Abstract
Background
Lung cancer is the leading cause of cancer‐related mortality worldwide. Studies have demonstrated that long noncoding RNA nicotinamide nucleotide transhydrogenase‐antisense RNA1 (NNT‐AS1) functioned as an oncogene in most malignancies, including non‐small cell lung cancer (NSCLC). This study aimed to investigate the underlying mechanisms of NNT‐AS1 in NSCLC progression.
Methods
The levels of NNT‐AS1, miR‐22‐3p and Yes‐associated protein (YAP1) were detected by qRT‐PCR in NSCLC tissues and cells. Kaplan‐Meier analysis was conducted to analyze the correlation between NNT‐AS1 expression and overall survival of NSCLC patients. Cell proliferation was evaluated by MTT assay. Cell migration and invasion were assessed using transwell assay. The protein levels of YAP1 and EMT‐related proteins were detected by western blot. The molecular mechanism was predicted by starBase2.0 and validated by dual‐luciferase reporter assay or RNA pull‐down assay. Xenograft analysis was carried out to analyze tumor growth in vivo.
Results
We found that the levels of NNT‐AS1 and YAP1 were enhanced, while miR‐22‐3p expression was decreased in NSCLC tissues and cells. High NNT‐AS1 expression was correlated with poor prognosis. NNT‐AS1 knockdown impeded proliferation, migration, invasion and EMT of NSCLC cells. NNT‐AS1 targeted miR‐22‐3p, and YAP1 was a target of miR‐22‐3p in NSCLC cells. Furthermore, NNT‐AS1 facilitated the progression of NSCLC by regulating miR‐22‐3p/YAP1 axis. NNT‐AS1 knockdown repressed tumor growth in vivo.
Conclusion
NNT‐AS1 facilitated proliferation, migration, invasion and EMT of NSCLC cells by sponging miR‐22‐3p and regulating YAP1 expression, which might provide a potential biomarker and therapeutic target for NSCLC.
Keywords: miR‐22‐3p, NNT‐AS1, non‐small cell lung cancer, progression, YAP1
Introduction
Lung cancer is the main origin of cancer mortality worldwide, and non‐small cell lung cancer (NSCLC) accounts for about 85% of lung cancer.1, 2 Despite great achievements in diagnosis and therapy, the five‐year overall survival rate for NSCLC patients is lesser than 16%.3 Therefore, exploring new molecular mechanisms of NSCLC is helpful for NSCLC treatment.
Long noncoding RNAs (lncRNAs) are highly conserved RNAs over 200 nucleotides in length.4 Accumulating evidence has suggested that lncRNAs occupy a vital position in various cancers, including NSCLC.5 For example, Hu et al. disclosed that LINC01296 triggers cell migration by competitively combining with miR‐5095 in NSCLC.6 Zhang et al. found that lncRNA PICART1 inhibits NSCLC progression through regulating AKT1 pathway.7 LncRNA nicotinamide nucleotide transhydrogenase‐antisense RNA1 (NNT‐AS1) is an oncogene in various cancers, such as cervical cancer,8 gastric cancer,9 and osteosarcoma.10 Li et al. revealed that NNT‐AS1 induces epithelial‐to‐mesenchymal transition (EMT) process in breast cancer by sponging miR‐142‐3p and upregulating ZEB1.11 Downregulation of NNT‐AS1 inhibits NSCLC progression via regulating NNT‐AS1/miR‐129‐5p axis.12 Nevertheless, the mechanism of NNT‐AS1 in NSCLC requires further investigation.
MicroRNAs (miRNAs) are a class of short noncoding RNAs composed of 19–24 nucleotides.13 Emerging evidence has certified that lncRNAs play a vital regulatory role in various biological processes by acting as miRNA sponges or competing endogenous RNAs (ceRNAs).14, 15 Besides, mature miRNAs have been reported to impede the expression of targets by targeting them.16 MicroRNAs occupy a crucial position in various biological processes, including tumor metastasis.17 Recent studies have shown that many miRNAs, such as microRNA‐612 and miRNA‐621, function as tumor suppressors in NSCLC by regulating target genes.18, 19 A previous study found that circFGFR3 facilitates NSCLC progression via targeting miR‐22‐3p.20 However, the relationship between NNT‐AS1 and miR‐22‐3p has not been studied.
In addition, Yes‐associated protein (YAP1) is the main effector of the Hippo pathway and contributes to the development of many cancers.21 EMT exerts a crucial role in tumor metastasis.22 Activation of the extracellular signal‐regulated kinase (ERK) signaling pathway contributes to EMT in lung cancer.23 YAP1 has been reported to be associated with EMT program in various cancers.24 YAP1 is conspicuously enhanced and promoted EMT by inducing Slug transcription in NSCLC.25 Therefore, we hypothesized that miR‐22‐3p and YAP1 may be involved in NNT‐AS1‐induced NSCLC progression. In our research, we investigated NNT‐AS1 expression in NSCLC and assessed the effect on NSCLC progression. In addition, we explored the relationship between NNT‐AS1, miR‐22‐3p and YAP1, which might be a biomarker for NSCLC therapy.
Methods
Specimen collection
A total of 37 NSCLC samples and adjacent normal tissues were collected from the Second Xiangya Hospital of Central South University. The study was ratified by the Ethics Committee of the Second Xiangya Hospital of Central South University. Written informed consents were signed by all participants.
Cell culture
Three NSCLC cell lines H1650, A549 and H1299, and human lung epithelial cells BEAS‐2B were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). NSCLC cell line PC‐9 was obtained from Tongpai (Shanghai, China). Cells were incubated in Dulbecco's Modified Eagle Medium (DMEM; Seebio, Shanghai, China) supplemented with 10% fetal bovine serum (Seebio) at 37°C with 5% CO2.
Cell transfection
Short hairpin RNA (shRNA) against NNT‐AS1 (sh‐NNT‐AS1), shRNA targeting YAP1 (sh‐YAP1), the negative control shRNA (sh‐NC), NNT‐AS1 overexpression vector (NNT‐AS1), YAP1 overexpression vector (YAP1) and the empty overexpression vector (pcDNA) were obtained from GenePharma (Shanghai, China). MiR‐22‐3p mimic (miR‐22‐3p) and mimic control (miR‐NC) were obtained from RiboBio (Guangzhou, China). Cell transfection was performed by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative real‐time PCR
RNA extraction was carried out using Trizol (15596026, Invitrogen). cDNA was then synthesized by High‐Capacity cDNA Reverse Transcription Kits (1725037, Bio‐Rad, Hercules, CA, USA) or Reverse Transcription Kit (4366596, Thermo Fisher Scientific, Waltham, MA, USA). The expression was detected using SYBR Green Mixture (DRR041A, Takara, Dalian, China). Primers were presented below: NNT‐AS1 (F 5′‐ACGTGCAGACAACATCTACCT‐3′; R 5′‐TACAACACCTTCCCGCAT‐3′), miR‐22‐3p (F 5′‐AAGCTGCCAGTTGAAGAACTGTA‐3′; R 5′‐GCTGTCAACGATACGCTACGTAAC‐3′), YAP1 (F 5′‐TAGCCCTGCGTAGCCAGTTA‐3′; R 5′‐TCATGCTTAGTCCACTGTCTGT‐3′), GAPDH (F 5′‐AATGGGCAGCCGTTAGGAAA‐3′; R 5′‐GCGCCCAATACGACCAAATC‐3′), U6 (F 5′‐CTCGCTTCGGCAGCACA‐3′; R 5′‐AACGCTTCACGAATTTGCGT‐3′). GAPDH or U6 was taken as internal control.
MTT assay
Cells were injected into 96‐well plates, and MTT solution (M1025, Solarbio, Beijing, China) was added into each well after incubation for 24 hours, 48 hours, 72 hours and 96 hours. After incubation for another four hours, dimethyl sulfoxide (DMSO; Solarbio) was added into each well. The absorbance was monitored by Microplate Reader (Bio‐Rad, Hercules, CA, USA) at 490 nm.
Transwell assay
For cell invasion assay, the upper chamber was supplemented with Matrigel (354234, Corning, Corning, NJ, USA), while the upper chamber without Matrigel coating was used for the migration assay. First, cells were resuspended in serum‐free medium and placed into the upper chamber. Complete medium in the lower chamber was considered as a chemical attractant. After incubating for 48 hours, the migrated or invaded cells attached to the lower surface were fixed with formaldehyde and stained with crystal violet, and then photographed and counted under a microscope.
Western blot assay
RIPA buffer (R0010, Solarbio) was used to extract total protein. Protein samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) and then transferred onto polyvinylidene fluoride membranes (Millipore, Bradford, MA, USA). The membranes were blocked by 5% defatted milk for two hours at room temperature and then incubated with primary antibodies against E‐cadherin (1:1000; ab15148, Abcam, Cambridge, UK), N‐cadherin (1:1000; ab76057, Abcam), Vimentin (1:1000; ab137321, Abcam), YAP1 (1:1000; ab81183, Abcam) or GAPDH (1:500; ab9485, Abcam) at 4°C overnight. Next, the membranes were incubated with HRP‐labeled secondary antibody (1:4000; ab7090, Abcam) at room temperature for two hours. The chemiluminescence intensity was detected by ECL kit (32109, Pierce, Rockford, IL, USA).
Dual‐luciferase reporter assay
The sequence of NNT‐AS1 containing the putative binding site (WT‐NNT‐AS1) or mutant (MUT‐NNT‐AS1) of miR‐22‐3p was inserted into pGL3 luciferase reporter plasmids (Promega, Madison, WI, USA). Similarly, the fragments of wild‐type (YAP1‐3'UTR‐WT) or mutant YAP1 3' UTR (YAP1‐3'UTR‐MUT) was cloned into pGL3 plasmids. Then, miR‐22‐3p or miR‐NC was cotransfected with the corresponding vectors into H1299 and A549 cells using Lipofectamine 2000 (Invitrogen). After transfection for 48 hours, Dual‐Lucy Assay Kit (D0010, Solarbio) was carried out to monitor the luciferase activity.
RNA pull‐down assay
H1299 and A549 cells were transfected with biotinylated miR‐22‐3p (Bio‐miR‐22‐3p) or the control (Bio‐miR‐NC). After transfection for 48 hours, the cells were lysed by lysis buffer. The cell lysates were incubated with Dynabeads M‐280 Streptavidin (Thermo Fisher Scientific) at 37°C for two hours. The enrichment of NNT‐AS1 was examined by qRT‐PCR.
Xenograft tumor experiment
BALB/c nude male mice (5‐week‐old) were maintained under pathogen‐free conditions and randomized into two groups (n = 5 per group). A549 cells infected with lentivirus harboring sh‐NC or sh‐NNT‐AS1 were subcutaneously injected into the back of nude mice. Tumor volume was estimated every seven days. Tumors were removed after 35 days, and tumor weight was measured. Tumor tissues were snap‐frozen for RNA extraction. The protein levels were examined using western blot assay. The xenograft analysis was authorized by the Animal Ethics Committee of the Second Xiangya Hospital of Central South University.
Statistical analysis
Data were tested at least three times independently and represented as mean ± standard deviation. The correlation was evaluated using Spearman's correlation method. Graphpad Prism 7.0 software (GraphPad, San Diego, CA, USA) was applied for all data. Student's t‐test or one‐way ANOVA was performed to assess the differences. P < 0.05 was identified as statistically significant.
Results
LncRNA NNT‐AS1 is upregulated in NSCLC and associated with poor prognosis
First, we detected the expression of NNT‐AS1 in NSCLC tissues and cells using qRT‐PCR. The results revealed that NNT‐AS1 expression in NSCLC tissues was overtly higher than that in adjacent normal tissues (Fig 1a and b). Furthermore, Kaplan‐Meier survival analysis and log‐rank test exhibited that higher NNT‐AS1 expression is significantly associated with overall survival of NSCLC patients compared with lower NNT‐AS1 expression (Fig 1c). Additionally, NNT‐AS1 expression in NSCLC cell lines was detected using qRT‐PCR, and the results suggested that NNT‐AS1 expression in NSCLC cells (H1650, PC‐9, A549 and H1299) was dramatically increased in comparison with human lung epithelial cells BEAS‐2B (Fig 1d). From these data, we speculated that NNT‐AS1 might play roles in NSCLC tumorigenesis and progression.
Figure 1.
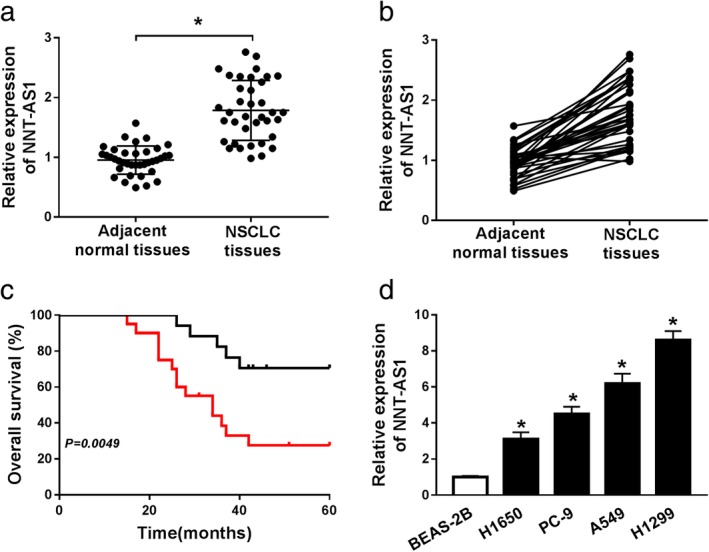
LncRNA NNT‐AS1 is upregulated in NSCLC and associated with poor prognosis. (a and b) The expression of NNT‐AS1 was examined in 37 paired NSCLC tissues and adjacent normal tissues by qRT‐PCR. (c) Kaplan‐Meier survival analysis was conducted to detect the correlation between NNT‐AS1 expression and overall survival of NSCLC patients. (d) The expression of NNT‐AS1 was measured in normal lung epithelial cell line (BEAS‐2B) and NSCLC cell lines (H1650, PC‐9, A549 and H1299) by qRT‐PCR. *P < 0.05.  sh‐NC,
sh‐NC,  sh‐NNT‐AS1.
sh‐NNT‐AS1.
Knockdown of NNT‐AS1 represses proliferation, migration, invasion and EMT of NSCLC cells
To investigate the effects of NNT‐AS1 on the progression of NSCLC, H1299 and A549 cells were transfected with sh‐NC or sh‐NNT‐AS1. The results of qRT‐PCR showed that NNT‐AS1 expression was dramatically reduced in the sh‐NNT‐AS1 group compared to the sh‐NC group (Fig 2a and b). MTT assay revealed that cell viability was distinctly inhibited in H1299 and A549 cells transfected with sh‐NNT‐AS1 compared to the sh‐NC group (Fig 2c and d). Transwell assay showed that cell migration and invasion were remarkably suppressed in H1299 and A549 cells transfected with sh‐NNT‐AS1 compared with the sh‐NC group (Fig 2e‐h). In addition, EMT‐related proteins were measured using western blot assay, and the results showed that NNT‐AS1 knockdown distinctly increased the level of epithelial marker E‐cadherin, and obviously decreased the levels of mesenchymal markers (N‐cadherin and Vimentin) (Fig 2i and j). All these data demonstrated that NNT‐AS1 depletion hinders NSCLC progression.
Figure 2.
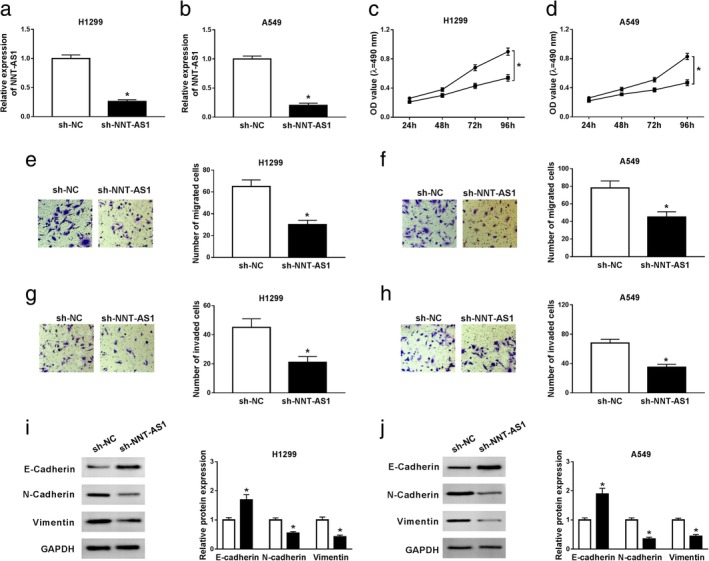
Knockdown of NNT‐AS1 represses proliferation, migration, invasion and EMT of NSCLC cells. (a–j) H1299 and A549 cells were transfected with sh‐NC or sh‐NNT‐AS1. (a and b) NNT‐AS1 expression was examined in transfected cells by qRT‐PCR. (c and d) MTT assay was performed to evaluate cell viability. (e–h) Transwell assay was carried out to detect migration and invasion of NSCLC cells. (i and j) The protein levels of E‐cadherin, N‐cadherin and Vimentin were detected using western blot assay. *P < 0.05.  sh‐NC,
sh‐NC,  sh‐NNT‐AS1.
sh‐NNT‐AS1.
LncRNA NNT‐AS1 directly binds to miR‐22‐3p in NSCLC cells
First, starBase2.0 predicted the targets of NNT‐AS1, and miR‐22‐3p was selected as the research target. Prediction showed that NNT‐AS1 had putative binding sites with miR‐22‐3p (Fig 3a). To validate whether NNT‐AS1 targeted miR‐22‐3p, dual‐luciferase reporter assay was performed. The results suggested that miR‐22‐3p mimic significantly decreased the luciferase activity of WT‐NNT‐AS1 in H1299 and A549 cells, but did not affect the luciferase activity of MUT‐NNT‐AS1 (Fig 3b and c). In addition, RNA pull‐down assay confirmed that NNT‐AS1 was strikingly enriched in H1299 and A549 cells transfected with Bio‐miR‐22‐3p (Fig 3d and e). Furthermore, NNT‐AS1 expression was effectively elevated in the NNT‐AS1 group compared with the pcDNA group (Fig 3f and g). Transfection with sh‐NNT‐AS1 prominently upregulated miR‐22‐3p expression, whereas NNT‐AS1 overexpression obviously reduced miR‐22‐3p expression in H1299 and A549 cells (Fig 3h and i). Additionally, miR‐22‐3p expression was notably decreased in NSCLC tissues and cells (Fig 3j‐l). Finally, NNT‐AS1 expression was negatively correlated with miR‐22‐3p expression in NSCLC tissues (Fig 3k). These data indicated that NNT‐AS1 negatively targets miR‐22‐3p in NSCLC cells.
Figure 3.
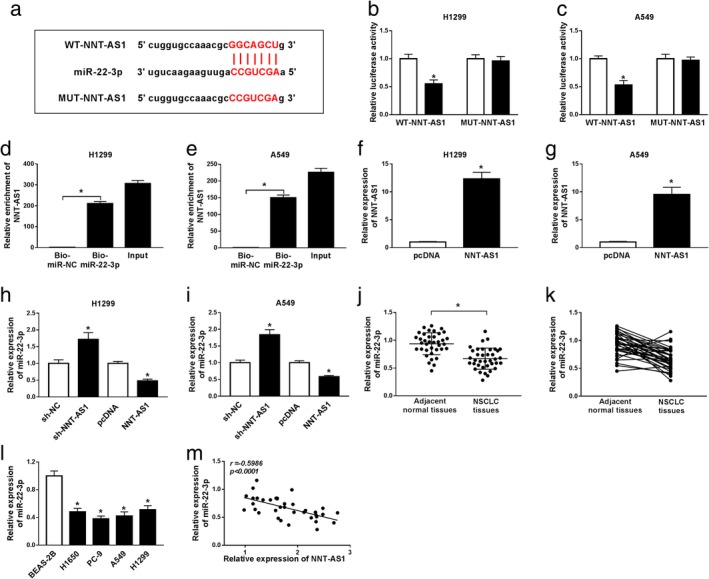
LncRNA NNT‐AS1 directly binds to miR‐22‐3p in NSCLC cells. (a) The putative binding sites of NNT‐AS1 and miR‐22‐3p were shown. (b and c) Dual‐luciferase reporter assay was conducted to detect the luciferase activity in H1299 and A549 cells cotransfected with WT‐NNT‐AS1 or MUT‐NNT‐AS1 and miR‐22‐3p mimic (miR‐22‐3p) or miR‐NC. (d and e) RNA pull‐down assay was performed to validate NNT‐AS1 targeted miR‐22‐3p. (f and g) The overexpression efficiency of NNT‐AS1 was measured in H1299 and A549 cells by qRT‐PCR analysis. (h and i) The expression of miR‐22‐3p was measured in H1299 and A549 cells transfected with sh‐NC, sh‐NNT‐AS1, pcDNA or NNT‐AS1. (j and k) The expression of miR‐22‐3p was detected in 37 paired NSCLC tissues and adjacent normal tissues using qRT‐PCR. (l) The expression of miR‐22‐3p was examined in BEAS‐2B cells and NSCLC cell lines (H1650, PC‐9, A549 and H1299) by qRT‐PCR. (m) The correlation between NNT‐AS1 and miR‐22‐3p in NSCLC tissues was analyzed. *P < 0.05.  miR‐NC,
miR‐NC,  miR‐22‐3p.
miR‐22‐3p.
MiR‐22‐3p reverses the promotion of NNT‐AS1 on the progression of NSCLC
To investigate the role of miR‐22‐3p in NSCLC progression, H1299 and A549 cells were transfected with pcDNA, NNT‐AS1, NNT‐AS1+miR‐NC or NNT‐AS1+miR‐22‐3p. The results exhibited that transfection of NNT‐AS1 markedly reduced miR‐22‐3p expression, while miR‐22‐3p expression was restored after transfection with miR‐22‐3p (Fig 4a and b). Moreover, NNT‐AS1 overexpression strikingly promoted cell viability, while miR‐22‐3p mimic abrogated the effects (Fig 4c and d). Transwell assay suggested that miR‐22‐3p mimic abrogated the promoting effcet of NNT‐AS1 overexpression on NSCLC cell migration and invasion (Fig 4e‐h). Furthermore, overexpression of NNT‐AS1 resulted in an obvious decrease in E‐cadherin levels and a distinct increase in N‐cadherin and Vimentin levels, while miR‐22‐3p mimic abolished the effects (Fig 4i and j). All these data indicated that NNT‐AS1 promotes the progression of NSCLC by regulating miR‐22‐3p.
Figure 4.
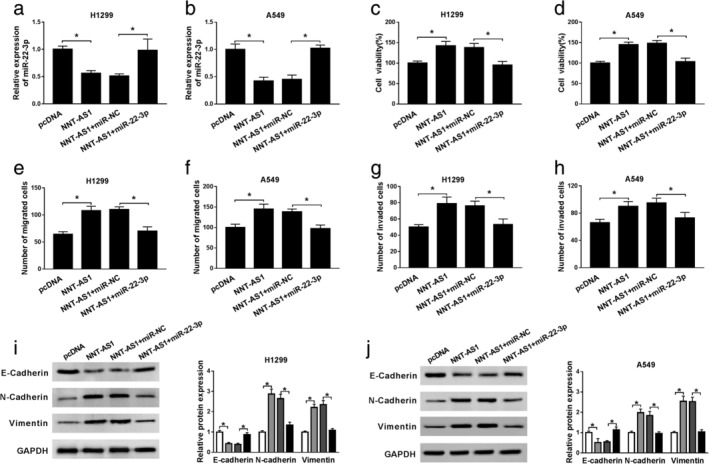
MiR‐22‐3p reverses the promotion of NNT‐AS1 on NSCLC progression. (a–h) H1299 and A549 cells were transfected with pcDNA, NNT‐AS1, NNT‐AS1+miR‐NC or NNT‐AS1+miR‐22‐3p. (a and b) The level of miR‐22‐3p was detected by qRT‐PCR. (c and d) Cell proliferation was evaluated by MTT assay. (e–h) The migrated and invaded cells were detected using transwell assay. (i and j) The protein levels of E‐cadherin, N‐cadherin and Vimentin were examined by western blot assay. *P < 0.05.  pcDNA,
pcDNA,  NNT‐AS1,
NNT‐AS1,  NNT‐AS1+miR‐NC,
NNT‐AS1+miR‐NC,  NNT‐AS1+miR‐22‐3p.
NNT‐AS1+miR‐22‐3p.
YAP1 is a target of miR‐22‐3p in NSCLC cells
StarBase v2.0 predicted that miR‐22‐3p might bind to YAP1 3'UTR (Fig 5a). Dual‐luciferase reporter assay showed that mature miR‐22‐3p notably decreased the luciferase activity in H1299 and A549 cells transfected with YAP1‐3'UTR‐WT, whereas did not affect the luciferase activity of YAP1‐3'UTR‐MUT (Fig 5b and c). In addition, the overexpression efficiency was confirmed by qRT‐PCR, indicating that miR‐22‐3p was evidently upregulated in H1299 and A549 cells transfected with miR‐22‐3p mimic (Fig 5d). The results of qRT‐PCR and western blot assay then showed that the mRNA and protein levels of YAP1 were significantly reduced in the miR‐22‐3p group compared with the miR‐NC group (Fig 5e and f). In addition, the mRNA and protein levels of YAP1 were remarkably increased in NSCLC tissues compared with adjacent normal tissues (Fig 5g–i). Meanwhile, the levels of miR‐22‐3p and YAP1 were negatively correlated in NSCLC tissues (Fig 5j). The mRNA and protein levels of YAP1 were strikingly higher in NSCLC cells than that in BEAS‐2B cells (Fig 5k and l). These data indicated that YAP1 is a target of miR‐22‐3p in NSCLC cells.
Figure 5.
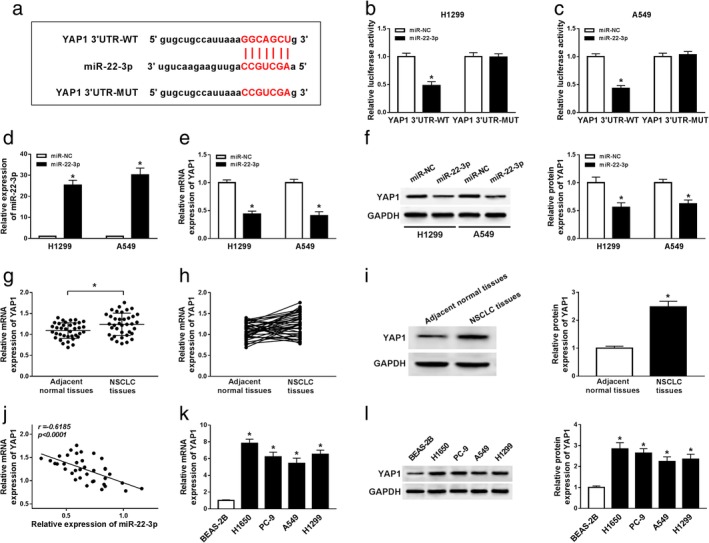
YAP1 is a target of miR‐22‐3p in NSCLC cells. (a) The putative binding sites of miR‐22‐3p and YAP1 3'UTR were exhibited. (b and c) Luciferase activity was detected in H1299 and A549 cells cotransfected with YAP1‐3'UTR‐WT or YAP1‐3'UTR‐MUT and miR‐22‐3p or miR‐NC. (d) The level of miR‐22‐3p was measured in H1299 and A549 cells transfected with miR‐NC or miR‐22‐3p by qRT‐PCR. (e and f) The levels of YAP1 were measured in H1299 and A549 cells transfected with miR‐NC or miR‐22‐3p by qRT‐PCR or western blot assay. (g–i) The mRNA and protein levels of YAP1 were detected in NSCLC tissues and adjacent normal tissues. (j) The correlation between miR‐22‐3p and YAP1 in NSCLC tissues was identified. (k and l) The levels of YAP1 were examined in normal lung epithelial cell line (BEAS‐2B) and NSCLC cell lines (H1650, PC‐9, A549 and H1299) by qRT‐PCR or western blot assay. *P < 0.05.  miR‐NC,
miR‐NC,  miR‐22‐3p.
miR‐22‐3p.
YAP1 reverses the inhibitory effects of miR‐22‐3p on the progression of NSCLC
Firstly, transfection with sh‐YAP1 markedly downregulated the protein level of YAP1, and YAP1 overexpression greatly upregulated the protein level of YAP1 (Fig 6a and b). To explore the role of YAP1 in NSCLC progression, H1299 and A549 cells were transfected with miR‐NC, miR‐22‐3p, miR‐22‐3p+pcDNA or miR‐22‐3p+YAP1. MTT assay revealed that YAP1 overexpression abrogated the inhibitory effects of miR‐22‐3p mimic on cell viability in NSCLC cells (Fig 6c and d). Transwell assay showed that miR‐22‐3p mimic suppressed cell migration and invasion, and the inhibition was restored after overexpression of YAP1 (Fig 6e‐h). Western blot assay suggested that miR‐22‐3p mimic hindered EMT of NSCLC cells, whereas YAP1 overexpression abolished the effects (Fig 6i and j). All these data reflected that miR‐22‐3p inhibits NSCLC progression by regulating YAP1.
Figure 6.
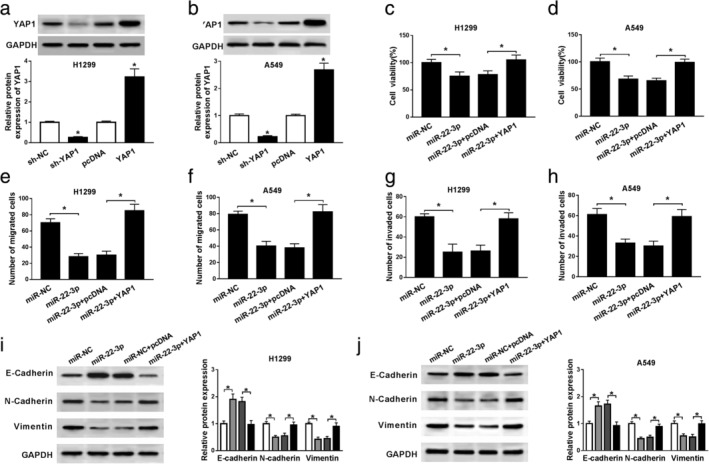
YAP1 reverses the inhibitory effects of miR‐22‐3p on NSCLC progression. (a and b) The protein level of YAP1 was detected in H1299 and A549 cells transfected with sh‐NC, sh‐YAP1, pcDNA or YAP1. (c–j) H1299 and A549 cells were transfected with miR‐NC, miR‐22‐3p, miR‐22‐3p+pcDNA or miR‐22‐3p+YAP1. (c and d) Cell proliferation was estimated by MTT assay. (e–h) The migrated and invaded cells were examined by transwell assay. (i and j) The levels of EMT‐related proteins were detected by western blot assay. *P < 0.05. miR‐NC,
miR‐NC,  miR‐22‐3p,
miR‐22‐3p,  miR‐22‐3p+pcDNA,
miR‐22‐3p+pcDNA,  miR‐22‐3p+YAP1
miR‐22‐3p+YAP1
YAP1 knockdown abrogates the promotion effects of NNT‐AS1 overexpression on NSCLC progression
To investigate the effects of NNT‐AS1 and miR‐22‐3p on YAP1 expression, H1299 and A549 cells were transfected with miR‐NC, miR‐22‐3p, miR‐22‐3p + pcDNA or miR‐22‐3p + NNT‐AS1. Western blot assay showed that miR‐22‐3p mimic restrained YAP1 expression, and NNT‐AS1 overexpression alleviated the inhibitory effects of miR‐22‐3p on YAP1 expression (Fig 7a and b). In addition, H1299 and A549 cells were transfected with pcDNA, NNT‐AS1, NNT‐AS1+sh‐NC or NNT‐AS1+sh‐YAP1 to investigate the effects of NNT‐AS1 and YAP1 on NSCLC progression. MTT assay showed that suppression of YAP1 reversed the promotion of cell viability induced by NNT‐AS1 overexpression (Fig 7c and d). Transwell assay suggested cell migration and invasion were apparently enhanced by overexpression of NNT‐AS1, which were counteracted after transfection with sh‐YAP1 (Fig 7e‐h). Western blot assay showed that interference of YAP1 undermined the promotion effect of NNT‐AS1 overexpression on EMT of NSCLC cells (Fig 7i and j). Furthermore, overexpression of NNT‐AS1 promoted phosphorylation of ERK in A549 and H460 cells, and the effect was overturned after transfection with sh‐YAP1 (Fig 7i and j). These data reflected that NNT‐AS1 facilitates NSCLC progression by modulating YAP1 and YAP1‐mediated ERK pathway.
Figure 7.
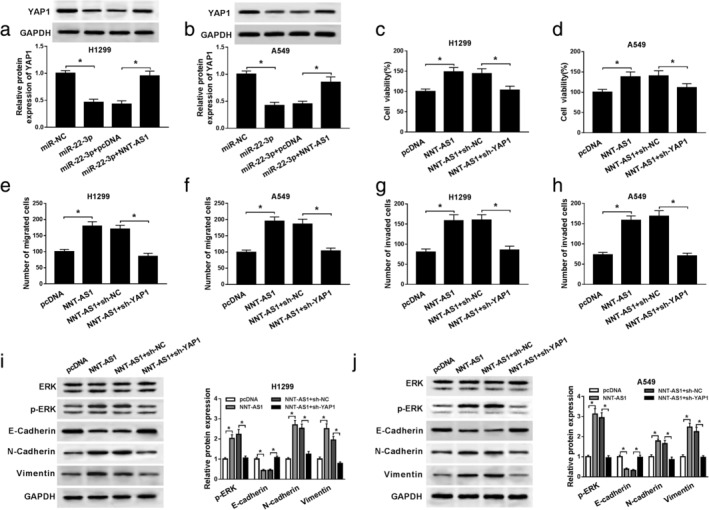
YAP1 knockdown abrogates the promotion effects of NNT‐AS1 overexpression on NSCLC progression. (a and b) The protein level of YAP1 was detected in H1299 and A549 cells transfected with miR‐NC, miR‐22‐3p, miR‐22‐3p+pcDNA or miR‐22‐3p+NNT‐AS1. (c–j) H1299 and A549 cells were transfected with pcDNA, NNT‐AS1, NNT‐AS1+sh‐NC or NNT‐AS1+sh‐YAP1. (c and d) Cell viability was detected by MTT assay. (e–h) The migrated and invaded cells were measured by transwell assay. (i and j) The protein levels of p‐ERK and EMT‐related proteins were detected by western blot assay. *P < 0.05.  pcDNA,
pcDNA,  NNT‐AS1,
NNT‐AS1,  NNT‐AS1+sh‐NC,
NNT‐AS1+sh‐NC,  NNT‐AS1+sh‐YAP1.
NNT‐AS1+sh‐YAP1.
Depletion of NNT‐AS1 blocks tumor growth in vivo
To explore the effect of NNT‐AS1 on tumor growth in vivo, A549 cells were transfected with sh‐NC or sh‐NNT‐AS1 and subcutaneously injected into the back of nude mice. The results revealed that tumor volume in the sh‐NNT‐AS1 group was strikingly decreased compared with the sh‐NC group (Fig 8a). Tumor weight in the sh‐NNT‐AS1 group was distinctly lower than that in sh‐NC group (Fig 8b). After the mice were sacrificed, the levels of NNT‐AS1, miR‐22‐3p, YAP1 and EMT‐related proteins were examined by qRT‐PCR or western blot assay. The results showed that the levels of NNT‐AS1 and YAP1 were overtly reduced, and the level of miR‐22‐3p was clearly increased in the sh‐NNT‐AS1 group compared with the sh‐NC group (Fig 8c and d). Moreover, interference of NNT‐AS1 markedly inhibited EMT of NSCLC cells (Fig 8d). Thus, the xenograft assay indicated that NNT‐AS1 knockdown suppresses tumor growth in vivo.
Figure 8.
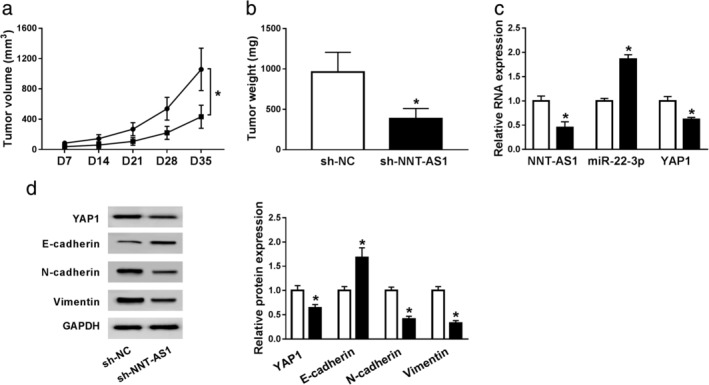
Depletion of NNT‐AS1 blocks tumor growth in vivo. (a–d) A549 cells were transfected with sh‐NC or sh‐NNT‐AS1. (a) Tumor volume was measured every seven days. (b) Tumor weight was measured after mice were sacrificed. (c) The levels of NNT‐AS1, miR‐22‐3p and YAP1 were examined using qRT‐PCR analysis. (d) The protein levels of YAP1 and EMT‐related proteins were detected by western blot assay. *P < 0.05.  sh‐NC,
sh‐NC,  sh‐NNT‐AS1,
sh‐NNT‐AS1,  sh‐NC,
sh‐NC,  sh‐NNT‐AS1.
sh‐NNT‐AS1.
Discussion
Recent studies have suggested that lncRNAs regulate the biological processes in lung cancer and act as potential therapeutic targets for NSCLC.5 LncRNA NNT‐AS1 serves as tumor promotor in various cancers. For example, higher NNT‐AS1 expression was strongly associated with poor prognosis, and NNT‐AS1 knockdown restrained osteosarcoma progression and blocked tumor growth.26 NNT‐AS1 functioned as an oncogene in gastric cancer progression via sponging miR‐363.9 NNT‐AS1 facilitated proliferation, metastasis and EMT of breast cancer cells via targeting miR‐142‐3p and upregulating ZEB1 expression.11 NNT‐AS1 silencing suppressed drug‐resistant cell proliferation and induced apoptosis via MAPK/Slug pathway.27 Additionally, Shen et al. corroborated that higher NNT‐AS1 expression was closely related to poor prognosis in patients with NSCLC, and NNT‐AS1 facilitated NSCLC progression by inhibiting miR‐129‐5p.12 In the current study, lncRNA NNT‐AS1 was significantly elevated in NSCLC. Furthermore, aberrantly expressed NNT‐AS1 could regulate NSCLC progression in H1299 and A549 cells.
Accumulating evidence indicated that lncRNAs could sponge miRNAs and regulate target genes via acting as ceRNAs.28 Besides, studies have demonstrated the mutual inhibition between lncRNA and miRNA.29, 30 In this study, we mainly analyzed the possible NNT‐AS1‐mediated ceRNA pathway. We first confirmed that NNT‐AS1 bound to miR‐22‐3p. In addition, recent studies manifested that miR‐22‐3p was a tumor inhibitor in various cancers. For instance, Chen et al. found that berberine suppressed cell proliferation via upregulating miR‐22‐3p expression in hepatocellular carcinoma.31 Sha et al. showed that LINC00858 induced colorectal cancer development via acting as a ceRNA of miR‐22‐3p.32 In lung adenocarcinoma, lncRNA DGCR5 contributed to cancer progression through sponging miR‐22‐3p.33 A recent study suggested that miR‐22 suppressed lung cancer cell EMT and invasion via targeting Snail.34 Ling et al. found that miR‐22 inhibited cell progression by regulating ErbB3 expression.35 However, the role of miR‐22‐3p in NSCLC remains unclear. Consistent with the above research, miR‐22‐3p was low expressed in NSCLC. Further, we demonstrated that NNT‐AS1 contributed to NSCLC progression by sponging miR‐22‐3p.
Some previous studies have demonstrated that yes‐associated protein (YAP1) acted as an oncogene in most malignancies. Zhang et al. found that miR‐345 impeded cell migration and invasion in NSCLC through binding to YAP1.36 Previous research demonstrated that miR‐497 suppressed NSCLC cell proliferation via targeting YAP1.37 Pu et al. found that COPB2 overexpression contributed to cell proliferation, tumorigenesis and restrained cell apoptosis by translocating YAP1 from cytoplasm to nuclear in lung adenocarcinoma.38 In the current study, miR‐22‐3p hindered NSCLC progression by regulating YAP1. Moreover, NNT‐AS1 expedited the progression of NSCLC by modulating YAP1, and YAP1‐mediated ERK pathway regulated EMT in NSCLC cells.
In conclusion, this study indicated that NNT‐AS1 facilitated proliferation, migration, invasion and EMT of NSCLC cells through sponging miR‐22‐3p and elevating YAP1 expression. Therefore, the discovery of the NNT‐AS1/miR‐22‐3p/YAP1 axis provides a potential therapeutic target for lung cancer treatment.
Disclosure
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
References
- 1. Kaderbhai C, Tharin Z, Ghiringhelli F. The role of molecular profiling to predict the response to immune checkpoint inhibitors in lung cancer. Cancers (Basel) 2019; 11 10.3390/cancers11020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Radovic M, Kanesvaran R, Rittmeyer A et al Multidisciplinary treatment of lung cancer in older patients: A review. J Geriatr Oncol 2019; 10: 405–10. [DOI] [PubMed] [Google Scholar]
- 3. Testa U, Castelli G, Pelosi E. Lung cancers: Molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel) 2018; 10 10.3390/cancers10080248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non‐coding rnas in cancer. Trends Mol Med 2018; 24: 257–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential clinical application of lncRNAs in non‐small cell lung cancer. Onco Targets Ther 2018; 11: 8045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu X, Duan L, Liu H, Zhang L. Long noncoding RNA LINC01296 induces non‐small cell lung cancer growth and progression through sponging miR‐5095. Am J Transl Res 2019; 11: 895–903. [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang C, Su C, Song Q, Dong F, Yu S, Huo J. LncRNA PICART1 suppressed non‐small cell lung cancer cells proliferation and invasion by targeting AKT1 signaling pathway. Am J Transl Res 2018; 10: 4193–201. [PMC free article] [PubMed] [Google Scholar]
- 8. Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non‐coding RNA NNT‐AS1 promotes cell proliferation and invasion through Wnt/beta‐catenin signaling pathway in cervical cancer. Biomed Pharmacother 2017; 92: 1128–34. [DOI] [PubMed] [Google Scholar]
- 9. Wang X, Ren M, Li Y et al Long noncoding RNA NNT‐AS1 promotes gastric cancer proliferation and invasion by regulating microRNA‐363 expression. J Cell Biochem 2019; 120: 5704–12. [DOI] [PubMed] [Google Scholar]
- 10. Li C, Zhang S, Qiu T, Wang Y, Ricketts DM, Qi C. Upregulation of long non‐coding RNA NNT‐AS1 promotes osteosarcoma progression by inhibiting the tumor suppressive miR‐320a. Cancer Biol Ther 2019; 20: 413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M. Long non‐coding RNA NNT‐AS1 affects progression of breast cancer through miR‐142‐3p/ZEB1 axis. Biomed Pharmacother 2018; 103: 939–46. [DOI] [PubMed] [Google Scholar]
- 12. Shen Q, Jiang Y. LncRNA NNT‐AS1 promotes the proliferation, and invasion of lung cancer cells via regulating miR‐129‐5p expression. Biomed Pharmacother 2018; 105: 176–81. [DOI] [PubMed] [Google Scholar]
- 13. Subramaniam S, Jeet V, Clements JA, Gunter JH, Batra J. Emergence of microRNAs as key players in cancer cell metabolism. Clin Chem 2019; 65: 1090–1101. [DOI] [PubMed] [Google Scholar]
- 14. Bayoumi AS, Sayed A, Broskova Z et al Crosstalk between long noncoding RNAs and microRNAs in health and disease. Int J Mol Sci 2016; 17 (3): 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med 2015; 8 (10): 17110–6. [PMC free article] [PubMed] [Google Scholar]
- 16. Takahashi RU, Prieto‐Vila M, Kohama I, Ochiya T. Development of miRNA‐based therapeutic approaches for cancer patients. Cancer Sci 2019; 110: 1140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 2019; 11: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang X, Kong F, Wu S et al microRNA‐612 suppresses the malignant development of non‐small‐cell lung cancer by directly targeting bromodomain‐containing protein 4. Onco Targets Ther 2019; 12: 4167–79. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Zhang M, Shi H, Zhang C, Zhang SQ. MiRNA‐621 inhibits the malignant progression of non‐small cell lung cancer via targeting SIX4. Eur Rev Med Pharmacol Sci 2019; 23: 4807–14. [DOI] [PubMed] [Google Scholar]
- 20. Qiu BQ, Zhang PF, Xiong D et al CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non‐small cell lung cancer by regulating Galectin‐1‐AKT/ERK1/2 signaling. J Cell Physiol 2019; 234: 11256–64. [DOI] [PubMed] [Google Scholar]
- 21. Lv X, He C, Huang C et al Timely expression and activation of YAP1 in granulosa cells is essential for ovarian follicle development. FASEB J 2019; 33: 10049–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aiello NM, Kang Y. Context‐dependent EMT programs in cancer metastasis. J Exp Med 2019; 216: 1016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo W, Liu Q, Jiang N, Li M, Shi L. Isorhamnetin inhibited migration and invasion via suppression of Akt/ERK‐mediated epithelial‐to‐mesenchymal transition (EMT) in A549 human non‐small cell lung cancer cells. Biosci Rep 2019; 39 10.1042/BSR20190159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Overholtzer M, Zhang J, Smolen GA et al Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A 2006; 103: 12405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu M, Chen Y, Li X et al YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co‐factor TEAD. Cell Death Dis 2018; 9: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of Long Non‐Coding RNA NNT‐AS1 Correlates with Tumor Progression and Poor Prognosis in Osteosarcoma. Cell Physiol Biochem 2018; 45: 1904–14. [DOI] [PubMed] [Google Scholar]
- 27. Cai Y, Dong ZY, Wang JY. LncRNA NNT‐AS1 is a major mediator of cisplatin chemoresistance in non‐small cell lung cancer through MAPK/Slug pathway. Eur Rev Med Pharmacol Sci 2018; 22: 4879–87. [DOI] [PubMed] [Google Scholar]
- 28. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci 2018; 19 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Z, Zhu Z, Watabe K et al Negative regulation of lncRNA GAS5 by miR‐21. Cell Death Differ 2013; 20 (11): 1558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146 (3): 353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J, Wu FX, Luo HL et al Berberine upregulates miR‐22‐3p to suppress hepatocellular carcinoma cell proliferation by targeting Sp1. Am J Transl Res 2016; 8: 4932–41. [PMC free article] [PubMed] [Google Scholar]
- 32. Sha QK, Chen L, Xi JZ, Song H. Long non‐coding RNA LINC00858 promotes cells proliferation, migration and invasion by acting as a ceRNA of miR‐22‐3p in colorectal cancer. Artif Cells Nanomed Biotechnol 2019; 47: 1057–66. [DOI] [PubMed] [Google Scholar]
- 33. Dong HX, Wang R, Jin XY, Zeng J, Pan J. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa‐mir‐22‐3p. J Cell Physiol 2018; 233: 4126–36. [DOI] [PubMed] [Google Scholar]
- 34. Zhang K, Li XY, Wang ZM, Han ZF, Zhao YH. MiR‐22 inhibits lung cancer cell EMT and invasion through targeting Snail. Eur Rev Med Pharmacol Sci 2017; 21: 3598–604. [PubMed] [Google Scholar]
- 35. Ling B, Wang GX, Long G, Qiu JH, Hu ZL. Tumor suppressor miR‐22 suppresses lung cancer cell progression through post‐transcriptional regulation of ErbB3. J Cancer Res Clin Oncol 2012; 138: 1355–61. [DOI] [PubMed] [Google Scholar]
- 36. Zhang MY, Lin J, Kui YC. MicroRNA‐345 suppresses cell invasion and migration in non‐small cell lung cancer by directly targeting YAP1. Eur Rev Med Pharmacol Sci 2019; 23: 2436–43. [DOI] [PubMed] [Google Scholar]
- 37. Huang C, Ma R, Yue J, Li N, Li Z, Qi D. MiR‐497 suppresses YAP1 and inhibits tumor growth in non‐small cell lung cancer. Cell Physiol Biochem 2015; 37: 342–52. [DOI] [PubMed] [Google Scholar]
- 38. Pu X, Wang J, Li W et al COPB2 promotes cell proliferation and tumorigenesis through up‐regulating YAP1 expression in lung adenocarcinoma cells. Biomed Pharmacother 2018; 103: 373–80. [DOI] [PubMed] [Google Scholar]


