pain. Due to these disadvantages, cancer researchers have been trying to address whether body liquids (e.g., blood and saliva) contain tumor-derived biomolecules that reflect the genetic characteristics of solid tumor tissues of cancer patients. Circulating cell-free DNA (cfDNA) has proven not only to be useful in terms of overcoming the disadvantages of standard biopsy listed above but has also provided the opportunity to track genetic changes through multiple, periodical blood sampling and determine the emergence of therapy-resistant clones several months earlier than currently used methods in routine clinical settings (Crowley et al., 2013).
Several recently reported elegant studies have revealed the feasibility of cfDNA genotyping for diagnosis, prognosis, and monitoring chemotherapy response in major subtypes of B cell lymphoma. Spina et al. evaluated whether circulating cell-free tumor DNA can be used for cHL genotyping (Spina et al., 2018). By applying a custom-designed, ultradeep targeted NGS panel, which consisted of 77 genes recurrently mutated in B cell tumors, on DNA samples from 80 diagnosed and 32 refractory cHL cases, the authors observed that 87.5% of the somatic mutations detected in tumor biopsies were also present in the circulating cell-free tumor DNA (ctDNA) of the same patients’ plasma. The same study also revealed that decreased plasma ctDNA concentrations are associated with markedly better survival for cHL patients treated with a combination of chemotherapeutic drugs (i.e. adriamycin, bleomycin, vinblastine, and dacarbazine). Another recent study, by Rossi et al., showed that the sensitivity for detection of mutations in cfDNA is 83% if tumor tissue gDNA is regarded as the gold standard for mutation detection, suggesting that noninvasive blood sampling and subsequent plasma cfDNA genotyping may have diagnostic value (Rossi et al., 2017). The authors also observed the changes in variant allele frequency (VAF) before and after R-CHOP therapy, which showed that patients responding to R-CHOP have decreased VAF, whereas others did not. Importantly, new variants emerged in different patients such as those of PIM1, BIRC3, and FBXW7. This observation suggests that mutations responsible for relapse or refractory disease in individual DLBCL patients can potentially be determined noninvasively using a blood-based test. From a precision medicine perspective, cfDNA genotyping may assist clinicians in choosing the most suitable option for targeted therapy through facilitated detection of individual genetic subclones responsible for relapse in DLBCL patients. There are other recent studies that evaluated plasma cfDNA levels in DLBCL (Kurtz et al., 2018), addressing whether it can predict or monitor therapy response or if it can predict tumor burden or prognosis of FL cases (Delfau-Larue et al., 2018). Using droplet digital PCR, Alcaide et al. detected previously known mutations (e.g., EZH2 Y641 and STAT6 D419) in circulating tumor DNA in major B cell NHL subtypes (Alcaide et al., 2016).
7. Conclusion
HL and B cell NHL, which represent most lymphoid cancers, include mutations related to malfunctioning immunogenetic mechanisms during germinal center reactions. These genetic aberrations may provide us with novel biomarkers of noninvasive diagnosis or prognosis if they distinguish B cell lymphoma subsets from similar B cell lymphomas or benign diseases and predict patient survival, respectively. Furthermore, biological signaling pathways dysregulated by activating mutations in B cell lymphomas offer new opportunities for targeted therapies.
Acknowledgments
This study was supported by the Young Scientists Award Program of the Turkish Academy of Sciences (TÜBA GEBİP 2017). A part of this review was presented in Turkish in Hematolog, which is a members-only periodical magazine of the Turkish Society of Hematology.
References
- Agarwal R Chan YC Tam CS Hunter T Vassiliadis D Dynamic molecular monitoring reveals that SWI–SNF mutations mediate resistance to ibrutinib plus venetoclax in mantle cell lymphoma. Nature Medicine. 2019;25:119. doi: 10.1038/s41591-018-0243-z. [DOI] [PubMed] [Google Scholar]
- Alcaide M Yu S Bushell K Fornika D Nielsen JS Multiplex droplet digital PCR quantification of recurrent somatic mutations in diffuse large b-cell and follicular lymphoma. Clinical Chemistry. 2016;62:1238. doi: 10.1373/clinchem.2016.255315. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA Eisen MB Davis RE Ma C Lossos IS Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Al-Tourah AJ Gill KK Chhana Bhai M Hoskins PJ Klasa RJ Population-based analysis of incidence and outcome of transformed non-Hodgkin’s lymphoma. Journal of Clinical Oncology. 2008;26:5165. doi: 10.1200/JCO.2008.16.0283. [DOI] [PubMed] [Google Scholar]
- Andor N Simonds EF Czerwinski DK Chen J Grimes SM Single-cell RNA-Seq of follicular lymphoma reveals malignant B-cell types and coexpression of T-cell immune checkpoints. Blood. 2019;133:1119. doi: 10.1182/blood-2018-08-862292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth TFE Martin-Subero JI Joos S Menz CK Hasel C Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood. 2003;101:3681. doi: 10.1182/blood-2002-08-2577. [DOI] [PubMed] [Google Scholar]
- Beà S Valdés-Mas R Navarro A Salaverria I Martín-Garcia D Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18250. doi: 10.1073/pnas.1314608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MJ Helping the CD8(+) T-cell response. Nature Reviews Immunology. 2004;4:595. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- Bouska A McKeithan TW Deffenbacher KE Lachel C Wright GW Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood. 2014;123:1681. doi: 10.1182/blood-2013-05-500595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouska A Zhang W Gong Q Iqbal J Scuto A Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia. 2017;31:83. doi: 10.1038/leu.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaccioli S Quattrone A Schiavone N Calastretti A Copreni E A bcl-2/IgH antisense transcript deregulates bcl-2 gene expression in human follicular lymphoma t(14;18) cell lines. Oncogene. 1996;13:105. [PubMed] [Google Scholar]
- Compagno M Lim WK Grunn A Nandula SV Brahmachary M Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C Schneider PA Dai H Dogan A Maurer MJ BCL2 mutations are associated with increased risk of transformation and shortened survival in follicular lymphoma. Blood. 2015;125:658. doi: 10.1182/blood-2014-04-571786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortelazzo S Ponzoni M Ferreri AJM Dreyling M Mantle cell lymphoma. Critical Reviews in Oncology/Hematology. 2012;82:78. doi: 10.1016/j.critrevonc.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Crowley E Di Nicolantonio F Loupakis F Bardelli A Liquid biopsy: monitoring cancer-genetics in the blood. Nature Reviews Clinical Oncology. 2013;10:472. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- Davids MS Roberts AW Seymour JF Pagel JM Kahl BS Phase I first-in-human study of venetoclax in patients with relapsed or refractory non-Hodgkin lymphoma. Journal of Clinical Oncology. 2017;35:826. doi: 10.1200/JCO.2016.70.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE Brown KD Siebenlist U Staudt LM Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. Journal of Experimental Medicine. 2001;194:1861. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE Ngo VN Lenz G Tolar P Young RM Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbacher KE Iqbal J Sanger W Shen Y Lachel C Molecular distinctions between pediatric and adult mature B-cell non-Hodgkin lymphomas identified through genomic profiling. Blood. 2012;119:3757. doi: 10.1182/blood-2011-05-349662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfau-Larue MH van der Gucht A Dupuis J Jais JP Nel I Total metabolic tumor volume, circulating tumor cells, cell-free DNA: distinct prognostic value in follicular lymphoma. Blood Advances. 2018;2:871. doi: 10.1182/bloodadvances.2017015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois S Jardin F Novel molecular classifications of DLBCL. Nature Reviews Clinical Oncology. 2018;15:474. doi: 10.1038/s41571-018-0041-z. [DOI] [PubMed] [Google Scholar]
- Einerson RR Kurtin PJ Dayharsh GA Kimlinger TK Remstein ED - FISH is superior to PCR in detecting t(14;18)(q32;q21)-IgH/bcl-2 in follicular lymphoma using paraffin-embedded tissue samples. American Journal of Clinical Pathology. 2005;24:421. doi: 10.1309/BLH8-MMK8-5UBQ-4K6R. [DOI] [PubMed] [Google Scholar]
- Eskelund CW Dahl C Hansen JW Westman M Kolstad A TP53 mutations identify younger mantle cell lymphoma patients who do not benefit from intensive chemoimmunotherapy. Blood. 2017;130:1903. doi: 10.1182/blood-2017-04-779736. [DOI] [PubMed] [Google Scholar]
- Falini B Flenghi L Aversa F Barbabietola G Martelli MF Response of refractory Hodgkin’s disease to monoclonal anti-CD30 immunotoxin. Lancet. 1992;339:1195. doi: 10.1016/0140-6736(92)91135-u. [DOI] [PubMed] [Google Scholar]
- Farrell K Jarrett RF The molecular pathogenesis of Hodgkin lymphoma. Histopathology. 2011;58:15. doi: 10.1111/j.1365-2559.2010.03705.x. [DOI] [PubMed] [Google Scholar]
- Fernàndez V Salamero O Espinet B Solé F Royo C Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Research. 2010;70:1408. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- Gaidano G Ballerini P Gong JZ Inghirami G Neri A p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:5413. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande BM Gerhard DS Jiang A Griner NB Abramson JS Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood. 2019;133:1313. doi: 10.1182/blood-2018-09-871418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner TC Moynihan MJ Chan WC Lytle DM Pedersen A p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood. 1996;87:4302. [PubMed] [Google Scholar]
- Halldórsdóttir AM Lundin A Murray F Mansouri L Knuutila S Impact of TP53 mutation and 17p deletion in mantle cell lymphoma. Leukemia. 2011;25:1904. doi: 10.1038/leu.2011.162. [DOI] [PubMed] [Google Scholar]
- Hans CP Weisenburger DD Greiner TC Gascoyne RD Delabie J Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Hoster E Dreyling M Klapper W Gisselbrecht C van Hoof A A new prognostic index (MIPI) for patients with advanced-stage mantle cell lymphoma. Blood. 2008;111:558. doi: 10.1182/blood-2007-06-095331. [DOI] [PubMed] [Google Scholar]
- Hu S Xu-Monette ZY Tzankov A Green T Wu L MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from the International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X Baytak E Li J Akman B Okay K The relationship of REL proto-oncogene to pathobiology and chemoresistance in follicular and transformed follicular lymphoma. Leukemia Research. 2017;54:30. doi: 10.1016/j.leukres.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Hüllein J Słabicki M Rosolowski M Jethwa A Habringer S MDM4Is targeted by 1q gain and drives disease in Burkitt lymphoma. Cancer Research. 2019;79:3125. doi: 10.1158/0008-5472.CAN-18-3438. [DOI] [PubMed] [Google Scholar]
- Ichikawa A Kinoshita T Watanabe T Kato H Nagai H Mutations of the p53 gene as a prognostic factor in aggressive B-cell lymphoma. New England Journal of Medicine. 1997;337:529. doi: 10.1056/NEJM199708213370804. [DOI] [PubMed] [Google Scholar]
- Italiano A Soria JC Toulmonde M Michot JM Lucchesi C Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncology. 2018;19:649. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- Jain N Hartert K Tadros S Fiskus W Havranek O Targetable genetic alterations of TCF4 ( E2-2 ) drive immunoglobulin expression in diffuse large B cell lymphoma. Science Translational Medicine. 2019;11 doi: 10.1126/scitranslmed.aav5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos S Falk MH Lichter P Haluska FG Henglein B Variable breakpoints in Burkitt lymphoma cells with chromosomal t(8;14) translocation separate c-myc and the IgH locus up to several hundred kb. Human Molecular Genetics. 1992;1:625. doi: 10.1093/hmg/1.8.625. [DOI] [PubMed] [Google Scholar]
- Juskevicius D Jucker D Klingbiel D Mamot C Dirnhofer S Mutations of CREBBP and SOCS1 are independent prognostic factors in diffuse large B cell lymphoma: mutational analysis of the SAKK 38/07 prospective clinical trial cohort. Journal of Hematology & Oncology. 2017;10:70–70. doi: 10.1186/s13045-017-0438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler H Küppers R Hansmann ML Rajewsky K Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. Journal of Experimental Medicine. 1996;184:1495. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kridel R Meissner B Rogic S Boyle M Telenius A Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119:1963. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- Kridel R Sehn LH Gascoyne RD Pathogenesis of follicular lymphoma. Journal of Clinical Investigation. 2012;122:3424. doi: 10.1172/JCI63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers R Mechanisms of B-cell lymphoma pathogenesis. Nature Reviews Cancer. 2005;5:251. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Küppers R The biology of Hodgkin’s lymphoma. Nature Reviews Cancer. 2009;9:15. doi: 10.1038/nrc2542. [DOI] [PubMed] [Google Scholar]
- Kurşun D Küçük C Systematic analysis of the frequently amplified 2p15-p16.1 locus reveals PAPOLG as a potential proto-oncogene in follicular and transformed follicular lymphoma. Turkish Journal of Biology. 2019;43:124. doi: 10.3906/biy-1810-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz DM Scherer F Jin MC Soo J Craig AFM Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. Journal of Clinical Oncology. 2018;36:2845. doi: 10.1200/JCO.2018.78.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen MB Reinholdtz L Schönherz AA Due H Jespersen DS High CXCR4 expression impairs rituximab response and the prognosis of R-CHOP-treated diffuse large B-cell lymphoma patients. Oncotarget. 2019;10:717. doi: 10.18632/oncotarget.26588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G Davis RE Ngo VN Lam L George TC Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Lenz G Nagel I Siebert R Roschke AV Sanger W Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. Journal of Experimental Medicine. 2007;204:633. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G Wright GW Emre NCT Kohlhammer H Dave SS Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13520. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY Leboeuf C Shi JY Li JM Wang L Rituximab plus CHOP (R-CHOP) overcomes PRDM1-associated resistance to chemotherapy in patients with diffuse large B-cell lymphoma. Blood. 2007;110:339. doi: 10.1182/blood-2006-09-049189. [DOI] [PubMed] [Google Scholar]
- Lo Coco F Gaidano G Louie DC Offit K Chaganti RS p53 mutations are associated with histologic transformation of follicular lymphoma. Blood. 1993;82:2289. [PubMed] [Google Scholar]
- Lossos IS Gascoyne RD Transformation of follicular lymphoma. Best Practice & Research: Clinical Haematology. 2011;24:147. doi: 10.1016/j.beha.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C Sun Z Jima D Li G Zhang J The genetic landscape of mutations in Burkitt lymphoma. Nature Genetics. 2012;44:1321. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matolcsy A Casali P Warnke RA Knowles DM Morphologic transformation of follicular lymphoma is associated with somatic mutation of the translocated Bcl-2 gene. Blood. 1996;88:3937. [PubMed] [Google Scholar]
- Minard-Colin V Brugières L Reiter A Cairo MS Gross TG Non-Hodgkin lymphoma in children and adolescents: progress through effective collaboration, current knowledge, and challenges ahead. Journal of Clinical Oncology. 2015;33:2963. doi: 10.1200/JCO.2014.59.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A Sandoval N Das M Pillai R Chen L CCND1 mutations increase protein stability and promote ibrutinib resistance in mantle cell lymphoma. Oncotarget. 2016;7:73558. doi: 10.18632/oncotarget.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil M Callanan M Dascalescu C Sotto JJ Leroux D Molecular diagnosis of t(11;14) in mantle cell lymphoma using two-colour interphase fluorescence in situ hybridization. British Journal of Haematology. 1996;93:656. doi: 10.1046/j.1365-2141.1996.d01-1675.x. [DOI] [PubMed] [Google Scholar]
- Morin RD Johnson NA Severson TM Mungall AJ An J Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genetics. 2010;42:181. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier N Briere J Gisselbrecht C Emile JF Lederlin P Rituximab plus CHOP (R-CHOP) overcomes bcl-2-associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL) Blood. 2003;101:4279. doi: 10.1182/blood-2002-11-3442. [DOI] [PubMed] [Google Scholar]
- Ngo VN Young RM Schmitz R Jhavar S Xiao W Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hayre M Inoue A Kufareva I Wang Z Mikelis CM Inactivating mutations in GNA13 and RHOA in Burkitt’s lymphoma and diffuse large B-cell lymphoma: a tumor suppressor function for the Gα13/RhoA axis in B cells. Oncogene. 2016;35:3771. doi: 10.1038/onc.2015.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J Bödör C Wang J Araf S Yang CY Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nature Genetics. 2014;46:176. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L Migliazza A Basso K Houldsworth J Chaganti RSK Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101:2914. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- Pastore A Jurinovic V Kridel R Hoster E Staiger AM Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: a retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncology. 2015;16:1111. doi: 10.1016/S1470-2045(15)00169-2. [DOI] [PubMed] [Google Scholar]
- Piccaluga PP De Falco G Kustagi M Gazzola A Agostinelli C Gene expression analysis uncovers similarity and differences among Burkitt lymphoma subtypes. Blood. 2011;117:3596. doi: 10.1182/blood-2010-08-301556. [DOI] [PubMed] [Google Scholar]
- Portis T Dyck P Longnecker R Epstein-Barr virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood. 2003;102:4166. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- Quesada AE Medeiros LJ Desai PA Lin P Westin JR Increased MYC copy number is an independent prognostic factor in patients with diffuse large B-cell lymphoma. Modern Pathology. 2017;30:1688. doi: 10.1038/modpathol.2017.93. [DOI] [PubMed] [Google Scholar]
- Rossi D Diop F Spaccarotella E Monti S Zanni M Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017;129:1947. doi: 10.1182/blood-2016-05-719641. [DOI] [PubMed] [Google Scholar]
- Schaffner C Idler I Stilgenbauer S Döhner H Lichter P Mantle cell lymphoma is characterized by inactivation of the ATM gene. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2773. doi: 10.1073/pnas.050400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R Wright GW Huang DW Johnson CA Phelan JD Genetics and pathogenesis of diffuse large B-cell lymphoma. New England Journal of Medicine. 2018;378:1396. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R Young RM Ceribelli M Jhavar S Xiao W Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell R Staak O Borchmann P Schwartz C Matthey B A phase I study with an anti-CD30 ricin a-chain immunotoxin (ki-4.dgA) in patients with refractory CD30+ Hodgkin’s and non-Hodgkin’s lymphoma. Clinical Cancer Research. 2002;8:1779. [PubMed] [Google Scholar]
- Schuetz JM Johnson NA Morin RD Scott DW Tan K BCL2 mutations in diffuse large B-cell lymphoma. Leukemia. 2012;26:1383. doi: 10.1038/leu.2011.378. [DOI] [PubMed] [Google Scholar]
- Shanbhag S Ambinder RF Hodgkin lymphoma: A review and update on recent progress. CA: A Cancer Journal for Clinicians. 2018;68:116. doi: 10.3322/caac.21438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankland KR Armitage JO Hancock BW Non-Hodgkin lymphoma. Lancet. 2012;380:848. doi: 10.1016/S0140-6736(12)60605-9. [DOI] [PubMed] [Google Scholar]
- Shannon-Lowe C Rickinson AB Bell AI Epstein–Barr virus-associated lymphomas. Philosophical Transactions of the Royal Society B Biological Sciences. 2017;372:20160271–20160271. doi: 10.1098/rstb.2016.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert R Matthiesen P Harder S Zhang Y Borowski A Application of interphase fluorescence in situ Hybridization for the detection of the Burkitt translocation t(8;14)(q24;q32) in B-cell lymphomas. Blood. 1998;91:984. [PubMed] [Google Scholar]
- Slifka MK Antia R Whitmire JK Ahmed R Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- Spina V Bruscaggin A Cuccaro A Martini M Trani M Di Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131:2413. doi: 10.1182/blood-2017-11-812073. [DOI] [PubMed] [Google Scholar]
- Swerdlow SH Campo E Pileri SA Harris NL Stein H The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiacci E Ladewig E Schiavoni G Penson A Fortini E Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood. 2018;131:2454. doi: 10.1182/blood-2017-11-814913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y Finger LR Yunis J Nowell PC Croce CM Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- Vaandrager JW Schuuring E Raap T Philippo K Kleiverda K Interphase FISH detection of BCL2 rearrangement in follicular lymphoma using breakpoint-flanking probes. Genes Chromosomes Cancer. 2000;27:85. [PubMed] [Google Scholar]
- Van den Neste E André M Gastinne T Stamatoullas A Haioun C A phase II study of the oral JAK1/JAK2 inhibitor ruxolitinib in advanced relapsed/refractory Hodgkin lymphoma. Haematologica. 2018;103:840. doi: 10.3324/haematol.2017.180554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gijn J Gijselhart JP Thomas Hodgkin and his disease. Nederlands Tijdschrift voor Geneeskunde. 2012;156:A4332–A4332. [PubMed] [Google Scholar]
- Vivier E Tomasello E Baratin M Walzer T Ugolini S Functions of natural killer cells. Nature Immunology. 2008;9:503. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- Weniger MA Küppers R NF-κB deregulation in Hodgkin lymphoma. Seminars in Cancer Biology. 2016;39:32. doi: 10.1016/j.semcancer.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Weniger MA Melzner I Menz CK Wegener S Bucur AJ Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25:2679. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- Weniger MA Tiacci E Schneider S Arnolds J Rüschenbaum S Human CD30+ B cells represent a unique subset related to Hodgkin lymphoma cells. Journal of Clinical Investigation. 2018;128:2996. doi: 10.1172/JCI95993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiestner A Tehrani M Chiorazzi M Wright G Gibellini F Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood. 2007;109:4599. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis TG Dyer MJ The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96:808. [PubMed] [Google Scholar]
- Wilson WH Young RM Schmitz R Yang Y Pittaluga S Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nature Medicine. 2015;21:922. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska I Nooyen P Maes B Martin-Subero JI Siebert R Frequent occurrence of BCL6 rearrangements in nodular lymphocyte predominance Hodgkin lymphoma but not in classical Hodgkin lymphoma. Blood. 2003;101:706. doi: 10.1182/blood-2002-05-1592. [DOI] [PubMed] [Google Scholar]
- Wu C de Miranda NF Chen L Wasik AM Mansouri L Genetic heterogeneity in primary and relapsed mantle cell lymphomas: impact of recurrent CARD11 mutations. Oncotarget. 2016;7:38180. doi: 10.18632/oncotarget.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P Liu X Ouyang J Chen B TP53 mutation predicts the poor prognosis of non-Hodgkin lymphomas: evidence from a meta-analysis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0174809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB Chu J Berg T Schapira M Cheng SWG Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan WX Gui YX Na WN Chao J Yang X Circulating microRNA-125b and microRNA-130a expression profiles predict chemoresistance to R-CHOP in diffuse large B-cell lymphoma patients. Oncology Letters. 2016;11:423. doi: 10.3892/ol.2015.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]