Abstract
Retinoic acid-inducible gene-I (RIG-I) is a cytosolic pathogen sensor that is crucial against a number of viral infections. Many viruses have evolved to inhibit pathogen sensors to suppress host innate immune responses. In the case of influenza, nonstructural protein 1 (NS1) suppresses RIG-I function, leading to viral replication, morbidity, and mortality. We show that silencing NS1 with in-vitro-transcribed 5′-triphosphate containing NS1 short hairpin RNA (shRNA) (5′-PPP-NS1shRNA), designed using the conserved region of a number of influenza viruses, not only prevented NS1 expression but also induced RIG-I activation and type I interferon (IFN) expression, resulting in an antiviral state leading to inhibition of influenza virus replication in vitro. In addition, administration of 5′-PPP-NS1shRNA in prophylactic and therapeutic settings resulted in significant inhibition of viral replication following viral challenge in vivo in mice with corresponding increases of RIG-I, IFN-β, and IFN-λ, as well as a decrease in NS1 expression.
Keywords: Influenza, antiviral, NS1, RIG-I, Interferon, 5'PPP-RNA, shRNA
Graphical Abstract

Introduction
Influenza epidemics cause significant morbidity and mortality every year in older adults and those with underlying medical conditions. Influenza infections resulted in 48.8 million illnesses, more than 22.7 million medical visits, 959,000 hospitalizations, and 79,400 deaths during the 2017–2018 season in the US alone.1 Although vaccination is the most cost-effective preventive strategy against influenza infections, antiviral agents, specifically neuraminidase inhibitors, are the drugs of choice to treat influenza viral infections.2 To have the antiviral effects, oseltamivir, the neuraminidase inhibitor, needs to be administered within 48 h of diagnosis and given twice a day for 5 days or once daily for 10 days for those who come in contact with flu-infected patients. Recently, a new drug, baloxavir marboxil, marketed as Xofluza, which inhibits influenza viral polymerase, was approved to treat influenza infections, almost two decades after the approval of oseltamivir.3 Similar to oseltamivir, Xofluza needs to be administered within 48 h of influenza diagnosis. However, drug resistance has been a major issue with neuraminidase inhibitors, and a majority of circulating viruses during 2008 became resistant to oseltamivir prior to the 2009 pandemic.4, 5, 6, 7 Circulation of drug-resistant strains of H1N1 and H3N2 against not only neuraminidase inhibitors but also against the newly introduced polymerase subunit inhibitor, Xofluza, have been reported.8, 9, 10, 11, 12, 13 The emergence of drug-resistant strains is of major concern because a severe epidemic or a pandemic with a drug-resistant strain can result in severe morbidity and a large number of deaths.14,15 Hence developing novel antiviral strategies is an immediate public health need.
Innate immunity is the first line of defense. Cells recognize molecular patterns associated with pathogens with a conserved set of pathogen recognition receptors located on the cell surface, in vesicles, or in the cytosol.16 Among cytosolic pathogen sensors, we and others have shown that retinoic acid-inducible gene-I (RIG-I) is a major pathogen sensor for influenza virus, and its activation is inhibited by nonstructural protein 1 (NS1) of influenza virus.17, 18, 19, 20 Short single-stranded RNAs (ssRNAs) containing a 5′-triphosphate (5′-PPP) end or longer ssRNAs without a 5′-PPP end from viruses have been shown to activate RIG-I.21 We have shown earlier that activation of the RIG-I pathway confers protection against not only drug-sensitive and -resistant strains of influenza independent of their hemagglutinin (HA) and neuraminidase (NA) type, pathogenicity, and pandemic potential, but also against Ebola virus.22, 23, 24 The RIG-I ligands can be delivered via nanoparticles to activate antiviral defenses.24 In addition, activation of the RIG-I pathway did not generate resistant strains. Because a significant proportion of the human population has defects in type 1 interferon (IFN) induction, as well as activation pathways, the RIG-I-mediated antiviral pathway will not work in that population.25,26 Hence we have developed a dual strategy that silences NS1 (needed for the population that has defects in type 1 IFN induction/activation pathways) and at the same time it activates the antiviral RIG-I pathway (in type 1 IFN-sufficient population), and demonstrate that it is efficacious against influenza virus infection in prophylactic and therapeutic settings, both in vitro and in vivo.
Results
Generation of 5′-PPP-NS1shRNA Using T7 Pol-Based In Vitro Transcription
In this study, we investigated the antiviral potential of 5′-PPP-NS1shRNA, which not only suppresses the NS1 gene of influenza virus but also activates RIG-I-mediated antiviral responses by virtue of its 5′-PPP moiety, therefore serving as a dual-function shRNA. In order to find small stretches of conserved NS1 sequence that can serve as a target sequence in 5′-PPP-NS1shRNAs, we performed sequence alignment of the NS gene segment of 12 different influenza viruses. Sequence alignment results revealed nine short stretches of sequence that were the most conserved among the 12 different influenza viruses and that served as the putative target (NS1) sense sequences in the 5′-PPP-NS1shRNAs tested (Table S1). In addition, we performed an in silico analysis of the A/PR/8/34 NS1 sequence utilizing a multi-parameter prediction of RNA accessibility (mppRNA) analysis paradigm. This analysis determines domains in an RNA sequence that are accessible to shRNA attack by predicting stretches with minimal to no secondary structure that would impede shRNA binding (Figure 1A).27 The double-stranded DNA (dsDNA) templates used for generating the 5′-PPP-NS1shRNAs were composed of a T7 promoter followed by the NS1 target sense sequence, stem loop, target antisense sequence, and hepatitis delta virus (HDV) ribozyme (Figure 1B). The purpose of using a T7 RNA polymerase (T7 Pol)-based transcription was that T7 Pol not only results in a triphosphate moiety at the 5′ end of its transcripts but also has high transcriptase activity with very strict specificity for its promoter.28 Therefore, it has been extensively used for expressing target proteins in eukaryotic cells.29,30 However, one major issue with the use of T7 Pol-based transcription is the heterogeneity at the 3′ end of its transcripts, which may interfere with RNAi phenomenon in mammalian cells.31 In order to circumvent this problem, we incorporated a HDV ribozyme sequence at the 3′ end of the dsDNA template (Figure 1B). Among all nine shRNAs, only three shRNAs, namely, 1, 4, and 9, were found to be effective and functional (data not shown). We also established an optimum dose of 3 μg for these shRNAs to be used in transfection studies by doing a dose-response study (data not shown).
Figure 1.
Design and Characterization of 5′-PPP-NS1shRNA
(A) Determining optimal shRNA sequences by multi-parameter prediction of RNA accessibility (mppRNA) analysis. Peaks indicate domains along the influenza A/PR/8/34 sequence with a higher probability of access for shRNA attack based on secondary structure. Orange shaded regions indicate domains with ≥97% homology with the consensus sequence of 12 influenza A strains. #1, #4, and #9 sequence domains from which shRNAs were derived that resulted in anti-influenza activity. *Location of shRNA species that was utilized in subsequent detailed in vitro and in vivo analysis. (B). The dsDNA template to generate 5′-PPP-NS1shRNA by T7 polymerase (7 Pol)-based in vitro transcription (IVT) was composed of a T7 Pol promoter region (in yellow) at the 5′ end of the dsDNA template followed by the target sense sequence (in blue), loop (in green), target antisense (in gray), and hepatitis delta virus (HDV) ribozyme (in pink). Both ends of this dsDNA template were flanked by restriction enzyme sites (in red). 5′-PPP-NS1shRNAs were synthesized using a T7 RNA Pol-based IVT kit. A549 cells (1 × 106/well) were dispensed into six-well plates and then mock transfected (cont) or transfected with 3 μg of IVT 5′-PPP shRNAs (with the indicated sense sequence or a scrambled control) or their m7G-capped counterparts, as indicated, and cells were harvested 24 h post-transfection for RNA analysis by quantitative real-time PCR. (C–E) In a separate set of experiments, these transfected cells were also infected with 1.0 MOI of A/Brisbane/59 (H1N1) influenza virus and were harvested 24 h post-infection for mRNA analysis of (C) RIG-I, (D) IFN-β, and (E) NS1 expression. mRNA levels are expressed as fold increase over controls in shRNA-transfected cells. Error bars represent the mean ± SD from three independent experiments. One-way analysis of variance (ANOVA) with multiple comparisons analysis was used to analyze differences among treatment groups and control group.
5′-PPP-NS1shRNA Enhances RIG-I and IFN-β, and Suppresses NS1 Expression in A549 Cells following Influenza Virus Infection
We and others have shown the antiviral potential of 5′-PPP-RNA for influenza and other viruses.21, 22, 23,32,33 In this study, we have designed a dual-function shRNA that can activate the RIG-I-mediated antiviral responses and also knock down the NS1 protein of influenza virus. In order to assess the functionality of our dual-function shRNAs, A549 cells transfected with all 5′-PPP-NS1shRNAs or their capped 5′ 7-methylguanosine (m7G)-NS1shRNAs lead to significant upregulation of RIG-I (up to 40-fold; Figure 1C) and IFN-β (up to 200-fold; Figure 1D), as compared with transfection control and capped counterparts of NS1shRNAs and capped scrambled shRNA, where the triphosphate moiety at the 5′ end was blocked by a synthetic cap analog, m7G. We also assessed the ability of these dual-function NS1shRNAs to suppress production of NS1 by the influenza virus. A549 cells were mock transfected or transfected with 5′-PPP-shRNAs or their m7G-capped counterparts, and 24 h later infected with A/Brisbane/9/2007 influenza virus. 80%–90% knockdown in NS1 mRNA levels was achieved with all NS1shRNAs with 5′-PPP or without 5′-PPP (m7G-capped), as compared with transfection control (Figure 1E). Interestingly, scrambled shRNA with a 5′-PPP also suppressed NS1 to the similar extent as other 5′-PPP-NS1shRNAs by virtue of the triphosphate moiety at the 5′ end, which activates RIG-I-mediated antiviral responses. However, scrambled shRNA without the 5′-PPP moiety (m7G-capped) behaved in a manner similar to the transfection control because of the lack of NS1 silencing ability, as well as the absence of the 5′-PPP moiety (Figure 1E).
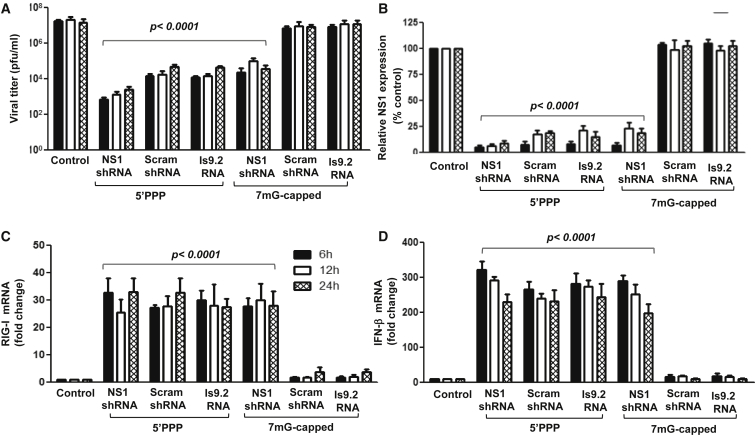
Differentiating NS1 Silencing versus RIG-I Activation
To distinguish the antiviral responses mediated by RIG-I activation and induction of type 1 IFN and its interaction with type 1 IFN receptors due to the presence of the 5′-PPP moiety on the shRNA versus those mediated by the NS1 silencing effect of the shRNA, we used Vero cells that lack the IFN-α and IFN-β genes.34 Vero cells were transfected with shRNAs with or without 5′-PPP groups and then infected with A/Brisbane/59/2007 at a multiplicity of infection (MOI) of 1.0 at 6 and 24 h post-transfection. RNA and cell lysates for NS1 mRNA and protein expression were collected. A 75% reduction in the levels of NS1 transcripts (Figure 2A), as well as NS1 protein (Figure 2B), was observed only in cells treated with NS1shRNA uncapped or m7G-capped followed by influenza virus infection as compared with controls. However, no reduction in NS1 expression either at message or protein level was observed in cells treated with 5′-PPP-scrambled shRNA or 5′-PPP control RNA (Is9.2). These results coupled with those described in Figure 1 support the hypothesis that NS1shRNA mediates antiviral effects both by suppression of NS1 expression and by induction of type 1 IFN-mediated antiviral effects.
Figure 2.
Differentiating NS1 Silencing and Type 1 IFN-Mediated Antiviral Effects
Vero cells (1 × 106) were mock transfected or transfected with 3 μg of shRNAs or their capped counterparts as shown. Six (empty bars) or 24 h (filled bars) later, the transfected cells were infected with 1 MOI of A/Brisbane/59/2007 influenza virus. The cells were harvested 24 h post-infection for NS1 mRNA and protein analysis by quantitative real-time PCR (A) and western blot (B), respectively. Error bars represent the mean ± SD from three independent experiments. One-way ANOVA with multiple comparisons analysis was used to analyze differences among NS1shRNA, scram shRNA, and Is9.2 RNA groups.
5′-PPP-NS1shRNA Exerts Its Antiviral Effect in a Prophylactic Setting
Having demonstrated the functionality of NS1shRNAs, we assessed the potential prophylactic utility of the dual-function shRNAs. We first transfected A549 cells with 3 μg of 5′-PPP-shRNA constructs and their m7G-capped counterparts, as well as control shRNA. At 6, 12, and 24 h post-transfection of shRNAs, A549 cells were infected with 1.0 MOI of A/Brisbane/59/2007 virus. Twenty-four hours post-infection, supernatants and cell lysates were collected to determine viral titers in plaque assay using Madin Darby canine kidney (MDCK) cells and RNA to determine NS1, RIG-I, and IFN-β transcript levels, respectively. All shRNA constructs with the exception of m7G-capped scrambled and m7G-capped control constructs significantly reduced viral titers when compared with control, with inhibitions ranging from 4 logs to 2 logs (Figure 3A). For example, NS1shRNA containing a 5′-PPP moiety significantly reduced viral titers by about 4 logs at the 6-h time point. Although the antiviral effect appears to be very high at 6 h when compared with 24 h, there were no statistically significant differences between 6, 12, and 24 h. The inhibition was mediated by RIG-I activation, as well as direct inhibition of NS1 transcripts. m7G-capped NS1shRNA inhibited viral titers by about 3 logs, and this inhibition was mediated by direct inhibition of NS1 by shRNA. RIG-I activation and NS1 suppression work synergistically in dual-functioning 5′-PPP-NS1shRNA. 5′-PPP-containing scrambled shRNA construct did reduce viral titers through RIG-I activation; however, the reduction in viral titers hovered around 3 logs for all time points (Figure 3A). Similarly, the positive control shRNA, Is9.2, also reduced viral titers by about 3 logs, and this inhibition was mediated through RIG-I activation. The inhibition of viral titers correlated with the significant inhibition (80%–90%) of NS1 transcription (Figure 3B), with corresponding increases of about 20- to 30-fold in RIG-I (Figure 3C) and 250- to 300-fold in IFN-β (Figure 3D) induction.
Figure 3.
Antiviral Activity Mediated by NS1shRNA in a Prophylactic Setting
A549 cells (1 × 106) were mock transfected or transfected with 3 μg of IVT shRNAs or their 7mG-capped counterparts, as indicated. The transfected cells were infected with 1.0 MOI of A/Brisbane/59/2007 influenza virus at indicated time points (i.e., 6 [filled bars], 12 [empty bars], and 24 h [hatched bars]) post-transfection and harvested for mRNA analysis by quantitative real-time PCR. Cell supernatants were collected for detecting viral titers by plaque assay. (A–D) Viral titer (A), relative mRNA expression of (B) NS1, (C) RIG-I, and (D) IFN-β measured by plaque assay and qRT-PCR analysis, respectively. mRNA expressions are displayed as fold increase over controls. Error bars represent the mean ± SD from three independent experiments. One-way ANOVA with multiple comparisons analysis was used to analyze differences among treatment groups and control group.
5′-PPP-NS1shRNA Exerts Its Antiviral Effect in Therapeutic Settings
To evaluate the therapeutic potential of our dual-functional shRNA, A549 cells were first infected with 1.0 MOI of A/Brisbane/59/2007 influenza virus and at 6, 12, and 24 h post-infection, the cells were treated with shRNAs (Figure 4). Viral titers were significantly lower (>3 logs reduction) in infected cells treated with NS1shRNA with or without 5′-PPP, or scrambled shRNA with 5′-PPP and Is9.2 with 5′-PPP at 6 and 12 h post-infection (Figure 4A). However, the reduction was about 1 log at 24 h post-infection. Neither m7G-capped scrambled shRNA nor m7G-capped Is9.2 RNA had any impact on viral titers. NS1shRNAs with or without 5′-PPP were able to suppress NS1 levels ranging from 30% to 70% as compared with controls in cells at 6, 12, and 24 h post-infection, unlike 5′-PPP-scramshRNA and 5′-PPP-Is9.2 RNA, which suppressed NS1 by 45%–50% at 6 h post-infection and marginally, 5%–15% at 12 and 24 h post-infection (Figure 4B). m7G-capped-scrambled shRNA and m7G-capped Is9.2shRNA failed to suppress NS1 expression. There was a 20- to 30-fold increase in the levels of RIG-I as compared with transfection control in virus-infected cells that were treated with 5′-PPP-NS1shRNA 6 h post-infection. However, this induction of RIG-I was not seen at 12- or 24-h time points during infection (Figure 4C). Interestingly, treatment of infected cells with NS1shRNA with or without the 5′-PPP moiety, 5′-PPP-scramshRNA, and 5′-PPP-Is9.2 RNA induced significantly higher levels of IFN-β (>100-fold) when cells were treated with them at 6 h post-infection (Figure 4D). Although NS1shRNA with or without the 5′-PPP group induced about a 70-fold increase in IFN-β at 12 h post-infection, none of the other constructs induced significant levels of IFN-β.
Figure 4.
Therapeutic Potential of 5′-PPP-NS1shRNA
A549 cells (1 × 106) were mock infected or infected with 1.0 MOI of A/Brisbane/59 influenza virus, and 6 (filled bars), 12 (empty bars), or 24 h (hatched bars) later the cells were transfected with 3 μg of IVT shRNAs or their 7mG-capped counterparts, as indicated. After 24 h, the cells were analyzed for mRNA by quantitative real-time PCR, and supernatants were collected for detecting viral titers by plaque assay. (A–D) Viral titers in the supernatants measured by plaque assay in MDCK cells (A): NS1 (B), RIG-I (C), and IFN-β (D). Error bars represent the mean ± SD from three independent experiments. One-way ANOVA with multiple comparisons analysis was used to analyze differences among treatment groups and control group.
Prophylactic and Therapeutic Potential of NS1shRNA In Vivo
To assess the ability of NS1shRNA and its capped counterparts, as well as scrambled shRNA, to reduce viral titers both in a prophylactic and a therapeutic model in vivo, we administered one dose of RNA constructs formulated with in vivo-jetPEI, intranasally, on days −1, 0, 1, 2, and 3, and infected all mice on day 0 with A/PR/8/34 virus. The control consisted of in vivo-jetPEI alone with PBS. We harvested lungs on day 4 post-infection to measure viral titers by plaque assay and RIG-I, NS1, IFN-β, IFN-λ, and RANTES by quantitative real-time PCR (Figure 5). A single-dose administration of NS1shRNA, scrambled shRNA, and m7G-capped NS1shRNA on day −1 (24 h prior to virus infection) in a prophylactic model reduced viral titers by 2 logs (or 90% reduction) (Figure 5A) with a corresponding 3 log decrease of NS1 expression (Figure 5B), 50- to 60-fold upregulation of RIG-I (Figure 5C), 150- to 250-fold increase in IFN-β expression (Figure 5D), 70- to 75-fold increase in IFN-λ expression (Figure 5E), and 30- to 35-fold increase in RANTES expression (Figure 5F). These findings clearly indicate that a single dose of 5′-PPP-shRNA results in NS1 suppression and RIG-I activation, and suppresses influenza viral replication significantly. However, single-dose administration on the same day of infection or 24 h post-infection, albeit less effective than day −1 administration, did result in a significant decrease in viral replication and NS1 expression with corresponding increases in RIG-I, IFN-β, IFN-λ, and RANTES in a therapeutic setting. However, delayed administration on day 2 or 3 was less effective, with modest or no desirable protective response.
Figure 5.
Prophylactic and Therapeutic Potential of NS1shRNA in Mice
Mice were intranasally administered 50 μg of 5′-PPP-NS1shRNA (□), m7G-capped-NS1shRNA (▲), 5′-PPP-scrambled shRNA (X), or m7G-capped-scrambled shRNA (Δ) mixed with in vivo transfection reagent in vivo-jetPEI or in vivo-jetPEI alone (▪). All shRNA constructs were administered on day −1, 0, 1, 2, or 3. Mice were infected with mouse-adapted A/PR/8/1934 (H1N1) on day 0, and mouse lungs were harvested on day 4. (A–F) Lungs were analyzed for (A) viral titers by plaque assay, and mRNA expression of (B) NS1, (C) RIG-I, (D) IFN-β, (E) IFN-λ, and (F) RANTES by quantitative real-time PCR and expressed as fold increase over controls. Error bars represent the mean ± SD from three independent experiments. One-way ANOVA with multiple comparisons analysis was used to analyze differences among treatment groups and PBS group.
Discussion
Despite being a vaccine-preventable disease, influenza continues to remain a major public health problem worldwide. Influenza viruses infect 5%–15% of the global population annually, resulting in between 291,000 and 646,000 influenza-associated deaths.35 In the United States alone, influenza viruses are estimated to infect more than 50 million people every year, resulting in more than 200,000 hospitalizations and 30,000–50,000 deaths.1 Influenza affects people of all age groups, but the highest risk of complications occurs among children under the age of 2 years, adults over 65 years old, pregnant women, and people with certain medical conditions, such as cancer, chronic lung disease, heart disease, diabetes, and blood, lung, or kidney disorders.
Current influenza vaccine formulations typically generate strain-specific immune responses and are reformulated every year to match the circulating virus strains. The effectiveness of influenza vaccines is largely dependent on the antigenic closeness of the vaccine virus strain with that of the circulating virus, as well as the attack rate. A meta-analysis showed a pooled vaccine effectiveness rate of 70% (95% confidence interval [CI]: 55%–80%) for matched viruses and 55% (95% CI: 42%–65%) for unmatched strains. The overall vaccine effectiveness of the 2018–2019 influenza vaccine against both influenza A and B viruses is estimated to be 40%.1 However, the effectiveness of influenza vaccines in young children and older adults is comparatively lower due to weaker immune systems in these populations. Hence, apart from prophylactic intervention strategies, therapeutic interventions with antiviral drugs play a central part in the treatment of influenza viral infections. Three classes of drugs are licensed to treat against influenza. They include M2-ion channel inhibitors (amantadine approved in 1966), neuraminidase inhibitors (oseltamivir approved in 2000), and a viral polymerase inhibitor (baloxavir marboxil [Xofluza] approved in 2018).2,3,36 However, amantadine and its derivatives are no longer used because 100% of circulating influenza viruses are resistant to them.37 The majority of circulating H1N1 viruses prior to the 2009 pandemic became resistant to oseltamivir, and the incidence of oseltamivir-resistant viruses of currently circulating pandemic H1N1 strains has been reported.4,6, 7, 8,10,38 Isolation of baloxavir marboxil-resistant viruses during clinical trial indicates the potential development of drug-resistant strains.9,11,12 Hence it is important to develop next-generation antivirals that confer protection against viruses that are resistant against currently licensed drugs. Instead of targeting viral components to inhibit viral replication, we targeted host innate immune components to create an antiviral state to suppress viral growth. We have shown earlier that activating the RIG-I pathway inhibited the growth of influenza viruses irrespective of their genetic makeup, drug sensitivity status, and pathogenicity both in vitro and in vivo.22 In addition, we demonstrated that activation of the RIG-I pathway also inhibited Ebola viral replication.23 Furthermore, the viruses did not develop resistance to RIG-I-mediated antiviral defenses.
We chose 5′-PPP-shRNA against NS1 to perform the dual function of NS1 silencing and RIG-I activation rather than other viral structural components because NS1 is the viral virulence factor that suppresses host innate immune responses to facilitate viral replication. A well-conserved NS1 sequence, along with appropriate controls, was selected to distinguish RIG-I-mediated effects versus NS1 silencing effects by m7G-capping of the 5′-PPP group and scrambling the NS1 sequence. Any antiviral strategy should be applicable to the entire population. A significant number of individuals have defects in type 1 IFN pathways resulting in lack of induction of type 1 IFNs and/or absence or suboptimal responses to type 1 IFN-mediated signaling. In this population, RIG-I-mediated antiviral effects via the 5′-PPP group for type 1 IFN induction will not work. Hence we exploited NS1shRNA-mediated silencing of NS1 to protect the population.25,26 This has been clearly shown in Figure 2 using Vero cells that are defective in type 1 IFN signaling cascade. Only NS1shRNAs capped or uncapped were able to suppress NS1 expression, and none of the other constructs were able to suppress NS1 expression through RIG-I-mediated effects via 5′-PPP. In a prophylactic model in cell lines, there was a 3.5–4 log reduction in A/Brisbane/59/2007 viral titers when the cells were infected 6, 12, and 24 h post-transfection of shRNA constructs (Figure 3A). Although the dual-functioning RNA, 5′-PPP-NS1shRNA, is most effective, 7mG-capped NS1shRNA and 5′-PPP-scrambled shRNA are also very effective against viral replication. 7mG-capped control 9.2 RNA and 7mG-capped scrambled NS1shRNA lost their effect, indicating the importance of the 5′-PPP group for RIG-I activation. Using small interfering RNA against nucleoprotein (NPsiRNA) of influenza virus, a reduction of 1–2 logs in viral titer has been reported depending on the amount of 5′-PPP-NPsiRNA used.32 In a therapeutic setting,33 there was 3.5–4 log, 3.5 log, and 1–1.5 log reduction if 5′-PPP-NS1shRNA is transfected 6, 12, and 24 h post-infection. A similar trend is seen with m7G-capped-NS1shRNA (Figure 4A). Corresponding increases in IFN-β and RIG-I, and decreases in NS1 mRNA were also observed. An earlier report utilized 5′-PPP-NS1 silencing RNA and demonstrated only 40%–45% reduction in A/PR/8/34 viral titers in contrast with our 3.5 to 4 log reduction in viral titers.33 The observed poor effectiveness of their approach may be caused by the shRNA sequence, time of transfection of shRNA, and/or the viruses and cell lines (MDCK versus A549) used.
In a prophylactic setting in vivo, administration of 5′-PPP-NS1shRNA 24 h prior to infection resulted in a 2 log reduction in viral titers for 5′-PPP-NS1shRNA, m7G-capped-NS1shRNA, and 5′-PPP-scrambled NS1shRNA (Figure 5A) with corresponding increases in RIG-I, IFN-β, IFN-λ, and RANTES and a concomitant decrease in NS1 mRNA. In a therapeutic setting, when NS1shRNA constructs were administered on the day of viral infection and 24 h post-infection, there was a reduction of viral titers by 2 and 1.5 logs, respectively. However, the therapeutic effect was only a 1.5 to 1 log reduction in viral titers when the shRNAs were administered on days 2 and 3. When Svancarova et al.33 used NS1siRNA in a similar therapeutic model with A/PR/8/34 challenge, reduction of viral titers was not statistically significant, which may be because of the dose of the virus they used 4 times the viral dose that kills 50% of mice (4LD50). Similarly, using 5′-PPP-NPsiRNA, a 1 log reduction in viral titer was observed.32 Unlike the earlier observations, our study extended our initial findings demonstrating that activation of the RIG-I pathway results in an antiviral state in cells and showed that a single administration of dual-acting 5′-PPP-NS1shRNA suppresses viral replication >3 logs in vitro and 2 logs in vivo in a prophylactic setting. In a therapeutic setting, treatment can be delayed up to 24–48 h to have a beneficial effect in viral titer reduction. Furthermore, the ease of administration by the intranasal route, unlike the intravenous route used by others, facilitated targeted delivery and distribution to have the maximal antiviral effects in the target organ. RIG-I activation can be exploited to prevent or treat not only influenza, as shown by us earlier and in this report, but also to treat other viral infectious diseases, including Ebola, and cancer.17,22, 23, 24
Materials and Methods
Cell Lines
Human lung epithelial cells (A549), African green monkey kidney cells (Vero), and MDCK cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Influenza Viruses
A/Brisbane/59/2007 (H1N1) and mouse-adapted A/Puerto Rico/8/34 (H1N1) viruses were obtained from the influenza division Centers for Disease Control and Prevention (CDC) repository and were propagated in 10-day-old embryonated chicken eggs as previously described.39 Pooled allantoic fluid was clarified by centrifugation, aliquoted, and stored at −80°C until use. Both viruses were sequenced at the CDC for HA and NA, and titered for plaque-forming units (PFUs) in MDCK cells. The titer for A/Brisbane/59/2007 is 1.55 × 108 PFUs/mL and A/Puerto Rico/8/34 is 2 × 109 PFUs/mL.
mppRNA
NS1 RNA was subjected to a bioinformatics approach (mppRNA) to predict regions of accessibility in target RNAs that are subject to post-transcriptional gene silencing therapeutics. mppRNA uses three algorithms: MFold, SFold, and Oligowalk. MFold identifies the most stable secondary structure and a neighborhood of less stable structures. SFold uses a Boltzmann energy sampling algorithm to assess the access probability across the entire secondary structural folding space. OligoWalk assesses the local folding energy along the target RNA. Details of the approach and the underlying algorithms can be found elsewhere.40, 41, 42, 43 The accessibility vector output from each algorithm was combined with equal weighting, and the final access probability plotted versus nucleotide along the NS1 RNA. Strong peaks of accessibility were identified in the map that were used as small interfering RNA (siRNA) targeting sites.
In Vitro Synthesis of 5′-PPP-NS1shRNA
The sequences of the NS1 gene segment from 12 different influenza A viruses were aligned using NCBI tblastn to find the most conserved regions to serve as the universal target sense sequence in DNA templates. shRNAs based on these target sequences were designed with the guidance of mppRNA.27,44 Nine sequences were tested for knockdown efficacy. Sequences #1 (5′-GTGATGCCCCATTCCTTGA-3′), #4 (5′-TGAGGATGTCAAAAATGCA-3′), and #9 (5′-ATAAAAAACACCCTTGTTTCTACT-3′) were found to be the most effective in knocking down NS1 message and protein of influenza viruses used in this study, and #4 was used subsequently for the rest of the experiments. As a control, we used Is9.2, a 19-mer shRNA (5′-AGCUUAACCUGUCCUUCAA-3′), with 5′-PPP that did not target NS1 and its m7G-capped counterpart.45
DNA templates, composed of a T7 promoter followed by desired target sense sequence, loop, target antisense, HDV ribozyme, and ends flanked with a restriction enzyme site (Figure 1B), were created by primer annealing using two separate oligos (25 pmol each) with a 40+ bp complementary overlap region and DNA polymerase extension. PCR conditions consisted of: 94°C for 5 min followed by 3 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, and final extension at 72°C for 10 min. The DNA oligos were custom-made in an in-house CDCDSR/BIOS Oligo service center. 5′-PPP-NS1shRNAs were synthesized using a T7 Pol-based in vitro transcription kit (Ambion MEGAscript T7 High yield transcription kit; Austin, TX, USA), as per the manufacturer’s instructions. The in vitro transcription reaction was carried out for 16 h followed by treatment with DNase I (NEB, Ipswich, MA, USA) to digest the DNA template, and the RNA was purified using TRIzol (Invitrogen, Carlsbad, CA, USA), followed by ethanol precipitation. m7G-capped NS1shRNAs were prepared by replacing the guanosine triphosphate (GTP) in the in vitro transcription reaction with a 12:1 ratio of m7G(5′)PPP(5′)G cap analog:GTP. The 5′-PPP-RNA and capped RNA used in this study were prepared as previously described.21,22
Differentiating RIG-I-Mediated Effects and NS1 Silencing Effects
Vero cells that lack type 1 IFN genes were grown in six-well tissue culture plates and transfected with 3 μg of shRNAs containing the 5′-PPP group, as well as their m7G-capped counterparts using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA), as per the manufacturer’s instructions, and infected with A/Brisbane/59/2007 virus at an MOI of 1.0 at 6 and 24 h post-transfection. RNA and cell lysates were collected 24 h later for analysis of NS1 message expression by qRT-PCR and NS1 protein expression by western blot analysis.
Assessing Functionality of shRNA Constructs In Vitro
Prophylactic Settings
A549 (six-well tissue culture plates) were transfected with 3 μg/well (as optimized by dose-response study) of in-vitro-transcribed 5′-PPP-NS1shRNA, 5′-PPP-scramshRNA, control Is9.2 5′-PPP-RNA, or their 7mG-capped counterparts using Lipofectamine 2000 (Life Technologies, Grand Island, NY, USA), as per manufacturer’s instructions. At 6, 12, or 24 h post-transfection, cells were infected with A/Brisbane/59/2007 influenza virus at an MOI of 1.0 with trypsin supplementation. At 24 h post-infection, cells were harvested for RNA and protein analysis, and supernatants were collected for determination of viral titer by plaque assay using MDCK cells, as described previously.17
Therapeutic Settings
To study the therapeutic potential of 5′-PPP-NS1shRNA, A549 cells were first infected with A/Brisbane/59/2007 influenza virus at an MOI of 1.0 with trypsin supplementation for 6, 12, or 24 h, followed by transfection with in-vitro-transcribed 5′-PPP-NS1shRNA, 5′-PPP-scramshRNA, Is9.2 5′-PPP-RNA, or their m7G-capped counterparts. Cell lysates and supernatants were harvested 24 h post-transfection to perform various assays. Three independent experiments were performed under both prophylactic and therapeutic settings with three time points (6, 12, or 24 h), and each treatment was done in duplicate cultures.
Western Blot
Total protein was separated on a 4%–20% SDS-PAGE gel (Precise; Thermo Fisher Scientific, Waltham, MA, USA) at a constant current of 20 mA. The proteins on the gel were then transferred onto a nitrocellulose membrane using the iBlot 7-Minute Blotting System (Life Technologies). NP and NS1 antibodies used for the western blot were obtained from Influenza Reagents Repository and Dr. Adolfo Garcia-Sastre, and the antibody against β-actin was purchased from Millipore Sigma (St. Louis, MO, USA).
Real-Time RT-PCR
Total RNA was isolated from cells or lung tissue using the RNAeasy plus kit (QIAGEN, Valencia, CA, USA), as per the manufacturer’s protocol. The relative amount of mRNA for RIG-I, IFN-β, NS1, and β-actin was measured by real-time RT-PCR using the Superscript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen) according to the manufacturer’s protocol on a Stratagene Mx3000P PCR machine (Stratagene, La Jolla, CA, USA). The PCR protocol consisted of: 94°C for 15 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s for a total of 45 cycles, and relative mRNA levels were expressed as fold change. The human primers used in these studies were:22 IFN-β, forward 5′-TGG GAG GCT TGA ATA CTG CCT CAA-3′ and reverse 5′-TCT CAT AGA TGG TCA ATG CGG CGT-3′; RIG-I, forward 5′-AAA CCA GAG GCA GAG GAA GAG CAA-3′ and reverse 5′-TCG TCC CAT GTC TGA AGG CGT AAA-3′; NS1, forward 5′-AGA AAG TGG CAG GCC CTC TTT GTA-3′ and reverse 5′-TGT CCT GGA AGA GAA GGC AAT GGT-3′; β-actin, forward 5′-ACC AAC TGG GAC GAC ATG GAG AAA-3′ and reverse 5′-TAG CAC AGC CTG GAT AGC AAC GTA-3′. Mouse primer sets used in this study were: mRIG-I, forward 5′-GCCCTGTACCATGCAGGTTAC-3′ and reverse 5′ -AGTCCCAACTTTCGATGGCTT-3′; mIFN-β, forward 5′-CCAGCTCCAAGAAAGGACGA-3′ and reverse 5′-CGCCCTGTAGGTGAGGTTGAT-3′; mβ-actin, forward 5′-ATGCTCCCCGGGCTGTAT-3′ and reverse 5′-CATAGGAGTCCTTCTGACCCATTC-3′.
In Vivo Studies
Female BALB/c mice, 6–12 weeks old (Jackson Laboratories, Bar Harbor, ME, USA), were anesthetized with isoflurane and administered (5 mice/group) 25 μg of 5′-PPP-NS1shRNA, capped-NS1shRNA, Is9.2 synthetic RNA, or PBS complexed with in vivo-jetPEI according to the manufacturer’s protocol (6 μL/mouse; Polyplus-transfection, San Diego, CA, USA), intranasally, in a volume of 40 μL. Control mice were immunized with in vivo-jetPEI alone. in vivo-jetPEI is a linear polyethylenimine that mediates efficient nucleic acid delivery into animal models. All shRNA constructs were administrated on day −1, 0, 1, 2, or 3. Mice were challenged on day 0, intranasally, with ten 50% mouse infectious doses (MID50) of mouse-adapted A/PR/8/34 virus in a final volume of 50 μL under anesthesia. Mice were observed for any clinical signs, such as ruffled fur, lethargy, huddling together, or labored breathing, which are indicative of morbidity or sickness. Mouse lungs were harvested on day 4 postchallenge and immediately frozen and stored at −80°C in a freezer. Animal research was conducted under the guidance of the CDC’s Institutional Animal Care and Use Committee in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility.
IFN-β ELISA
IFN-β levels were measured in lung homogenates using a mouse ELISA kit (PBL Assay Science, Piscataway, NJ, USA) according to the manufacturer’s protocol. In brief, lung homogenates and assay standards were added to antibody-coated microtiter strips, followed by a biotinylated-labeled antibodies solution, streptavidin-HRP conjugate, and finally, a substrate and stop solution. The plates were read using a BioTek Synergy 4 plate reader (BioTek, Winooski, VT, USA). Absorbance at 450 nm values were converted to concentrations (pg/mL) from a standard curve.
Statistical Methods
Statistical analysis was performed using one-way analysis of variance (GraphPad Prism 5.0; GraphPad Software, La Jolla, CA, USA). The data were presented as mean ± SD. The differences were considered statistically significant when p <0.05.
Author Contributions
N.S., P.R., W.C., J.P., and S.G. performed experiments and analyzed data. J.M.S. has analyzed the constructs used in this study. B.A.D., P.N.P., P.R.K., and S.S. have planned and coordinated the studies and have written the manuscript.
Acknowledgments
This study was supported by National Institutes of Health grant AI084410 (awarded to P.R.K., P.N.P., and S.S.) and intramural research support from the Centers for Disease Control and Prevention (Atlanta, GA, USA). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency or Centers for Disease Control and Prevention.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.025.
Contributor Information
Paul R. Knight, Email: pknight@buffalo.edu.
Suryaprakash Sambhara, Email: ssambhara@cdc.gov.
Supplemental Information
References
- 1.Centers for Disease Control and Prevention Estimated influenza illnesses, medical visits, hospitalizations, and deaths in the United States—2017–2018 influenza season. 2018. https://www.cdc.gov/flu/about/burden/2017-2018.htm
- 2.Hayden F.G., Atmar R.L., Schilling M., Johnson C., Poretz D., Paar D., Huson L., Ward P., Mills R.G. Use of the selective oral neuraminidase inhibitor oseltamivir to prevent influenza. N. Engl. J. Med. 1999;341:1336–1343. doi: 10.1056/NEJM199910283411802. [DOI] [PubMed] [Google Scholar]
- 3.Heo Y.A. Baloxavir: First Global Approval. Drugs. 2018;78:693–697. doi: 10.1007/s40265-018-0899-1. [DOI] [PubMed] [Google Scholar]
- 4.Influenza Project Team Oseltamivir resistance in human seasonal influenza viruses (A/H1N1) in EU and EFTA countries: an update. Euro Surveill. 2008;13:8032. [PubMed] [Google Scholar]
- 5.Lackenby A., Hungnes O., Dudman S.G., Meijer A., Paget W.J., Hay A.J., Zambon M.C. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 2008;13:8026. doi: 10.2807/ese.13.05.08026-en. [DOI] [PubMed] [Google Scholar]
- 6.Hauge S.H., Dudman S., Borgen K., Lackenby A., Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 2009;15:155–162. doi: 10.3201/eid1502.081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moscona A. Global transmission of oseltamivir-resistant influenza. N. Engl. J. Med. 2009;360:953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 8.Okomo-Adhiambo M., Fry A.M., Su S., Nguyen H.T., Elal A.A., Negron E., Hand J., Garten R.J., Barnes J., Xiyan X., 2013–14 US Influenza Antiviral Working Group Oseltamivir-resistant influenza A(H1N1)pdm09 viruses, United States, 2013-14. Emerg. Infect. Dis. 2015;21:136–141. doi: 10.3201/eid2101.141006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omoto S., Speranzini V., Hashimoto T., Noshi T., Yamaguchi H., Kawai M., Kawaguchi K., Uehara T., Shishido T., Naito A., Cusack S. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018;8:9633. doi: 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tandel K., Sharma S., Dash P.K., Parida M. Oseltamivir-resistant influenza A(H1N1)pdm09 virus associated with high case fatality, India 2015. J. Med. Virol. 2018;90:836–843. doi: 10.1002/jmv.25013. [DOI] [PubMed] [Google Scholar]
- 11.Gubareva L.V., Fry A.M. Baloxavir and Treatment-Emergent Resistance: Public Health Insights and Next Steps. J. Infect. Dis. 2020;221:337–339. doi: 10.1093/infdis/jiz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubareva L.V., Mishin V.P., Patel M.C., Chesnokov A., Nguyen H.T., De La Cruz J., Spencer S., Campbell A.P., Sinner M., Reid H. Assessing baloxavir susceptibility of influenza viruses circulating in the United States during the 2016/17 and 2017/18 seasons. Euro Surveill. 2019;24:1800666. doi: 10.2807/1560-7917.ES.2019.24.3.1800666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takashita E., Kawakami C., Ogawa R., Morita H., Fujisaki S., Shirakura M., Miura H., Nakamura K., Kishida N., Kuwahara T. Influenza A(H3N2) virus exhibiting reduced susceptibility to baloxavir due to a polymerase acidic subunit I38T substitution detected from a hospitalised child without prior baloxavir treatment, Japan, January 2019. Euro Surveill. 2019;24:1900170. doi: 10.2807/1560-7917.ES.2019.24.12.1900170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N.X., Zheng J.J. Computational studies of H5N1 influenza virus resistance to oseltamivir. Protein Sci. 2009;18:707–715. doi: 10.1002/pro.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiso M., Iwatsuki-Horimoto K., Yamayoshi S., Uraki R., Ito M., Nakajima N., Yamada S., Imai M., Kawakami E., Tomita Y. Emergence of Oseltamivir-Resistant H7N9 Influenza Viruses in Immunosuppressed Cynomolgus Macaques. J. Infect. Dis. 2017;216:582–593. doi: 10.1093/infdis/jix296. [DOI] [PubMed] [Google Scholar]
- 16.Ranjan P., Bowzard J.B., Schwerzmann J.W., Jeisy-Scott V., Fujita T., Sambhara S. Cytoplasmic nucleic acid sensors in antiviral immunity. Trends Mol. Med. 2009;15:359–368. doi: 10.1016/j.molmed.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z., Chen L.M., Zeng H., Gomez J.A., Plowden J., Fujita T., Katz J.M., Donis R.O., Sambhara S. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 2007;36:263–269. doi: 10.1165/rcmb.2006-0283RC. [DOI] [PubMed] [Google Scholar]
- 18.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 19.Mibayashi M., Martínez-Sobrido L., Loo Y.M., Cárdenas W.B., Gale M., Jr., García-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opitz B., Rejaibi A., Dauber B., Eckhard J., Vinzing M., Schmeck B., Hippenstiel S., Suttorp N., Wolff T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 2007;9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 21.Davis W.G., Bowzard J.B., Sharma S.D., Wiens M.E., Ranjan P., Gangappa S., Stuchlik O., Pohl J., Donis R.O., Katz J.M. The 3′ untranslated regions of influenza genomic sequences are 5'PPP-independent ligands for RIG-I. PLoS ONE. 2012;7:e32661. doi: 10.1371/journal.pone.0032661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjan P., Jayashankar L., Deyde V., Zeng H., Davis W.G., Pearce M.B., Bowzard J.B., Hoelscher M.A., Jeisy-Scott V., Wiens M.E. 5'PPP-RNA induced RIG-I activation inhibits drug-resistant avian H5N1 as well as 1918 and 2009 pandemic influenza virus replication. Virol. J. 2010;7:102. doi: 10.1186/1743-422X-7-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiropoulou C.F., Ranjan P., Pearce M.B., Sealy T.K., Albariño C.G., Gangappa S., Fujita T., Rollin P.E., Nichol S.T., Ksiazek T.G., Sambhara S. RIG-I activation inhibits ebolavirus replication. Virology. 2009;392:11–15. doi: 10.1016/j.virol.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 24.Chakravarthy K.V., Bonoiu A.C., Davis W.G., Ranjan P., Ding H., Hu R., Bowzard J.B., Bergey E.J., Katz J.M., Knight P.R. Gold nanorod delivery of an ssRNA immune activator inhibits pandemic H1N1 influenza viral replication. Proc. Natl. Acad. Sci. USA. 2010;107:10172–10177. doi: 10.1073/pnas.0914561107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jing H., Su H.C. New immunodeficiency syndromes that help us understand the IFN-mediated antiviral immune response. Curr. Opin. Pediatr. 2019;31:815–820. doi: 10.1097/MOP.0000000000000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin S., Hahn T. Interferon deficiency syndrome. Clin. Exp. Immunol. 1985;60:267–273. [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan J.M., Yau E.H., Taggart R.T., Butler M.C., Kolniak T.A. Relieving bottlenecks in RNA drug discovery for retinal diseases. Adv. Exp. Med. Biol. 2012;723:145–153. doi: 10.1007/978-1-4614-0631-0_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borkotoky S., Murali A. The highly efficient T7 RNA polymerase: a wonder macromolecule in biological realm. Int. J. Biol. Macromol. 2018;118(Pt A):49–56. doi: 10.1016/j.ijbiomac.2018.05.198. [DOI] [PubMed] [Google Scholar]
- 29.Clarke D.K., Sidhu M.S., Johnson J.E., Udem S.A. Rescue of mumps virus from cDNA. J. Virol. 2000;74:4831–4838. doi: 10.1128/jvi.74.10.4831-4838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieber A., Kiessling U., Strauss M. High level gene expression in mammalian cells by a nuclear T7-phase RNA polymerase. Nucleic Acids Res. 1989;17:8485–8493. doi: 10.1093/nar/17.21.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin L., Liu Q., Berube N., Detmer S., Zhou Y. 5′-Triphosphate-short interfering RNA: potent inhibition of influenza A virus infection by gene silencing and RIG-I activation. J. Virol. 2012;86:10359–10369. doi: 10.1128/JVI.00665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svancarova P., Svetlikova D., Betakova T. Synergic and antagonistic effect of small hairpin RNAs targeting the NS gene of the influenza A virus in cells and mice. Virus Res. 2015;195:100–111. doi: 10.1016/j.virusres.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Juang Y.T., Lowther W., Kellum M., Au W.C., Lin R., Hiscott J., Pitha P.M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., Global Seasonal Influenza-associated Mortality Collaborator Network Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendel H.A., Snyder M.T., Pell S. Trial of amantadine in epidemic influenza. Clin. Pharmacol. Ther. 1966;7:38–43. doi: 10.1002/cpt19667138. [DOI] [PubMed] [Google Scholar]
- 37.Centers for Disease Control and Prevention Influenza antiviral medications: summary for clinicians. 2019. https://www.cdc.gov/flu/professionals/antivirals/summary-clinicians.htm
- 38.Bouvier N.M., Lowen A.C., Palese P. Oseltamivir-resistant influenza A viruses are transmitted efficiently among guinea pigs by direct contact but not by aerosol. J. Virol. 2008;82:10052–10058. doi: 10.1128/JVI.01226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szretter K.J., Balish A.L., Katz J.M. Influenza: propagation, quantification, and storage. Curr. Protoc. Microbiol. 2006;3 doi: 10.1002/0471729256.mc15g01s3. Unit 15G.1.1–15G.1.22. [DOI] [PubMed] [Google Scholar]
- 40.Abdelmaksoud H.E., Yau E.H., Zuker M., Sullivan J.M. Development of lead hammerhead ribozyme candidates against human rod opsin mRNA for retinal degeneration therapy. Exp. Eye Res. 2009;88:859–879. doi: 10.1016/j.exer.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Froebel B.R., Trujillo A.J., Sullivan J.M. Effects of Pathogenic Variations in the Human Rhodopsin Gene (hRHO) on the Predicted Accessibility for a Lead Candidate Ribozyme. Invest. Ophthalmol. Vis. Sci. 2017;58:3576–3591. doi: 10.1167/iovs.16-20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yau E.H., Butler M.C., Sullivan J.M. A cellular high-throughput screening approach for therapeutic trans-cleaving ribozymes and RNAi against arbitrary mRNA disease targets. Exp. Eye Res. 2016;151:236–255. doi: 10.1016/j.exer.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yau E.H., Taggart R.T., Zuber M., Trujillo A.J., Fayazi Z.S., Butler M.C., Sheflin L.G., Breen J.B., Yu D., Sullivan J.M. Systematic screening, rational development, and initial optimization of efficacious RNA silencing agents for human rod opsin therapeutics. Transl. Vis. Sci. Technol. 2019;8:28. doi: 10.1167/tvst.8.6.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D.H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.