Abstract
Background
Breast cancer (BC) is a common cancer in women worldwide. Emerging evidence has indicated that circular RNA hsa‐circ_0007255 (circ_0007255) is a prognostic mediator in BC progression. However, the functional role of circ_0007255 needs to be determined.
Methods
The expression of circ_0007255, microRNA (miR)‐335‐5p, and SIX Homeobox 2 (SIX2) was evaluated using quantitative real‐time polymerase chain reaction (qRT‐PCR) or western blot assay. Actinomycin D and RNase R treatment was performed to analyze the stability of circ_0007255. Additionally, Seahorse extracellular flux, colony formation and transwell analyses were carried out to detect oxygen consumption ratio (OCR), colony formation and cell mobility, respectively. The interaction between miR‐335‐5p and circ_0007255 or SIX2 was confirmed via dual‐luciferase reporter assay. A xenograft tumor model was established to explore the role of circ_0007255 in vivo.
Results
Circ_0007255 and SIX2 were overexpressed, but miR‐335‐5p was diminished in BC tissues and cells. Circ_0007255 absence inhibited oxygen consumption, colony formation, cell migration and invasion, and these effects were particularly abrogated via miR‐335‐5p upregulation in BC cells. Moreover, SIX2 deficiency eliminated the promotion effects of miR‐335‐5p inhibitor on oxygen consumption, colony formation, and cell mobility in BC cells. Importantly, circ_0007255 inhibited tumor growth in vivo. Mechanically, circ_0007255 was a sponge of miR‐335‐5p to regulate SIX2 expression in BC progression.
Conclusion
Circ_0007255 functioned as a novel oncogene in the progression of BC by regulating miR‐335‐5p/SIX2 axis, and might be a promising biomarker for BC treatment.
Key points
Significant findings of the study: Levels of circ_0007255 and SIX2 were upregulated, but miR‐335‐5p was diminished in BC tissues and cells. Circ_0007255 was an oncogene in BC development and exerted its function via miR‐335‐5p/SIX2 axis in BC. Tumor growth was reduced by circ_0007255 absence.
What this study adds: Circ_0007255 functioned as a novel oncogene in the progression of BC by regulating miR‐335‐5p/SIX2 axis, and might be a promising biomarker for BC treatment.
Keywords: BC, circ_0007255, miR‐335‐5p, SIX2
Introduction
Breast cancer (BC) is the most common cancer in women with the second highest mortality worldwide.1 According to the definition, BC can be classified into multiple subtypes, and the most general type is deemed as hormone receptor (HR) positive.2 The occurrence of BC is related to numerous factors, such as age, overweight status, physical inactivity, and radiation exposure.3 Over the past decades, estrogen signaling acted as a master function in the onset and progression of HR‐positive BC,4, 5 and available antihormone therapy was regarded as the standard treatment. However, curative molecular‐targeted treatments of any type have been reported to be unsatisfactory.6, 7 Thus, it is necessary to search for novel therapeutic targets for the treatment of BC patients.
Circular RNAs (circRNAs) are a class of non‐coding RNAs (ncRNAs) with covalently closed‐loop characteristics.8 They have been verified to derive from pre‐mRNA transcripts.9 Over the past decades, with the continuous development of high throughput sequencing, multiple circRNAs have been discovered and identified in mammalian cells.10, 11, 12 Emerging findings have indicated that circRNAs are associated with the regulation of gene transcription, suggesting that circRNAs play vital roles in diverse diseases, including cancers. Specifically, in the study by Chen et al. hsa_circRNA_000479 (circEPSTI1) modulated cell proliferation and apoptosis of triple‐negative breast cancer by sponging microRNA (miRNA/miR)‐4753 and miR‐6809.13 Hsa_circ_0008717 (circABCB10) triggered aggressive progression of breast cancer by targeting miR‐1271 in the study by Liang et al.14 In addition, hsa_circ_0007255 (circ_0007255, also known as circKIF4A) has been reported to be a sponge of miR‐375 in the initiation and process of triple‐negative breast cancer.15 Nevertheless, further work is needed in this regard.
Previous studies have indicated that the identified circRNAs harbor multiple common fragments for miRNAs, thereby acting as competitive inhibitors and miRNAs sponges.16 Earlier records showed that a circRNA contained diverse miRNA‐binding sites and was sufficiently excavated in the cytoplasm.17 In addition, the relationship between the ectopic expression of miRNAs and the pathogenesis of multiple cancers has been researched.18, 19 MiR‐335 has been confirmed to serve as a master suppressor in BC.20 Also, miR‐335‐5p, an aberrantly expressed miRNA, could be a major monitor in the progression of gastric cancer.21 However, there is little knowledge of the molecular mechanism by which miR‐335‐5p regulates biological development. In several studies, MiRNAs have been reported to bind to the 3′‐untranslated region (3'‐UTR) of the target mRNA, thereby leading to translational reduction or mRNA degradation.22, 23 SIX Homeobox 2 (SIX2) contributed to cell metastasis of BC,24 and we hypothesized that SIX2 was a direct target of miR‐335‐5p in tumorigenesis in BC.
In this study, we explored the expression of circ_0007255, miR‐335‐5p, and SIX2 in BC tissues and cell lines. In addition, we investigated the potential regulatory mechanism among them in BC progression.
Methods
Clinical specimens
BC patients (n = 50) and healthy volunteers (n = 48) were recruited from East Hospital, Xiamen University. Serum from BC patients and healthy individuals was collected. For tissue collections, the BC tissues (n = 50) and peritumor samples (n = 50) were obtained from BC patients. All available tissues and serum wasmaintained in −80°C until use. Written informed consents were given by the enrolled patients and volunteers, and our study was ratified by the Ethics Committee of the East Hospital, Xiamen University.
Cell culture
BC cell lines (T47D, MCF‐7, MB231, and MB468) and normal human breast epithelial cells (MCF‐10A) were purchased from Be Na collection (Beijing, China). T47D and MCF‐10 cells grown in Roswell Park Memorial Institute‐1640 (RPMI‐1640; Gibco, Carlsbad, CA, USA). MCF‐7 and MB231 cells were maintained in Dulbecco's modified eagle medium (DMEM; Gibco). Leibovitz's L‐15 (Thermo Fisher Scientific, Rockford, IL, USA) was employed to incubate MB468 cells. Ten percent fetal bovine serum (FBS; Gibco) was added to the medium. MB468 cells were incubated at 37°C with moist air and the same conditions with the addition of 5% CO2 was used to culture the other cell lines at 37°C.
Cell transfection
To knockdown circ_0007255, small interfering RNA (siRNA) or short hairpin (shRNA) targeting the back‐splice junction sites of circ_0007255 (si‐circ_0007255 or sh‐circ_0007255), and siRNA and shRNA were scrambled (si‐NC and sh‐NC) and synthesized by Geneseed (Guangzhou, China). In addition, siRNA against SIX2 (si‐SIX2) was also constructed. To overexpress circ_0007255, the full‐length cDNA of circ_0007255 was cloned into a basic vector (pLCDH‐ciR, Geneseed), and the blank control was similarly formed. With regard to miR‐335‐5p, the mimic (miR‐335‐5p) and inhibitor (anti‐miR‐335‐5p), as well as their controls (miR‐NC and anti‐miR‐NC) were obtained from GenePharma (Shanghai, China). Cell transfection was implemented using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) as per the protocols. ShRNA was used to create stably transfected cells via lentivirus‐mediation.
Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay
Total RNA was extracted from tissues and cells using Trizol reagent (Invitrogen). The PARIS Kit (Thermo Fisher Scientific) was applied to isolate the nuclear and cytoplasmic fractions according to the manufacturer's protocol. After that, the complementary DNA (cDNA) was synthesized using PrimeScript RT Reagent Kit (Takara, Dalian, China), and real‐time polymerase chain reaction was administrated on a Quantstudio DX system (Applied Biosystems, Foster City, CA, USA) after mixing with cDNA and the reagent of TB Green Premix Ex Taq II (Takara). Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH; for circ_0007255, SIX2, and KIF4A) and U6 (for miR‐335‐5p) served as the internal controls. Relative levels were calculated via the 2−ΔΔCt method. Primer information was listed: Circ_0007255 (Forward: 5′‐GTATTAATATTAACCGAGG‐3′, Reverse: 5′‐GTTATAGATCCAGGCAGGGT‐3′); miR‐335‐5p (Forward: 5′‐GTCAAGAGCAATAACGAAAAATG‐3′, Reverse: 5′‐GAGGTCAGGAGCAATAATGAA‐3′); SIX2 (Forward: 5′‐AAGGCACACTACATCGAGGC‐3′, Reverse: 5′‐CACGCTGCGACTCTTTTCC‐3′); KIF4A (Forward: 5′‐TACTGCGGTGGAGCAAGAAG‐3′, Reverse: 5′‐CATCTGCGCTTGACGGAGAG‐3′); GAPDH (Forward: 5′‐ACTCCTCCACCTTTGACGC‐3′, Reverse: 5′‐GCTGTAGCCAAATTCGTTGTC‐3′). U6 (Forward: 5′‐CTCGCTTCGGCAGCACA‐3′, Reverse: 5′‐AACGCTTCACGAATTTGCGT‐3′).
Actinomycin D and RNase R treatment
For circ_0007255 stability assay, actinomycin D (2 mg/mL; Sigma, St. Louis, MO, USA) or dimethyl sulfoxide (DMSO; Sigma) was added to constrain transcription. Total RNA (2 μg) was incubated with RNase R (Epicenter Technologies, Madison, WI, USA). After that, the reactive solution from RNase R treatment was purified using the RNeasy MinElute Cleaning Kit (Qiagen, Valencia, CA, USA). Next, the corresponding levels were determined using qRT‐PCR.
Seahorse extracellular flux analysis
For oxygen consumption, 2 × 104 cells were plated and cultured for 24 hours. Next, oxygen consumption ratio (OCR) was measured adopting the Seahorse XF24 analyzer25 (Seahorse Bioscience, Santa Clara, CA, USA) under corresponding conditions, including basic condition and supplement of ATP synthase inhibitor (oligomycin), the mitochondrial uncoupler (carbonyl cyanide 4‐(trifluoromethoxy) phenylhydrazone; FCCP) and the complex I+II inhibitors (rotenone+antimycin A) following the manuals. All the chemical reagents were obtained from Sigma.
Colony formation
Cells were trypsinized and plated into a 6 cm dish and cultured for two weeks. The medium was replaced every five days, and formed colonies were stained with crystal violet after methanol fixation. Only colonies with over 50 numbers of cells were available. Each assay was carried out in triplicate.
Transwell assay
For cell invasion assay, transwell inserts (8 μm pore‐size, Corning Costar, Corning, NY, USA) precoated with Matrigel in the upper chamber (Corning) were employed. After transfection for 24 hours, MB231 and MB468 cells with serum‐free media (200 μL) were seeded in the upper chamber. At the same time, 600 μL medium was added to the lower chamber. Cells on the lower surface of the membrane were then stained and photographed. The cell migration assay procedure was similar to the protocol of cell invasion except that the upper chamber was not precoated with Matrigel.
Dual‐luciferase reporter assay
The wild‐type sequences of circ_0007255 and the 3'‐UTR of SIX2 containing the binding sites of miR‐335‐5p were subcloned into the basic vector (pmirGLO; Promega, Madison, WI, USA), thereby generating WT‐CIRC_0007255 and WT‐SIX2, respectively. Similarly, the mutant sequences were also designed and inserted into the pmirGLO vector, and the constructed plasmids named MUT‐CIRC_0007255 and MUT‐SIX2. Briefly, the above reporters were cotransfected with miR‐335‐5p or miR‐NC into MB231 and MB468 cells, respectively. The cells were lysed at 48 hours post‐transfection, and dual‐luciferase assay system kit (Promega) was used to measure the luciferase activity.
Western blot assay
As previously described,26 proteins were segregated on the sodium dodecyl sulfate‐polyacrylamide gels (10%) and then transfected onto polyvinylidene fluoride membranes (PVDF; Millipore, Bedford, MA, USA). The primary antibodies (Abcam, Cambridge, MA, USA), including SIX2 (ab111827, 1:6000) and GAPDH (ab8245, 1:7500), were employed to incubate with the membranes, and then relative secondary antibodies were supplemented after washing with tris‐buffered saline Tween‐20 (TBST; Solarbio, Beijing, China). Finally, the protein levels were assessed utilizing an enhanced chemiluminescence reagent (Millipore).
Xenograft tumor model
A total of 10 female nude mice (n = 5/group, 4–6 weeks old) were purchased from Vital River Laboratory Animal Technology (Beijing, China) and then divided randomly into two groups. This in vivo experiment was approved by the Institutional Animal Care and Use Committee of East Hospital, Xiamen University. Stably transfected MB231 cells (sh‐circ_0007255 or sh‐NC) were subcutaneously injected into one side of the posterior flank. Tumor size (length and width) was measured every seven days for a total of five times. The tumor volume was calculated as follows: volume = length×width2 × 0.5. After five weeks, the mice were sacrificed, and the subcutaneous tumors were excised and weighed.
Statistical analysis
The data from each test were demonstrated as the mean ± standard deviation (SD). Student's t‐test or one‐way analysis of variance with Tukey's test was recruited to perform the statistical analysis. P < 0.05 indicated statistical significance.
Results
Level of circ_0007255 was upregulated in BC serum, tissues and cell lines
To investigate the biological role of circ_0007255 in BC, circ_0007255 expression was measured by qRT‐PCR in serum (n = 50) from BC patients (n = 50) and healthy volunteers (n = 48). We found that circ_0007255 was upregulated in BC patients’ serum in comparison to that in the matched controls (Fig 1a). Relative operating characteristic curves (ROC) and the area under the ROC curves (AUC) were applied to investigate the profile of circ_0007255 in BC. From all subjects, the ROC curves exhibited apparent separation between BC tissues and the healthy donators, with an AUC of 0.7748 for circ_0007255 (Fig 1b). Moreover, high expression of circ_0007255 was observed in BC tissues and cell lines compared with the corresponding controls (Fig 1c,d). The ectopic expression of circ_0007255 might be a promising marker for BC therapy.
Figure 1.
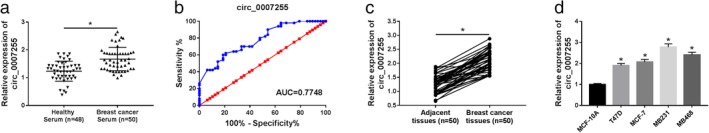
The level of circ_0007255 was upregulated in BC serum, tissues and cell lines. (a) QRT‐PCR assay was used to detect the level of circ_0007255 in breast cancer serum (n = 50) and healthy controls (n = 48). (b) ROC curve in which breast cancer serum (n = 50) was compared with matched healthy serum (n = 48). (c and d) Circ_0007255 expression was significantly overexpressed in breast cancer tissues and cells compared with relative controls. The data are presented as mean ± SD of three independent experiments. *P < 0.05.
Circ_0007255 existed in cytoplasm with stable characteristics
As referenced in the circBank database (http://www.circbank.cn), circ_0007255 is derived from the kinesin family member 4A (KIF4A). To research the structure of circ_0007255, transcription assay was performed using random primers or oligo (dT)18 primers, and our data suggested that circ_0007255 was almost undetectable when Oligo (dT)18 primers were used, confirming that circ_0007255 was a circRNA with circular structure (Fig 2a). Meanwhile, qRT‐PCR analysis of nuclear and cytoplasmic RNAs demonstrated that circ_0007255 existed predominantly in the cytoplasm in MB231 and MB468 cells (Fig 2b). Furthermore, we explored the characteristic of circ_0007255 via RNase R and Actinomycin D treatment. Results from qRT‐PCR confirmed that circ_0007255 was resistant to RNase R digestion in vitro (Fig 2c). Also, the half‐life of circ_0007255 transcript exceeded 24 hours, but that of the KIF4A mRNA was less than 12 hours, suggesting that the circ_0007255 was stable in MB231 and MB468 cells (Fig 2d). Collectively, these results indicated that circ_0007255 had a stable closed‐loop structure without poly‐A tail, and generally resided in cytoplasm.
Figure 2.
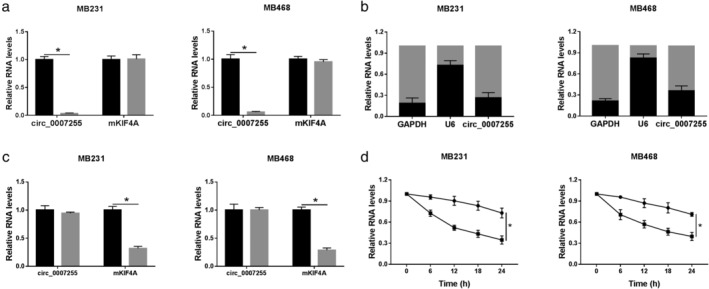
Circ_0007255 existed in cytoplasm with stable characteristics. (a) Back‐splice junction was detected in cDNA synthesized using random primers but not Oligo dT(18) primed RNA. MB231 ( ) random primers and (
) random primers and ( ) oligo (dT)18 primers. MB468 (
) oligo (dT)18 primers. MB468 ( ) random primers and (
) random primers and ( ) oligo (dT)18 primers. (b) QRT‐PCR assay was performed to analyze the levels of circ_0007255, GAPDH, and U6 in the cytoplasm and nucleus in MB231 and MB468 cells. MB231 (
) oligo (dT)18 primers. (b) QRT‐PCR assay was performed to analyze the levels of circ_0007255, GAPDH, and U6 in the cytoplasm and nucleus in MB231 and MB468 cells. MB231 ( ) cytoplasm and (
) cytoplasm and ( ) nuclear. MB468 (
) nuclear. MB468 ( ) cytoplasm and (
) cytoplasm and ( ) nuclear. (c and d) The levels of circ_0025202 and GAPDH expression in BC cells were reckoned after treating with RNase R or Actinomycin D. Data are presented as mean ± SD of three independent tests. *P < 0.05. (c) MB231 (
) nuclear. (c and d) The levels of circ_0025202 and GAPDH expression in BC cells were reckoned after treating with RNase R or Actinomycin D. Data are presented as mean ± SD of three independent tests. *P < 0.05. (c) MB231 ( ) RNase R− and (
) RNase R− and ( ) RNase R+. MB468 (
) RNase R+. MB468 ( ) RNase R− and (
) RNase R− and ( ) RNase R+. (d) MB231 (
) RNase R+. (d) MB231 ( ) circ_0007255 and (
) circ_0007255 and ( ) mKIF4A. MB468 (
) mKIF4A. MB468 ( ) circ_0007255 and (
) circ_0007255 and ( ) mKIF4A.
) mKIF4A.
Knockdown of circ_0007255 diminished cell growth and metastasis in BC cells
Next, we explored the functional role of circ_0007255 in BC progression. After transfection with si‐circ_0007255, the level of circ_0007255 was downregulated in MB231 and MB468 cells (Fig 3a). Functional assays were subsequently performed. As shown in Figure 3b, OCR analysis demonstrated that oxygen consumption was inhibited in circ_0007255‐silenced BC cells. Next, colony formation analysis verified that circ_0007255 deficiency decreased the capacity of colony formation in BC cells (Fig 3c). Moreover, a significant inhibition of cell mobility (cell migration and invasion) was observed in MB231 and MB468 cells (Fig 3d,e). These findings suggested that circ_0007255 might play an oncogenic role in BC progression.
Figure 3.
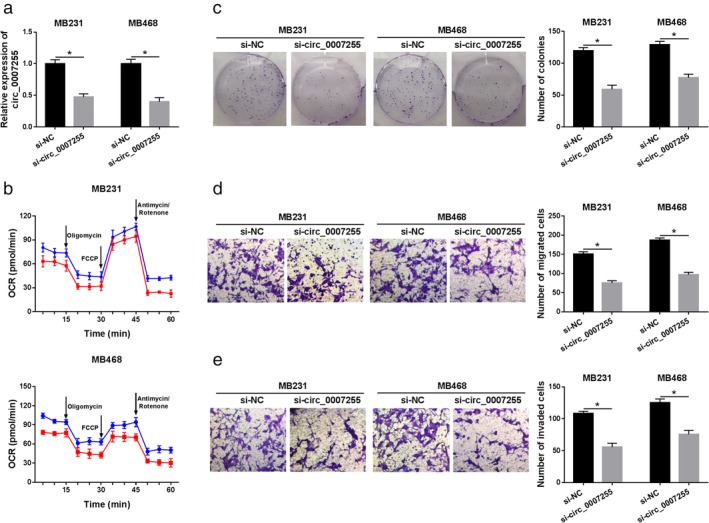
Knockdown of circ_0007255 diminished cell growth and metastasis in BC cells. (a) QRT‐PCR analysis of circ_0007255 in MB231 and MB468 cells with si‐circ_0007255 and si‐NC transfection was shown. (b) Oxygen consumption ratio was analyzed by Seahorse extracellular flux analyzer in BC cells transfected with si‐circ_0007255 and its control. MB231 ( ) si‐NC and (
) si‐NC and ( ) si‐circ_0007255. MB468 (
) si‐circ_0007255. MB468 ( ) si‐NC and (
) si‐NC and ( ) si‐circ_0007255. (c) Colony formation was shown in MB231 and MB468 cells under circ_0007255 absence. (d and e) The effect of circ_0007255 deficiency on cell migration and invasion in vitro was shown. Data are presented as mean ± SD of three different experiments. *P < 0.05.
) si‐circ_0007255. (c) Colony formation was shown in MB231 and MB468 cells under circ_0007255 absence. (d and e) The effect of circ_0007255 deficiency on cell migration and invasion in vitro was shown. Data are presented as mean ± SD of three different experiments. *P < 0.05.
Circ_0007255 was a sponge of miR‐335‐5p
As shown in Fig 4a, Interactome software predicted that miR‐335‐5p was a candidate target of circ_0007255. The relationship between miR‐335‐5p and circ_0007255 was then determined. Briefly, luciferase reporters (WT‐circ_0007255 and MUT‐circ_0007255) were cotransfected with miR‐335‐5p or miR‐NC, and we found that miR‐335‐5p decreased the luciferase activity in both MB231 and MB468 cells transfected with wild‐type reporter (WT‐circ_0007255) (Fig 4b). For miR‐335‐5p analysis, the low expression of miR‐335‐5p was observed in BC tissues and cells in comparison with corresponding controls (Fig 4c,d). To determine the regulatory mechanism between miR‐335‐5p and circ_0007255, the efficiency of circ_0007255 vector was confirmed. qRT‐PCR analysis indicated that circ_0007255 showed a successful efficiency in BC cells (Fig 4e). Moreover, si‐circ_0007255 or circ_0007255 was transfected into MB231 and MB468 cells, and the level of miR‐335‐5p was inversely regulated by circ_0007255 in vitro (Fig 4f). Taken together, circ_0007255 directly targeted miR‐335‐5p.
Figure 4.
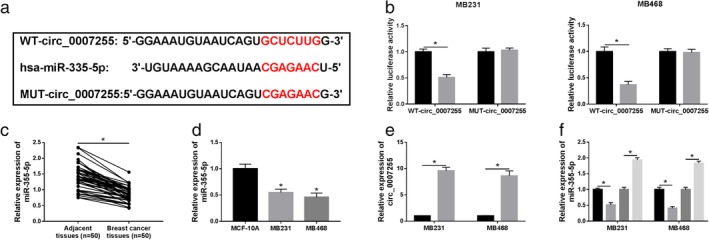
Circ_0007255 was a sponge of miR‐335‐5p. (a) The predictive binding sites between circ_0007255 and miR‐335‐5p were presented. (b) The luciferase activities of WT‐circ_0007255 and MUT‐circ_0007255 in MB231 and MB468 cells were evaluated after transfection with miR‐335‐5p. MB231 ( ) miR‐NC and (
) miR‐NC and ( ) miR‐335‐5p. MB468 (
) miR‐335‐5p. MB468 ( ) miR‐NC and (
) miR‐NC and ( ) miR‐335‐5p. (c and d) QRT‐PCR assay was conducted to assess the relative level of miR‐335‐5p in BC specimens and cells in comparison to the matched controls. (e) The overexpressed efficiency of circ_0007255 was assessed in BC cells. (
) miR‐335‐5p. (c and d) QRT‐PCR assay was conducted to assess the relative level of miR‐335‐5p in BC specimens and cells in comparison to the matched controls. (e) The overexpressed efficiency of circ_0007255 was assessed in BC cells. ( ) Vector and (
) Vector and ( ) circ_0007255. (f) Relative level of miR‐335‐5p was determined in MB231 and MB468 cells under circ_0007255 or si‐circ_0007255 transfection. Data are expressed as mean ± SD of three independent experiments. *P < 0.05. (
) circ_0007255. (f) Relative level of miR‐335‐5p was determined in MB231 and MB468 cells under circ_0007255 or si‐circ_0007255 transfection. Data are expressed as mean ± SD of three independent experiments. *P < 0.05. ( ) Vector and (
) Vector and ( ) circ_0007255. (
) circ_0007255. ( ) si‐NC and (
) si‐NC and ( ) si‐circ_0007255.
) si‐circ_0007255.
Circ_0025202 regulated cell behavior via miR‐335‐5p in BC cells
Owing to the interaction between miR‐335‐5p and circ_0007255, rescue assays were carried out to uncover the regulatory mechanism of circ‐0007255 in BC. MB231 and MB468 cells were transfected with miR‐NC, miR‐335‐5p, miR‐335‐5p+vector or miR‐335‐5p+circ‐0007255. QRT‐PCR analysis indicated that miR‐335‐5p was remarkably upregulated by miR‐335‐5p overexpression, and circ_0007255 could repress the promotion effect of miR‐335‐5p mimic on miR‐335‐5p expression in BC cells (Fig 5a). Also, circ_0007255 overexpression could reverse the inhibition effect of miR‐335‐5p upregulation on oxygen consumption in MB231 and MB468 cells (Fig 5b). Moreover, the number of colonies was diminished by miR‐335‐5p overexpression, while circ_0007255 upregulation reversed the inhibition effect of miR‐335‐5p overexpression (Fig 5c). In addition, circ_0007255 blocked the inhibitory effects of miR‐335‐5p overexpression on cell migration and invasion in BC cells (Fig 5d,e). The evidence proved that circ_0007255 exerted its effects by targeting miR‐335‐5p in BC cells.
Figure 5.
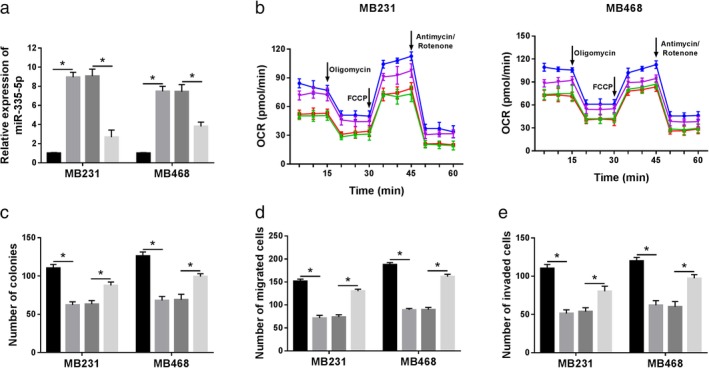
Circ_0025202 regulated cell behaviors via miR‐335‐5p in BC cells. MiR‐NC, miR‐335‐5p, miR‐335‐5p+vector, or miR‐335‐5p+circ_0007255 was transfected into MB231 and MB468 cells. (a) Relative level of miR‐335‐5p was measured in vitro. ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) miR‐335‐5p+vector and (
) miR‐335‐5p+vector and ( ) miR‐335‐5p+circ_0007255 (b) Oxygen consumption ratio was determined in MB231 and MB468 cells at indicated time after treatment with Oligomycin, FCCP, or Antimycin/Rotenone. MB231 (
) miR‐335‐5p+circ_0007255 (b) Oxygen consumption ratio was determined in MB231 and MB468 cells at indicated time after treatment with Oligomycin, FCCP, or Antimycin/Rotenone. MB231 ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) miR‐335‐5p+vector and (
) miR‐335‐5p+vector and ( ) miR‐335‐5p+circ_0007255. MB468 (
) miR‐335‐5p+circ_0007255. MB468 ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) miR‐335‐5p+vector and (
) miR‐335‐5p+vector and ( ) miR‐335‐5p+circ_0007255. (c) The effect of miR‐335‐5p and circ_0007225 on colony formation in BC cells was detected. (
) miR‐335‐5p+circ_0007255. (c) The effect of miR‐335‐5p and circ_0007225 on colony formation in BC cells was detected. ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) miR‐335‐5p+vector and (
) miR‐335‐5p+vector and ( ) miR‐335‐5p+circ_0007255. (d and e) Transwell assay was employed to examine the ability of cell migration and invasion in MB231 and MB460 cells. Data are presented as mean ± SD, and each experiment was repeated three times. *P < 0.05. (d) (
) miR‐335‐5p+circ_0007255. (d and e) Transwell assay was employed to examine the ability of cell migration and invasion in MB231 and MB460 cells. Data are presented as mean ± SD, and each experiment was repeated three times. *P < 0.05. (d) ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) miR‐335‐5p+vector and (
) miR‐335‐5p+vector and ( ) miR‐335‐5p+circ_0007255. (e) (
) miR‐335‐5p+circ_0007255. (e) ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) miR‐335‐5p+vector and (
) miR‐335‐5p+vector and ( ) miR‐335‐5p+circ_0007255
) miR‐335‐5p+circ_0007255
SIX2 was a direct target of miR‐335‐5p
As depicted in Fig 6a, StarBase software predicted that there were complementary binding sites between miR‐335‐5p and SIX2. Next, dual‐luciferase reporter assay was performed, and the results implied that miR‐335‐5p could reduce the luciferase activity of the wild‐type reporter in BC cells, but had no effect on the mutant reporter system (Fig 6b). High mRNA and protein expression levels of SIX2 were observed in BC tissues and cell lines in comparison to the matched controls (Fig 6c–f). Meanwhile, anti‐miR‐335‐5p could markedly inhibit the level of miR‐335‐5p in MB231 and MB468 cells (Fig 6g). The molecular mechanism between miR‐335‐5p and SIX2 was then identified. As described in Fig 6h,i, miR‐335‐5p mimic decreased the level of SIX2, whereas miR‐335‐5p inhibitor promoted the level of SIX2 in vitro. Overall, SIX2 was directly targeted by miR‐335‐5p.
Figure 6.
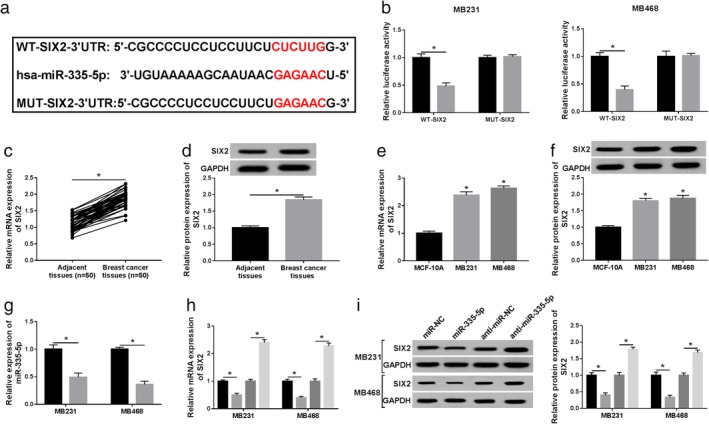
SIX2 was a direct target of miR‐335‐5p. (a) The complementary sequences between miR‐335‐5p and SIX2 were predicted by StarBase. (b) Dual‐luciferase reporter assay was conducted to test the luciferase activity of WT‐SIX2 and MUT‐SIX2 reporters in MB231 and MB468 cells with miR‐335‐5p or miR‐NC transfection. MB231 ( ) miR‐NC and (
) miR‐NC and ( ) miR‐335‐5p. MB468 (
) miR‐335‐5p. MB468 ( ) miR‐NC and (
) miR‐NC and ( ) miR‐335‐5p. (c–f) The mRNA and protein expression of SIX2 in BC tissues and cells was determined. (g) Knockdown efficiency of anti‐miR‐335‐5p was identified using qRT‐PCR assay. (
) miR‐335‐5p. (c–f) The mRNA and protein expression of SIX2 in BC tissues and cells was determined. (g) Knockdown efficiency of anti‐miR‐335‐5p was identified using qRT‐PCR assay. ( ) Anti‐miR‐NC and (
) Anti‐miR‐NC and ( ) anti‐miR‐335‐5p. (h and i) The effect of miR‐335‐5p or anti‐miR‐335‐5p transfection on the level of SIX was analyzed in BC cells. All data are presented as mean ± SD, and each test was repeated three times. *P < 0.05. (h) (
) anti‐miR‐335‐5p. (h and i) The effect of miR‐335‐5p or anti‐miR‐335‐5p transfection on the level of SIX was analyzed in BC cells. All data are presented as mean ± SD, and each test was repeated three times. *P < 0.05. (h) ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) anti‐miR‐NC and (
) anti‐miR‐NC and ( ) anti‐miR‐335‐5p. (i) (
) anti‐miR‐335‐5p. (i) ( ) miR‐NC, (
) miR‐NC, ( ) miR‐335‐5p, (
) miR‐335‐5p, ( ) anti‐miR‐NC and (
) anti‐miR‐NC and ( ) anti‐miR‐335‐5p.
) anti‐miR‐335‐5p.
MiR‐335‐5p axis regulated the progression of BC by targeting SIX2
As mentioned above, we next investigated the functional mechanism between miR‐335‐5p and SIX2. The mRNA and protein expression levels of SIX2 were decreased in BC cells transfected with si‐SIX2, and miR‐335‐5p inhibitor reversed the repressive effect of si‐SIX2 on the level of SIX2 (Fig 7a,b). The inhibitory effects of SIX2 knockdown on OCR were reversed by miR‐335‐5p overexpression (Fig 7c). Simultaneously, the number of colonies was notably weakened in SIX2 deficient BC cells, and miR‐335‐5p inhibitor blocked this effect of SIX2 deletion in vitro (Fig 7d). Also, cell migration and invasion were distinctly impeded by deficiency of SIX2, which were blocked by miR‐335‐5p inhibitor in MB231 and MB468 cells (Fig 7e,f). Moreover, the results confirmed that circ_0007255 increased on the level of SIX2 by targeting miE‐335‐5p in BC cells (Fig 8a,b). All tdata clarified that circ_0007255/miR‐335‐5p axis participated in the aggressive progression by modulating SIX2 in BC.
Figure 7.
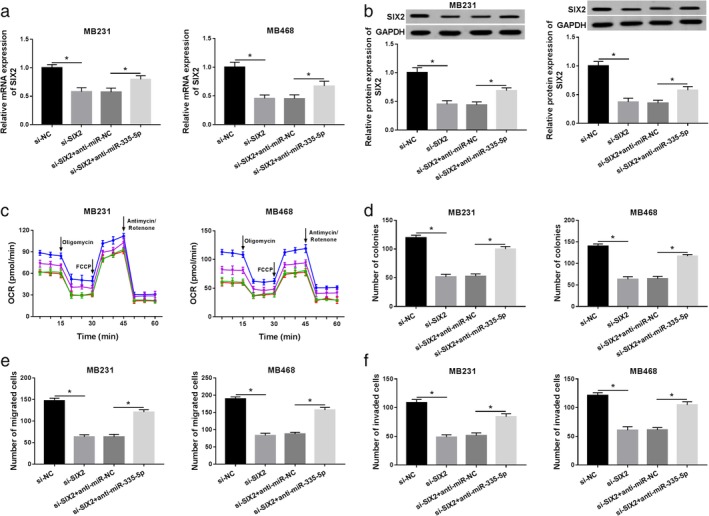
MiR‐335‐5p regulated the progression of BC by targeting SIX2. Si‐NC, si‐SIX2, si‐SIX2+miR‐NC, or si‐SIX2+miR‐335‐5p was transfected into MB231 and MB468 cells, respectively. (a and b) Relative level of SIX2 in MB231 and MB468 cells transfected with SIX2 and miR‐335‐5p is presented. (c) Oxygen consumption ratio was detected by Seahorse extracellular flux analysis in BC cells. MB231 ( ) si‐NC, (
) si‐NC, ( ) si‐SIX2, (
) si‐SIX2, ( ) si‐SIX2+anti‐miR‐NC and (
) si‐SIX2+anti‐miR‐NC and ( ) si‐SIX2+anti‐miR‐335‐5p. MB468 (
) si‐SIX2+anti‐miR‐335‐5p. MB468 ( ) si‐NC, (
) si‐NC, ( ) si‐SIX2, (
) si‐SIX2, ( ) si‐SIX2+anti‐miR‐NC and (
) si‐SIX2+anti‐miR‐NC and ( ) si‐SIX2+anti‐miR‐335‐5p. (d) The capacity of colony formation was exhibited in MB231 and MB468 cells. (e and f) The effect of SIX2 and circ_0007245 on cell migration and invasion was assayed in BC cells. Data are presented as mean ± SD of three independent assays. *P < 0.05.
) si‐SIX2+anti‐miR‐335‐5p. (d) The capacity of colony formation was exhibited in MB231 and MB468 cells. (e and f) The effect of SIX2 and circ_0007245 on cell migration and invasion was assayed in BC cells. Data are presented as mean ± SD of three independent assays. *P < 0.05.
Figure 8.
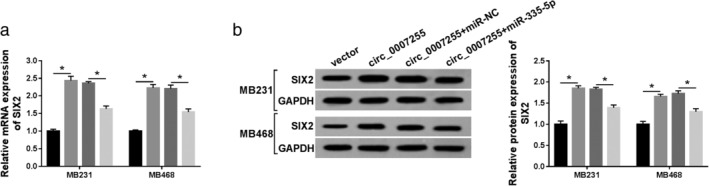
Circ_0007255 regulated SIX2 expression by sponging miR‐335‐5p. MB231 and MB468 cells were transfected with vector, circ_0007255, circ_0007255+miR‐NC, or circ_0007255+miR‐335‐5p, respectively. (a and b) The mRNA and protein levels of SIX2 are shown. Three independent experiments were conducted, and the data were expressed as mean ± SD. *P < 0.05. (a) ( ) Vector, (
) Vector, ( ) circ_0007255, (
) circ_0007255, ( ) circ_0007255+miR‐NC and (
) circ_0007255+miR‐NC and ( ) circ_0007255+miR‐335‐5p. (b) (
) circ_0007255+miR‐335‐5p. (b) ( ) Vector, (
) Vector, ( ) circ_0007255, (
) circ_0007255, ( ) circ_0007255+miR‐NC and (
) circ_0007255+miR‐NC and ( ) circ_0007255+miR‐335‐5p
) circ_0007255+miR‐335‐5p
The absence of circ_0007255 inhibited xenograft tumor growth
We then investigated the biological role of circ_0007255 in vivo. Stably transfected MB231 cells (lentivirus‐mediated sh‐circ_0007255 or sh‐NC) were injected into female nude mice, and tumor volume and weight recorded. We found that tumor volume and weight were inhibited in sh‐circ_0007255 group (Fig 9a,b). Moreover, circ_0007255 deletion induced the decrease of circ_0007255 and increase of miR‐335‐5p in excised tumors (Fig 9c,d). The level of SIX2 was also analyzed, and qRT‐PCR and western blot analyses indicated that the mRNA and protein levels of SIX2 were forcefully hindered in circ_0007255‐silenced specimens (Fig 9e,f). In summary, circ_0007255 might be an oncogene in tumor growth in BC.
Figure 9.
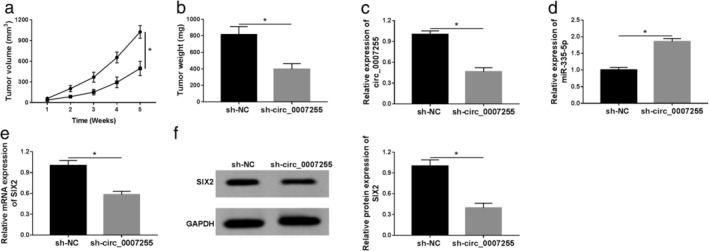
The absence of circ_0007255 inhibited tumor growth. (a and b) The data of tumor volume and weight were recorded and graphed. ( ) sh‐NC and (
) sh‐NC and ( ) sh‐circ_0007255. (c–e) QRT‐PCR assay was used to detect the levels of circ_0007255, miR‐335‐5p, and XIS2 in excised tumors. (f) Relative level of SIX2 was analyzed and exhibited. Three independent experiments were conducted, and the data were expressed as mean ± SD. *P < 0.05.
) sh‐circ_0007255. (c–e) QRT‐PCR assay was used to detect the levels of circ_0007255, miR‐335‐5p, and XIS2 in excised tumors. (f) Relative level of SIX2 was analyzed and exhibited. Three independent experiments were conducted, and the data were expressed as mean ± SD. *P < 0.05.
Discussion
A growing number of circRNAs have to date been identified using bioinformatics analysis and high‐throughput sequencing. In addition, circRNAs have also been reported to be a type of promising biomarker.27 Currently, circRNAs have attracted more attention as they have been reported to alter the aggressive phenotypes of different human cancers.28, 29, 30 For example, circ_ciRS‐7‐a facilitated cell‐cycle, and might be a promising biomarker in colorectal cancer.28 However, the role of circRNAs in BC still requires further investigation.
In this study, we focused on a novel identified circRNA, circ_0007255, in carcinogenesis of BC. Circ_0007255, derived from chrX: 69549254‐69553539, was upregulated in BC tissues and cell lines. Next, the characteristic of circ_0007255 was determined and we found that circ_0007255 with closed‐loop structure, was primarily located in the cytoplasm and resisted the degradation of RNase R. A previous study showed that circ_0007255 acted as the main monitor to modulate the progression of BC.15 We then analyzed the function role of circ_0007255 in BC and aimed to explore the molecular mechanism of circ_0007255 in BC. In our study, high expression of circ_0007255 was observed in BC serum, tissues, and cell lines compared with the matched controls, suggesting that circ_0007255 might be an oncogenic regulator in BC. To confirm this hypothesis, si‐circ_0007255 and its scramble were designed and constructed. We then found that cell growth and metastasis were significantly inhibited by circ_0007255 deletion in BC cells, as shown by the repression of oxygen consumption, colony formation, cell migration, and invasion. Similarly, the deficiency of circ_0007255 led to the repression of the tumor growth in vivo.
Accumulating records confirm that circRNAs, a type of endogenous ncRNAs, are the competitive endogenous RNAs (ceRNAs) and sponges of miRNAs.16, 27, 31 For example, circ_0007874 (circMTO1) could prevent tumor growth in hepatocellular carcinoma by inhibiting miR‐9.32 Furthermore, previous reports have suggested that the cytoplasmic localization of circRNAs was strictly implicated in miRNA sponging.33 In the present research, the possible targets of circ_0007255 were predicted by Interactome software, and the results indicated that miR‐335‐5p was a target of circ‐0007255. Also, the interaction between miR‐335‐5p and circ_0007255 was confirmed by dual‐luciferase reporter assay. A previous study reported that low expression of miR‐335‐5p was a common profile in human BC.34 MiR‐335 was found to diminish cell viability, mobility, and promote apoptosis in BC cells.35 Currently, a significant low level of miR‐335‐5p was found in BC tissues and cells, implying that miR‐335‐5p served as a tumor suppressor in BC. In our study, the aggressive progression, including cell oxygen consumption, colony formation, and mobility, was strongly weakened by miR‐335‐5p overexpression in BC cells. Also, the regulatory effect of circ_0007255 on cell behaviors was partially eliminated after cotransfection with miR‐335‐5p in vitro. All the data revealed that circ_0007255 exerted its oncogenic role via repressing miR‐335‐5p in BC progression.
Accumulating evidence has demonstrated that miRNAs exerted their roles via repression or deregulation of targeted mRNA.36 We then aimed to determine the target genes of miR‐335‐5p. Results from StarBase prediction confirmed that SIX2 was a potential candidate gene of miR‐335‐5p. As a potent oncogene, SIX2 has previously been demonstrated to accelerate tumorigenesis through regulating the proliferation of tumor cells.37 Similarly, the higher expression of SIX2 was validated in BC tissues and cells relative to the corresponding controls. Moreover, the absence of SIX2 declined cellular aggression, also abrogated miR‐335‐5p inhibitor‐mediated inhibition effects on cell growth and metastasis in BC cells and the level of SIX2 was coregulated by the circ_0007255/miR‐335‐5p axis in vitro.
In conclusion, circ_0007255 and SIX2 were upregulated, whereas miR‐335‐5p was decreased in BC tissues and cells. Circ_0007255 regulated cell behaviors, including oxygen consumption, colony formation, cell migration, and invasion by miR‐335‐5p/SIX2 axis. Hence, circ_0007255 might serve as a novel diagnostic and prognostic biomarker for the therapy of BC (Fig 10).
Figure 10.
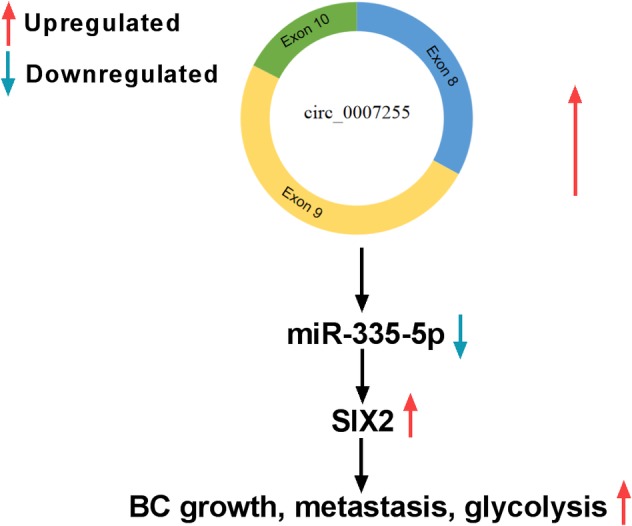
The mechanism schematic model by circ_0007255/miR‐335‐5p/SIX2 in BC. Circ_0007255 regulated the number of colonies, migrated and invaded BC cells, as well as glycolysis via targeting miR‐335‐5p/SIX2 axis.
Disclosure
The authors declare that they have no financial conflicts of interest.
Acknowledgment
None.
Contributor Information
Qianxin Jia, Email: ajeozv@163.com.
Ziqian Chen, Email: vwafdq@163.com.
References
- 1. Bray F, Ferlay J, Soerjomataram I et al Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Ojo D, Wei F, Liu Y et al Factors promoting tamoxifen resistance in breast cancer via stimulating breast cancer stem cell expansion. Curr Med Chem 2015; 22: 2360–74. [DOI] [PubMed] [Google Scholar]
- 3. Abdel‐Mohsen MA, Ahmed OA, El‐Kerm YM. BRCA1 gene mutations and influence of chemotherapy on autophagy and apoptotic mechanisms in Egyptian breast cancer patients. Asian Pac J Cancer Prev 2016; 17: 1285–92. [DOI] [PubMed] [Google Scholar]
- 4. Tryfonidis K, Zardavas D, Katzenellenbogen BS, Piccart M. Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat Rev 2016; 50: 68–81. [DOI] [PubMed] [Google Scholar]
- 5. Osborne CK, Schiff R, Fuqua SA, Shou J. Estrogen receptor: Current understanding of its activation and modulation. Clin Cancer Res 2001; 7: 4338s–42s; discussion 411s‐412s. [PubMed] [Google Scholar]
- 6. Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR‐34a microRNA. Breast Cancer Res Treat 2011; 130: 663–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao S, Li X, Ding X, Qi W, Yang Q. Cepharanthine induces autophagy, apoptosis and cell cycle arrest in breast cancer cells. Cell Physiol Biochem 2017; 41: 1633–48. [DOI] [PubMed] [Google Scholar]
- 8. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013; 340: 440–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ashwal‐Fluss R, Meyer M, Pamudurti NR et al circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 2014; 56: 55–66. [DOI] [PubMed] [Google Scholar]
- 10. Memczak S, Jens M, Elefsinioti A et al Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–8. [DOI] [PubMed] [Google Scholar]
- 11. Salzman J, Chen RE, Olsen MN et al Cell‐type specific features of circular RNA expression. PLOS Genet 2013; 9: e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeck WR, Sorrentino JA, Wang K et al Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19: 141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen B, Wei W, Huang X et al circEPSTI1 as a prognostic marker and mediator of triple‐negative breast cancer progression. Theranostics 2018; 8: 4003–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liang HF, Zhang XZ, Liu BG et al Circular RNA circ‐ABCB10 promotes breast cancer proliferation and progression through sponging miR‐1271. Am J Cancer Res 2017; 7: 1566–76. [PMC free article] [PubMed] [Google Scholar]
- 15. Tang H, Huang X, Wang J et al circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple‐negative breast cancer. Mol Cancer 2019; 18: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hansen TB, Jensen TI, Clausen BH et al Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–8. [DOI] [PubMed] [Google Scholar]
- 17. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32: 453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol 2014; 24: R762–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302: 1–12. [DOI] [PubMed] [Google Scholar]
- 20. Tavazoie SF, Alarcon C, Oskarsson T et al Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008; 451: 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang LL, Zhang LF, Guo XH et al Downregulation of miR‐335‐5p by long noncoding RNA ZEB1‐AS1 in gastric Cancer promotes tumor proliferation and invasion. DNA Cell Biol 2018; 37: 46–52. [DOI] [PubMed] [Google Scholar]
- 22. Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol 2013; 14: 205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. [DOI] [PubMed] [Google Scholar]
- 24. Wang CA, Drasin D, Pham C et al Homeoprotein Six2 promotes breast cancer metastasis via transcriptional and epigenetic control of E‐cadherin expression. Cancer Res 2014; 74: 7357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karedath T, Ahmed I, Al Ameri W et al Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer 2019; 19: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng J, Liu X, Wang P et al CRNDE promotes malignant progression of glioma by attenuating miR‐384/PIWIL4/STAT3 axis. Mol Ther 2016; 24: 1199–215. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016; 238: 42–51. [DOI] [PubMed] [Google Scholar]
- 28. Weng W, Wei Q, Toden S et al Circular RNA ciRS‐7‐a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 2017; 23: 3918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang M, Zhao K, Xu X et al A peptide encoded by circular form of LINC‐PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 2018; 9: 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xie F, Li Y, Wang M et al Circular RNA BCRC‐3 suppresses bladder cancer proliferation through miR‐182‐5p/p27 axis. Mol Cancer Res 2018; 17: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhong Y, Du Y, Yang X et al Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018; 17: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han D, Li J, Wang H et al Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology 2017; 66: 1151–64. [DOI] [PubMed] [Google Scholar]
- 33. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016; 17: 205–11. [DOI] [PubMed] [Google Scholar]
- 34. Png KJ, Yoshida M, Zhang XH et al MicroRNA‐335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev 2011; 25: 226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heyn H, Engelmann M, Schreek S et al MicroRNA miR‐335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer 2011; 129: 2797–806. [DOI] [PubMed] [Google Scholar]
- 36. Afonso‐Grunz F, Muller S. Principles of miRNA‐mRNA interactions: Beyond sequence complementarity. Cell Mol Life Sci 2015; 72: 3127–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pierce J, Murphy AJ, Panzer A et al SIX2 effects on Wilms tumor biology. Transl Oncol 2014; 7: 800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]


